Abstract
Adult hair follicle stem cells (HFSCs) undergo dynamic and periodic molecular changes in their cellular states throughout the hair homeostatic cycle. These states are tightly regulated by cell-intrinsic mechanisms as well as by extrinsic signals from the microenvironment. HFSCs are essential not only for fueling hair growth, but also for skin wound healing. Increasing evidence suggests an important role of HFSCs in organizing multiple skin components around the hair follicle, thus functioning as an organizing center during adult skin homeostasis. Here we focus on recent findings on cell-intrinsic mechanisms of HFSC homeostasis, which include transcription factors, histone modifications, DNA regulatory elements, non-coding RNAs, cell metabolism, cell polarity and post-transcriptional mRNA processing. Several transcription factors are now known to participate in well-known signaling pathways that control hair follicle homeostasis, as well as in super-enhancer activities to modulate HFSC and progenitor lineage progression. Interestingly, HFSCs have been shown to secrete molecules that are important in guiding the organization of several skin components around the hair follicle, including nerves, arrector pili muscle, and vasculature. Finally, we discuss recent technological advances in the field such as single cell RNA-sequencing and live imaging, which revealed HFSC and progenitor heterogeneity and brought new light to understanding crosstalking between HFSCs and the microenvironment. The field is well on its way to generate a comprehensive map of molecular interactions that should serve as a solid theoretical platform for application in hair and skin disease and aging.
Keywords: skin stem cells, transcription factors, epigenetics, metabolism, stem cell niche
Introduction
Adult stem cell (SC) identity and potential to regenerate tissues are defined by specific combinations of cell-intrinsic molecular mechanisms that control gene expression, in particular transcription (1,2). These mechanisms include chromatin structure, transcription factors, DNA regulatory elements such as enhancers, and non-coding RNAs. Cell metabolism is another emergent cell-intrinsic mechanism of SC regulation (3,4). Furthermore, SCs respond to microenvironmental (or niche) signals as well as signal themselves to the surrounding microenvironment for reciprocal control of cell behavior and tissue homeostasis (1,5). All these mechanisms collectively govern cell growth and proliferation, cell adhesion, migration, and differentiation, and maintain SC regenerative potential throughout life. Deregulation of these molecular mechanisms affect normal adult SC behavior and leads to diseased or aged tissues (6,7). Whereas each tissue SC presents its own unique combination of regulatory factors, similar molecular mechanisms and principles controlling these functions are emerging from diverse adult mammalian tissues (8,9).
Mouse hair follicle stem cells (HFSCs) constitute an excellent model system that helped deepen our global understanding of tissue SC regulation (10). The HFSCs are largely quiescent (11) but periodically undergo dynamic and coordinated activities that involve cell migration, proliferation, and differentiation (12), which occur sequentially and somewhat synchronously on the adult mouse back skin. These activities occur in distinct and tractable location along the hair follicle, enabling examination of genetic and epigenetic perturbations effects on SC behavior. Quiescent HFSCs located in the bulge generate the early progenitor cells or ‘primed SCs’, in the hair germ (Figure 1). The latter in turn generate the rapidly proliferating progenitor cells located in the hair matrix, which subsequently terminally differentiate and move upwards to form the inner hair follicle lineages, which include the inner root sheath and the hair shaft (13). As the matrix cells undergo differentiation, the bulge SCs pass through a brief phase of symmetric self-renewal at anagen, remain confined to the bulge, and finally return to quiescence once again (14,15). From here, some quiescent HFSCs migrate out of the ‘old’ bulge to generate a new set of primed stem cells in a ‘new’ hair germ as well as a ‘new’ bulge, and the cycle begins again. These cyclic processes occur during distinct phases of hair remodeling, known as telogen, anagen, and catagen (Figures 1 and 2) (16).
Figure 1.
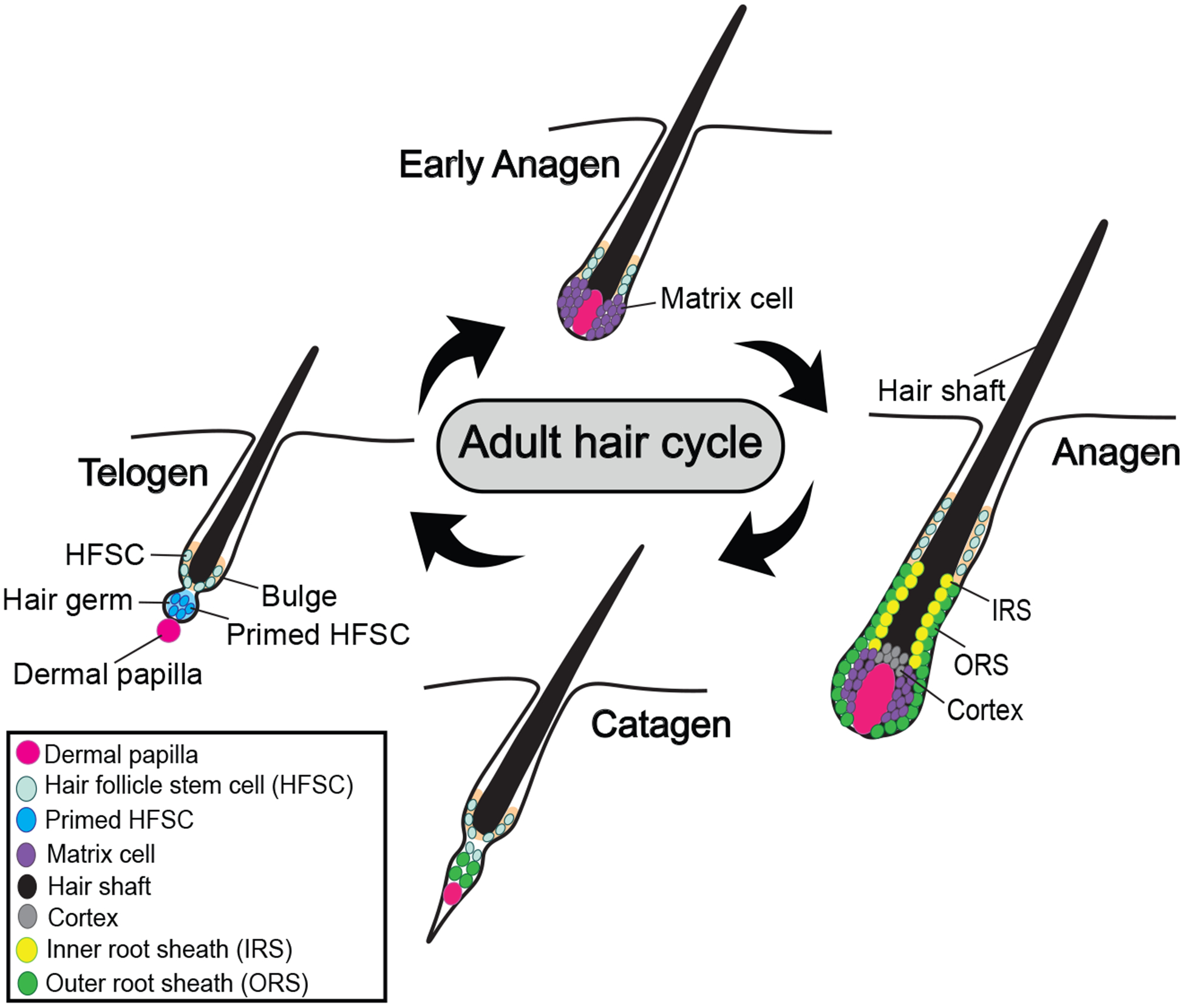
Adult hair cycle. Adult hair follicles are maintained via repeated cycles of growth (anagen), regression (catagen), and quiescence (telogen). In telogen, primed HFSCs form a tight, ball-like structure in hair germ, underneath quiescent HFSCs in the bulge. Mesenchymal cells in DP send signals to activate primed HFSCs during telogen-anagen transition, and primed HFSCs undergo rapid proliferation and expansion to give rise to matrix progenitors during early anagen. Matrix cells further differentiate to form IRS, cortex, and hair shaft during anagen, after which differentiated cells undergo systematic cell death during subsequent catagen.
Figure 2.
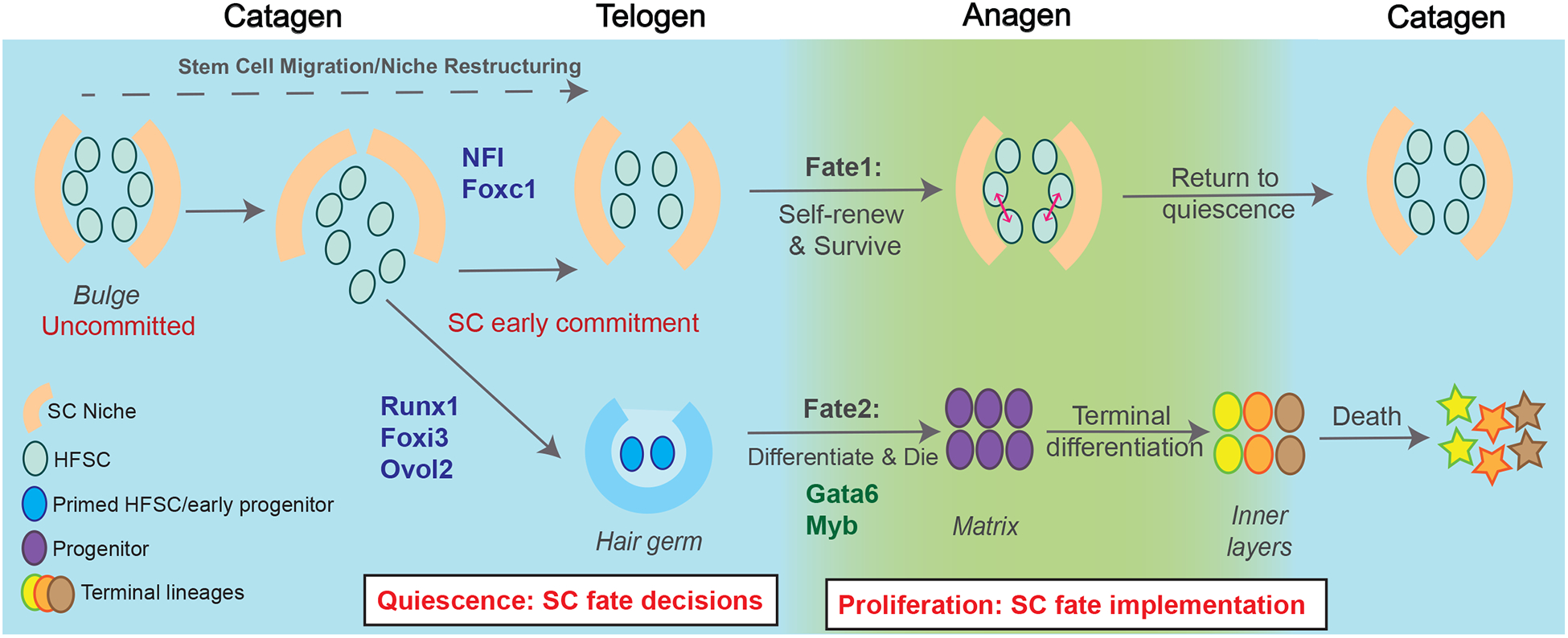
Changes in HFSC states during hair cycle. Bulge HFSCs in catagen are yet uncommitted to one of two subsequent fates during anagen. At this stage a fraction of bulge HFSCs will take residence below the bulge to form the primed HFSCs in the hair germ. Recently studied TFs discussed in text are shown on the arrows corresponding to their described function.
As with other tissue SCs, HFSCs function is driven by both cell-intrinsic mechanisms as well as by cell-extrinsic regulation from the microenvironment, the SC niche. This review focuses on the SC-intrinsic molecular regulation of dynamic adult HFSC states of proliferation and quiescence, prior to differentiation along the matrix/inner lineage axis. It also briefly summarizes how HFSCs provide a source of signaling molecules and extracellular matrix components, thus organizing their own microenvironment or niche. The reverse signaling, comprising instructions received from the niche towards the HFSCs has been considered in detail elsewhere (5,6,9,17–20). Here we focus on recent studies of intrinsic HFSC mechanisms of adult skin homeostasis published in the past 5 years. We only briefly refer here to earlier work previously reviewed (13,21,22), which we concisely summarized here in Table 1.
Table 1.
