Abstract
Introduction
Apolipoprotein E (APOE) alleles are associated with cognitive decline, mild cognitive impairment (MCI), and Alzheimer's disease in Whites, but have weaker and inconsistent effects reported in Latinos. We hypothesized that this heterogeneity is due to ancestry‐specific genetic effects.
Methods
We investigated the associations of the APOE alleles with significant cognitive decline and MCI in 4183 Latinos, stratified by six Latino backgrounds, and explored whether the proportion of continental genetic ancestry (European, African, and Amerindian) modifies these associations.
Results
APOE ε4 was associated with an increased risk of significant cognitive decline (odds ratio [OR] = 1.15, P‐value = 0.03), with the strongest association in Cubans (OR = 1.46, P‐value = 0.007). APOE‐ε2 was associated with decreased risk of MCI (OR = 0.37, P‐value = 0.04) in Puerto Ricans. Amerindian genetic ancestry was found to protect from the risk conferred by APOE ε4 on significant cognitive decline.
Discussion
Results suggest that APOE alleles' effects on cognitive outcomes differ across six Latino backgrounds and are modified by continental genetic ancestry.
Keywords: admixture, Alzheimer's disease, ancestry, apolipoprotein E, cognitive decline, genetic epidemiology, Hispanics/Latinos, mild cognitive impairment
1. BACKGROUND
Cognitive decline, mild cognitive impairment (MCI), and Alzheimer's disease and related dementias (ADRD) are a growing worldwide epidemic and one of the leading causes of death in the elderly population. 1 MCI is a prodromal cognitive impairment state preceding the more serious cognitive dysfunction characteristic of dementia. It can involve problems with memory, language, thinking, and judgment that are greater than normal age‐related changes. 2 , 3 Cognitive decline is a normal process of aging; however, in ADRD patients it begins many years before dementia is diagnosed and accelerates during the course of the disease. 3 Self‐reported cognitive decline has lately been introduced to the field, to extend ADRD risk diagnosis to an earlier stage before MCI. 4 Hispanics/Latinos (Latinos henceforth) are the fastest‐growing ethnic group in the United States, 5 and suffer from higher rates of ADRD, compared to Whites. 6 , 7
The apoplipoprotein E (APOE) ε4 allele is the strongest known genetic risk factor for ADRD, 8 and it has also been linked to more rapid cognitive aging, such as increased cognitive decline and MCI. 9 , 10 , 11 The APOE gene exists as three polymorphic allelesε2, ε3, and ε4which are determined by two single nucleotide polymorphisms (SNPs: rs429358 and rs7412), that substitute amino acids in the protein, resulting in functional changes. In general, APOE ε4 confers increased risk for cognitive decline, MCI, and ADRD compared to the more common APOE ε3, whereas the APOE ε2 is considered neuroprotective. 12 However, most of these findings are based on studies of individuals of European ancestry. Several population‐based studies have shown ancestry heterogeneity of the APOE ε4ADRD association. 13 , 14 , 15 By comparing haplotype ε4/ε4 to haplotype ε3/ε3, Farrer et al. 8 reported strong effects in Japanese (odds ratio [OR] = 33.1), and Whites (OR = 12.5), and weaker effects among African Americans (OR = 5.7) and Latinos (OR = 2.2). Other studies investigating the association of APOE ε4 with MCI and dementia in Latinos have produced inconsistent results. 16 , 17 , 18 , 19 , 20 A study of Mexican Americans indicated that haplotype ε4/ε4, compared to haplotype ε3/ε3, was associated with lower cognitive scores and higher dementia, though not significant (risk ratio [RR] = 2.04, confidence interval [CI] = 0.88–4.72). 16 Similarly, two other studies suggested that the APOE ε4 allele is both less common and confers less risk for MCI or ADRD in Mexican Latinos compared to Whites. 17 , 18 However, another study in Caribbean Latinos (ie, Dominicans and Puerto Ricans) showed that late‐onset familial Alzheimer's disease (AD) is strongly associated with APOE ε4, with the APOE ε4 allele more likely to be transmitted among affected individuals than unaffected relatives. 19 These inconsistent results may be due, in part, to small sample sizes, different study designs and samples, and definitions of cognitive outcome, 20 as well as to the heterogeneity of the Latino groups in which the studies were conducted.
The Hispanic Community Health Study/Study of Latinos (HCHS/SOL) is a population‐based longitudinal cohort study of 16,415 U.S. Hispanic/Latino adults that enrolled participants from Cuban, Central American, Dominican, Mexican, Puerto Rican, and South American backgrounds. 21 , 22 Previous characterization of the genetic diversity in the HCHS/SOL cohort has shown that Latino individuals have admixed genomes consisting of three predominant continental ancestries: Amerindian, European, and African with varying proportions among and within each background group. 23 Furthermore, it was shown that the APOE alleles have different distributions among the six Latino background groups, 20 consistent with different APOE allele frequencies among the ancestral populations.
Previous association analysis of the APOE alleles with MCI in the HCHS/SOL ancillary Study of Latinos‐Investigation of Neurocognitive Aging (SOL‐INCA) did not detect significant associations. 24 We hypothesized that there are differential association effects of the APOE alleles in the six Latino background groups on significant cognitive decline and MCI, which could potentially explain the inconsistent reported literature results of the association of APOE alleles with cognitive function, MCI, and dementia in Latinos. To address this, we tested the association of APOE alleles with significant cognitive decline and MCI and stratified the analyses by the six Latino background groups. Next, we hypothesized that differences in proportions of continental ancestries in the six Latino background groups will explain the heterogeneous APOE alleles' effects on significant cognitive decline and MCI. The purpose of this study was to determine whether genetic ancestry modifies the effect of APOE alleles on the significant cognitive decline and MCI by testing interaction effects between the three Latino continental ancestries and the APOE alleles.
