Abstract
The spinal cord is a fascinating structure responsible for coordinating movement in vertebrates. Spinal motor neurons control muscle activity by transmitting signals from the spinal cord to diverse peripheral targets. We profiled 43,890 single-nucleus transcriptomes from the adult mouse spinal cord using fluorescence-activated nuclei sorting to enrich for motor neuron nuclei. We identified 16 sympathetic motor neuron clusters, which are distinguishable by spatial localization and expression of neuromodulatory signaling genes. We found surprising skeletal motor neuron heterogeneity in the adult spinal cord, including transcriptional differences that correlate with electrophysiologically and spatially distinct motor pools. We also provide evidence for a novel transcriptional subpopulation of skeletal motor neuron (γ*). Collectively, these data provide a single-cell transcriptional atlas (http://spinalcordatlas.org) for investigating the organizing molecular logic of adult motor neuron diversity, as well as the cellular and molecular basis of motor neuron function in health and disease.
The central nervous system (CNS) receives sensory input from its surroundings, integrates that information, and then communicates with muscles and organs throughout the body to enact an appropriate response. While a vast, interconnected network of neurons is responsible for processing information and planning motor behaviors, the transmission of signals from the CNS to peripheral muscles is controlled by one special and rare cell population: the spinal motor neuron.
Spinal motor neurons are unique because they reside in the CNS yet extend their axons far into the periphery in order to reach their innervation targets. Their activity is essential for virtually all skeletal and smooth muscle contractions in the body, controlling actions that range from the regulation of blood pressure and sweat secretion to the contraction of fast-twitch muscle fibers. Because of the vast diversity of muscular function within the body, motor neurons must tune their synaptic connections and electrophysiological properties to the unique features of the effector cells they control1. The intricate process of establishing motor neuron identity requires both cell-intrinsic and extrinsic signaling during spinal cord development, culminating in the tightly controlled expression of transcription factors and cell signaling molecules that define motor circuits2–4.
Considering their physiological importance, it is not surprising that spinal motor neuron dysfunction underlies many human neuromuscular diseases. Indeed, spinal motor neuron degeneration is causally responsible for amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA), and numerous other rare neuromuscular disorders. In many of these diseases, certain populations of spinal motor neurons are selectively affected, while others are spared5. Defining the ground state for transcriptional differences between motor neuron subtypes in the adult will empower the analysis of molecular mechanisms that separate susceptible and resistant populations, leading to a better understanding of the molecular underpinnings of cell-type vulnerability.
Despite their critical functional role, spinal motor neurons make up less than 0.4% of the total cells in the mammalian spinal cord. This rarity has made them notoriously difficult to transcriptionally characterize6. To overcome this challenge, we developed a motor neuron enrichment strategy using a fluorescent reporter mouse (see Methods). This yielded an ~100-fold increase in representation of motor neuron nuclei over past efforts7, enabling us to assess the full transcriptional diversity of motor neurons in the adult spinal cord.
We performed single-nucleus RNA sequencing on 43,890 nuclei from the adult mouse spinal cord, providing unprecedented single-cell resolution of the spinal motor system. This method allowed us to transcriptionally distinguish spinal motor neurons of the autonomic nervous system (visceral motor neurons) and somatic nervous system (skeletal motor neurons) based on numerous newly discovered marker genes. Foundational work in the field of spinal cord development has established the molecular logic of spinal neuron differentiation and target specification3,8. These efforts set the stage for the present study, where we performed an unbiased analysis and validation of motor neuron diversity in the adult mouse spinal cord. We demonstrate that single-nucleus transcriptional profiling can provide key insights into the link between the brain and the body by defining the neuropeptides, transmitters, and receptors that motor neurons use to communicate. Furthermore, this detailed characterization will enable the development of a broad range of molecular tools, including transgenic mice and a roadmap for reprogramming stem cells into specific motor neuron subtypes, which will unlock genetic access to previously uncharacterized spinal motor neuron populations.
Single-nucleus transcriptional profiling of the adult mouse spinal cord
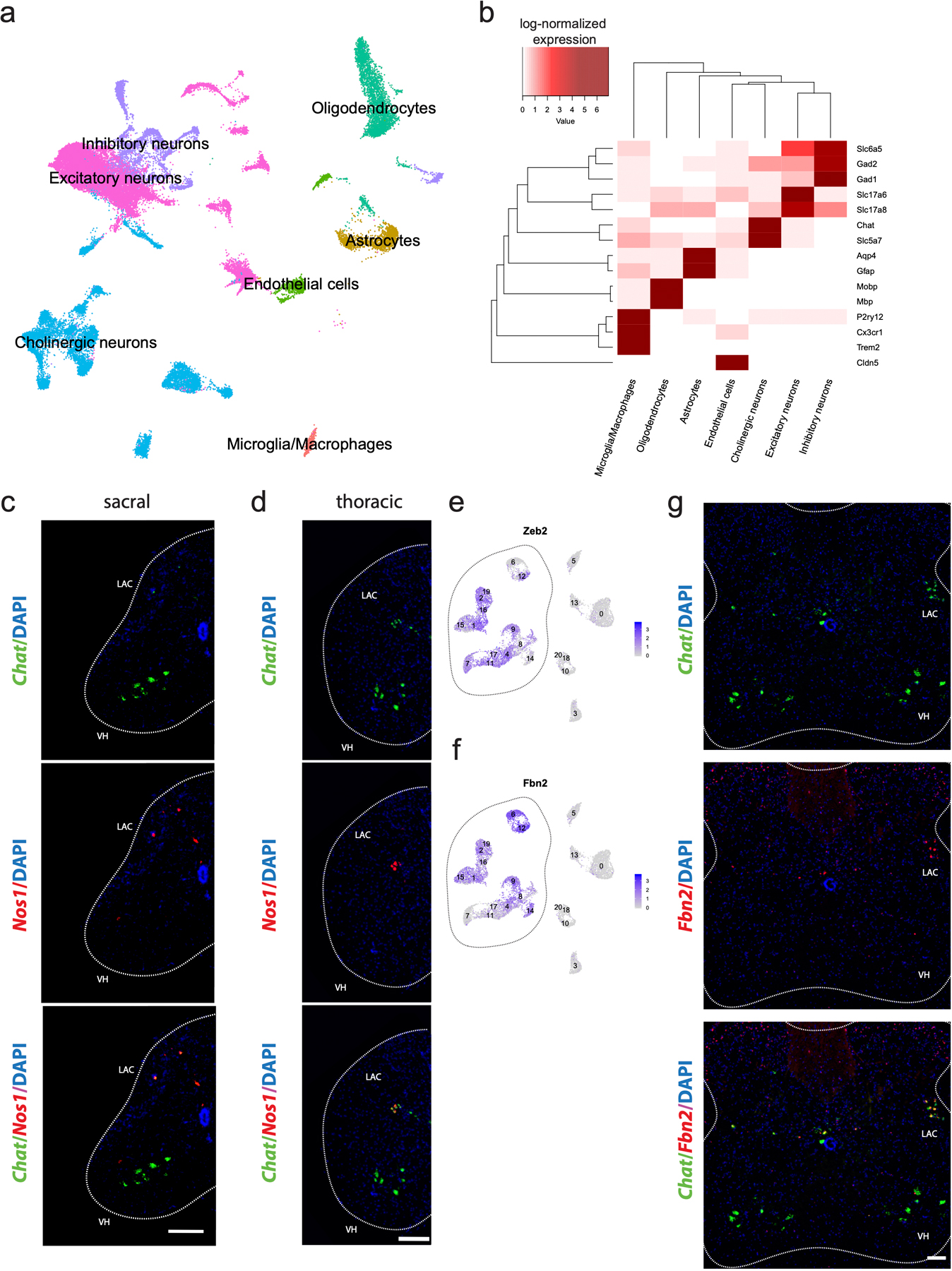
Because spinal motor neurons are so scarce, we enriched for motor neuron nuclei using a transgenic fluorescent reporter mouse9. This technique enabled us to selectively isolate cholinergic nuclei (Chat+), a population that encompasses all motor neurons and several interneuron subtypes in the adult mouse spinal cord (see Methods). Given the important role of non-cell autonomous mechanisms in neurodegeneration, we also isolated non-motor neuron cells – including interneurons, astrocytes, microglia, and oligodendrocytes. In total, we transcriptionally profiled 43,890 nuclei from a collection of male, female, and mixed cohorts of wild-type adult mice, with 20–40% of nuclei coming from motor neurons and 60–80% from other cells in the spinal cord (Fig. 1a). We used graph-based methods to cluster nuclei and then annotate cell types across clusters based on averaged expression of common marker genes, including genes encoding neurotransmitter signaling machinery (Fig. 1b,c, Methods). This approach enabled us to simultaneously characterize motor neurons, while also comparing their transcriptomes with other cells within the spinal cord, to find marker genes that are exclusively expressed in cell populations of interest. We assigned all profiled nuclei into seven broad categories: excitatory interneurons, inhibitory interneurons, cholinergic neurons, astrocytes, microglia, oligodendrocytes, and endothelial cells (Fig. 1c, Extended Data Fig. 1a,b, Supplementary Table 1). Based on these categories, we estimate that ~30% (13,589 cells) of the profiled single-nucleus transcriptomes correspond to cholinergic neurons. This is a considerable improvement in representation over prior efforts7 and provides unparalleled access to the transcriptional heterogeneity of spinal motor neurons. We provide an interactive web portal to access and search all of the spinal cord transcriptome data: http://spinalcordatlas.org.
Fig. 1: Motor neuron enrichment and single-nucleus transcriptional analysis of the adult mouse spinal cord uncovers novel skeletal and visceral motor neuron markers.