Intrinsic regulators of HFSCs homeostasis. Notable TFs and proteins involved in intrinsic regulation of HFSCs homeostasis and their sources are listed. Proteins found in the top half of the table in bold letters indicate regulators that have been studied in recent 5 years, while those listed in the bottom half indicate notable HFSC regulators from earlier studies. denotes expressions observed during development.
| Proteins | Expression location | HC stage | Function | Associated signaling pathway | Targets | Source |
|---|---|---|---|---|---|---|
| NFI | Bulge, hair germ, ORS | Telo, Ana | SC Identity & lineage specification | Chromatin remodeling | SC super-enhancers | Adam et al., 2020 |
| Myb | bulge, ORS | Telo, Ana | SC proliferation & differentiation | Bmp | HF-lineage genes | Veselá et al., 2014; Hu et al., 2019 |
| Runx1 | bulge, hair germ, ORS | All | SC activation & proliferation | Stat3, Wnt | p21, p15 | Osorio et al., 2008; Osorio et al., 2011; Scheitz et al., 2012; Lee et al., 2013 |
| Foxc1 | bulge, IRS | All | SC quiescence | Bmp | Nfatc1, Bmp2 | Wang et al„ 2016; Lay et al., 2016 |
| Foxi3 | hair germ | Telo | SC proliferation & bulge specification | Shh | HFSC genes | Shirokova et al., 2016 |
| Gata6 | hair germ, IRS | Telo, Ana | Proliferation & DNA damage protection | NF-kB | DNA damage repair genes | Wang et al., 2017; Krieger et al., 2018 |
| Ovol2 | bulge, hair germ | Telo, Ana | SC migration & proliferation | Ovol2-Zeb1 | Zeb1 | Haensel et al., 2019 |
| Lgr4 | bulge, hair germ, ORS, IRS | All | SC activation & proliferation | mTOR, Wnt, Bmp | R-spondin | Ren et al., 2020 |
| Hes1 | bulge, hair germ | Telo, Ana | HF growth | Shh | Gli1, Ptch1 | Suen et al., 2020 |
| Axin2 | bulge | Telo, Ana | SC quiescence | Wnt | Dkks, Sfrp1 | Lim et al., 2016 |
| Sirt7 | bulge, hair germ | Telo, Ana | SC activation | Bmp/Nfatc1 | Nfatc1 | Li et al., 2020 |
| Msi2 | bulge, hair germ, ORS, IRS | All | SC quiescence | Shh | Shh | Ma et al., 2017 |
| Acer1 | bulge | Ana? | Lineage specification | unspecified | unspecified | Lin et al., 2017; Liakath-Ali et al., 2016 |
| Par3 | unspecified | unspecified | Lineage specification | unspecified | unspecified | Liakath-Ali et al., 2016 |
| Endoglin | unspecified | All | SC quiescence & activation | Wnt/β-catenin, Bmp | Bmp | Calvo-Sánchez et al., 2019 |
| Lgr5 | bulge, hair germ, ORS | All | SC proliferation & self-renewal; lineage specification | Wnt, Shh | R-spondin, HF-lineage genes | Jaks et al., 2008 |
| Tcf3/4 | bulge, hair germ, ORS | All | Lineage specification | Wnt/β-catenin | HF-lineage genes | Merrill et al., 2001; Lien et al., 2014 |
| Lef1 | hair germ, IRS | Ana | Lineage specification | Wnt/β-catenin, Notch | HF-lineage genes | Zhou et al., 1995; Merrill et al., 2001; Niemann et al., 2002; Zhang et al., 2013; Lien et al., 2014; Adam et al., 2018 |
| Lhx2 | bulge, hair germ, ORS | All | Bulge organization & maintenance | Wnt | HFSC genes, cytoskeletal & adhesion protein genes | Rhee et al., 2006; Folgueras et al., 2013; Mardaryev et al., 2011 |
| Sox9 | bulge, hair germ, ORS | All | Lineage specification | Tgfβ/Activin, Wnt, Bmp, Shh | HF-lineage genes | Vidal et al., 2005; Kadaja et al., 2014; Adam et al., 2015 |
| Nfatc1 | bulge | All | SC quiescence | Bmp | CDK4 | Horsley et al., 2008 |
| Ctip2 | bulge, hair germ, ORS | All | SC quiescence & maintenance | Notch†, Egfr† | Nfatc1, Nhx2 | Bhattacharya et al., 2015 |
| Foxp1 | bulge | All | SC quiescence | p19/p53 | Fgf18, p57 | Leishman et al., 2013; Zhao et al., 2015 |
| p63 | bulge, hair germ, ORS | Telo | Lineage specification | Wnt/β-catenin†, Shh† | Wnt5a†, Tcf3/Tcf4†, Lef1† | Romano et al., 2010 |
| Gab1 | bulge, hair germ, ORS | Telo, Ana | SC quiescence & maintenance | Ras/Mapk | Mapk | Ozturk et al., 2015 |
| Hopx | bulge, hair germ, ORS, IRS | All | unspecified | unspecified | unspecified | Takeda et al., 2013 |
| aPKCλ | unspecified | unspecified | SC quiescence; lineage specification | unspecified | Bmp6, Fgf18 | Niessen et al., 2013; Osada et al., 2015 |
It is well known that HFSCs not only drive hair and skin homeostasis but they can also rapidly respond to injury signals by exiting quiescence and contributing to wound repair (23,24). Nonetheless, during homeostasis maintaining quiescence is a vital self-preservation mechanism for HFSCs, preventing SC depletion and reduced cell-fate plasticity (25,26). Failure to exit quiescence in a timely manner disrupts homeostatic tissue regeneration and wound healing (24,27,28). While we focus here on mechanisms of control in normal (un-injured) skin homeostasis, where known we briefly summarize implications of these mechanisms in wound healing, disease, and aging. The complexity of these latter aspects of skin biology surely warrants them a separate in depth account elsewhere.
Transcription factors and signaling pathways acting in quiescent bulge HFSCs
Transcription factors (TFs) are proteins that bind to specific DNA sequences or TF motifs and regulate the rate of gene transcription (29). Embryonic stem cells (ESCs) have high levels of specific TFs, most famous of which are the Yamanaka factors (Oct4, Sox2, Klf4, c-Myc). TFs maintain ESC pluripotency by regulating different signaling pathways, such as Notch, Wnt, and Sonic Hedgehog (Shh) (30,31). Likewise, HFSCs also upregulate specific sets of TFs to maintain their stemness, defined by quiescence, self-renewal, or differentiation into specific cell-type lineages throughout the hair cycle (Figure 2). TFs may act downstream or upstream of select signaling pathways to modulate expressions of other cell-intrinsic regulators and coordinate timely HF growth and regression in homeostasis or during wound healing (32–35). Several signaling pathways have been intensively studied in hair follicles over the past few decades, and were extensively reviewed (9,10,16,22,36–38). Of these, essential are Bmp and Wnt signaling, which govern HFSC cell fates during hair cycle (16,39–41). Briefly, Bmp signaling attenuates Wnt/β-catenin signaling to inhibit HFSC activation and proliferation, keeping HFSCs in quiescence (42,43). Bmp inhibition leads to premature bulge HFSCs activation, whereas inhibition of Wnt signaling leads to premature HF regression and eventual hair loss (43,44). Other signaling pathways such as Shh and Notch are also needed for HFSC homeostasis (40), as they are required for the differentiation and proliferation of progenitor cells, which in turn regulate HFSCs (discussed latter in this review).
Canonical Wnt/β-catenin signaling especially, plays a central role during both homeostatic hair growth and wound healing, and many TFs are known to take part or are downstream of this pathway (45). Tcf/Lef family of TFs are well-known effectors and can act as either Wnt target suppressors or transactivators (46). During postnatal hair growth, Tcf3/4 repress Wnt target genes to inhibit HFSC activation and differentiation, effects of which are reversed upon β-catenin binding (47). Lef1 acts largely as a transactivator and require β-catenin to drive the expression of HF-lineage genes in progenitor cells, without which sebum containing vacuoles and sebaceous tumors start to form (48–51). Tcf/Lef family of TFs and others such as Notch effector Rbpj and Bmp effector pSmad1 bind to super-enhancers (SEs), or large genomic regions enriched with clusters of enhancers, in a coordinated manner to suppress or activate different lineage-specific genes and to drive lineage progression in hair follicle progenitors (52,53). Thus, transient and coordinated activation of different signaling pathways is required for proper HFSC activity and hair growth during anagen.
Lgr5 is a well-known Wnt signaling target gene, highly expressed not only by HFSCs, but also by SCs of small intestine, mammary glands, and other organs. Lgr5 acts as an enhancer of Wnt-signaling and promotes SC self-renewal (54,55). Lgr5 expressing bulge HFSCs upregulate Shh signaling to facilitate rapid proliferation and differentiation during anagen, without which the hair fail to regenerate (56,57). Another family member, Lgr4, is also important for HFSC homeostasis, as its loss leads to failed HFSC activation and delayed hair growth, caused by decreased mTOR and Wnt activities and increased Bmp activity (58). Lgr5 is often used as a HFSC marker for imaging as well as in lineage tracing experiments (56,59,60). Recently Joost and colleagues used the Lgr5 promoter to genetically label HFSCs and study their transcriptomic changes in respond to skin injury using single-cell RNA-sequencing (scRNA-seq) (59). This study revealed at single cell resolutions how HFSCs and their progeny quickly migrate to the wound and gradually adopt a new fate as interfollicular epidermis (IFE), advancing the field’s understanding of HFSC contribution to wound healing since its first observation more than a decade ago (24).
Axin2 is another Wnt target gene expressed in HFSCs (61). Using Axin2-lacZ mice, Lim and colleagues showed that the outer bulge cells continuously secrete Wnt ligands throughout the hair cycle to maintain SC potency. Conversely, HFSC secretion of Wnt inhibitors such as Dkk and Sfrp1 keeps inner bulge cells quiescent during telogen (61). These findings highlight the importance of Wnt/β-catenin signaling control in bulge HFSC maintenance, without which premature HFSC activation and bulge depletion occur.
Surface glycoprotein endoglin (Eng) is a receptor for the Tgf-β pathway known to modulate the crosstalk between Bmp and Wnt signaling in the hematopoietic system during early development (62,63). Recently, it has been found to serve a similar function in HFSC during adult homeostasis, where mice with reduced whole-body expression of Eng exhibited dissonant hair growth patterns and out-of-sync expressions of hair cycle regulators such as Bmp4, Noggin and Shh (64). Further analysis revealed Wnt effector β-catenin and Bmp effector Smad4 forming heterodimers to drive Eng expression, suggesting the communication between Wnt/β-catenin and Tgf-β/Bmp/Smad signaling pathways in HFSCs during homeostasis (65).
Since the first molecular characterization of bulge HFSCs as label retaining cells almost two decades ago (11), many developmental TFs have been identified as important regulators of HFSC quiescence and activation. These include notable examples such as Runx1, Lhx2, Nfatc1, Sox9 and etc., studied in depth over the past couple of decades (Table 1 and (66–75)). Recent studies of HFSCs TFs expressed by bulge cells uncovered new facets of their regulation as well as new TFs, which we summarize here and depict in Figure 2. Foxc1 is a newly uncovered TF, loss of which results in shortened telogen, premature hair growth, and club hair loss (76,77). Foxc1-depleted HFSCs undergo premature activation and loss of quiescence. Foxc1 promotes expressions of quiescence genes such as Nfatc1 and Bmp2, and maintains the old bulge cells, which secrete Bmp5 and Fgf18 and induce HFSC quiescence in the new bulge. Upon Foxc1 loss, the old bulge is lost due to downregulation of E-cadherin (76). These findings indicate that Foxc1 can mediate HFSC quiescence by controlling relevant cell-intrinsic target genes, as well as indirectly through microenvironment maintenance.
A recent study of the TF Ovol2 reinforced the notion that not only HFSC activation and quiescence, but also migration is crucial for HFSC homeostasis and wound healing (78). Ovol2 is a TF that modulates epithelial-to-mesenchymal transition (EMT), whose expression has been observed in the basal layer of IFE as well as in the hair bulge, HG, and matrix cells in murine skin during homeostasis. Lack of Ovol2 in skin epithelium leads to delayed anagen progression and shorter HFs, as well as delayed wound healing due to abnormal cell adhesion and migration. In vitro and ex vivo explant cultures revealed that Ovol2-deficient HFSCs exhibited highly reduced clonal growth and displayed abnormal cell morphology under microenvironmental changes. Subsequent live cell imaging and cell cycle analysis indicated Ovol2 functions in HFSC migration directionality and cell cycle progression, part of which may be by inhibiting EMT promoting Zeb1 (78). This study suggests a novel role of Ovol2-Zeb1 EMT-regulatory circuit in HF homeostasis as well as during wound healing.
Nuclear Factor I (NFI) transcription factors NFIB and NFIX are known for their roles regulating progenitor differentiations during embryonic nervous system development (79,80). Recent findings from the Fuchs laboratory revealed that NFI TFs also play vital roles in preserving the HFSC identity, the loss of which leads to human scarring alopecia-like phenotypes in Nfib and Nfix double knockout (NFI dKO) mice (81). Further analysis using ATAC-seq and ChIP-seq indicated NFI dKO HFSCs had altered chromatin structure. This allowed increased accessibility to binding motifs of wound-induced TFs such as AP1 and Foxo1, previously discovered by the group to promote HF-to-epidermis lineage infidelity (82). Subsequent scRNA-seq analysis also confirmed highly reduced bulge HFSC signature gene expression in purified NFI dKO HFSCs as well (81). Their findings specify NFI TFs as a previously unknown mechanism that HFSCs employ against epidermal fate conversion during homeostasis. Another intriguing role NFIB seems to play is regulating melanocyte SCs located adjacent to HFSCs through Edn2/EdnrB signaling (83), which we will discuss in more depth elsewhere (Li and Tumbar, in review).