2. METHODS
2.1. Study population
The HCHS/SOL is a multisite, prospective cohort study of diverse Latinos enrolled at four field centers (Bronx, New York; Chicago, Illinois; Miami, Florida; and San Diego, California). The sampling design details have been previously described. 21 , 22 A total of 16,415 self‐identified Hispanic/Latino adults, 18 to 74 years old, were enrolled at HCHS/SOL baseline visit 1 (2008–2011). Anthropometry, biospecimens, and health information about risk/protective factors were collected. The baseline cognitive battery included four tests: Six‐Item Screener (SIS; mental status); 25 Brief‐Spanish English Verbal Learning Test (B‐SEVLT; verbal episodic learning and memory); 26 Word Fluency; 27 and Digit Symbol Substitution test (DSS; processing speed, executive function). HCHS/SOL visit 2 occurred between 2014 and 2017 with an abbreviated protocol. SOL‐INCA is an HCHS/SOL ancillary study that occurred at visit 2, and included the oversampled middle‐aged and older participants (50 years and older) who were administered the visit 1 cognitive battery, plus additional complementing cognitive tests to determine self‐reported measures of cognitive decline (Everyday Cognition‐12 [E‐Cog12]) 28 and functional status (Instrumental Activities of Daily Living Scale [IADL]), among other cognitive tests. 29 Overall, 6377 participants were re‐examined in SOL‐INCA after an average of 7 years since their visit 1 cognitive assessment. Of the 6377 participants, 2127 were excluded from analyses (1688 did not consent for genetic data, 420 failed APOE genotyping, 67 had missing cognitive outcomes, and 19 had missing covariates), totaling an analytic sample of 4183 individuals (mean age = 62.1 years and 52.5% were women). All participants in this analysis signed informed consent in their preferred language (Spanish/English) to use their genetic and non‐genetic data. The study was reviewed and approved by the institutional review boards at all collaborating institutions.
RESEARCH IN CONTEXT
Systematic review: The authors reviewed the literature using traditional sources. Several publications have shown weak and inconsistent results for the association of the apolipoprotein E (APOE) alleles with cognitive decline, mild cognitive impairment (MCI), and Alzheimer's disease‐related dementias in Latinos, despite higher rates of these disorders compared to non‐Latino Whites. Publications are appropriately cited.
Interpretation: Our findings suggest a differential association between the APOE ε2 and APOE ε4 alleles and risk for significant cognitive decline and MCI in the six Latino groups defined by country of origin. We also found that Amerindian genetic ancestry protects from the risk conferred by APOE ε4 on significant cognitive decline. Inconsistent estimated associations in Latinos may be due to different admixture patterns of continental ancestry across Latino groups.
Future directions: Future studies are needed to (a) identify enriched Amerindian genetic variants interacting with the APOEε4 allele, and the mechanism of interactions and (b) develop genetic measures for predicting significant cognitive decline and MCI in admixed individuals, potentially taking into account proportion ancestry or specific genetic variants.
2.2. Cognitive outcomes
We analyzed two binary cognitive variables that were previously constructed 30 based on cognitive tests and self‐report: significant cognitive decline, and MCI. Cognitive decline measures the cognitive decline between HCHS/SOL visit 1 and the SOL‐INCA exam, based on a latent factor model taking into account cognitive test scores. Individuals were classified as meeting significant cognitive decline criteria if they had a change in global cognitive performance between the two exams that exceeded –0.055 standard deviation (SD) yearly. 30
Individuals were classified as MCI according to National Institute on Aging‐Alzheimer's Association if they fit the following three criteria: 31 (1) a cognitive test score below –1SD based on SOL‐INCA robust internal norms, (2) significant cognitive decline (described above), and (3) self‐reported cognitive decline based on the E‐Cog12. 28 Also, individuals that had both a cognitive deficit < –2SD in any neurocognitive test at the SOL‐INCA exam and more than minimal cognitive impairment in the IADL scale, were classified as MCI. Additional information about the SOL‐INCA cognitive assessment approach is provided in detail in Appendix 1 of a previous publication. 30
2.3. Genetic data
Genotyping, quality control, and continental ancestry inference were previously described. 23 , 32 In brief, genotyping was performed using Illumina custom array and genome‐wide imputation was conducted with the 1000 Genome Project reference panel. 32 Principal components (PCs) were estimated using PC‐Relate 33 and continental‐ancestry proportions were calculated using ADMIXTURE software. 23 , 34 “Genetic analysis groups” were constructed based on a combination of self‐identified Hispanic/Latino background and genetic similarity, and are classified as Cuban, Dominican, and Puerto Rican (Caribbean groups); and Mexican, Central American, and South American (Mainland groups). 23 APOE genotyping was performed using commercial TaqMan assays previously described. 20 APOE variants were in Hardy‐Weinberg equilibrium (P > 0.05).
2.4. Statistical analysis
We provided descriptive statistics to characterize the demographic and cognitive outcomes and APOE allele distributions in the full analytic dataset and by background groups. We tested the associations between APOE ε2 and ε4 alleles with cognitive outcomes in the same model (APOE ε3 used as the reference allele) using a complex survey design from the R “survey” package, 35 with a “quasipoisson” family for binary traits. This method accounts for the stratification, clustering, and probability weighting in HCHS/SOL to allow correct generalizations to the target population of U.S. Latinos. Models were adjusted for age, sex, education, center, first five genetic PCs, and “genetic analysis groups.” We tested both an additive and a dominant inheritance mode of the APOE alleles. For a given APOE allele, additive inheritance mode counts the number of the alleles for each individual (0, 1, or 2); dominant mode 1 if an individual had at least one allele, 0 otherwise. Carriers of both alleles ie, APOE ε2/ε4 were included in the model by giving a value 1 for the APOE ε2 variable and a value of 1 for the APOE ε4 variable (for both modes of inheritance).