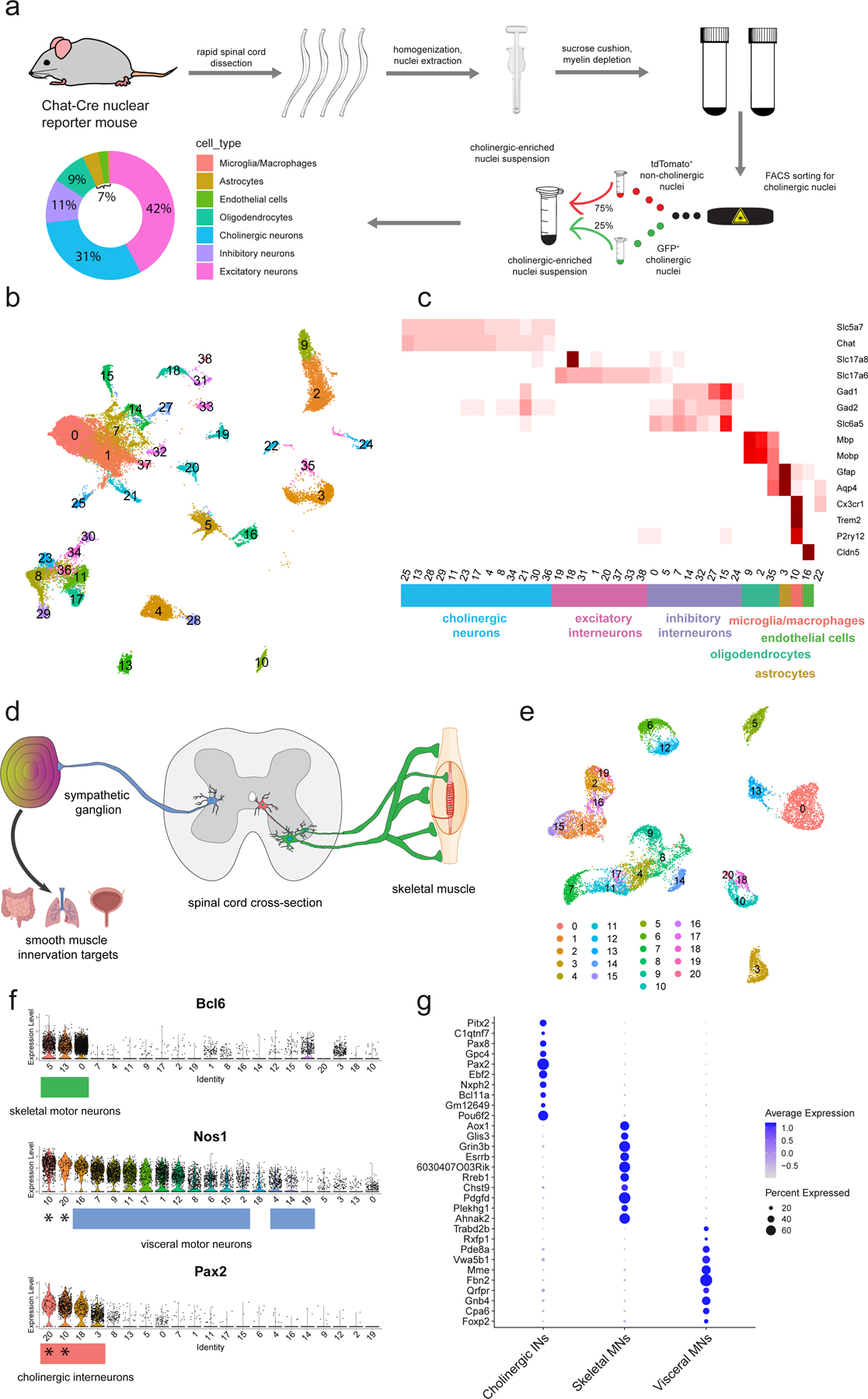
a, Workflow for cholinergic nucleus enrichment and single-nucleus RNA sequencing (snRNAseq)—GFP+ and TdTomato+ cells were mixed at a ratio between 1:3 and 2:3. Plot shows the distribution of canonical cell types with their proportional representation. FACS was used to select appropriate proportion of singlet, DAPI+/GFP+/tdTomato− nuclei. b, UMAP of clustered snRNAseq data from 43,890 transcriptomes. c, Average expression levels per cluster for marker genes of each canonical cell population. Cell type labels based on expression patterns of marker genes. d, Schematic depicting expected cholinergic cell types in the spinal cord. Visceral motor neurons (blue) innervate sympathetic ganglia, skeletal motor neurons (green) directly innervate muscle fibers, and cholinergic interneurons (red) innervate motor neurons and other cells. e, UMAP with graph-based clustering of all cholinergic neurons reveals 21 clusters. f, Ranked expression of known marker genes Pax2 (interneurons) and Nos1 (visceral motor neurons), as well as novel marker gene Anxa4 (skeletal motor neurons) by cluster. Cell labels were assigned hierarchically by expression levels of Pax2, Nos1, and Bcl6 and are reported below each plot. Cholinergic interneuron clusters that also express Nos1 are denoted with a (*). g, Enriched differentially expressed genes for cholinergic interneurons, skeletal motor neurons, and visceral motor neurons. Dot size is proportional to the percent of each cluster expressing the marker gene, while blue color intensity is correlated with expression level. All expression values were log-normalized in Seurat50.
Novel genetic markers distinguish autonomic and skeletal motor neurons
We next asked whether the observed transcriptional diversity of spinal motor neurons corresponds to functionally defined cell types, as in development (Fig. 1d). We computationally isolated and used graph-based clustering (see Methods) to segregate all cholinergic neurons into 20 clusters (Fig. 1e). We annotated these subpopulations as skeletal motor neurons, cholinergic interneurons, and visceral motor neurons (Fig. 1f) based on expression of known marker genes as well as expression patterns of uncharacterized genes in the publicly available Allen Mouse Spinal Cord Atlas10. Specifically, we identified cholinergic interneurons based on their expression of Pax211 and visceral motor neurons (which are part of the autonomic nervous system) by their expression of neuronal nitric oxide synthase (Nos1+)12. We performed double label in situ hybridization with Chat and Nos1 to confirm that Nos1 is expressed specifically in the lateral autonomic columns of the thoracic and sacral spinal cord—where visceral motor neurons are located (Extended Data Fig. 1c,d).
We hypothesized that the remaining three clusters represented skeletal motor neurons—a broad cell population with no known genetic markers that distinguish them from visceral motor neurons. To test this hypothesis, we examined the Allen Mouse Spinal Cord Atlas10 for expression of two genes that are highly expressed in those clusters—Bcl6 and Tns1. Indeed, in contrast to ubiquitous (Actb) and pan-neuronal (Syn1) transcripts, which show broad expression in transverse sections, it is apparent that Bcl6 and Tns1 are strongly expressed in small and large diameter neurons in the ventral horn of the spinal cord (Extended Data Fig. 2a–d). Future studies will investigate the expression patterns of these putative markers in greater depth, but this pattern is consistent with skeletal motor neurons. Our classification of skeletal and visceral motor neurons differs from a previous study, which postulated that two other genes, Fbn2 and Zeb2 were novel markers specifically expressed in alpha (α) motor neurons13. Instead, our transcriptional data indicate that that Zeb2 and Fbn2 are absent from skeletal motor neuron clusters (Extended Data Fig. 1e,f), the latter of which we confirm by multiplexed Chat/Fbn2 in situ hybridization (Extended Data Fig. 1g). Thus, these new transcriptional profiles reveal dozens of possible new marker genes that reliably distinguish skeletal motor neurons from other cells in the spinal cord. (Fig. 1g; Supplementary Table 2a).
Single-nucleus transcriptomics reveals diversity within the autonomic nervous system
Unlike skeletal motor neurons, which control voluntary movement, visceral motor neurons in the spinal cord control the activity of involuntary smooth muscles responsible for regulating homeostatic processes throughout the body. These cells are part of the sympathetic nervous system, while comparable cells in the brain stem are part of the parasympathetic nervous system12. These sympathetic visceral motor neurons are very closely related, developmentally, to skeletal motor neurons14,15, but do not innervate muscle fibers directly. Instead, they project from the lateral autonomic column of the spinal cord and synapse onto the peripheral ganglia in the sympathetic chain that control smooth muscle contraction (Fig. 2a)12. Neurons of the autonomic nervous system innervate nearly every organ in the body and thus have different functional requirements to fit the needs of each peripheral target (Fig. 2b). Could these functional differences be encoded by transcriptional heterogeneity?
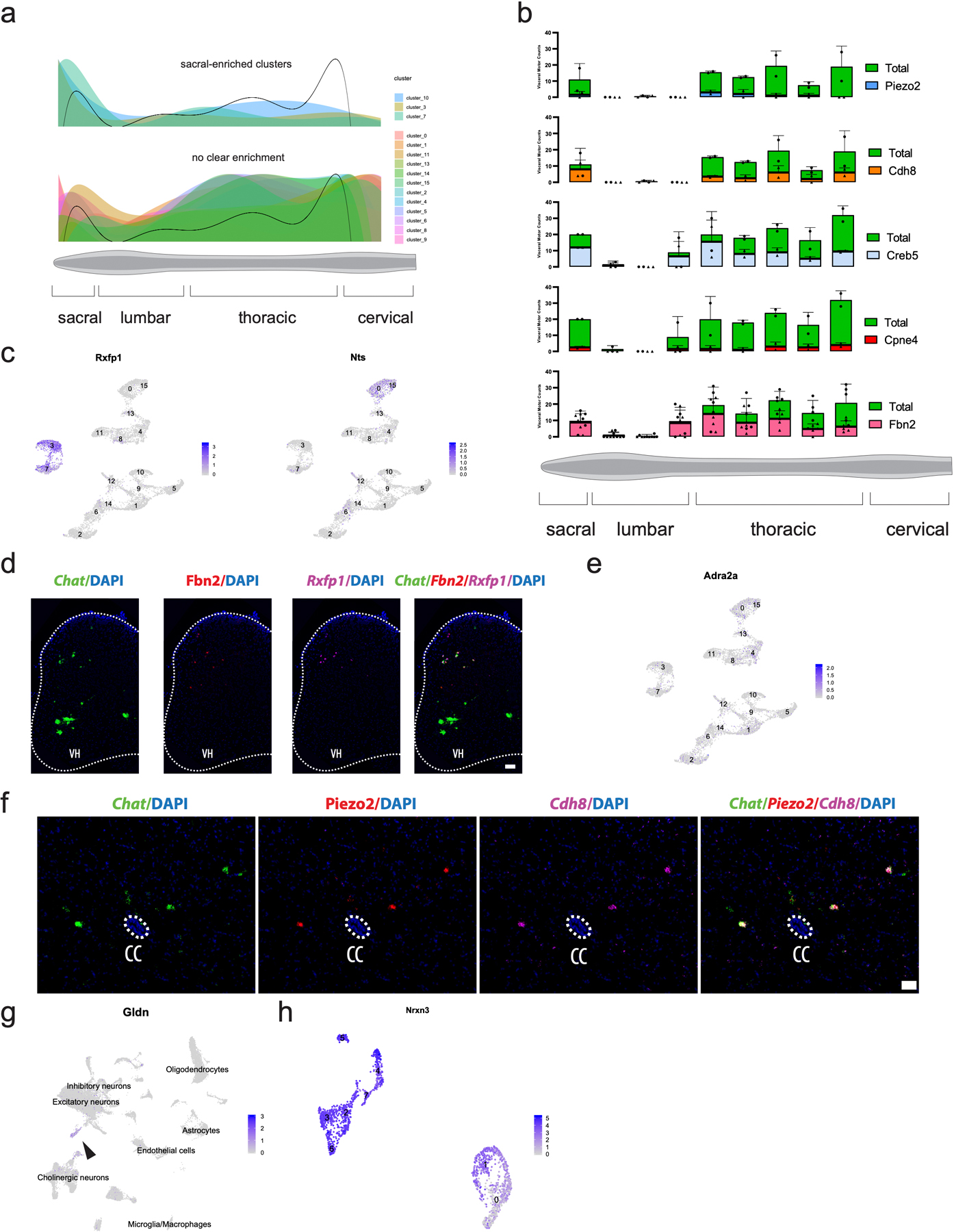
Fig. 2: Single-nucleus transcriptomics reveals immense diversity within the autonomic nervous system and partition cells.