Overall, these recent findings emphasize the importance of coordination between Wnt and other signaling pathways for HFSC maintenance during homeostasis, in which various TFs and target genes are activated to fine-tune strengths of these signals. Several TFs including NFI have been found to cause epigenetic changes to further modulate crosstalk between different pathways. Epigenetic changes will be further discussed in a latter section of this review.
Other cell-intrinsic regulators of bulge HFSC function
Apart from the above-discussed TFs and signaling pathways, SC intrinsic factors involved in other regulatory mechanisms such as deacetylation, RNA processing, and cell polarity have also been found in recent years to take part in HFSC maintenance. While histone deacetylases (HDAC) and certain members of the Sirtuin family silence gene expressions transcriptionally (84), ribonucleoprotein (RNP) complexes modulate those expressions post-transcriptionally during RNA processing (85). Cell polarity, or asymmetrical organization within a cell, affects cell morphology and cell division (86). Here, we discuss regulators of these mechanisms that have recently surfaced during HFSC homeostasis.
Sirt7 is a deacetylase commonly associated with ribosomal RNA transcription and cell senescence (87), and its expression in the bulge is necessary for timely anagen progression (88). Upon Sirt7 loss, HFSC activation and HF growth were delayed, while inducing Sirt7 expression during telogen led to significantly earlier anagen entry. Further in vitro experiments revealed Sirt7 mediated degradation of Nfatc1, a HFSC quiescence TF, as one of the mechanisms behind proper telogen-anagen transition. Intriguingly, increasing Sirt7 expression in aged HFs can alleviate aging-related HF growth defects (88). Thus, Sirt7 may be useful as a potential therapeutic target for hair loss due to aging.
Msi2 is a RNA binding protein that forms RNP to process mRNAs for post-transcriptional regulation (89). Msi2 has been suggested to promote SC self-renewal in ESCs and murine hematopoietic stem cells, with its dysfunction often found in malignant tumors of brain, liver, and other organs (89,90). Ma and colleagues discovered Msi2 functions in HFSC quiescence maintenance using gain and loss-of-function mouse models, where its overexpression led to prolonged telogen-anagen transition and impaired hair growth, and its loss resulted in faster hair growth and proliferative HFSCs post-depilation (91). Transcriptomic analysis indicated Shh pathway downstream of Msi2, with RNA crosslinking immunoprecipitation (CLIP)-PCR assay confirming Msi2 binding to Shh mRNA for translation repression. Therefore, Msi2-mediated suppression of Shh signaling is necessary for proper HFSC quiescence maintenance, highlighting the importance of post-transcriptional regulation in HF homeostasis.
Polarity protein Par3 is involved in processes such as cell polarization and asymmetric cell division, and it often forms a complex with Par6 and aPKC proteins to determine cell polarity in epithelial cells (92–95). In a recent study, skin epithelium specific Par3 KO mice showed unaltered hair cycle progression despite gradual HFSC depletion and hypertrophic sebaceous glands (96), suggesting Par3’s role in HFSC and sebaceous maintenance independent of hair cycle regulating pathways like Wnt and Bmp. The group speculated a possible role of Par3 in HFSC differentiation, and it will be interesting to learn how mitotic spindle orientation may affect HFSC fate decisions and self-renewal.
Thus, while HFSC proliferation and quiescence are largely governed by Wnt and Bmp signaling pathways, other regulatory mechanisms such as deacetylation, RNA processing, and cell polarity are also required for adult HFSC maintenance during homeostasis. De-regulation of these mechanisms leads to aberrant hair growth and eventual HFSC depletion and may explain the alopecia phenotype that often accompanies aging organisms. These studies suggest that HFSCs employ different and somewhat independent intrinsic mechanisms to preserve their pool in adult homeostasis.
Transcription factors of primed HFSCs in the hair germ
Whereas much attention has been given to regulation of quiescent HFSCs in the bulge, proper activation and proliferation of primed HFSCs in the hair germ is essential for the initiation of the new growth cycle and for the generation of a new hair shaft (16,97). Furthermore, Shh signaling from transit-amplifying cells (TACs) in the matrix, which are committed HF progenitors, is required for the activation of quiescent HFSCs in the bulge and for their self-renewal at anagen, without which bulge SC depletion occurs (98). Thus, primed SCs and their immediate matrix derivatives use progeny-related feedback mechanisms that are essential to maintain the HFSC pool. Several recent studies have uncovered novel TFs whose expression is found in these cell populations during homeostasis, which we will discuss next.
Foxi3, a developmental TF whose dysfunction leads to hair and toothless phenotypes in certain dog breeds (99), is primarily expressed by primed HFSCs in HG, where its absence in mice leads to severe bulge and HG reduction along with HF malformation and eventual hair loss during homeostasis (100). Without Foxi3 primed HFSCs fail to expand and to give rise to TACs, eventually leading to bulge SC depletion and niche loss due to lack of Shh signal from TACs (100). Transcriptome analysis also confirmed downregulation of HFSC genes such as Lhx2, Runx1, Klf4, and Nfatc1 upon Foxi3 loss, further implying its function in maintaining the HFSCs pool.
The importance of the Runx1 TF in SC activation and proliferation has been well-demonstrated by our laboratory in earlier studies, where Runx1 promotes HFSC activation and proliferation of primed stem cells in the hair germ, as well as the rates of bulge-cell divisions at anagen (101,102). This regulation occurs in part via inhibiting cyclin-dependent kinase inhibitors (CDKi), such as p21 and p15 (101–103). Runx1 also activates Stat3 signaling to promote tumorous growth in skin and oral epithelial SCs (104). Our recent work indicates yet another mechanism by which Runx1 mediates SC proliferation, where it regulates the amount of specific fatty acids in cell membranes by targeting lipid enzymes Scd1 and Soat1, which in turn may attenuate Wnt signal transduction (105). Both Scd1 and Soat1 are expressed in the bulge as well as in HG, suggesting Runx1 mediated lipid metabolism as another possible intrinsic mechanism involved in HFSC activation. In addition to its cell-autonomous roles, recent finding from our laboratory revealed that Runx1 also exerts non cell-autonomous regulation on the HFSC vascular niche (106), as we discussed in more detail in a follow up section in this review.
Gata6 is another developmental TF whose expression is primarily found in tissues originating from mesoderm and endoderm, such as lung and heart (107). In adult murine skin, we found that Gata6 expression is in HG during telogen-anagen transition, which then spreads to proliferative matrix cells during anagen (28). HFs become growth arrested upon Gata6 loss during telogen, and its loss during anagen causes HFs to degenerate into telogen-like structures. In vivo and in vitro data from our laboratory show evidence of DNA damage accumulation in proliferating matrix cells and in keratinocytes without Gata6 activity, suggesting that it plays a role in mitigating replication stress in rapidly dividing cells. Activation of NF-kB pathway by Gata6 target Edaradd might be the mechanism promoting DNA damage repair and cell survival in these keratinocytes, which is further supported by the robust NF-kB activity observed in HG and its proliferating progeny in vivo. However, HFs lacking NF-kB activity do not become growth arrested and in fact progress into anagen, albeit in a delayed manner (108). Thus, Gata6 may have other downstream mechanisms contributing to HG activation and progenitor expansion that remain unknown.
Hes1 is a Notch signaling transcriptional repressor that promotes stemness in stem and progenitor cells of organs such as brain and small intestines (109,110). It is found robustly expressed in the lower bulge and HG during telogen-anagen transition, and epithelial specific deletion of Hes1 has been found to cause delay in hair growth and primed HFSC activation (111). Furthermore, Hes1 conditional knockout mice exhibited decreased HF regeneration upon repetitive depilation challenge, suggesting exhaustion of the HFSC potential to self-renew. Comparative transcriptome analysis revealed compromised Shh signaling in Hes1-depleted HFSCs, indicating Hes1’s role in Shh signal induction for proper primed HFSCs activation. These findings show a close relationship between Notch and Shh signaling in anagen progression mediated by Hes1.
Myb is a proto-oncogene whose upregulation is associated with cancers such as leukemia and colon carcinoma (112). In hair follicle Myb is expressed in the lower bulge as well as ORS and matrix during homeostasis (113,114). Upon its overexpression in murine skin, hair follicle progenitors undergo impaired differentiation, leading to hair loss and eventual bulge cell depletion. Subsequent transcriptome analysis revealed that Bmp signaling genes were downregulated in the transgenic skin, which reiterated the importance of Bmp signaling in HFSC homeostasis (114). The group also noted increased regulatory T-cell infiltration to the bulge upon Myb overexpression, suggesting quiescence as a possible mechanism by which HFSCs escape immune surveillance, though further studies will be needed to confirm this model.
In conclusion, primed HFSCs have a specific set of TFs regulating their function, which differs from the bulge HFSCs. This may be responsible for their unique behavior, since unlike bulge HFSCs the primed HFSCs do not self-renew and are essentially lost to differentiation towards matrix and further inner lineages (Figure 2). The list of primed HFSCs TFs and other intrinsic factors controlling activation and initiation of anagen seems to be continuously expanding, suggesting potential synergistic and redundant functions that remain to be explored.
DNA regulatory elements and super-enhancers
DNA regulatory elements, such as promoters and enhancers, are regions in the genome where various proteins are recruited for regulating gene transcription (115). Promoters are TATA box containing regions near the gene’s transcription start site (TSS), where transcription initiation complex and RNA polymerase are recruited to begin gene transcription. Enhancers, on the other hand, are located further away from TSS and harbor multiple binding motifs of different TFs. TF binding to enhancers often forms DNA loops with the promoters, thereby regulating the rate of transcription. Because many TFs are cell type specific, enhancer activities give rise to specific gene expression patterns of a distinct cell lineage (116). SEs are regions containing multiple enhancers clustered together with high enrichment for transcriptional coactivator binding. They have been initially found essential for maintaining the pluripotent state of ESCs by fine-tuning expressions of various genes (116,117), and have recently been studied in HFSC as well.
Promoter regions have been intensively used to generate transgenic tools, such as the K14-CreER mice (118–120), which allowed close investigation of SC behavior and lineage progression in vivo as well as identification of different signaling pathways involved in HFSC homeostasis throughout the hair cycle (121,122). However, the exact mechanism by which HFSCs maintain or alter their gene expression patterns during lineage progression have remained unknown until recently, due to the lack of tools to monitor enhancer activities. Recent identification of HFSC SE regions and their use to generate SE reporter mice allowed the monitoring of activities of dense TF binding site clusters, or epicenters in HFSCs during telogen (52). These results showed evidence of SE activities driving HFSC stemness and quiescence, where the master TF Sox9 (123,124) was observed to act as a pioneer factor to de-compress chromatin and allow bindings of other HF TFs such as Nfatc1 and Lhx2. Upon microenvironmental changes such as in vitro or skin wounding however, these SEs underwent structural changes to activate different epicenters or new SEs to upregulate IFE-signature genes rather than HF lineage genes. A subsequent study by the group also revealed that coordinated switching of different Wnt and Bmp effectors binding to SE drove further lineage progression of HFSCs progenitors as well, where Tcf3/4 binding switch to Lef1 binding activated expression of HF differentiation genes in matrix cells, and Rbpj or pSmad1 co-binding with Lef1 at lineage specific enhancers led to activation of inner root sheath (IRS) or hair shaft gene expressions, respectively (53).
Taken together, these results indicate SE activities as a main mechanism by which HFSCs maintain their homeostasis or adapt to acquire new IFE fate in response to injuries. They suggest that TFs promote SC identity in HFSC via SE accessibility changes, further corroborating the importance of SE activities during HFSC homeostasis. It seems likely the list of TFs involved in these SE activities will continue to grow in coming years.
Epigenetic modifications and chromatin structure
While TFs modulate expression of HF lineage genes by direct binding to the DNA, epigenetic modifications such as histone methylation or acetylation remodel the chromatin structure to modulate DNA accessibility of large regions of the genome. Specifically, trimethylation of histone H3 at lysine 9 (H3 K9me3) and 27 (H3 K27me3) suppress transcription presumably by compacting chromatin structure, while trimethylation of histone H3 at lysine 4 (H3 K4me3) does the opposite to promote active transcription (125,126). Mechanisms such as DNA methylation, histone acetylation and others also participate in modulating epigenetic profile changes in HFSCs, and were discussed in detail in our recent review last year (13). Thus, in this review we discuss only work in this area related to the most recent publications from the past year.