Further association analyses of APOE ε4 and ε2 alleles with cognitive outcomes were done separately for each of the six genetic analysis groups and the significance of the results was evaluated through 10,000 permutations for each group, to protect from potential high type 1 error due to the low proportion of APOE variants. Effect modification by genetic analysis groups was tested by including multiplicative interaction terms of these groups with the APOE alleles followed by a Cochran's Q heterogeneity test accounting for correlations between effect estimates. Continental‐ancestry proportion interaction with APOE alleles effects was tested separately for each of the three ancestries (European, African, and Amerindian), by including the continental‐ancestry proportion variable together with a multiplicative interaction term of ancestry with the APOE allele (analytic dataset n = 3618). 23
Power calculation analysis for the two cognitive outcomes (cognitive decline and MCI) was calculated using population risk allele frequencies. 36 For APOE ε4 association with cognitive decline, the effect size was estimated based on the Cuban group and for APOE ε4 association with MCI, the effect size was estimated based on the Puerto Rican group, because these groups showed significant effects. This calculation did not account for age distribution in the background groups.
3. RESULTS
Table 1 presents summary statistics of the SOL‐INCA analytic sample used in this article, including sample size, distribution of sex, age, education, measures of cognitive function, distribution of the APOE alleles, and genetic proportion ancestries by genetic background groups. Overall, our dataset included 4183 participants (1568 men; 2615 women), with a mean age of 62.1 years. Figure S1 in supporting information illustrates the overlap between the two dichotomous cognitive outcomes. Thirty percent of the population was classified with significant cognitive decline (n = 1250); of these, one third were also classified as MCI (n = 430). Twelve more participants have solely MCI.
TABLE 1.
Demographics, neurocognitive, and genetic characteristics of SOL‐INCA by genetic background groups
| Cuban | Dominican | Mexican | Puerto Rican | South American | Central American | Overall | ||
|---|---|---|---|---|---|---|---|---|
| N | 875 | 424 | 1411 | 734 | 313 | 417 | 4183 | |
| Sex (%) | Female | 490 (48.1) | 300 (60.5) | 902 (53.1) | 447 (50.9) | 192 (56) | 277 (58.4) | 2,615 (52.5) |
| Age in years | Mean (SD) | 62.6 (8.58) | 61.65 (8.19) | 61.7 (7.57) | 62.9 (8.12) | 61.96 (8.13) | 61.35 (7.39) | 62.08 (8.18) |
| Age (%) | ||||||||
| 50–59 | 342 (31.6) | 192 (40.9) | 642 (45.7) | 281 (35.2) | 139 (39.3) | 182 (42.4) | 1782 (38.4) | |
| 60–69 | 364 (32.3) | 162 (37.3) | 547 (35.5) | 306 (35.6) | 123 (35.7) | 184 (39.6) | 1690 (35) | |
| 70+ | 169 (36.1) | 70 (21.8) | 222 (18.8) | 147 (29.2) | 51 (25.1) | 51 (18) | 711 (26.6) | |
| Education (%) | ||||||||
| < 12 | 208 (24) | 191 (43) | 686 (46.1) | 305 (42.1) | 65 (18.3) | 162 (39.5) | 1622 (36) | |
| 12 | 226 (24.5) | 84 (20.1) | 283 (21.4) | 164 (20.7) | 64 (18.2) | 83 (17.8) | 906 (21.7) | |
| > 12 | 441 (51.6) | 149 (36.9) | 442 (32.5) | 265 (37.3) | 184 (63.5) | 172 (42.7) | 1655 (42.3) | |
| Neurocognitive traits (%) | ||||||||
| Significant cognitive decline | 232 (27.9) | 140 (32) | 436 (32.5) | 241 (37.4) | 87 (27.4) | 113 (28.6) | 1250 (31.2) | |
| MCI | 85 (11.1) | 47 (11.8) | 148 (10.6) | 87 (13.4) | 28 (8.3) | 46 (12.3) | 442 (11.3) | |
| APOE genotype (%) | ||||||||
| ε2/ε2 | 2 (0.1) | 4 (1) | 4 (0.2) | 2 (0.3) | 0 (0) | 1 (0) | 14 (0.3) | |
| ε2/ε3 | 96 (10) | 56 (13.8) | 70 (4.1) | 60 (8.2) | 20 (4.9) | 27 (6.5) | 330 (7.8) | |
| ε2/ε4 | 14 (1.2) | 14 (4) | 7 (0.8) | 11 (1.3) | 2 (0.7) | 3 (0.9) | 51 (1.3) | |
| ε3/ε3 | 570 (66.3) | 231 (56) | 1059 (76.2) | 490 (68.1) | 235 (78.4) | 303 (72.3) | 2893 (69.5) | |
| ε3/ε4 | 178 (20.6) | 107 (22.6) | 253 (17.2) | 163 (21.3) | 46 (13.4) | 75 (18.1) | 824 (19.4) | |
| ε4/ε4 | 15 (1.7) | 12 (2.5) | 18 (1.5) | 8 (0.9) | 10 (2.6) | 8 (2.2) | 71 (1.7) | |
| APOE allele (%) | ||||||||
| ε2 | 114 (0.06) | 78 (0.10) | 85 (0.03) | 75 (0.05) | 22 (0.03) | 32 (0.04) | 409 (0.05) | |
| ε3 | 1414 (0.82) | 625 (0.74) | 2441 (0.87) | 1203 (0.83) | 536 (0.88) | 708 (0.85) | 6940 (0.83) | |
| ε4 | 222 (0.13) | 145 (0.16) | 296 (0.11) | 190 (0.12) | 68 (0.10) | 94 (0.12) | 1017 (0.12) | |
| Proportion genetic ancestry (mean [SD]) | ||||||||
| N | 780 | 358 | 1194 | 641 | 272 | 364 | 3618 | |
| African | 0.16 (0.20) | 0.46 (0.16) | 0.04 (0.02) | 0.22 (0.13) | 0.07 (0.08) | 0.10 (0.06) | 0.16 (0.18) | |
| European | 0.80 (0.21) | 0.48 (0.15) | 0.46 (0.20) | 0.65 (0.12) | 0.50 (0.22) | 0.46 (0.15) | 0.60 (0.24) | |
| Amerindian | 0.05 (0.04) | 0.06 (0.02) | 0.49 (0.20) | 0.13 (0.03) | 0.43 (0.23) | 0.44 (0.15) | 0.24 (0.24) |
Note: (%) based on the sampling weights and complex survey design.