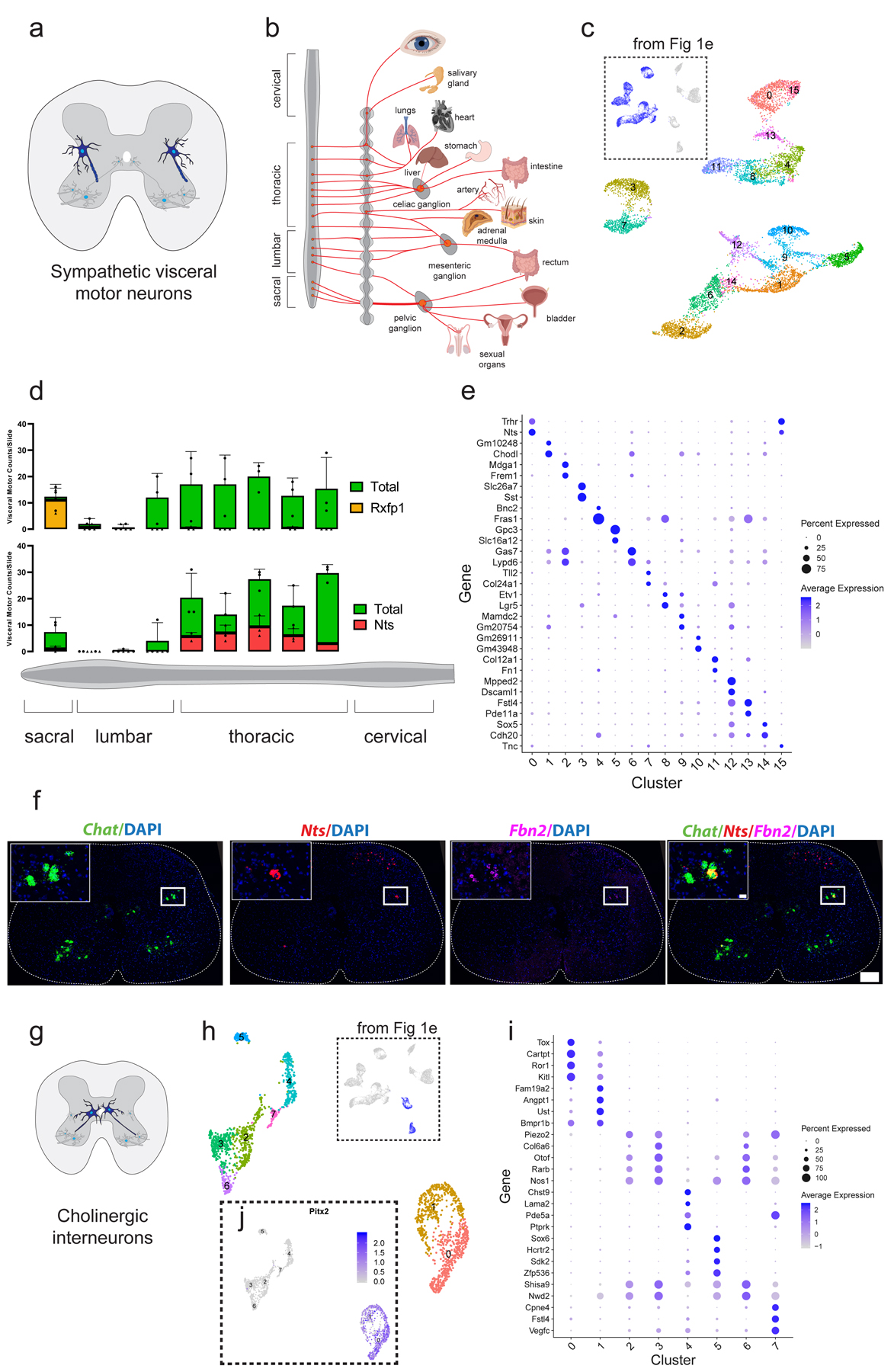
a, Schematic illustrating the position of sympathetic visceral motor neurons (blue) in the lateral autonomic column of the spinal cord. b, Diagram adapted from Espinosa-Medina 2016 showing innervation targets of the sympathetic nervous system12. c, UMAP with 16 visceral motor neuron subclusters. Inset shows all cells from Fig. 1e that were subclustered. d, Frequency of visceral motor neuron subpopulations (Rxfp1+ and Nts+) along the rostral-caudal axis of the spinal cord. Individual data points for total visceral motor neurons shown with filled circles, while marker gene-positive cell numbers shown with filled triangles. n=3 biologically independent animals. e, Novel marker genes for each cluster of visceral motor neurons. f, Representative in situ hybridization of Chat, Nts, and Fbn2 demonstrating Nts expression in visceral motor neurons. 200 µm scale bar in overview and 20 µm in inset. n=3 biologically independent animals. g, Schematic showing cholinergic interneuron innervation of skeletal motor neurons as demonstrated previously25. h, UMAP with graph-based clustering labels for cholinergic interneurons. Inset shows all cells from Fig. 1e that were subclustered. i, Novel marker genes for cholinergic interneuron clusters identified. j, Expression of Pitx2 in cholinergic interneuron populations, overlaid on UMAP projection from (h). Cluster 0 and 1 are Pitx2-positive. All expression values were log-normalized in Seurat50. Dot size is proportional to the percent of each cluster expressing the marker gene, while blue color intensity is correlated with expression level in (e,i).
The sympathetic nervous system is organized along the rostral-caudal axis of the spinal cord, such that visceral motor neurons that control the same organs are coarsely grouped near one another within the spinal cord (Fig. 2b). Intriguingly, past electrophysiological studies have uncovered heterogeneity among visceral motor neurons with respect to their membrane properties, responsiveness to neurotransmitters16, and sensitivity to hormones such as dopamine17 and noradrenaline18. This evidence strongly suggests that the visceral motor system contains physiologically distinct subpopulations, but the overarching molecular logic underlying the sympathetic nervous system remains unresolved.
We hypothesized that transcriptomic clusters may reflect distinct populations of visceral motor neurons that either innervate specific peripheral targets, are selectively responsive to hormones17, and/or utilize distinct classes of neuropeptides in transmitting signals to peripheral ganglia19. To test these hypotheses, we subclustered all visceral motor neurons. By limiting the diversity of cell types that are concurrently analyzed and subclustering along principal axes of variation specific to visceral motor neurons, we resolved 16 transcriptionally distinct subpopulations (Fig. 2c). We identified marker genes that are, according to the Allen Spinal Cord Atlas10, expressed in the adult mouse lateral autonomic column of the spinal cord and are significantly enriched in at least one of the 16 visceral motor neuron populations (Supplementary Table 3). For each marker gene we estimated a normalized spatial density of expressing cells along the rostral-caudal spinal cord using data from the Allen Spinal Cord Atlas (see Methods). We then estimated the positional distribution of cells within each visceral motor neuron cluster as an average of these spatial cell distributions, weighted by the relative expression levels of the marker genes within each cluster (see Methods). While most clusters showed no strong spatial bias along this axis (Extended Data Fig. 3a), clusters 3, 7, and 10 showed a clear enrichment in the sacral spinal cord.
To confirm these findings, we selected several genes that are enriched in distinct clusters (Rxfp1, Nts, Cdh8, Piezo2, Creb5, Fbn2) and performed in situ hybridizations on sections every 600 µm along the rostral-caudal axis of the adult spinal cord (Fig. 2d, Extended Data Fig. 3b). The results of this analysis were striking – we confirmed that Rxfp1 (Clusters 3 and 7) is expressed exclusively in the sacral spinal cord (Fig. 2d, Extended Data Fig. 3c,d). Among other roles, sacral visceral motor neurons modulate sexual function—an intriguing finding given that male and female sexual organs release relaxin family peptides20,21, which bind to Rxfp1. We provide Rxfp1 as an example for how these sequencing data can reveal functionally relevant receptor expression and there are other highly specific hormone receptor genes in distinct clusters (Table 1). We speculate that differentially expressed hormone receptor expression among visceral motor populations may be tuned to central presynaptic inputs and/or peripheral innervation targets.
Table 1:
Differentially expressed signaling machinery genes among visceral motor neuron clusters. All genes listed show differential enrichment in one cluster compared with all others (Wilcoxon rank sum test, p_adj<0.01 (Bonferroni), log2fc>0.5).
| Gene Product | Diff. expressed cluster | |
|---|---|---|
| Serotonergic | ||
| Htr2c | Serotonin receptor 2 | 2,4 |
| Htr1f | Serotonin receptor 1 | 3 |
| Htr2a | Serotonin receptor 2 | 13 |
| Adrenergic | ||
| Adra1a | Adrenergic receptor 1 | 3, 7 |
| Dopaminergic | ||
| Drd2 | Dopamine receptor 2 | 11 |
| Hormone signaling | ||
| Prlr | Prolactin receptor | 0,15 |
| Trhr | Thyrotropin releasing hormone receptor | 0,15 |
| Trhde | Thyrotropin releasing hormone degrading enzyme | 7,11 |
| Rxfp1 | Relaxin family peptide receptor 1 | 3,7 |
| Ghr | Growth hormone receptor | 11 |
| Gfra1 | GDNF Family Receptor | 3,5,14 |
| Qrfpr | Orexigenic neuropeptide QRFP receptor | 0,13,15 |
| Hcrtr2 | Orexin receptor type II | 3,5,15 |
| Tacr1 | Tachykinin (substance P) receptor 1 | 2,3 |
| Opioid signaling | ||
| Penk | Enkephalin | 3 |
| Oprm1 | Opioid receptor mu 1 | 9 |
| Neuropeptide (other) | ||
| Sst | Somatostatin | 3 |
| Nts | Neurotensin | 0,15 |
| Cartpt | Cart peptide | 1,9 |
| Misc. signaling | ||
| Lifr | Lif receptor | 4,5 |
| Tnc | Tenascin | 15 |
| Tnr | Tenascin receptor | 11,13,15 |
We found preganglionic clusters to be highly transcriptionally divergent — with numerous individual markers capable of distinguishing specific clusters (Fig. 2e). Strikingly, the most common subtype of visceral motor neurons (cluster 0) expresses high levels of Neurotensin (Nts) and are therefore neurotensinergic. These neurotensinergic motor neurons are distributed throughout the thoracic and sacral lateral autonomic columns (Fig. 2d). Nts expression was remarkably binary, with Ntson and Ntsoff visceral motor neurons frequently observed directly adjacent to one another in transverse sections (Fig. 2f). Neurotensin is a 13-amino acid peptide, which when injected into rats causes potent inhibition of sympathetic circuits that regulate blood pressure, heart rate, and inspiratory drive—among other effects on the sympathetic nervous system22. Remarkably, although Nts is clearly involved in regulating sympathetic nervous function, its role in spinal preganglionic motor neurons has not been studied. We present strong evidence that Nts expression is a defining feature of preganglionic motor identity and provide the transcriptional roadmap for more detailed future characterization.
Because several studies have shown that hormones, neuropeptides, and monoaminergic signaling play a crucial role in the sympathetic nervous system17–23, we were interested in differentially expressed genes that would affect those pathways. We observed remarkable specificity of neuropeptides and receptors—as well as neurotransmitter receptors—across visceral motor neuron clusters (Fig. 2f, Table 1). Adrenergic receptors, for example, show a remarkable degree of expression specificity in our single-nucleus sequencing data. Type I adrenergic receptor Adra1a is expressed in clusters 3 and 7, which we previously determined to correspond with sacral autonomic motor neurons (Fig. 2d). On the other hand, type II adrenergic receptor Adra2a is expressed at low levels among all visceral populations at (Extended Data Fig. 3e). We also found highly specific expression of neuroactive peptide precursors, such as Penk and Sst in cluster 3 (Table 1). Together, these findings suggest that the repertoire of neuropeptide and hormone receptor expression is an organizing logic within the sympathetic preganglionic motor system.
Transcriptional characterization of cholinergic inhibitory neurons
Cholinergic interneurons are a rare cell population marked by Pax2 expression11. They play key roles in the circuits underlying locomotor behaviors24 (Fig. 2g). Subclustering revealed 7 distinct transcriptional populations of cholinergic interneurons (Fig. 2h) —including clusters 2, 3, 5, 6, and 7 that express high levels of Nos1, which is traditionally considered to be a marker of the autonomic nervous system in the spinal cord12 (Fig. 2i). It remains to be determined whether these Nos1+ cholinergic cells are interneurons or instead a preganglionic motor neuron population that projects into the periphery but also expresses the interneuron marker Pax2. A subset of this population of cells (clusters 2, 3, and 6) also expresses higher levels of Piezo2, a mechanosensitive ion channel involved in proprioception, than any other population in the adult spinal cord (Supplementary Table 2a). We performed in situ hybridization to demonstrate that cholinergic, Piezo2-high cells have large cell bodies and predominantly localize just lateral of the central canal (CC) (Extended Data Fig. 3f). Notably, the localization and size of these cholinergic interneurons strongly resemble previously identified Pitx2- ‘C3’ cells25.