Global reduction of H3 K9/27me3 has been associated with high cell fate plasticity in quiescent SCs, disturbance of which leads to premature or compromised cell differentiation (127,128). Furthermore, global changes in H3 K27me3 have also been observed between quiescent and active HFSCs as well as their early progenitors previously, indicating different chromatin dynamics at each distinct cell state (129).
In our recent studies, global reduction of H3 K4/9/27me3 (global hypomethylation) have been shown to occur in quiescent late catagen HFSCs compared to proliferative early anagen HFSCs, suggesting increased HFSC cell plasticity during quiescence, prior to HFSC fate decision for self-renewal or differentiation (26,130). Follow up in vitro and in vivo experiments further strengthened our model, where quiescent HFSCs were found to have higher chromatin factor exchange rates and increased tendencies to reprogram upon Yamanaka factors exposure, compared to their proliferative counterparts (130). Data suggest that BMP signaling may be found both upstream and downstream of histone methylation in a negative feedback loop that remains to be directly tested in vivo (26,60). Results of our studies demonstrate the importance of histone methylation marks in adult HFSC function and suggest a link with quiescence and fate plasticity during homeostasis and injury repair. They also begin to explore the connection between epigenetics and essential signaling pathways known to control HFSC quiescence (Figure 3).
Figure 3.
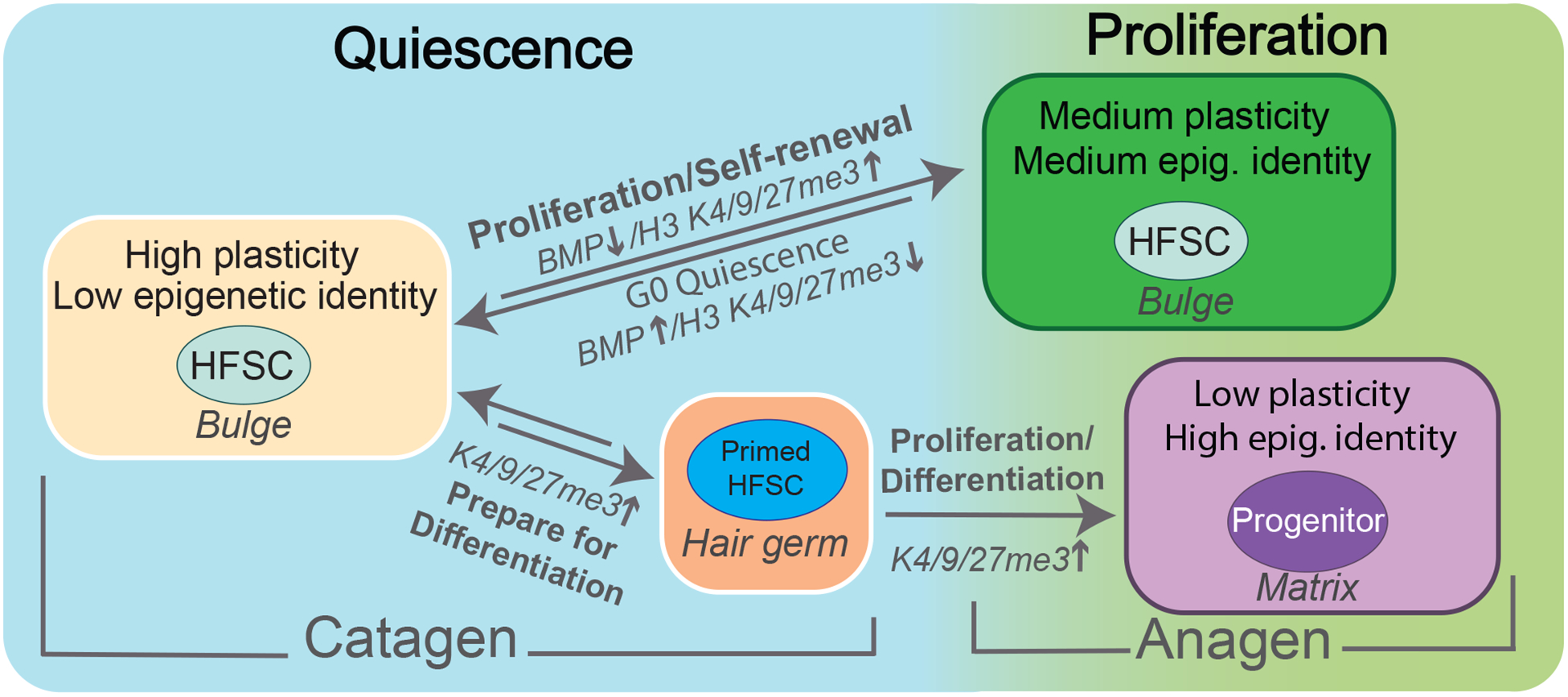
Model of epigenetic and cell plasticity states of HFSCs and progenitors. During quiescence bulge HFSCs maintain highest cell fate plasticity and lowest epigenetic identity as defined by low levels of histone H3 methylation marks. As bulge cells approach anagen for self-renewal, Bmp signal strength decrease and levels of histone H3 methylation marks increase, resulting in medium plasticity and epigenetic identity in proliferating bulge HFSCs. Matrix cells have high levels of histone H3 methylation marks associated with low cell plasticity and high epigenetic identity as progenitor cells.
Multiple SE studies mentioned in the earlier section further imply changes in HFSC chromatin structure throughout the hair cycle, where TFs such as Sox9 and NFI have been found to alter HFSC SE chromatin accessibility to drive expression of SC identity genes (52,81). Another recent study investigated HFSC chromatin structure changes induced by constant low-dose radiation (LDR) exposure, and examined subsequent HFSC behavior (131). The group observed high DNA damage accumulation in Lgr5+ HFSCs, which led to sustained chromatin structure changes and increased heterochromatin marks. HFSCs responded by either entering senescence or migrating out of the niche and differentiating, in which case cell proliferation was inadvertently induced to replace excluded SCs. Taken together, these findings suggest that the accumulation of chromatin structure changes in HFSCs induced by constant environmental genotoxin exposure may modify HFSC behavior.
Polycomb repressive complex 1 (PRC1) is an important player in epigenetic modifications, known to monoubiquitinate histone H2A (H2AK119Ub) and also facilitate PRC2-mediated methylation of H3 K27me3, repressing gene expression (132). Whereas previous studies have focused on the role of this complex in embryonic and early post-natal life (13,133), a recent study showed that PRC1 activity loss in Lgr5+ HFSCs leads to reduced level of H2AK119Ub and significantly delayed HF regeneration, likely due to reduced HFSC proliferation (134). The group noted that the absence of PRC1 activity leads to upregulation of genes associated with development and downregulation of HF lineage genes such as Shh in PRC1-null HFSCs. Comparative analysis with intestinal SC transcriptome profile indicated PRC1’s role in non-lineage specific TF suppression in both SC populations (134). These findings implied that in adult SCs, PRC1 regulates SC lineage specificity by inhibiting non-HF lineage TFs including epidermal-signature TFs. Whether this repression is due to PRC1 mediated H2AK119Ub or H3K27me3 or both remain to be investigated in future studies.
While current studies began to scratch the surface of epigenetic and chromatin control of adult HFSCs, much remains unknown. Most histone modifications deemed essential in ESCs are entirely unexplored in adult SCs, including the HF, and how these chromatin states might be involved in HFSC regulations is still unclear. Direct genetic approaches to target individual histone marks via loss of their positive and negative regulators in specific HF cell types to complement the chemical inhibition experiments are missing. Global changes of the chromatin structure and dynamics in adult HFSCs await further experimentation.
Non-coding RNA activities in HFSCs
Another important layer of HFSC intrinsic regulation comes from non-coding RNAs (ncRNAs). For earlier studies in this area we refer the reader to an excellent review by Dr. Botchkareva (135). ncRNAs or RNAs that are not translated into proteins, have been found to play vital roles in transcriptional and post-transcriptional regulation of genes (136,137). Micro-RNAs (miRNAs) are ncRNAs about 22-nucleotide long on average that mediate mRNA degradation and induce target gene silencing, whereas long ncRNAs (lncRNAs) are more than 200-nucleotide long and have more diverse functions such as miRNA regulation and chromatin remodeling, to name a few. Although many of their functions are studied in a disease-context, some studies have investigated their roles during homeostasis as well (138–140).
MiRNAs have been found in several studies to facilitate proper HF development as well as homeostasis (141–144). MiR-205 was found to be highly upregulated in HF placode, upon whose loss PI3K/Akt signaling was inhibited and HFSCs failed to expand (142). MiR-22 and miR-125b activities in HFSCs modulate cell differentiation and promote HFSC stemness (143–145). A recent study by Ge and colleagues showed that miR-29 was expressed the highest in telogen HFSCs. Its repression led to premature HG activation and anagen induction likely via Wnt and Bmp signaling genes, such as Lrp6 and Bmpr1a, which were identified as miR-29 targets (146). Similarly, miR-214 was recently shown to have an inhibitory effect on HFSC proliferation and differentiation and hair cycle progression partly by down-regulating Wnt signaling (147,148). Combining the covalent ligation of endogenous Argonaute-bound RNAs (CLEAR)-CLIP technique and in vivo experiments, Hoefert and colleagues also found that the miR-200 family plays various roles during HF development and homeostasis (149). miR-200 target mRNAs function in cell adhesion, actin cytoskeleton, and Hippo signaling, disturbance of which leads to defective HF development. Thus, these studies noted the importance of miRNA regulation of important signaling pathways and cellular processes, such as cell adhesion and cell junction formation in HFSCs.
Dicer is a riboendonuclease that regulates mature miRNA processing as well as facilitates post-transcriptional gene silencing via miRNA-induced silencing complex (miRISC) (150,151). Recently, bulge specific ablation of Dicer has been found to cause HF differentiation defects following depilation induced anagen, likely due to decreased keratin expressions in HF layers (151). Since ablation of another miRISC component Tarbp2 in bulge did not yield phenotypes observed in Dicer cKO HFs, the group speculated Dicer’s miRNA processing function is necessary for proper HF development during anagen, where more miRNAs than previously thought might be involved.
The role of long non-coding RNA in HFSCs also became more evident in the past few years. A study in vitro showed that PlncRNA-1 promotes proliferation and differentiation of cultured HFSCs through the up-regulation of TGF-β1 and the subsequent activation of Wnt signaling (152). In addition to directly targeting signaling pathways, lncRNA also interacts with miRNA to fine-tune HFSC activities. For example, lncRNA5322 functions as a competing endogenous RNA (ceRNA) to suppresses miR-19b-3p, thus promoting HFSC proliferation and antagonizing apoptosis (153). In vitro work also showed lncRNA5322 promotes proliferation and differentiation through the upregulation of miR-21, which is upstream of the phosphorylation of PI3K and AKT (154).
A deeper understanding of the non-coding RNAs in the HFSCs, and how this regulation interfaces with the more traditional regulatory molecules, such as transcription factors and chromatin structure, will add an important perspective on the complicated signaling mechanisms behind HFSC behavior. In addition, the de-regulation of non-coding RNAs in the skin has implications in various diseases such as cancer and psoriasis (135). In the clinical field, non-coding RNAs hold promising potential as targets of novel therapies (155).
Metabolism as a cell-intrinsic mechanism in HFSCs
Metabolism is a cell-intrinsic function that has been clearly linked with SC behavior (156). However, compared to the considerable amount of work done to understand the transcriptional and epigenetic regulation of the HFSC, less is known about metabolic pathways utilized by HFSCs and how they are involved in HFSC fate decisions.
Several metabolic pathways were found to be significant for HFSC activation. For instance, lactate production is indispensable for HFSC activation whereas promoting lactate production leads to accelerated activation (157). Glutamate signaling may be another pathway important in HFSC activation, as the glutamate transporter Slc1a3 is required in HFSCs for timely activation and anagen onset (158). In addition, lineage tracing experiments revealed that bulge HFSCs contain a subpopulation of Slc1a3+ cells that contributes to long-term hair follicle regeneration. Of note, Slc1a3 has been identified as a marker for non-label retaining stem cells in the basal layer in the IFE (159). Interestingly, ablation of Slc1a3 gene uncouples the expansion of sebaceous gland and IFE (158), pointing to a potential cross-talk between HFSC and SG/IFE niches mediated by Slc1a3. Whether the effect of Slc1a3 knockout on HFSCs comes from the niches or from HFSCs intrinsically is a question that can be answered by doing population-specific knockout of Slc1a3.