Abbreviations: APOE, apolipoprotein E; MCI, mild cognitive impairment; SD, standard deviation; SOL‐INCA, Study of Latinos‐Investigation of Neurocognitive Aging.
The distribution of the APOE alleles in our SOL‐INCA analytic sample by genetic background groups demonstrates the differential distribution, similar to the results published by González et al. 20 Overall, APOE ε3 is the most frequent allele in all Latino background groups, while ε4 and ε2 are relatively rare. Table 2 summarizes the associations between the APOE alleles and cognitive outcomes in the SOL‐INCA population based on an additive inheritance model. The OR for cognitive decline with one APOE ε4 allele, compared to individuals without APOE ε4 allele, was 1.15 (95% CI [1.01;1.32], P‐value = 0.03); restriction to older subsets of the SOL‐INCA population resulted in stronger association (ORs = 1.15‐1.31; Table S1 in supporting information). However, no significant associations were obtained between APOE ε4 allele and MCI, and between APOE ε2 allele and both cognitive outcomes in the total SOL‐INCA analytic sample.
TABLE 2.
APOE alleles association with neurocognitive function in the SOL‐INCA (additive inheritance mode)
| Trait | APOE allele | OR [95% CI] | P‐value |
|---|---|---|---|
| Significant cognitive decline | 2 | 1.04 [0.85;1.29] | 6.85E‐01 |
| 4 | 1.15 [1.01;1.32] | 3.35E‐02 | |
| MCI | 2 | 0.78 [0.51;1.17] | 2.30E‐01 |
| 4 | 0.94 [0.72;1.22] | 6.19E‐01 |
Notes: Models adjusted for sex, age, education, center, genetic background groups, and first five PCs. APOE ε3 used as the reference allele. Effect sizes and SEs were estimated based on the complex survey design method and were used to ORs and 95% CIs. OR values larger than 1 represent decreased neurocognitive function
Abbreviations: APOE, apolipoprotein E; CI, confidence interval; MCI, mild cognitive impairment; OR, odds ratio; PC, principal component; SOL‐INCA, Study of Latinos‐Investigation of Neurocognitive Aging; SE, standard error.
We further stratified and explored the association of the APOE alleles with cognitive outcomes in the six background groups: Cuban, Dominican, Mexican, Puerto‐Rican, South‐American, and Central‐American, summarized in Table 3 (additive model). The association between the APOE ε4 allele with cognitive decline remained significant only for Cubans showing a stronger effect compared to the effect in the total population (OR = 1.46, 95% CI [1.13;1.88], P‐value = 0.007). Additionally, stratification to Latino background groups also revealed a new nominally significant protective association between the APOE ε2 allele and MCI in the Puerto Rican group (OR = 0.37, 95% CI [0.16;0.84], P‐value = 0.04). Heterogeneity tests for differences between the background group association tests were not significant.
TABLE 3.
APOE alleles association with neurocognitive function in the SOL‐INCA by background groups (additive inheritance mode)
| Cuban (n = 875) | Dominican (n = 424) | Mexican (n = 1411) | Puerto Rican (n = 734) | South American (n = 313) | Central American (n = 417) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | APOE allele | OR [95% CI] | P‐value | OR [95% CI] | P‐value | OR [95% CI] | P‐value | OR [95% CI] | P‐value | OR [95% CI] | P‐value | OR [95% CI] | P‐value |
| Significant cognitive decline | 2 | 1.20 [0.80;1.80] | 3.96E‐01 | 0.98 [0.62;1.55] | 9.46E‐01 | 1.03 [0.69;1.54] | 8.79E‐01 | 1.06 [0.69;1.62] | 7.95E‐01 | 1.93 [1.03;3.62] | 7.17E‐02 | 0.65 [0.30;1.38] | 3.07E‐01 |
| 4 | 1.46 [1.13;1.88] | 7.00E‐03 | 0.95 [0.70;1.30] | 7.69E‐01 | 1.05 [0.77;1.43] | 7.69E‐01 | 1.04 [0.84;1.29] | 7.31E‐01 | 0.91 [0.55;1.50] | 7.39E‐01 | 1.05 [0.76;1.46] | 7.71E‐01 | |
| MCI | 2 | 0.98 [0.49;1.96] | 9.54E‐01 | 1.05 [0.40;2.72] | 9.33E‐01 | 0.54 [0.23;1.29] | 2.19E‐01 | 0.37 [0.16;0.84] | 4.41E‐02 | 0.74 [0.14;3.87] | 7.85E‐01 | 0.34 [0.05;2.19] | 3.23E‐01 |
| 4 | 0.80 [0.42;1.53] | 5.26E‐01 | 0.98 [0.55;1.72] | 9.43E‐01 | 0.90 [0.58;1.38] | 6.64E‐01 | 1.04 [0.65;1.67] | 8.64E‐01 | 0.83 [0.39;1.76] | 6.90E‐01 | 1.08 [0.58;2.01] | 8.34E‐01 | |
Notes: Models adjusted for sex, age, education, center, and first five PCs. Effect sizes and SEs were estimated based on the complex survey design method, and were used to compute ORs and 95% CIs. APOE ε3 used as the reference allele. OR values larger than 1 represent decreased neurocognitive function. P‐values were estimated based on 10,000 permutations for each genetic background group. Heterogeneity P‐values were not significant.