In contrast, clusters 0 and 1 do not express Nos1 but instead express Pitx2—an established marker of partition cells25,26 (Fig. 2j). Partition cells are a subset of cholinergic interneurons that make direct cholinergic synapses with motor neuron soma and proximal dendrites. These synapses, referred to as ‘C boutons,’ modulate motor neuron excitability during locomotor activity25. There is a robust transcriptional signature that separates partition cells from other cells in the spinal cord (Supplementary Table 2a,d). Of particular interest is Gldn, a gene that is selectively expressed in the cluster of cholinergic Pitx2+ cells that have previously been identified as partition cells (Extended Data Fig. 3g). Mutations in GLDN cause lethal congenital contracture syndrome, a crippling neurodegenerative disease in which joints become permanently fixed in a bent or straight position27. Future studies should seek to examine if GLDN mutations affect partition cells.
Elegant viral tracing studies have delineated partition cells into ipsilaterally and contralaterally projecting populations that make exquisitely specific synaptic connections with motor neurons28. We find a parallel transcriptional bifurcation in partition cells, which segregate into two main clusters that are genetically delineated by numerous differentially expressed genes. One example is Nrxn3, which encodes a cell adhesion molecule responsible for establishing synaptic specificity (Extended Data Fig. 3h). Further in vivo experiments will be necessary to definitively show whether these transcriptionally distinct populations correspond to ipsilateral and contralateral projecting populations, and whether Nrxn3 plays a functional role in partition circuit assembly/maintenance. By identifying these populations in our data, we present a detailed molecular characterization of partition cells and reveal several novel marker genes of cholinergic interneurons cell classes.
Identification of novel alpha and gamma motor neuron markers
Traditionally, skeletal motor neurons have been defined based on their muscle innervation target4,29, developmental lineage30, morphology, and electrophysiological properties31,32. They are classified as α, beta (β), and gamma (γ) spinal motor neurons (Fig. 3a). α motor neurons directly innervate extrafusal muscle fiber neuromuscular junctions (NMJs). In contrast, γ MNs innervate intrafusal muscle spindles33. We identified skeletal motor neurons by Tns1/Bcl6 expression (as above, see Methods), and then subclustered them (Fig. 3b). We utilized the few robust genetic markers of α and γ motor neurons that have been confirmed in adult animals33–36. Thus, we identified γ motor neurons by their expression of Htr1d and α motor neurons by their high level of Rbfox3 and Spp1 expression.
Fig. 3: Transcriptional differences between alpha (α) and gamma (γ) motor neurons.
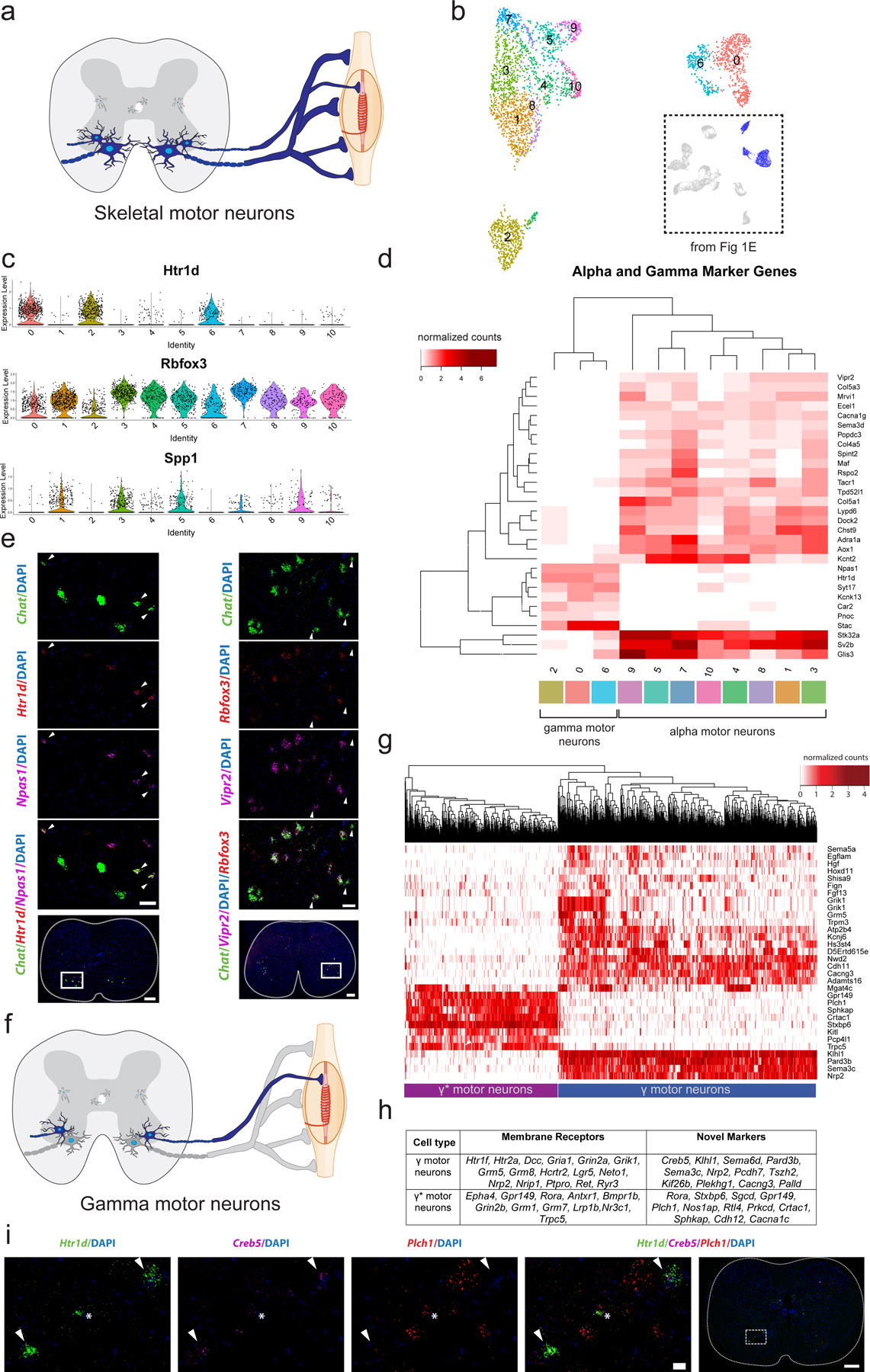
a, Transverse schematic illustrating position of skeletal motor neurons (blue) in the ventral horn of the spinal cord. Gamma motor neurons are small and innervate intrafusal muscle fibers. α motor neurons are large and innervate extrafusal fibers. b, UMAP with 11 subclustered skeletal motor neurons populations. Inset shows all cells from Fig 1e that were subclustered. c, Average expression of known γ marker Htr1d and α markers Rbfox3 and Spp1 by cluster. d, Heatmap with average expression by cluster of differentially expressed genes in α and γ populations. Differentially expressed genes between γ and α populations. e, Representative in situ hybridization against Chat/Htr1d/Npas1 and Chat/Rbfox3/Vipr2 in transverse lumbar spinal cord sections. Arrowheads indicate γ motor neurons in both images. Scale bars are 200 µm (overview) and 50 µm (inset). n=4 biologically independent animals. f, Transverse schematic illustrating γ motor neurons (blue) innervating intrafusal muscle fibers. Inset shows all cells from Fig 3b that were subsequently subclustered. g, Heatmap showing fundamental subdivision between γ and γ* motor neurons, hierarchically clustered by expression of highly variable genes among all classes of γ motor neurons (see Methods). h, Differentially expressed membrane receptors between two main populations of γ motor neurons, as well as novel markers that delineate them. i, Representative in situ hybridization against Htr1d/Plch1/Creb5 in transverse lumbar spinal cord. Plch1 and Creb5 are expressed reciprocally in Htr1d+ cells and represent γ and γ* motor neurons. Arrow heads demarcate Creb5+ γ motor neurons and * indicates γ* motor neurons. n=5 biologically independent animals. All differential expression calculated using Wilcoxon rank sum test and adjusted for multiple comparisons (Bonferroni method) (p_adj < 0.01, log2fc>0.5). All expression values were log-normalized in Seurat50. Scale bars=50 µm.
To identify putative α and γ motor neuron populations, we examined marker gene expression within each cluster (Fig. 3c). Clusters 0, 2, and 6 express high levels of Htr1d and low levels of Rbfox3 and Spp1 (Fig. 3c, Supplementary Table 2e), suggesting that they represent γ motor neurons. The remaining clusters minimally express Htr1d, suggesting that they represent α motor neurons (Fig. 3c). We calculated differential gene expression between putative α and γ motor neuron clusters, yielding a collection of novel markers of each population (Fig. 3d). To validate a putative marker of γ motor neurons (Npas1), we performed multiplexed in situ hybridization with the canonical γ marker Htr1d and Chat in the adult spinal cord. The resulting images demonstrate robust coexpression in cholinergic cells in the ventral horn (Fig. 3e). Similarly, we performed in situ hybridization comparing expression of a novel α marker (Vipr2), Chat, and an established marker of α motor neurons (Rbfox3) to confirm that Vipr2 is expressed solely in α motor neurons (Fig. 3e, Extended Data Fig. 4a,b). Additionally, Vipr2 and Htr1d have non-overlapping expression patterns in Chat+ cells (Extended Data Fig. 4c, as do Npas1 and Rbfox3 (Extended Data Fig. 4d). As a final confirmation, we show that Vipr2 and Npas1 are expressed in reciprocal populations of motor neurons in the ventral horn (Extended Data Fig. 4e). Notably, all existing α motor neuron markers are insufficient on their own to distinguish them from other cells in the spinal cord. In contrast, Vipr2 is expressed exclusively in α motor neurons. Together, these results provide a robust molecular basis for distinguishing α and γ motor neurons using newly described genetic markers.
Transcriptional profiles of gamma spinal motor neurons reveal two highly divergent motor neuron types
Gamma motor neurons innervate intrafusal muscle fibers, which maintain the tension required for skeletal muscle to function properly (Fig. 3f). Hierarchical clustering of γ motor neuron marker genes (Htr1d+) revealed two main clusters (Fig. 3g), which may be distinguished by the expression of numerous individual transcripts (Fig. 3h). Clustering may also be taken to an even more granular level, segmenting γ motor neurons into 4 distinct populations (Extended Data Fig. 5a,b)—however, the majority of variation is captured by dividing them into two populations. We noticed that many of the genes enriched in the Stxbp6+ population are also expressed more broadly by α motor neurons (Extended Data Fig. 5c,d). We named this new population γ* (pronounced gamma star), while all other Htr1d+ cells are canonical γ motor neurons. We can reliably distinguish γ from γ* by reciprocal expression of either Stxbp6 or Plch1 (γ*) and Creb5 or Pard3b (γ), both in our single cell dataset (Extended Data Fig. 5e–h) and by in situ hybridization (Fig. 3i, Extended Data Fig. 5i,j). Owing to the extensive number of differentially expressed genes that separate γ from γ*, as well as the lack of an intermediate population between them that would suggest that they are two functional ‘states’ of γ motor neurons, we hypothesize that these cell types represent a fundamental subdivision of the fusimotor system. However, future work will be necessary to conclusively determine if this subdivision corresponds to transient ‘activity states’ of γ motor neurons or developmentally/functionally distinct populations of cells.
One kind of skeletal motor neuron that has been defined physiologically and anatomically, but not yet transcriptionally, is the β motor neuron. This cell type innervates both intrafusal and extrafusal fibers, and therefore has properties of both α and γ motor neurons37. Could γ* actually correspond to this historically elusive and long-sought skeletal motor neuron subtype? We present a list of novel markers that differentiate the γ and γ* populations (Fig. 3h), which will enable a more detailed exploration in the future.