Previously, it has been shown that reactive oxygen species (ROS), besides their conventionally accepted role in cytotoxicity, have implications in regulating epidermis differentiation through Notch and Wnt signaling (160). Recent findings also tie ROS with HFSC proliferation. Zhao et al. proposed that ROS has an inhibitory effect on HFSC proliferation, as failure in ROS accrual by Foxp1 loss and anti-oxidant treatment promotes HFSC proliferation (161). However, the Espada group showed an opposite effect of ROS in HFSC proliferation through photoactivated transient ROS production in the bulge (162). In line with this result, similar photoactivation treatment in ex vivo cultured human hair follicles cause anagen onset in a Wnt-dependent manner (163). The discrepancy in these studies may suggest that long-term ROS has different effects from transient ROS or may be attributed to side effects from different ROS induction methods.
In anagen, differentiating matrix cells have elongated mitochondria filled with cristae (164). This morphological change is accompanied with higher mitochondrial activity compared to quiescent bulge HFSCs. Transcriptional profiling of enzyme expression further revealed that differentiating matrix cells utilize aerobic respiration whereas quiescent bulge HFSCs utilize anaerobic respiration (164). This finding highlights the differential metabolic needs at different points along the way of HFSC lineage progression.
Lipid metabolism contributes to various aspects of cellular activities including energy production and lipid membrane synthesis. How lipid metabolism relates to HFSCs behaviors has been explored recently. Overexpression of sPLA2-IIA, a phospholipase that hydrolyzes glycerophospholipids to fatty acids and lysophospholipids, was shown to promote the differentiation of HFSCs, likely by activating mitogens and pro-proliferating transcriptional factors such as c-Jun and FosB. This eventually leads to HFSC exhaustion and depletion over time (37). As mentioned in the TF section, Runx1 controls HFSC behavior from many aspects, among which lipid metabolism was newly identified (105). This finding provides a new perspective on the function of conserved TFs in SCs and how they regulate SC behavior.
Ceramide is an essential component of the skin barrier and an important metabolite that actively regulates cellular activities. Disruption of ceramide metabolism by inactivating Ceramide Synthase 4 leads to precocious HFSC activation by a stepwise shift from the inhibitory Bmp signaling to the activating Wnt signaling, thus depleting the HFSC pool and eventually causing hair loss (165). However, knockout of ceramidase Acer1 does not have an impact on the hair cycle except for disrupting the K15 bulge organization (96,166). Whether it is the ceramide level or the molecular signaling pathway controlled by those enzymes that regulate HFSC behavior needs further clarification.
Many of the recent findings highlight the active role of metabolism in dictating HFSC behavior, rather than being a mere cellular state. The intricate relationship between metabolic pathways and classic HFSC signaling pathways such as Bmp, Notch and Wnt pathways is also coming to light. In the future, further dissecting each metabolite’s function in regulating HFSC activities and its connections with other pathways will be of great scientific interest. In addition, HFSCs is often found to be the origin of skin cancers and many studies showed that the de-regulation of metabolic pathways can turn tissue stem cells into cancer cells (4). Therefore, exploring the link between HFSC and skin cancers from the perspective of metabolism may shed new light on cancer therapeutics.
Signaling from the hair follicle stem cells towards the surrounding skin environment
Cell-intrinsic mechanisms of HFSCs not only provide spatiotemporal control of cell-autonomous behaviors, but also generate outwards signaling to modulate the nearby microenvironment that is generally referred to as the HFSC niche. Here we briefly summarize evidence suggesting that signals emanating from the HFSCs can regulate the organization of surrounding skin vasculature, nerves, arrector pili muscle, melanocyte stem cells, adipose tissue and possibly the dermal sheet. A comprehensive review on this fascinating subject is currently from our laboratory is currently in review (Li and Tumbar, in review).
Most recent attention has been given to the dermal vasculature, which we recently showed that it dynamically reorganizes around the hair germ and bulb during the hair cycle (106). Three studies published in the last year, including our own, agreed in that signals from the HF epithelium are critical in the organization of vasculature around the hair follicle (106,167,168). First, loss of Runx1 TF from the hair epithelium alters the vasculature organization around and bellow the hair germ at telogen. This is coupled with a subsequent delay in HFSC activation and likely occurs through down-regulation of relevant Runx1 target genes. The latter are known to function as secreted molecules important for vasculature remodeling, such as Ntn4, among others (106). Significantly, Ntn4 was found important in HFSCs along with ANGPTLs and Wnt ligands to remodel the lymphatic capillary that sit alongside the hair bulge (167,168).
Furthermore, HFSCs are responsible for deposition of neurotrophins such as BDNF and of ECM proteins such as EGFL6, thus ensuring proper nerve patterning around the HF, which is critical for mechanosensory response and piloerection (169–171). Similarly, HFSCs deposit nephronectin on one side of the bulge, thus establishing a niche for the arrector pili muscle (APM), a muscle string that connects the epidermis with the hair follicle causing piloerection (171,172). HFSCs also secrete Edn2, Wnt ligands and antagonists, KIT ligand, CXCL12, and Col17a1 (83,121,173–176), which signal to the melanocyte SCs in the bulge to regulate coordinated responses during hair cycle. A recent study also sheds light on metabolites’ role on niche regulation, as retinoic acid is found to be upstream of the melanocyte SC differentiation program (176).
Other components of the HFSC environment include the skin adipose tissue, whose progenitor cells signal to HFSCs for proper activation whereas the HF epithelium signals back for adipocyte production (177). Last but not least are the mesenchymal cells, including the dermal papillae, which is essential in HFSC activation (18) and the dermal sheet, which was recently shown to function as a muscle important through each contractions for degradation of the epithelial hair bulb during catagen and for bringing the dermal papillae in close contact with the bulge (178). To our knowledge there is no direct evidence indicating that adult HFSCs regulate these two neighboring compartments, but our epithelial mutant mice that halt the hair cycle, Runx1 and Gata6 (101,179), also halt remodeling of the dermal sheet, suggesting a tight coupling between these compartments. Specific signals that might be responsible for the adult HFSC communication toward these mesenchymal compartments is awaiting.
Technological advances shed new light on our understanding of the HFSC field
In recent years novel technologies have been adopted and robustly used in the skin field to advance our current understanding of HFSCs behavior and regulation. Most notable of those technologies is single cell RNA sequencing (scRNA-seq), which allows identification of different cell populations based on individual cell’s transcriptome profile, as well as unbiased lineage trajectory progression among these populations (180–182). Its use in the skin field was first pioneered by the Watt laboratory to examine epidermal SC heterogeneity in human skin in 2006 (183). A decade later Joost and colleagues used scRNA-Seq to identify distinct interfollicular and hair follicle cell populations in adult murine skin at telogen and anagen (184,185). Their results revealed new transcriptomic heterogeneity within each skin compartment. The Kasper group also coupled the use of lineage tracing and scRNA-seq to demonstrate gradual transcriptional adaptation of Lgr5+ HFSCs to IFE-lineage upon wound healing (59). Recently, scRNA-seq has also been utilized by several groups to investigate hair follicle progenitor heterogeneity and lineage progressions (53,81,186), wound-induced fibroblast conversion to dermal papilla (34), the hair follicle dermal niche formation during development (187), and age-related HFSCs and microenvironment changes in murine skin (188).
Despite all this progress, scRNA-seq data on proliferating or self-renewing bulge HFSCs at anagen, a small cell population that is under-represented in studies of whole skin, have been missing so far. In this issue of Experimental Dermatology, our laboratory presents high resolution scRNA-seq data of self-renewing bulge cells, which were sorted from the anagen skin to enrich in this rare population (Chovatya et al., in revision). Unbiased clustering and GO analysis of differentially expressed genes suggested that each anagen-bulge subpopulation may have distinct functions within the bulge, providing a platform for future functional studies.
In addition to the advancement in genomic technologies, another technological breakthrough during the past decade or so is intravital imaging, which allows direct visualization of cellular events including cell division, differentiation, migration, and other physical changes in real time. The Greco laboratory pioneered this technique in the skin and their earlier work revealed remarkable details on HFSC biology, which can be achieved only by revisiting the same hair follicle over time. The spatiotemporal information provided by intravital imaging uncovered dynamic cellular processes including HFSC morphological reorganization during HF regeneration and epithelial phagocytosis during HF regression, both regulating HFSC behavior (189,190). In addition, tracking individual HFSC at different locations over time revealed that HFSC position dictates subsequent HFSC fate (191). For the first time they also demonstrated high plasticity of epithelial cells even under homeostasis, as they can reconstitute the HFSC pool after HFSC ablation (191). While in the past five years there has been no new finding that used live imaging to further our understanding on the bulge HFSCs specifically, recent work that employed this technique brought new clarity to the process of progenitor matrix cell differentiation to inner lineages. Although our review here focuses on the bulge cell regulation and does not deal in depth with the progenitor cells, this work raised new questions and challenges our understanding of fate determination in the hair follicle, and is hence worth discussing in this context here.
By tracking the starting position of the differentiating SC in the hair germ through live imaging, Xin et al. found that the location is determinant of the subsequent fate of the descending matrix cells to differentiate to the inner hair follicle lineages (192). Moreover, this fate decision appears to be flexible at later stages, when progenitor matrix cells change position along the DP vertical axis and also switch the fate of their descendants according to their new location. Turning on mutations that block differentiation in some progenitor cells does not have an effect, as the remaining progenitors adapt and compensate, all attesting to the flexible fate of matrix progenitor cells. This work also demonstrated the continuous replenishment of the matrix progenitor by lower ORS cells at the base of the bulb (192,193).
This live imaging work attests to the flexibility of fate acquisition of progenitor cells in the matrix and corroborated well with scRNA-seq from the Kasper laboratory, in which progenitor cells were found to be largely transcriptionally uncommitted (185). These findings contradicted previous models in which the matrix progenitors were deemed unipotent based on evidence from single-cell lineage tracing, in which results are inferred from indirect observations (179,186,194–196). A recent article reported a novel technology that allows both single cell ATACseq and single cell RNAseq within the same single cell (200). When applying this new technology to sequencing cells of the mammalian skin, the authors found that the early matrix progenitors, although transcriptionally uncommitted, already have distinct chromatin changes that may bias them towards specific fates (200), attesting one more time to the notion that fate commitment may be a more fluid process than initially thought.
These studies represent an excellent example of how emerging technologies can shed new light on old models, and re-shape our understanding of fundamental biological mechanisms of cell fate determination. In future, as new technologies emerge, it will be critical to harness their power to shine new light on HFSC biology.
Conclusion and future directions
HFSCs undergo cycles of activation and quiescence to replenish progenitor pools during homeostasis. During cycles, Wnt and Bmp signaling strengths are modulated by various intrinsic mechanisms such as TFs, SE activity, ncRNAs, and epigenetic remodeling via histone modifications, with progenitors of HFSCs also partaking HFSC regulation through Shh and Notch signaling. Recently, new findings in the field shed light on novel roles of TFs in coordinating activities between HFSCs and the niche, such as Runx1 involvement in vasculature reorganization, and Rbpj role in melanocyte stem cell differentiation (106,176). Knowledge of HFSC capabilities to remodel the surrounding microenvironment through secreted molecules provides a new angle to view the function of HFSCs, as not only providers of the building blocks necessary for production of differentiated tissue (the hair shaft), but also as an organizing center for proper SC niche remodeling and adult skin homeostasis (Li and Tumbar, in review). These intrinsic mechanisms, along with HFSC crosstalk with niche cells, are all essential for proper tissue regeneration during homeostasis and wound healing.
Perhaps what is missing in the field is an understanding of how different epigenetic mechanisms and niche interactions all come together and function in a coordinated manner. An example is the correlations between global histone methylation levels and SE activities of HFSCs, clarification of which may reveal the underlying mechanism between HFSC quiescence and cell fate plasticity. Given the heterogeneity uncovered in bulge cells, this endeavor will undoubtedly require the development of novel techniques or innovative strategies to utilize existing tools. A recent example is the technique developed by Weinreb and colleague (182). The group generated random DNA barcoding constructs to uniquely label individual multipotent cells, enabling simultaneous fate mapping and transcriptional analysis of barcode-sharing progenitors of a specific cell.
Another considerable gap in our knowledge is an in-depth understanding of the global chromatin structure and nuclear organization in HFSCs during adult homeostasis, and how global changes in histone marks manifest in this context. Recent studies in systems other than skin suggest that biophysical measures of chromatin compaction are more accurate than those who rely on distribution of post-translational modification of histones, such as heterochromatin marks H3K9me3 and H3K27me3 (197). However, those approaches are largely missing from the skin, most likely due to technical hurdles related to small amount of materials available from sorted cells. Advanced technologies examining DNA accessibility and hence the compaction of chromatin include single cell ATAC-Seq (198), which has been applied to embryonic skin (199), but not yet to the adult bulge HFSCs during different stages of the hair cycle. The recent approach that enables simultaneous assessment of chromatin accessibility and transcriptome profile of single cells known as SHARE-seq will provide further insight into cell fate bias and determination in HFSCs (200). Development of technologies such as SHARE-seq will allow direct probing of chromatin structure in vivo, and permit the field to move to the next level of understanding chromatin structure in the context of epigenetic marks and cell fate decisions in a living tissue. Chromatin dynamics, and regulation of DNA-bound protein exchange rates, and how this affects HFSC plasticity is also in its infancy (130) and requires more investigation.