Abbreviations: APOE, apolipoprotein E; CI, confidence interval; MCI, mild cognitive impairment; OR, odds ratio; PC, principal component; SOL‐INCA, Study of Latinos‐Investigation of Neurocognitive Aging; SE, standard error.
Tables S2 and S3 in supporting information present similar associations between APOE alleles and cognitive outcomes in the SOL‐INCA analytic sample, based on a dominant inheritance mode. Power analysis for the cognitive traits is presented in Table S4 in supporting information. Power for the APOE allele associations with cognitive functions is substantially different among the six Latino backgrounds (ranges: MCI 0.34–1.00, cognitive decline 0.26–0.88).
Proportion ancestry interaction with APOE alleles' effects on cognitive outcomes is presented in Table 4. A significant interaction effect was found between Amerindian ancestry and APOE ε4 on cognitive decline (OR = 0.47, 95% CI [0.24;0.93], P‐value = 0.04), such that protection from the risk of cognitive decline in APOE ε4 carriers was associated with higher Amerindian proportion ancestry (Figure 1).
TABLE 4.
The interaction effect of proportion ancestry and APOE alleles on neurocognitive function in the SOL‐INCA (additive inheritance mode)
| Europe | Africa | Amerindian | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Interaction | APOE allele | Interaction | APOE allele | Interaction | APOE allele | ||||||||
| Trait | APOE allele | OR [95% CI] | P‐value | OR [95% CI] | P‐value | OR [95% CI] | P‐value | OR [95% CI] | P‐value | OR [95% CI] | P‐value | OR [95% CI] | P‐value |
| Significant cognitive decline | 2 | 1.76 [0.70;4.40] | 2.41E‐01 | 0.77 [0.42;1.40] | 4.06E‐01 | 0.60 [0.22;1.61] | 3.22E‐01 | 1.23 [0.90;1.67] | 2.03E‐01 | 1.14 [0.32;4.05] | 8.43E‐01 | 1.08 [0.79;1.46] | 6.48E‐01 |
| 4 | 1.58 [0.82;3.06] | 1.90E‐01 | 0.88 [0.58;1.33] | 5.47E‐01 | 1.20 [0.64;2.28] | 5.77E‐01 | 1.11 [0.93;1.34] | 2.63E‐01 | 0.47 [0.24;0.93] | 3.83E‐02 | 1.34 [1.11;1.63] | 3.70E‐03 | |
| MCI | 2 | 0.47 [0.05;4.71] | 5.50E‐01 | 1.20 [0.30;4.83] | 8.12E‐01 | 4.63 [0.66;32.36] | 1.57E‐01 | 0.49 [0.24;1.01] | 6.84E‐02 | 0.03 [0.00;1.38] | 1.07E‐01 | 1.16 [0.60;2.25] | 6.70E‐01 |
| 4 | 1.37 [0.38;4.92] | 6.59E‐01 | 0.81 [0.36;1.85] | 6.39E‐01 | 0.76 [0.16;3.68] | 7.50E‐01 | 1.05 [0.72;1.52] | 8.26E‐01 | 1.01 [0.34;3.01] | 9.92E‐01 | 0.99 [0.66;1.50] | 9.72E‐01 | |
Notes: Models adjusted for sex, age, education, center, and first five PCs. Effect sizes and SEs were estimated based on the complex survey design method and were used to compute ORs and 95% CIs. APOE ε3 used as the reference allele. Interaction P‐values were estimated based on 10,000 permutations for each ancestry separately. OR values larger than 1 represent decreased neurocognitive function.
Abbreviations: APOE, apolipoprotein E; CI, confidence interval; MCI, mild cognitive impairment; OR, odds ratio; PC, principal component; SOL‐INCA, Study of Latinos‐Investigation of Neurocognitive Aging.
FIGURE 1.
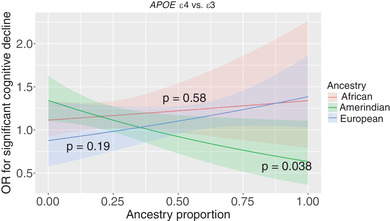
Interaction of proportion continental ancestry with apolipoprotein E(APOE) ε4 allele effects on significant cognitive decline. Interaction P‐values were estimated based on 10,000 permutations for each ancestry separately
Additional analyses of continuous cognitive decline are reported in Appendix 1 and show similar results to the significant cognitive decline outcome.
4. DISCUSSION
We performed an association study of APOE alleles and cognitive outcomes in a large cohort of diverse middle‐aged and older Latinos. Our main result showed an association between the APOE ε4 allele and the risk of significant cognitive decline. In the stratified analysis, this result remained significant only for Cubans. We also found an association between APOE ε2 and decreased risk of MCI only in Puerto Ricans, suggesting differential effects of the APOE alleles on cognitive function in the Latino background groups. We further discovered that an increased proportion of genetic Amerindian ancestry was associated with a protective effect from the risk of APOE ε4 on significant cognitive decline, compatible with the known low proportion of Amerindian ancestry in the Cuban background group. 23
We infer that ancestry‐specific genetic variants may explain the differential effects of APOE alleles in the six Latino backgrounds.