Transcriptional analysis of alpha motor neurons reveals motor pools
During development, motor neurons require cell-intrinsic and extrinsic cues that coordinate expression of transcription factors and cell adhesion molecules3,4. This molecular cascade enables α motor neurons to form into groups, known as motor pools, that cluster alongside one another in the spinal cord and collectively innervate the extrafusal fibers of distinct muscles34 (Fig. 4a). Owing to a vast heterogeneity in muscle location throughout the body and the types of muscle contractions required for coordinated movement, mature α motor neurons display substantial functional differences38–40. However, a priori, it was unknown whether or not electrophysiological subtypes of α motor neurons with specific innervation targets express different transcriptional programs.
Fig. 4: Alpha (α) motor neuron pool, position, and electrophysiological subtype reflect transcriptional differences.
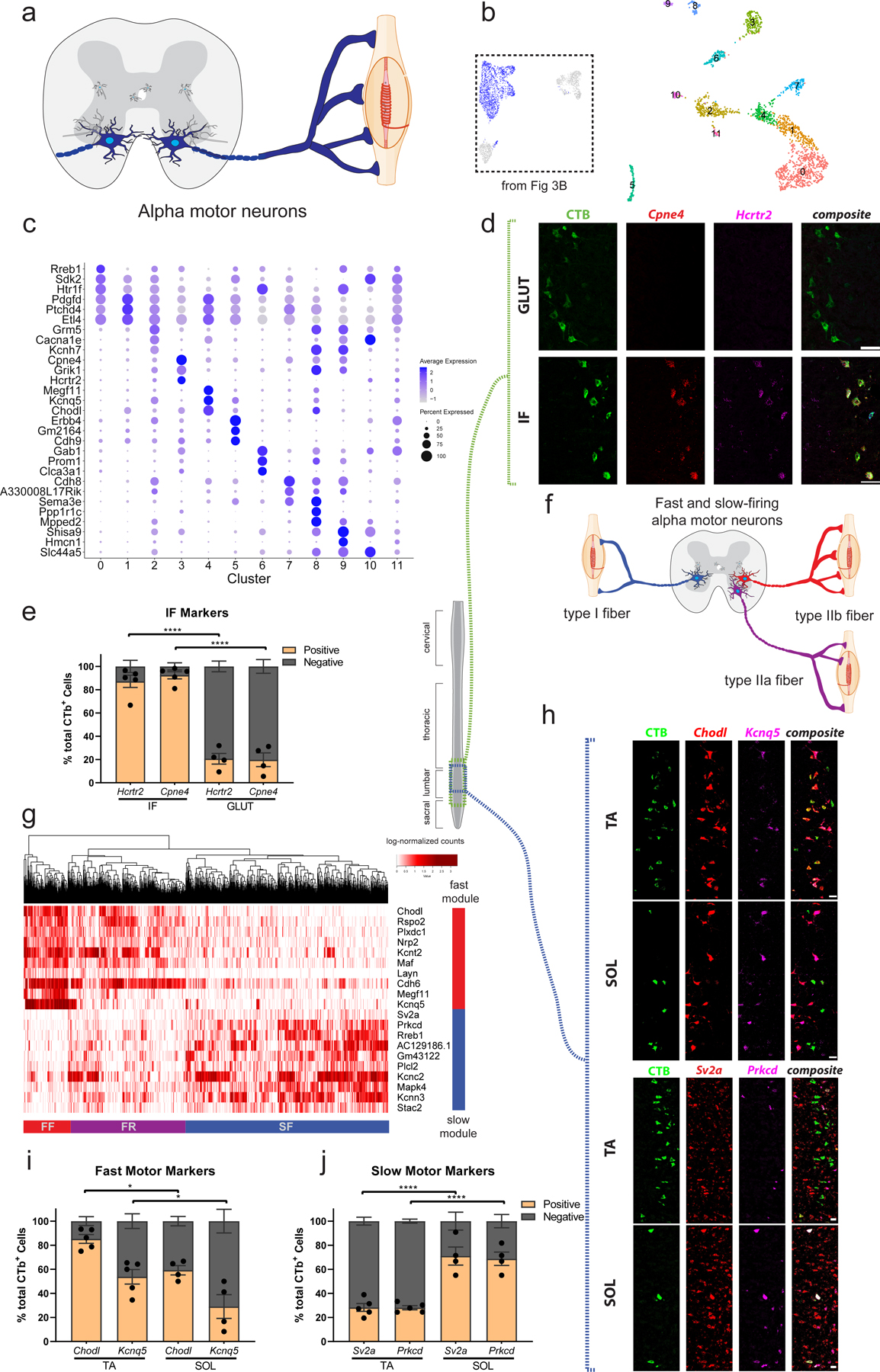
a, Transverse schematic shows α motor neurons (blue) innervating extrafusal muscle fibers. b, UMAP with 12 subclustered α motor neuron populations. Inset shows all α motor neurons from Fig. 3b that were subclustered. c, Novel marker gene expression across α motor neuron subpopulations. Dot size is proportional to the percent of each cluster expressing the marker gene, while blue color intensity is correlated with expression level. d, Representative in situ hybridization against novel (Hcrtr2) and known (Cpne4) intrinsic-foot (IF) motor pool markers in longitudinal sections, overlaid with CTb labeled cells that innervate the gluteus (GLUT) and IF. n=5 (IF) and 4 (GLUT) biologically independent animals. e, Proportion of CTB-labeled cells from GLUT and IF that are labeled with Hcrtr2 and Cpne4 shows novel and known markers selectively label the IF motor pool. n=5 (IF) and 4 (GLUT) biologically independent animals. f, Schematic illustrating slow-firing (SF, blue), fast fatigue-resistant (FR, purple) and fast fatigable (FF, red) α motor neuron populations innervating type I, IIa, and IIb fibers respectively. g, Heatmap showing all α motor neurons hierarchically clustered and colored by expression of differentially expressed genes between fast (Chodl+) and slow (Sv2a+) α motor neurons shows three cell populations corresponding to SF, FR, and FF motor neurons. Red and blue bars show gene modules enriched in fast- and slow-firing α motor neurons, respectively. h, Representative in situ hybridization in longitudinal sections against novel (Kcnq5, Prkcd) and known (Chodl, Sv2a) fast and slow-firing motor neuron markers, respectively. Images show CTb labeled cells that innervate a muscle with predominantly fast-twitch fibers (TA) and slow-twitch fibers (SOL). n=5 (TA) and 4 (SOL) biologically independent animals. i, Proportion of CTB-labeled cells from TA and SOL that are labeled with Chodl and Kcnq5. The TA has a significantly larger proportion of Chodl+ and Kcnq5+ cells than the SOL, though Kcnq5+ cells are significantly less frequently found in both populations. n=5 (TA) and 4 (SOL) biologically independent animals. Adjusted p-values=0.0197 (Chodl), 0.0262 (Kcnq5). j, Proportion of CTB-labeled cells from TA and SOL that are labeled with Sv2a and Prkcd. The SOL pool has a significantly larger proportion of Prkcd+ and Sv2a+ cells than the TA. n=5 (TA) and 4 (SOL) biologically independent animals. All differential expression calculated using Wilcoxon rank sum test and adjusted for multiple comparisons (Bonferroni method) (p_adj < 0.01, log2fc>0.5). All expression values were log-normalized in Seurat50. Scale bars=50µm (h), 75 µm (all others). One-way ANOVA with post-hoc Sidak multiple comparison test between same-gene conditions. *=p value<0.05, **=p value <0.01, ***=p value<0.001, ****=p value<0.0001. Error bars are SEM.
Foundational studies strongly suggest that motor neuron target specificity arises from transcriptional heterogeneity3 during development, but whether these differences persist into adulthood is unknown. We subclustered the α motor neuron transcriptomes (as above—see Methods; Fig. 4b). This analysis revealed 12 clusters. Most α motor neurons fall in one large population that consists of clusters 0, 1, and 4. Other clusters diverge transcriptionally from this main population, and express specific distinguishing markers (Fig. 4c). Cluster 3 expresses Cpne4 and Fign specifically (Fig. 4c, Supplementary Table 2f), genes which were recently shown to be highly expressed in intrinsic-foot (IF) motor neurons during development38. Furthermore, clusters 7 and 8 express high levels of Sema3e (Fig. 4c)—which within the developing lumbar spinal cord is a specific genetic marker for Gluteus maximus (Glut)-innervating motor neurons41, as well as shoulder-innervating motor neurons in the cervical spinal cord42. Sema3e encodes a protein that, together with its receptor PlexinD1, contributes to synaptic specificity between sensory and motor neurons in that pool41,42. These findings raise the hypothesis that single-nucleus transcriptomics is sufficient to distinguish subpopulations of α motor neurons that in the adult mouse specifically innervate unique muscle groups.
To test whether the α motor neuron clusters that we identified by transcriptomics correspond to functionally defined motor pools, which collectively innervate a specific muscle, we performed intramuscular injections of fluorescently conjugated cholera toxin beta subunit (CTB)—a recombinant protein that is taken up by motor neuron axon terminals and transported retrogradely to the cell body43. By performing simultaneous CTB labeling and in situ hybridization against candidate marker genes for a given population, we are able to perform whole-transcriptome characterization of functionally defined cell populations (akin to performing laser-capture microdissection on an anatomically distinct population of cells). In other words, the individual marker genes that we derive are not the end goal of these experiments, but rather a means to define the full transcriptome of functionally defined motor neuron populations. To accomplish this, we first demonstrated that IF adult motor neurons express the developmentally defined IF marker Cpne4 (Fig. 4d,e), while other populations of motor neurons (GLUT) do not. This experiment enabled the tentative conclusion that cluster 3 from our transcriptional data corresponds with IF motor neurons labeled by CTB, because Cpne4 was exclusively expressed in that population using orthogonal methods. However, to conclusively demonstrate this finding, we tested a novel, functionally intriguing marker that we found specifically expressed in cluster 3 of our single-nucleus experiment (Hcrtr2). Similarly, this gene was a robust marker of IF motor neurons (87% of IF MNs are Hcrtr2+) and not substantially expressed in GLUT motor neurons (21% of GLUT MNs are Hcrtr2+) (Fig. 4d,e). It is intriguing that a specific marker of IF motor neurons, Hcrtr2, encodes a membrane-bound hypocretin (orexin) receptor, a gene whose disruption causes muscle weakness and cataplexy in animal models44.
We next examined whether Sema3e+ clusters present in our dataset correspond to the GLUT motor pool. Indeed, we found an increase in proportion of both Sema3e+ and Cdh8+ cells in GLUT-innervating motor neurons when compared to the IF motor pool (Extended Data Fig. 6a). Overall, our results demonstrate that motor pools in the adult mouse have distinct transcriptional properties, which can be resolved by single-nucleus sequencing of α motor neurons. Still, there are far more motor pools (~fifty)4 than clusters (twelve). To explain this discrepancy, we propose that transcriptional differences among adult α motor neurons are more subtle than those that emerge during embryonic development, as is the case in the olfactory system45, and single-nucleus profiling is only sufficient to delineate more dramatic transcriptional differences. We show that the embryonic marker for GLUT innervating motor neurons (Sema3e) is less specific in the adult mouse, suggesting that future work should seek to utilize more robust, novel markers (like the ones identified here) to map transcriptional subpopulations onto their peripheral targets. The abundance of specifically expressed genes in each novel population will empower further functional study of the adult motor system.