Furthermore, we lack the appropriate tools to examine nascent or ongoing transcription in small populations of cells in vivo in tissue SCs, as it has been done in ESCs (201). Hence, we may completely mis-interpret the total (or the steady-state) level of mRNA detected in cells via RNA-seq. Currently, we presume this level as being transcriptionally controlled, thus overlooking possible contributions from RNA processing, RNA stability or degradation, which are likely to also play important roles. Along those lines, it is interesting that protein synthesis rates are very different between proliferative and quiescent HFSCs in vivo, and that this global regulation can affect HFSC function (202). Protein post-translational modification are now known to be intimately connected with cell metabolism, which in turn control transcriptional states and ultimately cell fate (203), another area of investigation that is currently lacking in the skin.
Perhaps the most exciting part of the current advances in the field is that now we are beginning to get a glimpse not only at individual players in the SC-intrinsic control of HFs, but also at the broader mechanisms and the myriad interplays amongst them that govern HFSC homeostasis. For instance, TF, SE, and signaling pathways have now been clearly linked in their orchestration of cell fate acquisition. Changes in HFSC epigenetic modifications at different cellular states may be modulated by signaling, and specific TF-SE interactions have been shown to alter HFSC chromatin accessibility patterns. These discoveries suggest the existence of cross-talks between multiple mechanisms throughout the hair cycle, regulating HFSC homeostasis. Elucidating these complex relationships will help us better understand associations among various cell-intrinsic mechanisms in controlling SC states, with generation of comprehensive molecular interaction networks. It is probably the deepest hope of every hair follicle biologist that this broad understanding will help in future to alleviate perturbed pathways in hair and skin disease and in aging. These findings will also provide a road map of investigation for other tissue systems, which are not as easily amenable to in-depth exploration as the hair follicle.
Acknowledgments
We thank all our colleagues for their seminal contributions to knowledge of skin and hair follicle biology and for making this an exciting and rewarding field of research. We apologize to those whose articles are not cited owing to space limitation. This work was supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR070157 and R01AR073806) to TT.
Footnotes
Conflict of Interests
The authors have declared no conflicting interests.
References
- 1.Watt FM, Hogan BLM. Out of eden: Stem cells and their niches [Internet]. Science (80-) 2000: 287: 1427–1430. [DOI] [PubMed] [Google Scholar]
- 2.Cho IJ, Lui PPW, Obajdin J et al. Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence [Internet]. Stem Cell Reports 2019: 12: 1190–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandel NS, Jasper H, Ho TT et al. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing [Internet]. Nat Cell Biol 2016: 18: 823–832. [DOI] [PubMed] [Google Scholar]
- 4.Shapira SN, Christofk HR. Metabolic Regulation of Tissue Stem Cells. Trends Cell Biol 2020: 30: 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rompolas P, Greco V. Stem cell dynamics in the hair follicle niche [Internet]. Semin Cell Dev Biol 2014: 25–26: 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji J, Ho BS-Y, Qian G et al. Aging in hair follicle stem cells and niche microenvironment [Internet]. J Dermatol 2017: 44: 1097–1104. [DOI] [PubMed] [Google Scholar]
- 7.So WK, Cheung TH. Molecular regulation of cellular quiescence: A perspective from adult stem cells and its niches. In: Methods in Molecular Biology. Humana Press Inc., 2018: 1–25. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: Stem cells and their niche. Cell 2004: 116: 769–778. [DOI] [PubMed] [Google Scholar]
- 9.Chacón-Martínez CA, Koester J, Wickström SA. Signaling in the stem cell niche: regulating cell fate, function and plasticity [Internet]. Development 2018: 145: dev165399. [DOI] [PubMed] [Google Scholar]
- 10.Houschyar KS, Borrelli MR, Tapking C et al. Molecular Mechanisms of Hair Growth and Regeneration: Current Understanding and Novel Paradigms [Internet]. Dermatology 2020:: 1–10. [DOI] [PubMed] [Google Scholar]
- 11.Tumbar T, Guasch G, Greco V et al. Defining the epithelial stem cell niche in skin. [Internet]. Science 2004: 303: 359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mesler AL, Veniaminova NA, Lull MV. et al. Hair Follicle Terminal Differentiation Is Orchestrated by Distinct Early and Late Matrix Progenitors. Cell Rep 2017: 19: 809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang S, Chovatiya G, Tumbar T. Epigenetic control in skin development, homeostasis and injury repair [Internet]. Exp Dermatol 2019: 28: 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang YV, Cheong J, Ciapurin N et al. Distinct Self-Renewal and Differentiation Phases in the Niche of Infrequently Dividing Hair Follicle Stem Cells [Internet]. Cell Stem Cell 2009: 5: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waghmare SK, Bansal R, Lee J et al. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. [Internet]. EMBO J 2008: 27: 1309–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Tumbar T. Hairy tale of signaling in hair follicle development and cycling. Semin Cell Dev Biol 2012: 23: 906–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang AB, Jain P, Tumbar T. The Hair Follicle Stem Cell Niche: The Bulge and Its Environment [Internet]. Springer, Cham, 2015: 1–26. [Google Scholar]
- 18.Morgan BA The dermal papilla: An instructive niche for epithelial stem and progenitor cells in development and regeneration of the hair follicle [Internet]. Cold Spring Harb Perspect Med 2014: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clevers H, Loh KM, Nusse R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control [Internet]. Science (80-) 2014: 346 [DOI] [PubMed] [Google Scholar]
- 20.Chen CL, Huang WY, Wang EHC et al. Functional complexity of hair follicle stem cell niche and therapeutic targeting of niche dysfunction for hair regeneration [Internet]. J Biomed Sci 2020: 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botchkarev VA, Mardaryev AN. Repressing the Keratinocyte Genome: How the Polycomb Complex Subunits Operate in Concert to Control Skin and Hair Follicle Development. J Invest Dermatol 2016: 136: 1538–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi R Concise Review: Mechanisms of Quiescent Hair Follicle Stem Cell Regulation [Internet]. Stem Cells 2017: 35: 2323–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plikus MV, Mayer JA, De La Cruz D et al. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration [Internet]. Nature 2008: 451: 340–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito M, Liu Y, Yang Z et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis [Internet]. Nat Med 2005: 11: 1351–1354. [DOI] [PubMed] [Google Scholar]
- 25.Van Velthoven C T J, Rando TA. Cell Stem Cell Review Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling [Internet]. Stem Cell 2019: 24: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, Kang S, Lilja KC et al. Signalling couples hair follicle stem cell quiescence with reduced histone H3 K4/K9/K27me3 for proper tissue homeostasis. [Internet]. Nat Commun 2016: 7: 11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myung PS, Takeo M, Ito M et al. Epithelial wnt ligand secretion is required for adult hair follicle growth and regeneration. J Invest Dermatol 2013: 133: 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang AB, Zhang YV, Tumbar T. Gata6 promotes hair follicle progenitor cell renewal by genome maintenance during proliferation [Internet]. EMBO J 2017: 36: 61–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matys V, Fricke E, Geffers R et al. TRANSFAC 1 : transcriptional regulation, from patterns to profiles [Internet]. [DOI] [PMC free article] [PubMed]
- 30.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006: 126: 663–676. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Huang J, Chen T et al. Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells. Cell Res 2008: 18: 1177–1189. [DOI] [PubMed] [Google Scholar]
- 32.Bielefeld KA, Amini-Nik S, Alman BA. Cutaneous wound healing: Recruiting developmental pathways for regeneration [Internet]. Cell Mol Life Sci 2013: 70: 2059–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito M, Yang Z, Andl T et al. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 2007: 447: 316–320. [DOI] [PubMed] [Google Scholar]
- 34.Lim CH, Sun Q, Ratti K et al. Hedgehog stimulates hair follicle neogenesis by creating inductive dermis during murine skin wound healing. Nat Commun 2018: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wier EM, Garza LA. Through the lens of hair follicle neogenesis, a new focus on mechanisms of skin regeneration after wounding [Internet]. Semin Cell Dev Biol 2019: 2020 April: 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akilli Öztürk Ö, Pakula H, Chmielowiec J et al. Gab1 and Mapk Signaling Are Essential in the Hair Cycle and Hair Follicle Stem Cell Quiescence [Internet]. Cell Rep 2015: 13: 561–572. [DOI] [PubMed] [Google Scholar]
- 37.Sarate RM, Chovatiya GL, Ravi V et al. sPLA2 -IIA Overexpression in Mice Epidermis Depletes Hair Follicle Stem Cells and Induces Differentiation Mediated Through Enhanced JNK/c-Jun Activation [Internet]. Stem Cells 2016: 34: 2407–2417. [DOI] [PubMed] [Google Scholar]
- 38.Rishikaysh P, Dev K, Diaz D et al. Signaling involved in hair follicle morphogenesis and development [Internet]. Int J Mol Sci 2014: 15: 1647–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Botchkarev VA Bone morphogenetic proteins and their antagonists in skin and hair follicle biology. J Invest Dermatol 2003: 120: 36–47. [DOI] [PubMed] [Google Scholar]
- 40.Veltri A, Lang C, Lien W-H. Concise Review: Wnt Signaling Pathways in Skin Development and Epidermal Stem Cells [Internet]. Stem Cells 2018: 36: 22–35. [DOI] [PubMed] [Google Scholar]
- 41.So W-K, Cheung TH. Molecular Regulation of Cellular Quiescence: A Perspective from Adult Stem Cells and Its Niches [Internet]. Humana Press, New York, NY, 2018: 1–25. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, He XC, Tong W-G et al. Bone Morphogenetic Protein Signaling Inhibits Hair Follicle Anagen Induction by Restricting Epithelial Stem/Progenitor Cell Activation and Expansion [Internet]. Stem Cells 2006: 24: 2826–2839. [DOI] [PubMed] [Google Scholar]
- 43.Kobielak K, Stokes N, De La Cruz J et al. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling [Internet]. Proc Natl Acad Sci U S A 2007: 104: 10063–10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi YS, Zhang Y, Xu M et al. Distinct functions for Wnt/β-Catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis [Internet]. Cell Stem Cell 2013: 13: 720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Hu W, Ma K et al. Are hair follicle stem cells promising candidates for wound healing? [Internet]. Expert Opin Biol Ther 2019: 19: 119–128. [DOI] [PubMed] [Google Scholar]
- 46.Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 2012: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lien WH, Polak L, Lin M et al. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat Cell Biol 2014: 16: 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merrill BJ, Gat U, DasGupta R et al. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev 2001: 15: 1688–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niemann C, Owens DM, Hülsken J et al. Expression of ΔNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development 2002: 129: 95–109. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Yu J, Shi C et al. Lef1 contributes to the differentiation of bulge stem cells by nuclear translocation and cross-talk with the notch signaling pathway [Internet]. Int J Med Sci 2013: 10: 738–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou P, Byrne C, Jacobs J et al. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate [Internet]. Genes Dev 1995: 9: 700–713. [DOI] [PubMed] [Google Scholar]
- 52.Adam RC, Yang H, Rockowitz S et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. [Internet]. Nature 2015: 521: 366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adam RC, Yang H, Ge Y et al. Temporal Layering of Signaling Effectors Drives Chromatin Remodeling during Hair Follicle Stem Cell Lineage Progression. Cell Stem Cell 2018: 22: 398–413.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leung C, Hui Tan S, Barker N. Recent Advances in Lgr5 + Stem Cell Research General Principles of Homeostatic Stem Cell Biology Gleaned from Lgr5 + ISCs [Internet]. Trends Cell Biol 2018: 28: 380–391. [DOI] [PubMed] [Google Scholar]
- 55.de Lau W, Peng WC, Gros P et al. The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength. Genes Dev 2014: 28: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaks V, Barker N, Kasper M et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet 2008: 40: 1291–1299. [DOI] [PubMed] [Google Scholar]
- 57.Hoeck JD, Biehs B, Kurtova AV. et al. Stem cell plasticity enables hair regeneration following Lgr5 + cell loss [Internet]. Nat Cell Biol 2017: 19: 666–676. [DOI] [PubMed] [Google Scholar]
- 58.Ren X, Xia W, Xu P et al. Lgr4 Deletion Delays the Hair Cycle and Inhibits the Activation of Hair Follicle Stem Cells [Internet]. J Invest Dermatol 2020: 140: 1706–1712.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joost S, Jacob T, Sun X et al. Single-Cell Transcriptomics of Traced Epidermal and Hair Follicle Stem Cells Reveals Rapid Adaptations during Wound Healing [Internet]. Cell Rep 2018: 25: 585–597.e7. [DOI] [PubMed] [Google Scholar]