The association of APOE ε4 allele with significant cognitive decline (OR = 1.15) is compatible with previous results presenting a relatively weaker association between APOE ε4 allele and ADRD in the Latino population compared to Whites (OR = 2.2). 8 The difference in effect sizes may be due to the difference in the tested phenotypes, significant cognitive decline versus ADRD. Cognitive decline modeling differs substantially between studies, thus a comparison of effect sizes of APOE alleles on significant cognitive decline is not feasible. 37 In the analysis stratified by “genetic analysis groups,” we anticipated some relationship between APOE ε4 and significant cognitive decline specifically among Cubans because they have higher degrees of European ancestry. 23 Cubans in our analytic dataset are also older (a risk factor for cognitive decline) and more educated (a protective factor for cognitive decline) compared to the other Latino backgrounds; however, our models controlled for age and education. A previous study conducted in Cubans from Cuba also reported an association between APOE ε4 and incident of dementia with a stronger effect in middle‐aged adults (<70 years) compared to older adults (>70 years). 38
The non‐significant results for the APOE ε4 allele association with significant cognitive decline in the five other Latino backgrounds are consistent with their lower percentage of European ancestry. 23 This could also result from the predominantly middle‐aged SOL‐INCA population (mean age 62.1 years), a population not fully presenting with significant cognitive decline, whereas most epidemiological studies on APOE and cognitive decline outcomes are conducted in individuals ≥65 years. This claim is supported by analysis restricted to older subsets of the population resulting in a stronger association between APOE ε4 allele and significant cognitive decline in the older subsets (Table S1). Alternatively, it may result from limited statistical power eg, power = 0.26 for the South American group for the cognitive decline trait (Table S4), or it could present a true differential effect size of APOE ε4 on significant cognitive decline in the different Latino backgrounds. The latter hypothesis is supported by the fact that the statistical power for the Mexican group for the association of APOE ε4 with cognitive decline status is 0.88, yet not significant. Three previous studies focusing on Mexican‐origin Latinos also showed the non‐significant result with a relatively weak effect of APOE ε4 allele on dementia, MCI, or AD. 16 , 17 , 18
Unique to this study, continental‐ancestry proportion, which captures genetic variation across the genomes, further revealed that an increased proportion of genetic Amerindian ancestry was associated with a protective effect from the risk of APOE ε4 on significant cognitive decline. This result is also compatible with the APOE ε4 significant risk effect we found in Latino Cubans, which were shown to have the lowest proportion of Amerindian ancestry among all Latino background groups (Table 1). By using all three ancestries of the admixed Latino population we could infer the protective effect of the Amerindian ancestry, rather than the European or African ancestries being harmful (if there were only two studied ancestries, we could not distinguish between the protective effect of one and the risk effect of the other). This result is inconsistent with a recent report in Peruvians (78 AD cases and 128 controls) suggesting that Amerindian local ancestry in the APOE region is contributing to a strong risk for AD in APOE ε4 carriers. 39 Two other studies in Caribbean Hispanics suggest a protective effect of the African local ancestry in the APOE region from the risk of APO ε4 on AD. 40 , 41 Therefore, we also studied whether local ancestry at the APOE region modifies the effects of APOE on significant cognitive decline (results not shown). While the Amerindian local ancestry association was protective, it was not statistically significant (P‐value >0.2). It is a topic of future research to perform a more comprehensive analysis of local ancestry at an expanded region around the APOE gene and potentially genome‐wide, and search for specific genetic variants explaining the observed interaction of global Amerindian genetic ancestry with APOE ε4 in its effect on significant cognitive decline.
Our study also highlights a protective association between APOE ε2 and MCI solely in the Puerto Rican background group (Table 3), compatible with the known neuroprotective effect of APOE ε2. 8 The direction of this effect was similarly protective in several other Latino background groups; however, they were not significant, despite high power estimations especially for the Cuban and Mexican background groups (Table S4). APOE ε2 is relatively rare and its effect on cognitive function is less studied compared to APOE ε4, 42 specifically in the Latino population. A meta‐analysis based predominantly on the Chinese population suggests that APOE ε2/ε3 genotype provides slight protection for MCI. 43
We did not observe the expected risk effects of the APOE ε4 alleles with MCI. This could be explained by our middle‐aged SOL‐INCA population, not fully presenting MCI. While both significant cognitive decline and MCI are markers for ADRD, our results might indicate that significant cognitive decline, which appears in an earlier stage before MCI, is an important risk marker for ADRD in this SOL‐INCA dataset. 44
This is the first study to examine Latino genetic diversity in the context of significant cognitive decline and MCI that used both background groups and continental genetic ancestry proportions, which is a major step forward in the field of cognitive aging and ADRD precision medicine. Second, this cohort is composed of Latino middle‐aged and older adults, thus there is less significant survival bias compared to studies of older adults. However, this dataset does not include biomarkers such as advanced imaging or fluid biomarkers, which could have validated the significant cognitive decline and MCI status. Also, we note that significant cognitive decline measure is not a clinical phenotype. To address this limitation, we report a similar analysis of APOE alleles with continuous cognitive decline (Appendix 1), and the results are similar to the results of significant cognitive decline. Because we examined six different Latino background groups, Winner's curse bias might also explain our significant result in the stratified analyses. Overall, our differential association results in the Latino background groups may suggest a true differential genetic association between APOE alleles and cognitive outcomes related to admixed genomes. However, they could alternatively represent the lifestyle and environmental factors differing between the Latino ancestries, such as smoking, nutrition, alcohol consumption, physical activity, sleep phenotypes, air pollution, and metal exposure, that may also interact with APOE alleles and cognitive function risk. 45 Further analysis in SOL‐INCA and other samples with older Latino populations, accounting for environmental characteristics, may further delineate the associations between the APOE alleles and cognitive outcomes.