Fast and slow-firing alpha motor neurons have divergent transcriptional signatures
Skeletal muscles innervated by motor neurons are composed of slow (type I), intermediate (type IIa), and fast (type IIx/IIb) twitch fibers, each of which requires different patterns of synaptic input (Fig. 4f)32. Likewise, α motor neurons innervating these fibers are divided into slow-firing (SF), fast-fatigue resistant (FR), and fast-fatigable (FF) cell types—each with specific electrophysiological and metabolic properties34. Importantly, these groups of α motor neurons have drastically different susceptibilities to degeneration in neuromuscular disorders such as ALS5,46. Thus, determining the transcriptional programs that define these classes of motor neurons may provide insight into divergent function and susceptibility to disease5,32. Past work has begun to address this question, leading to the discovery of several markers of FF/FR (Chodl, Mmp9)31 and SF (Sv2a) motor neurons. However, the broader transcriptional differences beyond these few validated markers has thus far remained elusive.
To identify the gene expression modules that underly differences between SF, FR, and FF motor neurons, we segmented all α motor neurons by their mutually exclusive expression of the known markers Chodl (fast-firing) and Sv2a (slow-firing)47. We identified differentially expressed genes between Chodl+ and Sv2a+ α motor neurons, and then hierarchically clustered cells based on expression of this gene set (Fig. 4g). This analysis was sufficient to segment α motor neurons into three main populations (Fig. 4g). It is clear that fast and slow gene modules are expressed in reciprocal populations of α motor neurons across virtually all motor pools, but in different proportions (Extended Data Fig. 6c,d). Importantly, hierarchical clustering of cells revealed three main populations of cells, two of which are Chodl+ and one that is Sv2a+. As Chodl is expressed in both FF and FR and Sv2a is expressed in SF motor neurons, this raises the possibility that the three populations may in fact correspond to FF, FR, and SF α motor neurons. We identified novel transcripts to distinguish each population, including a putative marker of FF α motor neurons (Kcnq5) and a specific marker of SF motor neurons (Prkcd). To confirm these findings, we demonstrated that Kcnq5 is expressed in a subpopulation of Chodl+ (FF/FR) and Mmp9+ (FF/FR) α motor neurons (Extended Data Fig. 6e,f, 7a). We also validated that Prkcd is expressed in Sv2a+ α motor neurons and is excluded from Mmp9+ motor neurons (Extended Data Fig. 7b,c).
To validate that the expression of the fast and slow-firing gene modules that we identified define electrophysiological subtypes of motor neurons, we leveraged unique properties of the soleus (Sol) and tibialis anterior (TA) muscles, which consist of predominantly slow and fast-twitch fibers, respectively31,48. We performed intramuscular CTB injections that label TA and Sol-innervating motor neurons and measured expression of novel FF and SF marker genes by in situ hybridization (Fig. 4h–j, Extended Data Figs. 6b, 7d,e). In Sol-innervating motor neurons, Prkcd was expressed in 69% of cells, while Kcnq5 was expressed in just 29%. This trend was opposite in the TA, which contained 54% Kcnq5+ cells and only 28% Prkcd+ cells (Extended Data Fig. 7d,e). We observed almost identical patterns of expression for Prkcd and Sv2a, however it was clear that within these motor pools Kcnq5 was only expressed in a subpopulation of fast-firing motor neurons. Intriguingly, the proportion of Kcnq5+ cells in the TA (54%) is quite close to the proportion of type IIb fibers in that muscle48. These data support the conclusion that Kcnq5 is expressed in FF motor neurons, while Chodl is more broadly expressed in FF and FR motor neurons—although future electrophysiological characterization will be required to demonstrate this conclusively. In addition, Prkcd is a robust and specific marker of SF motor neurons.
Both the FF and SF expression modules contain genes that encode subunits of voltage-gated potassium channels (Table 2). Potassium channel subtypes play a vital role in determining the resting membrane potential and basal firing rate of neurons. The differential expression of potassium channel isoforms suggests a mechanism through which α motor neuron electrophysiological properties are established32. Furthermore, we found that SF neurons specifically express Prkcd, which encodes a protein with a fundamental role in determining how cells respond to oxidative stress and DNA damage. In contrast, FF motor neurons do not express Prkcd, but instead express high levels of Prkcb (Table 2), which encodes a key regulator of autophagy. Dysregulation of each of these pathways is thought to be fundamental to ALS ontology and progression. Collectively, these data reveal a rich transcriptional basis for the functional diversity of fast- and slow-firing motor neurons.
Table 2:
Enriched GO terms among genes in fast and slow-firing expression modules.
| Voltage-gated K+ channels | Calmodulin binding | Calcium Ion Binding | Protein Kinase C Activity | |
|---|---|---|---|---|
| GO-TERM | GO:0005249 | GO:0005516 | GO:0005509 | GO:0019901 |
| GO-TERM adj p-value | 4.40E-05 | 1.50E-04 | 6.70E-04 | 4.10E-02 |
| fast-enriched | Kcnq5, Kcnt2 | Edil3, Cdh6, Dgkg, Mctp1, Mcc | Prkcb | |
| slow-enriched | Kcnq3, Kcnc2, Kcnd2 | Adcy8, Esrrg, Kcnn3, Kcnq3, Unc13c | Crtac1, Plch1, Fstl4, Dner | Prkcd |
Discussion
We report here a detailed molecular characterization of the adult mammalian motor system at single-cell resolution. Using a transgenic motor neuron enrichment strategy, we have greatly expanded the transcriptional characterization of these populations and discovered new divisions within the autonomic and somatic motor systems.
Within the autonomic motor system we discovered 16 subpopulations of sympathetic visceral motor neurons, including several clusters that localize to the sacral spinal cord. Visceral motor neuron subtypes express entirely different repertoires of neuromodulatory peptides, such as somatostatin, neurotensin, and proenkephalin. This suggests an underlying peptidergic logic that governs transmission between visceral motor neurons and their peripheral targets—a long-standing hypothesis that has never been comprehensively demonstrated. Together, our findings inspire the possibility that specific visceral motor neuron populations may someday be selectively targeted with therapies to treat autonomic dysfunction in humans.
Within the somatic motor system, we present novel α and γ motor neuron markers and have identified a new skeletal motor neuron population (γ*), which shares the expected features of the elusive β motor neuron population. Furthermore, we find that transcriptional subpopulations of α motor neurons correspond to previously described, distinct motor pools38,42. We propose that other transcriptionally distinct α motor neuron populations in our dataset may similarly correspond to specialized motor pools, and we offer numerous novel markers to facilitate testing this hypothesis.
Our analysis also reveals gene modules that are selectively expressed in electrophysiologically and metabolically distinct populations of fast and slow-firing α motor neurons. These differentially expressed genes offer insight into how motor neuron subtypes establish unique biophysical properties that are specifically matched to the properties of their muscle innervation targets32. Indeed, our analysis reveals that fast and slow-firing α motor neurons express a divergent collection of potassium channel subunits—which are specifically required to tune the resting membrane potential and firing rate in neurons. These expression profiles may also help suggest approaches to rescue aberrant function in fast-firing motor neurons, which specifically degenerate in ALS5,34,46,49.
We have identified new markers for motor neuron populations, unlocking unprecedented genetic access to these important cells in the adult spinal cord. Defining the transcriptomes of distinct motor neuron types introduces the possibility of engineering more refined stem cell models and provides a single-cell framework for characterizing their behavior in health and disease.
Methods
Data reporting
No statistical methods were used to predetermine sample size, but our sample sizes (such as minimum cluster size) are similar to those reported in previous publications7,13,50. No randomization was used in assigning experimental conditions to animals. All quantification of microscopy images was performed blinded. CTB experiments were included in analysis if, under blinded conditions, it was determined that CTB-labeled cells were easily distinguishable from background staining (e.g. successful retrograde labeling occurred). Additional information can be found in the Life Sciences Reporting Summary.
Statistics
For in situ hybridization quantifications, one-way ANOVA tests with Sidak and Tukey post-tests were used to determine statistical significance of differences among specific (Sidak) comparisons or all conditions (Tukey). For single-cell analysis, all pseudo-bulk differential expression was calculated using the Wilcoxon rank sum test and adjusted for multiple comparisons (Bonferroni method), implemented by default in Seurat50. These tests did not require assumptions of normally distributed data.
Mouse crosses
CHaT-IRES-Cre (Chat-CRE/Chat-CRE) mice were purchased from JAX (stock no. 006410m B6;129S6-Chattm2(cre)Lowl/J) and crossed with ROSAnT-nG/ROSAnT-nG (stock no. 023035, B6;129S6-Gt(ROSA)26Sortm1(CAG-tdTomato*,-EGFP*)Ees/J). F1 heterozygous reporter mice were aged to P100–150, and then sacrificed for subsequent sequencing experiments. All in situ hybridization was performed using wild-type male and female P100–150 B6 mice purchased from JAX (stock no. 000664). Mice were housed at ~55% humidity, 25° C, on a 12:12 light/dark cycle. Humane experiments were performed with ethical oversight by the Stanford Administrative Panel on Laboratory Testing (APLAC, protocol #30643).
Mouse nuclei collection
Four to 8 mice in five independent experiments were euthanized with CO2 and decapitated caudal to the brain stem. Their spinal columns were severed just caudal to the sacral spinal cord, and rapidly cut out as described7. Briefly, a blunt 18-gauge syringe containing ice-cold PBS was inserted into the caudal end of the spinal cord and used for rapid hydraulic extrusion of the entire, intact cord. Two spinal cords at a time were homogenized with a Dounce Homogenizer in 2 mL of nuclei extraction buffer (neB, Supplementary Table 4a). Spinal cords were homogenized with 10 strokes of Pestle A, followed by 5 strokes of Pestle B. The entire homogenate was transferred to a 25 mL, round-bottom plastic ultracentrifuge tube and 8 mL of nuclei spin buffer 1 (nsB1, Supplementary Table 4a) was added. Five mL of nuclei spin buffer 2 (nsB2, Supplementary Table 4a) were layered gently underneath the homogenate, and the gradient was spun for 15 minutes at 4000*g in a 4°C, benchtop swinging bucket centrifuge (Beckman 5810R). The supernatant was rapidly discarded, and the nuclei were gently resuspended in 5 mL nsB1. Five mL of nuclei spin buffer 3 (nsB3) were gently layered underneath, and the resulting gradient was spun for 15 minutes at 4000*g at 4° C. The supernatant was rapidly discarded, and the pellet was resuspended in 400 uL nuclei FACS buffer (nfB). Next, 0.5 uL DAPI was added to enable doublet discrimination on the sorter.
Fluorescence activated nuclei sorting (FANS)
FANS was performed on a BD Biosciences FACSAria II flow cytometer. Briefly, following calibration of the FACS machine or power-up of the flow cytometer, single nuclei were gated using forward scatter (FSC), side-scatter (SSC), and DAPI measurements to ensure that doublets were gated out. Following this initial gating, EGFP+/tdTomato− nuclei were identified using a 2-dimensional scatterplot. These nuclei stood out from the main population, enabling double-gating. To ensure that our samples would contain both cholinergic and non-cholinergic nuclei, we then sorted ~10,000–15,000 EGFP+/TdTomato− nuclei and 20,000–25,000 EGFP−/TdTomato+ nuclei into 400 uL 10X loading buffer (Supplementary Table 4a). After sorting was completed, this nuclei mixture was spun at 300xg for 5 minutes and resuspended in 10X loading buffer according to 10X loading criteria for the precise number of nuclei sorted—a number that varied.
Droplet-based snRNA-seq
For droplet-based snRNA-seq, libraries were prepared using the Chromium Single Cell 3′ Reagent Kits v.3 according to the manufacturer’s protocol (10x Genomics). The generated snRNA-seq libraries were sequenced using NextSeq 500/550 High Output v2 kits (150 cycles) to an average read depth of ~500,000 reads/cell.