- 60.Kang S, Long K, Wang S et al. Histone H3 K4/9/27 Trimethylation Levels Affect Wound Healing and Stem Cell Dynamics in Adult Skin. Stem Cell Reports 2020: 14: 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lim X, Tan SH, Lou Yu K et al. Axin2 marks quiescent hair follicle bulge stem cells that are maintained by autocrine Wnt/β-catenin signaling. Proc Natl Acad Sci U S A 2016: 113: E1498–E1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoonderwoerd MJA, Goumans MJTH, Hawinkels LJAC. Endoglin: Beyond the endothelium [Internet]. Biomolecules 2020: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baik J, Magli A, Tahara N et al. Endoglin integrates BMP and Wnt signalling to induce haematopoiesis through JDP2 [Internet]. Nat Commun 2016: 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Calvo-Sánchez MI, Fernández-Martos S, Carrasco E et al. A role for the Tgf-β/Bmp co-receptor Endoglin in the molecular oscillator that regulates the hair follicle cycle. J Mol Cell Biol 2019: 11: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calvo-Sánchez MI, Fernández-Martos S, Carrasco E et al. A role for the Tgf- β/Bmp co-receptor Endoglin in the molecular oscillator that regulates the hair follicle cycle [Internet]. J Mol Cell Biol 2018: 11: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Armiñán A, Gandía C, García-Verdugo JM et al. Cardiac transcription factors driven lineage-specification of adult stem cells. J Cardiovasc Transl Res 2010: 3: 61–65. [DOI] [PubMed] [Google Scholar]
- 67.Arnold K, Sarkar A, Yram MA et al. Sox2 + adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 2011: 9: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles [Internet]. Science (80-) 2006: 312: 1946–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Folgueras AR, Guo X, Pasolli HA et al. Architectural niche organization by LHX2 is linked to hair follicle stem cell function [Internet]. Cell Stem Cell 2013: 13: 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mardaryev AN, Meier N, Poterlowicz K et al. Lhx2 differentially regulates Sox9, Tcf4 and Lgr5 in hair follicle stem cells to promote epidermal regeneration after injury. Development 2011: 138: 4843–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kadaja M, Keyes BE, Lin M et al. SOX9: A stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev 2014: 28: 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horsley V, Aliprantis AO, Polak L et al. NFATc1 Balances Quiescence and Proliferation of Skin Stem Cells. Cell 2008: 132: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhattacharya S, Wheeler H, Leid M et al. Transcription Factor CTIP2 Maintains Hair Follicle Stem Cell Pool and Contributes to Altered Expression of LHX2 and NFATC1 [Internet]. J Invest Dermatol 2015: 135: 2593–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leishman E, Howard JM, Garcia GE et al. Foxp1 maintains hair follicle stem cell quiescence through regulation of Fgf18 [Internet]. Dev 2013: 140: 3809–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Romano RA, Smalley K, Liu S et al. Abnormal hair follicle development and altered cell fate of follicular keratinocytes in transgenic mice expressing DeltaNp63alpha. [Internet]. Development 2010: 137: 1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lay K, Kume T, Fuchs E. FOXC1 maintains the hair follicle stem cell niche and governs stem cell quiescence to preserve long-term tissue-regenerating potential. Proc Natl Acad Sci U S A 2016: 113: E1506–E1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, Siegenthaler JA, Dowell RD et al. Stem cells: Foxc1 reinforces quiescence in self-renewing hair follicle stem cells. Science (80-) 2016: 351: 613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haensel D, Sun P, MacLean AL et al. An Ovol2-Zeb1 transcriptional circuit regulates epithelial directional migration and proliferation [Internet]. EMBO Rep 2019: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matuzelski E, Bunt J, Harkins D et al. Transcriptional regulation of Nfix by NFIB drives astrocytic maturation within the developing spinal cord [Internet]. Dev Biol 2017: 432: 286–297. [DOI] [PubMed] [Google Scholar]
- 80.Fraser J, Essebier A, Brown AS et al. Common Regulatory Targets of NFIA, NFIX and NFIB during Postnatal Cerebellar Development [Internet]. Cerebellum 2020: 19: 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adam RC, Yang H, Ge Y et al. NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices [Internet]. Nat Cell Biol 2020: 22: 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ge Y, Gomez NC, Adam RC et al. Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. [Internet]. Cell 2017: 169: 636–650.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang CY, Pasolli HA, Giannopoulou EG et al. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche [Internet]. Nature 2013: 495: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bürger M, Chory J. Structural and chemical biology of deacetylases for carbohydrates, proteins, small molecules and histones [Internet]. Commun Biol 2018: 1: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hentze MW, Castello A, Schwarzl T et al. A brave new world of RNA-binding proteins [Internet]. Nat Rev Mol Cell Biol 2018: 19: 327–341. [DOI] [PubMed] [Google Scholar]
- 86.Butler MT, Wallingford JB. Planar cell polarity in development and disease [Internet]. Nat Rev Mol Cell Biol 2017: 18: 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blank MF, Grummt I. The seven faces of SIRT7 [Internet]. Transcription 2017: 8: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li G, Tang X, Zhang S et al. SIRT7 activates quiescent hair follicle stem cells to ensure hair growth in mice [Internet]. EMBO J 2020: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Minuesa G, Albanese SK, Xie W et al. Small-molecule targeting of MUSASHI RNA-binding activity in acute myeloid leukemia [Internet]. Nat Commun 2019: 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sutherland JM, McLaughlin EA, Hime GR et al. The Musashi Family of RNA Binding Proteins: Master Regulators of Multiple Stem Cell Populations [Internet]. Springer, Dordrecht, 2013: 233–245. [DOI] [PubMed] [Google Scholar]
- 91.Ma X, Tian Y, Song Y et al. Msi2 Maintains Quiescent State of Hair Follicle Stem Cells by Directly Repressing the Hh Signaling Pathway [Internet]. J Invest Dermatol 2017: 137: 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goldstein B, Macara IG. The PAR Proteins: Fundamental Players in Animal Cell Polarization [Internet]. Dev Cell 2007: 13: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Noguchi N, Hirose T, Suzuki T et al. Atypical protein kinase C isoforms differentially regulate directional keratinocyte migration during wound healing. J Dermatol Sci 2019: 93: 101–108. [DOI] [PubMed] [Google Scholar]
- 94.Niessen MT, Scott J, Zielinski JG et al. Apkcγ controls epidermal homeostasis and stemcell fate through regulation of division orientation [Internet]. J Cell Biol 2013: 202: 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Osada SI, Minematsu N, Oda F et al. Atypical Protein Kinase C Isoform, aPKCλ, Is Essential for Maintaining Hair Follicle Stem Cell Quiescence [Internet]. J Invest Dermatol 2015: 135: 2584–2592. [DOI] [PubMed] [Google Scholar]
- 96.Liakath-Ali K, Vancollie VE, Lelliott CJ et al. Alkaline ceramidase 1 is essential for mammalian skin homeostasis and regulating whole-body energy expenditure [Internet]. J Pathol 2016: 239: 374–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Panteleyev AA Functional anatomy of the hair follicle: The Secondary Hair Germ [Internet]. Exp Dermatol 2018: 27: 701–720. [DOI] [PubMed] [Google Scholar]
- 98.Hsu YC, Li L, Fuchs E. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration [Internet]. Cell 2014: 157: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Drögemüller C, Karlsson EK, Hytönen MK et al. A mutation in hairless dogs implicates FOXI3 in ectodermal development. Science (80-) 2008: 321: 1462. [DOI] [PubMed] [Google Scholar]
- 100.Shirokova V, Biggs LC, Jussila M et al. Foxi3 Deficiency Compromises Hair Follicle Stem Cell Specification and Activation [Internet]. Stem Cells 2016: 34: 1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Osorio KM, Lee SE, McDermitt DJ et al. Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development 2008: 135: 1059–1068. [DOI] [PubMed] [Google Scholar]
- 102.Lee J, Hoi CSL, Lilja KC et al. Runx1 and p21 synergistically limit the extent of hair follicle stem cell quiescence in vivo. Proc Natl Acad Sci U S A 2013: 110: 4634–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Osorio KM, Lilja KC, Tumbar T. Runx1 modulates adult hair follicle stem cell emergence and maintenance from distinct embryonic skin compartments. J Cell Biol 2011: 193: 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Scheitz CJF, Lee TS, McDermitt DJ et al. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J 2012: 31: 4124–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jain P, Nattakom M, Holowka D et al. Runx1 Role in Epithelial and Cancer Cell Proliferation Implicates Lipid Metabolism and Scd1 and Soat1 Activity [Internet]. Stem Cells 2018: 36: 1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li KN, Jain P, He CH et al. Skin vasculature and hair follicle cross-talking associated with stem cell activation and tissue homeostasis. Elife 2019: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu L, Zhao L, Yuan F et al. GATA6 loss-of-function mutations contribute to familial dilated cardiomyopathy [Internet]. Int J Mol Med 2014: 34: 1315–1322. [DOI] [PubMed] [Google Scholar]
- 108.Krieger K, Millar SE, Mikuda N et al. NF-κB Participates in Mouse Hair Cycle Control and Plays Distinct Roles in the Various Pelage Hair Follicle Types. J Invest Dermatol 2018: 138: 256–264. [DOI] [PubMed] [Google Scholar]
- 109.Liu ZH, Dai XM, Du B. Hes1: A key role in stemness, metastasis and multidrug resistance [Internet]. Cancer Biol Ther 2015: 16: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goto N, Ueo T, Fukuda A et al. Distinct roles of HES1 in normal stem cells and tumor stem-like cells of the intestine [Internet]. Cancer Res 2017: 77: 3442–3454. [DOI] [PubMed] [Google Scholar]
- 111.Suen WJ, Li ST, Yang LT. Hes1 regulates anagen initiation and hair follicle regeneration through modulation of hedgehog signaling [Internet]. Stem Cells 2020: 38: 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells [Internet]. Nat Rev Cancer 2008: 8: 523–534. [DOI] [PubMed] [Google Scholar]
- 113.Veselá B, Švandová E, Šmarda J et al. Mybs in mouse hair follicle development. Tissue Cell 2014: 46: 352–355. [DOI] [PubMed] [Google Scholar]
- 114.Hu Y, Song Z, Chen J et al. Overexpression of MYB in the Skin Induces Alopecia and Epidermal Hyperplasia. J Invest Dermatol 2019: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Doane AS, Elemento O. Regulatory elements in molecular networks [Internet]. Wiley Interdiscip Rev Syst Biol Med 2017: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Whyte WA, Orlando DA, Hnisz D et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes [Internet]. Cell 2013: 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pott S, Lieb JD. What are super-enhancers? [Internet]. Nat Genet 2015: 47: 8–12. [DOI] [PubMed] [Google Scholar]
- 118.Kretzschmar K, Watt FM. Lineage Tracing [Internet]. Cell 2012: 148: 33–45. [DOI] [PubMed] [Google Scholar]
- 119.Romagnani P, Rinkevich Y, Dekel B. The use of lineage tracing to study kidney injury and regeneration [Internet]. Nat Rev Nephrol 2015: 11: 420–431. [DOI] [PubMed] [Google Scholar]
- 120.Fan H, Liu X, Shen Y et al. In Vivo Genetic Strategies for the Specific Lineage Tracing of Stem Cells [Internet]. Curr Stem Cell Res Ther 2019: 14: 230–238. [DOI] [PubMed] [Google Scholar]
- 121.Rabbani P, Takeo M, Chou W et al. Coordinated activation of wnt in epithelial and melanocyte stem cells initiates pigmented hair regeneration [Internet]. Cell 2011: 145: 941–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Genander M, Cook PJ, Ramsköld D et al. BMP signaling and its pSMADS1/5 target genes differentially regulate hair follicle stem cell lineages [Internet]. Cell Stem Cell 2014: 15: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vidal VPI, Chaboissier MC, Lützkendorf S et al. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol 2005: 15: 1340–1351. [DOI] [PubMed] [Google Scholar]
- 124.Nowak JA, Polak L, Pasolli HA et al. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 2008: 3: 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hawkins RD, Hon GC, Lee LK et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 2010: 6: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wysocka J, Swigut T, Xiao H et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006: 442: 86–90. [DOI] [PubMed] [Google Scholar]
- 127.Efroni S, Duttagupta R, Cheng J et al. Global Transcription in Pluripotent Embryonic Stem Cells [Internet]. Cell Stem Cell 2008: 2: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu J, Magri L, Zhang F et al. Chromatin landscape defined by repressive histone methylation during oligodendrocyte differentiation [Internet]. J Neurosci 2015: 35: 352–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lien WH, Guo X, Polak L et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 2011: 9: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee J, Tumbar T. Linking chromatin dynamics, cell fate plasticity, and tissue homeostasis in adult mouse hair follicle stem cells [Internet]. Mol Life 2017: 1: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schuler N, Timm S, Rübe CE. Hair Follicle Stem Cell Faith Is Dependent on Chromatin Remodeling Capacity Following Low-Dose Radiation [Internet]. Stem Cells 2018: 36: 574–588. [DOI] [PubMed] [Google Scholar]