Overall, our study, together with other studies focusing on the Latino population, may lead to a better understanding of the role of APOE in the development of ADRD in Latinos and potentially in American Indians by extension thus advancing the development of personalized risk prediction and strategies to address Latinos’ health disparities in neurodegenerative aging and disorders.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
The authors thank the staff and participants of HCHS/SOL for their important contributions. Investigators website: http://www.cscc.unc.edu/hchs/. This work is supported by the National Institute on Aging (R01AG048642, RF1AG054548, RF1AG061022, and R21AG056952). Dr. González also receives additional support from P30AG062429 and P30AG059299. The Hispanic Community Health Study/Study of Latinos is a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (HHSN268201300001I/N01‐HC‐65233), University of Miami (HHSN268201300004I/N01‐HC‐65234), Albert Einstein College of Medicine (HHSN268201300002I/N01‐HC‐65235), University of Illinois at Chicago – HHSN268201300003I/N01‐HC‐65236 Northwestern Univ), and San Diego State University (HHSN268201300005I/N01‐HC‐65237). The following Institutes/Centers/Offices have contributed to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution‐Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03).
Granot‐Hershkovitz E, Tarraf W, Kurniansyah N, et al. APOE alleles’ association with cognitive function differs across Hispanic/Latino groups and genetic ancestry in the study of Latinos‐investigation of neurocognitive aging (HCHS/SOL). Alzheimer's Dement. 2021;17:466–474. 10.1002/alz.12205
REFERENCES
- 1. Heron M. Deaths: leading Causes for 2016. Natl Vital Stat Reports. 2016:67. [PubMed] [Google Scholar]
- 2. Boyle PA, Wilson RS, Aggarwal NT, Tang Y, Bennett DA. Mild cognitive impairment: risk of Alzheimer disease and rate of cognitive decline. Neurology. 2006;67:441‐445. [DOI] [PubMed] [Google Scholar]
- 3. Wilson RS, Segawa E, Boyle PA, Anagnos SE, Hizel LP, Bennett DA. The natural history of cognitive decline in Alzheimer's disease. Psychol Aging. 2012;27:1008‐1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng Y‐W, Chen T‐F, Chiu M‐J. From mild cognitive impairment to subjective cognitive decline: conceptual and methodological evolution. Neuropsychiatr Dis Treat. 2017;13:491‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vega IE, Cabrera LY, Wygant CM, Velez‐Ortiz D, Counts SE. Alzheimer's Disease in the Latino community: intersection of genetics and social determinants of health. J Alzheimer's Dis. 2017;58:979‐992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alzheimer's Association. 2018 Alzheimer's disease facts and figures. Alzheimer's Dement. 2018;14:367‐429. [Google Scholar]
- 7. Dilworth‐Anderson P, Hendrie HC, Manly JJ, Khachaturian AS, Fazio S. Diagnosis and assessment of Alzheimer's disease in diverse populations. Alzheimer's Dement. 2008;4:305‐309. [DOI] [PubMed] [Google Scholar]
- 8. Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E Genotype and Alzheimer disease. JAMA. 1997;278:1349‐1356. [PubMed] [Google Scholar]
- 9. Reas ET, Laughlin GA, Bergstrom J, Kritz‐Silverstein D, Barrett‐Connor E, McEvoy LK. Effects of APOE on cognitive aging in community‐dwelling older adults. Neuropsychology. 2019;33:406‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Makkar SR, Lipnicki DM, Crawford JD, et al. APOE ε4 and the influence of sex, age, vascular risk factors, and ethnicity on cognitive decline. J Gerontol A Biol Sci Med Sci. 2020:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ren D, Lopez OL, Lingler JH, Conley Y. The effect of the APOE ε2ε4 Genotype on the development of Alzheimer's disease (AD) and Mild Cognitive Impairment (MCI) in Non‐Latino Whites. J Am Geriatr Soc. 2020;68:1044‐1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu C‐C, Liu C‐C, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang M‐X, Stern Y, Marder K, et al. The APOE‐∊4 Allele and the Risk of Alzheimer Disease Among African Americans, Whites, and Hispanics. JAMA. 1998;279:751. [DOI] [PubMed] [Google Scholar]
- 14. Crean S, Ward A, Mercaldi CJ, et al. Apolipoprotein E 4 prevalence in Alzheimer's disease patients varies across global populations: a systematic literature review and meta‐analysis. Dement Geriatr Cogn Disord. 2011;31:20‐30. [DOI] [PubMed] [Google Scholar]
- 15. Maestre G, Ottman R, Stern Y, Gurland B, Chun M, ‐X TangM, et al. Apolipoprotein E and Alzheimer's disease: ethnic variation in genotypic risks. Ann Neurol. 1995;37:254‐259. [DOI] [PubMed] [Google Scholar]
- 16. Haan MN, Mungas DM, Gonzalez HM, Ortiz TA, Acharya A, Jagust WJ. Prevalence of dementia in older Latinos: the influence of type 2 diabetes mellitus, stroke and genetic factors. J Am Geriatr Soc. 2003;51:169‐177. [DOI] [PubMed] [Google Scholar]
- 17. O'Bryant SE, Johnson L, Reisch J, et al. Risk factors for mild cognitive impairment among Mexican Americans. Alzheimers Dement. 2013;9:622‐631.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Campos M, Edland SD, Peavy GM. An exploratory study of APOE‐ε4 Genotype and risk of Alzheimer's disease in Mexican Hispanics NIH public access. J Am Geriatr Soc. 2013;61:1038‐1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romas SN, Santana V, Williamson J, et al. Familial Alzheimer disease among Caribbean Hispanics. Arch Neurol. 2002;59:87‐91. [DOI] [PubMed] [Google Scholar]