Mouse CNS in situ hybridization
Mice were humanely euthanized using CO2 for 5 minutes prior to decapitation. The spinal cord was rapidly hydraulically extruded, as above, and briefly dried. For longitudinal sections, the spinal cord was placed into a plastic freezing mold and frozen briefly on dry ice, before adding O.C.T Compound freezing media (Tissue-Tek) to ensure that the spinal cord was frozen completely flat to the mold. For transverse sections, spinal cords were cut into three segments—roughly lumbosacral, thoracic, and cervical. Sections were mounted and cut to a thickness of 20 um using a Leica CM3050 S Cryostat. Sections were processed immediately or stored for 1–2 weeks at −80° C.
Sections were processed for RNAScope v2 (ACD Biosciences) according to manufacturer instructions for fresh frozen tissue. All in situ hybridization probes used are listed in Supplementary Table 4b. Briefly, tissue sections were fixed with 4% PFA at 4° C, and subsequently dehydrated with 5 minutes each of 50% EtOH, 70% EtOH, and finally 100% EtOH. Samples were dried and a hydrophobic barrier was drawn around each section. Samples were then incubated with Protease IV (ACD Biosciences) for 15 minutes at room temperature before removing and washing in 1X PBS. Before staining, probes were equilibrated to 40° C for 10 minutes and then cooled to room temperature. Mixtures of probes in channels 1–3 were diluted, and then incubated on samples for 2 hrs in a humidified 40° hybridization oven. Samples were washed twice in PBS, then incubated in the hybridization oven at 40° with Amp-1-FL, Amp-2-FL, Amp-3-FL, and Amp-4-FL for 30, 15, 30, and 15 minutes respectively (with 2X washes in between each incubation). Samples were then washed twice, briefly dried, and mounted with DAPI-containing VectaShield mounting media (Vector Laboratories, H-1200-10). Subsequent imaging was performed using a Zeiss LSM 710 confocal microscope. Image analysis and background correction was performed in ImageJ.
Analysis of snRNA-seq data
Counts per gene were generated by aligning reads to the mm10 genome (Mus_musculus.GRCm38—NCBI: GCA_000001635.2) using CellRanger software (v.3.0.0) (10x Genomics) running on the Sherlock Stanford Computing Cluster. To account for unspliced nuclear transcripts, reads mapping to pre-mRNA were counted. After this, the CellRanger aggr pipeline was used to aggregate all libraries and normalize the read depth between libraries before data merging (with the default parameters) to generate a gene-count matrix. We then used the cell-detection method employed by CellRanger to exclude doublets and empty droplets (10X genomics). One doublet-containing cluster was also removed subsequently.
Normalization, clustering and subtype annotation
Most subsequent analysis was performed using the Seurat R Package50 (https://satijalab.org, V3.0). All 43,890 transcriptomes were normalized for read-depth with the CellRanger aggr function, and then loaded into Seurat. The nuclei were batch-corrected using the Seurat Integrate function as previously described50, which enables cells from different experiments to be projected into the same high and low-dimensional spaces. Principal component analysis (PCA) was performed on the whole dataset, and the top 15 components were used to generate a uniform manifold approximation (UMAP). The same principal components were used to perform graph-based clustering via the FindClusters function, which identified 39 total clusters. To annotate clusters, a manually curated list of markers for major cell types was assembled from the literature – including excitatory interneurons, inhibitory interneurons, cholinergic neurons, oligodendrocytes, endothelial cells, astrocytes, and microglia. Average expression of these marker genes was calculated using Seurat (AverageExpression). Clusters that were positive for these marker genes were annotated by cell type (e.g. Aqp4+ clusters were classified as astrocytes).
To identify specific marker genes for each neuronal subtype, differential gene expression analysis was performed using the Seurat FindAllMarkers function, which leverages Wilcoxon rank sum test differential expression testing. Briefly, differentially expressed genes were identified for each subtype with respect to all remaining cells. Each subtype was down-sampled to 250 cells in order to compensate for stoichiometric biases. An FDR-corrected p-value threshold was set at p<0.01, and only markers with a log2-fold change>0.5 were included in subsequent analysis. To find robust markers, lists of differentially expressed marker genes were rank order based on the average expression level in all cells not included in the subtype of interest (low background expression yields high rank). Even though this occasionally resulted in the prioritization of markers that are not ubiquitously expressed in the subtype of interest, we determined this method empirically produces robust markers and view the loss of signal is some cells a likely result of a high dropout rate for single-nucleus sequencing. However, only genes detected in at least 25% of the cells within the given identity class were considered as candidate subtype markers. Skeletal and visceral motor neurons, as well as cholinergic interneurons, were each separately subsetted, subclustered, and umap-projected. Differential expression analysis was performed on clusters within each population, as above and explained in the main text.
To determine batch-to-batch variability and reproducibility of effects, all levels of clustering were plotted as UMAPs for each replicate, along with the age, sex, and number of mice that were pooled per replicate (Extended Data Fig. 8). This analysis demonstrated that cluster identity was largely invariant to sex, age, and number of mice pooled.
Retrograde Labeling
Motor neurons innervating the tibialis anterior (TA), soleus (SOL), or intrinsic muscles of the foot (IF) were retrogradely labeled by contralateral intramuscular injection of cholera toxin B subunit (CTb) conjugated to Alexa 488. Adult mice were deeply anesthetized with 2–3% isoflurane mixed with oxygen. To access the TA and SOL muscles, a small incision was made on the front of one hindlimb (TA) and on the lateral side of the contralateral hindlimb (SOL). For IF muscle injections CTb solution was directly injected into the footpad. Injections were performed with a 10 µl Hamilton syringe equipped with a 33-gauge needle. For all muscles were injected at three sites with 0.5 µl of 0.5–1% CTb-488 solution. Following each injection, the wound site was closed with 7mm Reflex clips. Spinal cords (and injected muscles) were harvested 5 days after injection. For the isolation – muscles were fixed for 20 min in 4% PFA at room temperature. They were washed in PBS, cryopreserved overnight in 30% sucrose, and embedded in O.C.T. compound. For the soleus and TA: longitudinal muscle sections were cut at 30 µm on the cryostat. Sections were immunostained with rabbit anti-CTb (abcam) and bungarotoxin (BTX) conjugated to CF55 (Biotium). Muscles were analyzed by microscopy to confirm the specificity of CTb labeling.
Tissue Preparation
Mice were transcardially perfused with phosphate-buffered saline (PBS) and spinal cords were removed by hydraulic extrusion. Spinal cords were fixed in 4% PFA overnight at 4°C. The following day tissues were washed with PBS prior to overnight cryoprotection in 30% sucrose. Tissues were embedded in O.C.T compound and 20 µm longitudinal sections were acquired on a Leica CM3050S cryostat.
Combined in situ hybridization and immunohistochemistry
Cryosections were rinsed in PBS and baked at 60°C for 45 min. Slides were then fixed in 4% PFA for 1 hour at room temperature. Ethanol dehydration was then performed with 50%, 70%, 100%, 100% ethanol for 5 minutes each. After air drying, sections were treated with RNAScope® hydrogen peroxide for 10 minutes, followed by antigen retrieval at 95°C in RNAScope® Target Retrieval Buffer. Slides were then baked at 60°C for 45 min. The remaining steps of the in situ hybridization procedure were performed using the RNAScope® Multiplex Fluorescent Reagent Kit v2 according to manufacturer guidelines. Upon completion of the in situ, CTb immunostaining was performed. Briefly, spinal cord sections were blocked in PBS containing 0.2% Triton-X and 10% donkey serum (Jackson ImmunoResearch). Rabbit anti-CTb (ab34992, Abcam) diluted at 1:8,000 in PBS containing 0.2% Triton-X was incubated overnight at 4°C. The following day, secondary antibody labeling was performed with donkey anti-rabbit-Alexa488 (Invitrogen).
Image Analysis
All sections with CTb retrograde labeling were imaged on a confocal microscope (Leica SP8, Leica Biosystems). 10x tile scans were acquired at high resolution (2048 × 2048) with a z-step size of 2.5 µm using LAS Navigator. Maximum intensity images were generated from merged tile scans (LAS X Software, Leica Biosystems). To prevent counting false-positive motor neurons that were artifactually labeled with CTb leakage, cells with high levels of CTb backfill were uniformly identified in each sample through thresholding. All CTb+ motor neurons were then scored cell for the presence or absence of a particular RNAScope® probe set.
Visceral motor neuron spatial localization
We determined where cells within each visceral motor neuron subcluster localize along this axis by the following method. Briefly, a list of approximately 100 genes that localize to the intermediolateral cell column (iMLC) was compiled from the Allen Spinal Cord Atlas10 (Supplementary Table 4c). Among these genes, we identified 29 marker genes differentially expressed in at least one visceral motor neuron subcluster. Differentially expressed genes among visceral clusters were identified as above. We then counted the number of iMLC cells that were positive for each gene in all available adult spinal cord sections in the Allen Spinal Cord Atlas and noted the relative position of each positive cell along the rostral-caudal spinal cord axis. For each marker gene, we divided the number of positive cells at each rostral-caudal section by the average positive cell count for that gene across all sections. We then normalized this cell density by the maximum number of cells in any section that were positive for that gene, to generate a normalized cell count representing the relative enrichment of positive cells in different sections of the spinal cord. Due to the sparsity of the data, we fit a polynomial function (order=8) to the normalized count, producing 29 independent polynomial functions (gene densities) that reflect the density of cells expressing each gene over the adult mouse spinal cord. We then estimated the positional density of each scRNA-seq visceral motor neuron cluster as the weighted average of the gene desities for which the gene is differentially expressed in the cluster. The averging weights were calculated as the log2 fold change of the gene expression level with a given cluster with respect to the mean expression of the gene across all clusters. This generates an estimated positional density for each cluster along the rostral-caudal spinal cord spatial atlas as reported.
If the log2-FC>0 and the FDR-adjusted p-value was <0.01, we considered a cluster enriched for expression of that gene.
Visceral motor neuron spatial validation by in situ hybridization
Spinal cords from wild-type mice were rapidly hydraulically extruded and cut into six uniform pieces, spanning from sacral to upper thoracic spinal cord, and frozen in cryopreservant. Four sequential cryostat sections were cut at 20 µm every 600 µm along the rostral-caudal axis to fully sample the thoracic and sacral autonomic columns. All subsequent tissue processing for RNAScope v2 (ACD Biosciences) was performed according to manufacturer instructions for fresh frozen tissue.
Background subtraction was performed on each channel of all images. For each image, masks of all cholinergic cells were made using the ChAT channel. To make the masks, a pixel value threshold and size cutoff were applied to remove noise, and the resulting images were converted into binary form. Using the binary images, masks corresponding to non-visceral MN cells were manually removed, and occasionally, visceral MN masks were manually split. Images without visceral MNs were not processed.
For each visceral MN mask, mean and maximum pixel values were obtained from the non-ChAT channels, which contain RNA scope data from the genes of interest (Rxfp1, Fbn2, Nts, Creb5, Piezo2, Cdh8, Cpne4). This was done by redirecting each binary image to the corresponding image/channel and analyzing particles. For each gene of interest, a threshold was set on the mean and maximum pixel values such that visceral MNs surpassing the threshold were counted as expressing the gene of interest. The total number of visceral MNs, number of visceral MNs expressing the gene of interest, and percentage of visceral MNs expressing the gene of interest were plotted as a function of distance along the rostral-caudal axis. The analysis was performed by a researcher who was blinded to the genes that were probed for.
Extended Data
Extended Data Fig. 1. Single-nucleus transcriptional analysis of the adult mouse spinal cord reveals canonical cell types.
a, Canonical cell class labels, visualized on UMAP. b, Average log-normalized marker gene expression across canonical cell classes. c–d, Representative in situ hybridization against Chat/Nos1 in transverse sacral (c) and thoracic (d) spinal cord hemi-sections. n=3 biologically independent animals. e-f, Average log-normalized expression of Zeb2 (e) and Fbn2 (f) across all cholinergic clusters (labeled), overlaid on UMAP. Dotted line surrounds clusters corresponding to visceral motor neurons. g, Representative in situ hybridization against Chat/Fbn2 in transverse thoracic spinal cord hemi-section. n=3 biologically independent animals. Scale bars=200 µm (c–d) and 100 µm (g). LAC=lateral autonomic column (c,d), VH=ventral horn (c,d).