- 132.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011: 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Miroshnikova YA, Cohen I, Ezhkova E et al. Epigenetic gene regulation, chromatin structure, and force-induced chromatin remodelling in epidermal development and homeostasis [Internet]. Curr Opin Genet Dev 2019: 55: 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pivetti S, Fernandez-Perez D, D’Ambrosio A et al. Loss of PRC1 activity in different stem cell compartments activates a common transcriptional program with cell type-dependent outcomes. Sci Adv 2019: 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Botchkareva NV The Molecular Revolution in Cutaneous Biology: Noncoding RNAs: New Molecular Players in Dermatology and Cutaneous Biology [Internet]. J Invest Dermatol 2017: 137: e105–e111. [DOI] [PubMed] [Google Scholar]
- 136.O’Brien J, Hayder H, Zayed Y et al. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018: 9: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics, Proteomics Bioinforma 2017: 15: 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang Y, Wang F, Wang R et al. 2A self-cleaving peptide-based multi-gene expression system in the silkworm Bombyx mori [Internet]. Sci Rep 2015: 5: 16273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ebert MS, Sharp PA. Roles for MicroRNAs in conferring robustness to biological processes. Cell 2012: 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Salviano-Silva A, Lobo-Alves SC, Almeida R C de et al. Besides pathology: Long non-coding RNA in cell and tissue homeostasis [Internet]. Non-coding RNA 2018: 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Amelio I, Lena AM, Bonanno E et al. miR-24 affects hair follicle morphogenesis targeting Tcf-3 [Internet]. Cell Death Dis 2013: 4: e922–e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang D, Zhang Z, O’Loughlin E et al. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway [Internet]. Nat Cell Biol 2013: 15: 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang L, Stokes N, Polak L et al. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment [Internet]. Cell Stem Cell 2011: 8: 294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yuan S, Li F, Meng Q et al. Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by miR-22 [Internet]. PLOS Genet 2015: 11: e1005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang L, Ge Y, Fuchs E. miR-125b can enhance skin tumor initiation and promote malignant progression by repressing differentiation and prolonging cell survival [Internet]. Genes Dev 2014: 28: 2532–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ge M, Liu C, Li L et al. miR-29a/b1 Inhibits Hair Follicle Stem Cell Lineage Progression by Spatiotemporally Suppressing WNT and BMP Signaling. Cell Rep 2019: 29: 2489–2504.e4. [DOI] [PubMed] [Google Scholar]
- 147.Ahmed MI, Alam M, Emelianov VU et al. MicroRNA-214 controls skin and hair follicle development by modulating the activity of the Wnt pathway [Internet]. J Cell Biol 2014: 207: 549–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Du KT, Deng JQ, He XG et al. MiR-214 Regulates the Human Hair Follicle Stem Cell Proliferation and Differentiation by Targeting EZH2 and Wnt/β-Catenin Signaling Way In Vitro. Tissue Eng Regen Med 2018: 15: 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hoefert JE, Bjerke GA, Wang D et al. The microRNA-200 family coordinately regulates cell adhesion and proliferation in hair morphogenesis. J Cell Biol 2018: 217: 2185–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Song MS, Rossi JJ. Molecular mechanisms of Dicer: Endonuclease and enzymatic activity [Internet]. Biochem J 2017: 474: 1603–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vishlaghi N, Lisse TS. Dicer- and Bulge Stem Cell-Dependent MicroRNAs During Induced Anagen Hair Follicle Development [Internet]. Front Cell Dev Biol 2020: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Si Y, Bai J, Wu J et al. LncRNA PlncRNA-1 regulates proliferation and differentiation of hair follicle stem cells through TGF-β-mediated Wnt/β-catenin signal pathway. Mol Med Rep 2018: 17: 1191–1197. [DOI] [PubMed] [Google Scholar]
- 153.Cai B, Wang X, Liu H et al. Up-regulated lncRNA5322 elevates MAPK1 to enhance proliferation of hair follicle stem cells as a ceRNA of microRNA-19b-3p. Cell Cycle 2019: 18: 1588–1600. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 154.Cai B, Zheng Y, Ma S et al. Long non-coding RNA regulates hair follicle stem cell proliferation and differentiation through PI3K/AKT signal pathway. Mol Med Rep 2018: 17: 5477–5483. [DOI] [PubMed] [Google Scholar]
- 155.Wang WT, Han C, Sun YM et al. Noncoding RNAs in cancer therapy resistance and targeted drug development [Internet]. J Hematol Oncol 2019: 12: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ly CH, Lynch GS, Ryall JG. A Metabolic Roadmap for Somatic Stem Cell Fate. Cell Metab 2020: 31: 1052–1067. [DOI] [PubMed] [Google Scholar]
- 157.Flores A, Schell J, Krall AS et al. Lactate dehydrogenase activity drives hair follicle stem cell activation [Internet]. Nat Cell Biol 2017: 19: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Reichenbach B, Classon J, Aida T et al. Glutamate transporter Slc1a3 mediates inter-niche stem cell activation during skin growth [Internet]. EMBO J 2018: 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sada A, Jacob F, Leung E et al. Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin [Internet]. Nat Cell Biol 2016: 18: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hamanaka RB, Glasauer A, Hoover P et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development [Internet]. Sci Signal 2013: 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhao J, Li H, Zhou R et al. Foxp1 Regulates the Proliferation of Hair Follicle Stem Cells in Response to Oxidative Stress during Hair Cycling [Internet]. PLoS One 2015: 10: e0131674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Carrasco E, Calvo MI, Blázquez-Castro A et al. Photoactivation of ROS Production in Situ Transiently Activates Cell Proliferation in Mouse Skin and in the Hair Follicle Stem Cell Niche Promoting Hair Growth and Wound Healing [Internet]. J Invest Dermatol 2015: 135: 2611–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Calvo-Sánchez MI, Fernández-Martos S, Montoya JJ et al. Intrinsic activation of cell growth and differentiation in ex vivo cultured human hair follicles by a transient endogenous production of ROS [Internet]. Sci Rep 2019: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tang Y, Luo B, Deng Z et al. Mitochondrial aerobic respiration is activated during hair follicle stem cell differentiation, and its dysfunction retards hair regeneration [Internet]. PeerJ 2016: 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Peters F, Vorhagen S, Brodesser S et al. Ceramide synthase 4 regulates stem cell homeostasis and hair follicle cycling [Internet]. J Invest Dermatol 2015: 135: 1501–1509. [DOI] [PubMed] [Google Scholar]
- 166.Lin CL, Xu R, Yi JK et al. Alkaline Ceramidase 1 Protects Mice from Premature Hair Loss by Maintaining the Homeostasis of Hair Follicle Stem Cells. Stem Cell Reports 2017: 9: 1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gur-Cohen S, Yang H, Baksh SC et al. Stem cell-driven lymphatic remodeling coordinates tissue regeneration [Internet]. Science (80-) 2019: 366: 1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Peña-Jimenez D, Fontenete S, Megias D et al. Lymphatic vessels interact dynamically with the hair follicle stem cell niche during skin regeneration in vivo. EMBO J 2019: 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cheng CC, Tsutsui K, Taguchi T et al. Hair follicle epidermal stem cells define a niche for tactile sensation. Elife 2018: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rutlin M, Ho CY, Abraira VE et al. The cellular and molecular basis of direction selectivity of Aδ-LTMRs [Internet]. Cell 2014: 159: 1640–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shwartz Y, Gonzalez-Celeiro M, Chen CL et al. Cell Types Promoting Goosebumps Form a Niche to Regulate Hair Follicle Stem Cells. Cell 2020: 182: 578–593.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fujiwara H, Ferreira M, Donati G et al. The basement membrane of hair follicle stem cells is a muscle cell niche [Internet]. Cell 2011: 144: 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guo H, Lei M, Li Y et al. Paracrine Secreted Frizzled-Related Protein 4 Inhibits Melanocytes Differentiation in Hair Follicle [Internet]. Stem Cells Int 2017: 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yamada T, Hasegawa S, Hasebe Y et al. CXCL12 regulates differentiation of human immature melanocyte precursors as well as their migration [Internet]. Arch Dermatol Res 2019: 311: 55–62. [DOI] [PubMed] [Google Scholar]
- 175.Tanimura S, Tadokoro Y, Inomata K et al. Hair follicle stem cells provide a functional niche for melanocyte stem cells [Internet]. Cell Stem Cell 2011: 8: 177–187. [DOI] [PubMed] [Google Scholar]
- 176.Lu Z, Xie Y, Huang H et al. Hair follicle stem cells regulate retinoid metabolism to maintain the self-renewal niche for melanocyte stem cells. Elife 2020: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zwick RK, Guerrero-Juarez CF, Horsley V et al. Anatomical, Physiological, and Functional Diversity of Adipose Tissue. Cell Metab 2018: 27: 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Heitman N, Sennett R, Mok KW et al. Dermal sheath contraction powers stem cell niche relocation during hair cycle regression. Science (80-) 2020: 367: 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang AB, Zhang YV, Tumbar T. Gata6 promotes hair follicle progenitor cell renewal by genome maintenance during proliferation. EMBO J 2017: 36: 61–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shin J, Berg DA, Zhu Y et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 2015: 17: 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines [Internet]. Exp Mol Med 2018: 50: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Weinreb C, Rodriguez-Fraticelli A, Camargo FD et al. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science (80-) 2020: 367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jensen KB, Watt FM. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regular of stem cell quiescence [Internet]. Proc Natl Acad Sci U S A 2006: 103: 11958–11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Joost S, Zeisel A, Jacob T et al. Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity [Internet]. Cell Syst 2016: 3: 221–237.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Joost S, Annusver K, Jacob T et al. The Molecular Anatomy of Mouse Skin during Hair Growth and Rest [Internet]. Cell Stem Cell 2020: 26: 441–457.e7. [DOI] [PubMed] [Google Scholar]
- 186.Yang H, Adam RC, Ge Y et al. Epithelial-Mesenchymal Micro-niches Govern Stem Cell Lineage Choices [Internet]. Cell 2017: 169: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gupta K, Levinsohn J, Linderman G et al. Single-Cell Analysis Reveals a Hair Follicle Dermal Niche Molecular Differentiation Trajectory that Begins Prior to Morphogenesis. Dev Cell 2019: 48: 17–31.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ge Y, Miao Y, Gur-Cohen S et al. The aging skin microenvironment dictates stem cell behavior [Internet]. Proc Natl Acad Sci U S A 2020: 117: 5339–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mesa KR, Rompolas P, Zito G et al. Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature 2015: 522: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rompolas P, Deschene ER, Zito G et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration [Internet]. Nature 2012: 487: 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate [Internet]. Nature 2013: 502: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xin T, Gonzalez D, Rompolas P et al. Flexible fate determination ensures robust differentiation in the hair follicle [Internet]. Nat Cell Biol 2018: 20: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Takeda N, Jain R, LeBoeuf MR et al. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to k6+ niche cells [Internet]. Dev 2013: 140: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Legué E, Nicolas JF. Hair follicle renewal: Organization of stem cells in the matrix and the role of stereotyped lineages and behaviors [Internet]. Development 2005: 132: 4143–4154. [DOI] [PubMed] [Google Scholar]
- 195.Legué E, Sequeira I, Nicolas JF. Hair follicle renewal: Authentic morphogenesis that depends on a complex progression of stem cell lineages [Internet]. Development 2010: 137: 569–577. [DOI] [PubMed] [Google Scholar]
- 196.Sequeira I, Nicolas JF. Redefining the structure of the hair follicle by 3D clonal analysis [Internet]. Dev 2012: 139: 3741–3751. [DOI] [PubMed] [Google Scholar]
- 197.Nicetto D, Zaret KS. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr Opin Genet Dev 2019: 55: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fiers MWEJ, Minnoye L, Aibar S et al. Mapping gene regulatory networks from single-cell omics data [Internet]. Brief Funct Genomics 2018: 17: 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fan X, Wang D, Burgmaier JE et al. Single Cell and Open Chromatin Analysis Reveals Molecular Origin of Epidermal Cells of the Skin [Internet]. Dev Cell 2018: 47: 21–37.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ma S, Zhang B, LaFave LM et al. Chromatin Potential Identified by Shared Single-Cell Profiling of RNA and Chromatin [Internet]. Cell 2020: 0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wissink EM, Vihervaara A, Tippens ND et al. Nascent RNA analyses: tracking transcription and its regulation. Nat Rev Genet 2019: 20: 705–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Blanco S, Bandiera R, Popis M et al. Stem cell function and stress response are controlled by protein synthesis [Internet]. Nature 2016: 534: 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tarazona OA, Pourquié O. Exploring the Influence of Cell Metabolism on Cell Fate through Protein Post-translational Modifications [Internet]. Dev Cell 2020: 54: 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]