- 20. González HM, Tarraf W, Jian X, et al. Apolipoprotein E genotypes among diverse middle‐aged and older Latinos: study of Latinos‐investigation of neurocognitive aging results (HCHS/SOL). Sci Rep. 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lavange LM, Kalsbeek WD, Sorlie PD, et al. Sample design and cohort selection in the Hispanic community health study/study of Latinos. Ann Epidemiol. 2010;20:642‐649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sorlie PD, Avilé S‐Santa LM, Wassertheil‐Smoller S, et al. Design and implementation of the Hispanic community health study/study of Latinos. Ann Epidemiol. 2010;20:629‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conomos MP, Laurie CA, Stilp AM, et al. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic community health study/study of Latinos. Am J Hum Genet. 2016;98:165‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. González HM, Tarraf W, Schneiderman N, et al. Prevalence and correlates of mild cognitive impairment among diverse Hispanics/Latinos: study of Latinos‐investigation of neurocognitive aging results. Alzheimer's Dement. 2019;15:1507‐1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Callahan Christopher M, Unverzagt Frederick W, Hui Siu L, Perkins Anthony J, Hendrie Hugh C. Six‐item screener to identify cognitive impairment among potential subjects for clinical research. Med Care. 2002;40(9):771–781. 10.1097/00005650-200209000-00007. [DOI] [PubMed] [Google Scholar]
- 26. González HM, Mungas D, Reed BR, Marshall S, Haan MN. A new verbal learning and memory test for English‐ and Spanish‐speaking older people. J Int Neuropsychol Soc. 2001;7:544‐555. [DOI] [PubMed] [Google Scholar]
- 27. Lezak MD, Howieson DB, Loring DW, Hannay HJ, Fischer JS. Neuropsychological Assessment, PsycNET. 4th ed. New York: Oxford University Press; 2004. [Google Scholar]
- 28. Tomaszewski Farias S, Mungas D, Harvey DJ, Simmons A, Reed BR, Decarli C. The measurement of everyday cognition: development and validation of a short form of the everyday cognition scales. Alzheimers Dement. 2011;7:593‐601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. González HM, Tarraf W, Gouskova N, et al. Neurocognitive function among middle‐aged and older Hispanic/Latinos: results from the Hispanic community health study/study of Latinos. Arch Clin Neuropsychol. 2015;30:68‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. González HM, Tarraf W, Fornage M, et al. A research framework for cognitive aging and Alzheimer's disease among diverse US Latinos: design and implementation of the Hispanic community health study/study of Latinos—investigation of neurocognitive aging (SOL‐INCA). Alzheimer's Dement. 2019;15:1624‐1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270‐279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sofer T, Wong Q, Hartwig FP, et al. Genome‐wide association study of blood pressure traits by Hispanic/Latino background: the Hispanic community health study/study of Latinos. Sci Rep. 2017;7:10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conomos MP, Reiner AP, Weir BS, Thornton TA. Model‐free estimation of recent genetic relatedness. Am J Hum Genet. 2016;98:127‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alexander DH, Novembre J, Lange K. Fast model‐based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655‐1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lumley T, Scott A. Tests for regression models fitted to survey data. Aust New Zeal J Stat. 2014;56:1‐14. [Google Scholar]
- 36. Nelson SC, Doheny KF, Pugh EW, Romm JM, Ling H, Laurie CA, et al. Imputation‐based genomic coverage assessments of current human genotyping arrays. G3 (Bethesda). 2013;3:1795‐1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karr JE, Graham RB, Hofer SM, Muniz‐Terrera G. When does cognitive decline begin? A systematic review of change point studies on accelerated decline in cognitive and neurological outcomes preceding mild cognitive impairment, dementia, and death. Psychol Aging. 2018;33:95‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodríguez JJL, Cepero AV, Gil IYS, et al. Incidence of dementia and association with APOE genotype in older Cubans. Dement Neuropsychol. 2014;8:356‐363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cornejo‐Olivas M, Rajabli F, Marca V, et al. Dissecting the role of Amerindian genetic ancestry and ApoE ε4 allele on Alzheimer disease in an admixed Peruvian population. BioRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rajabli F, Feliciano BE, Celis K, et al. Ancestral origin of ApoE ε4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet. 2018;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blue EE, Horimoto ARVR, Mukherjee S, Wijsman EM, Thornton TA. Local ancestry at APOE modifies Alzheimer ’ s disease risk in Caribbean Hispanics. Alzheimer's Dement. 2019:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu L, Zhao L. ApoE2 and Alzheimer's disease: time to take a closer look. Neural Regen Res. 2016;11:412‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang Y, He T, Deng W, Sun P. Association between apolipoprotein E gene polymorphism and mild cognitive impairment: a meta‐analysis. Clin Interv Aging. 2017;12:1941‐1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caselli RJ, Reiman EM, Locke DEC, et al. Cognitive domain decline in healthy apolipoprotein E ε4 Homozygotes before the diagnosis of mild cognitive impairment. Arch Neurol. 2007;64:1306. [DOI] [PubMed] [Google Scholar]
- 45. Eid A, Mhatre I, Richardson JR. Gene‐environment interactions in Alzheimer's disease: a potential path to precision medicine. Pharmacol Ther. 2019;199:173‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information