Extended Data Fig. 2. Novel markers of skeletal motor neurons confirmed by Allen Spinal Cord Atlas in situ hybridizations.
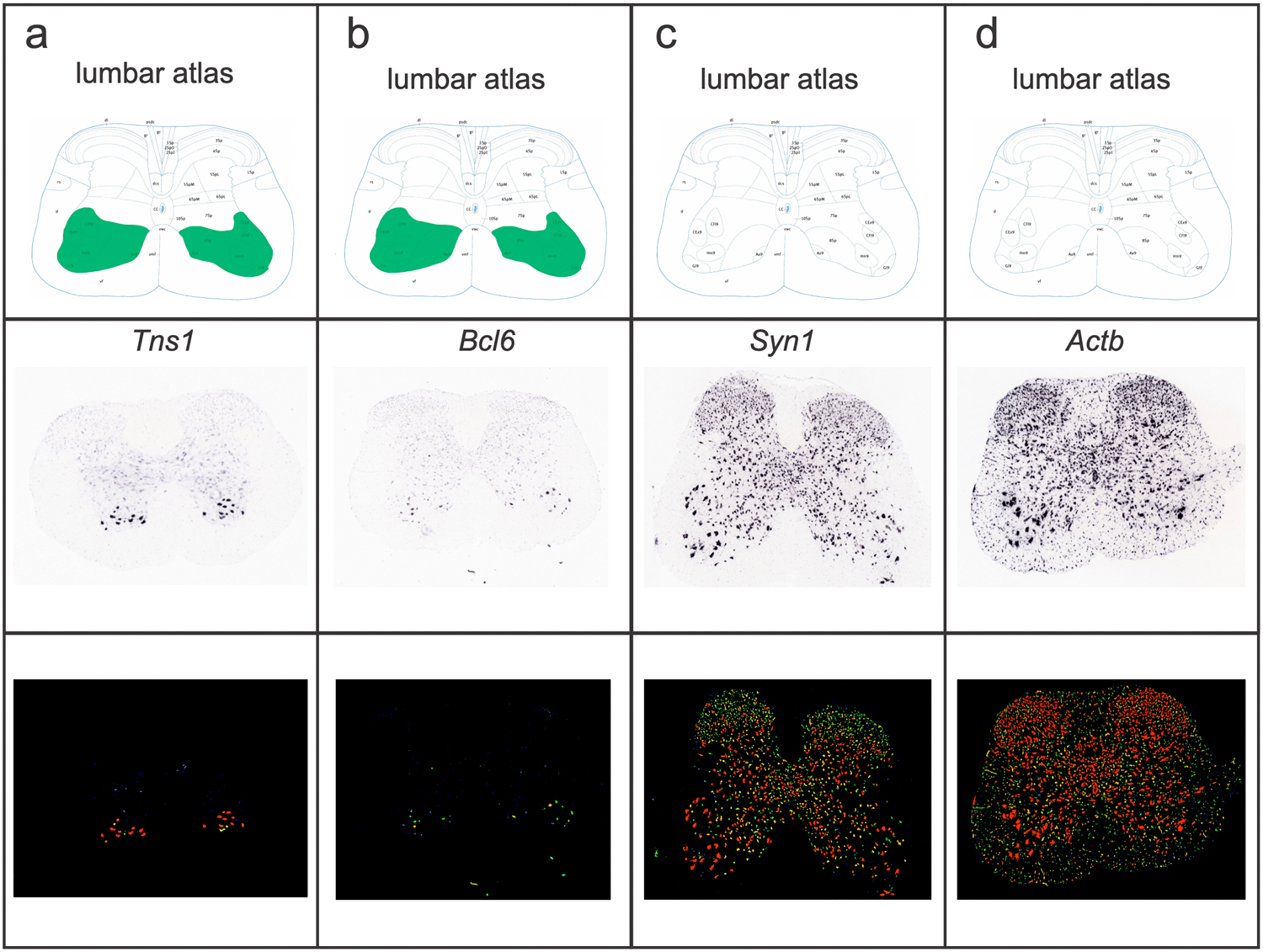
a-d, Transverse schematic illustrating expected positions of skeletal motor neurons in ventral horn (VH, green) in lumbar spinal cord. Second row—corresponding in situ hybridization against Tns1 (a), Bcl6 (b), Syn1 (c), and Actb (d). Third row—expression mask shows relative enrichment of Tns1 and Bcl6 in small and large cell bodies in the VH.
Extended Data Fig. 3. Visceral motor neuron populations express selective repertoires of neuropeptides and are spatially distinct.
a, Estimated relative density of visceral motor neurons along the rostral-caudal axis of the spinal cord based on Allen Spinal Cord Atlas10. Density functions are combined density estimates of marker genes for each cluster (see Methods). Clusters were grouped according to shape of density function, with clusters 3,7, and 10 clearly enriched in the sacral spinal cord. b, Validation of visceral spatial modeling from (a) via high-resolution in situ hybridization for Chat and visceral cluster markers (Piezo2, Cdh8, Creb5, Cpne4, Fbn2). Plots show number of motor neurons counted in the autonomic column, added across three counted slides in each region. Individual data points for total visceral motor neurons shown with filled circles, while marker gene-positive cell numbers shown with filled triangles. n=2 (Piezo2, Cdh8, Creb5, Cpne4) or 5 (Fbn2) biologically independent replicates. Error bars are SEM. c, Average log-normalized expression of Rxfp1 and Nts across all visceral motor neuron clusters (labeled), overlaid on UMAP. d, Representative in situ hybridization against Chat/Fbn2/Rxfp1 in transverse sacral spinal cord shows coexpression in the autonomic column but not in the ventral horn (VH). n=3 biologically independent animals. Scale bar=100 µm. e, Average log-normalized expression of Adra2a across all visceral clusters (labeled) shows that sporadic expression exists across populations, overlaid on UMAP. f, Representative in situ hybridization against Chat/Piezo2/Cdh8 in cholinergic cells around the central canal (CC). Scale bar=50 µm. n=2 biologically independent animals. g, Average log-normalized expression of Gldn across all cells in spinal cord shows clear enrichment in partition cell cluster (arrowhead), overlaid on UMAP. h, Average log-normalized expression of Nrxn3 across cholinergic interneurons shows that Nrxn3 expression is limited to half of partition cells (arrowhead), overlaid on UMAP.
Extended Data Fig. 4. Vipr2 and Npas1 are novel, robust, and specific markers of α and γ motor neurons in the spinal cord.
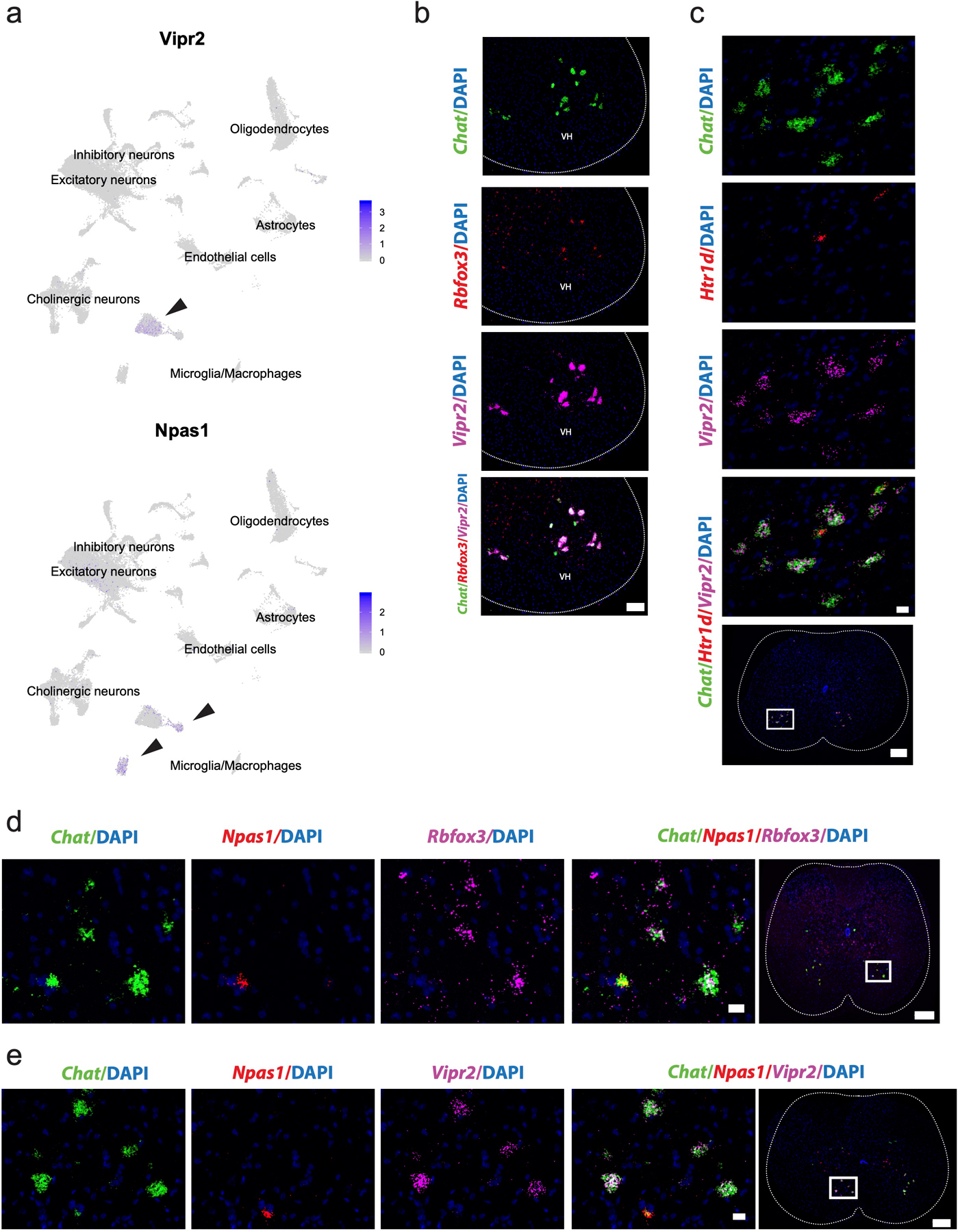
a, Average expression of Vipr2 and Npas1 across all spinal cord cell populations (labeled), overlaid on UMAP. Arrow points to α and γ motor neuron clusters, respectively. b, Representative in situ hybridization against Chat/Rbfox3/Vipr2 in transverse spinal cord shows coexpression in the ventral horn (VH). n=4 biologically independent animals. Scale bar=100 µm. c, Representative in situ hybridization against Chat/Htr1d/Vipr2 in transverse spinal cord shows mutual exclusion. Scale bar=50 µm (inset) and 200 µm (overview). n=4 biologically independent animals. d, Representative in situ hybridization against Chat/Npas1/Rbfox3 in transverse spinal cord shows mutual exclusion of Rbfox3 and Npas1 in Chat+ cells. n=5 biologically independent animals. Scale bar=20 µm (inset) and 200 µm (overview). e, Representative in situ hybridization against Chat/Npas1/Vipr2 in transverse spinal cord shows mutual exclusion of novel markers Vipr2 and Npas1 in Chat+ cells. n=4 biologically independent animals. Scale bar=20 µm (inset) and 200 µm (overview).
Extended Data Fig. 5. Discovery of a fundamental transcriptional bifurcation among γ motor neurons.
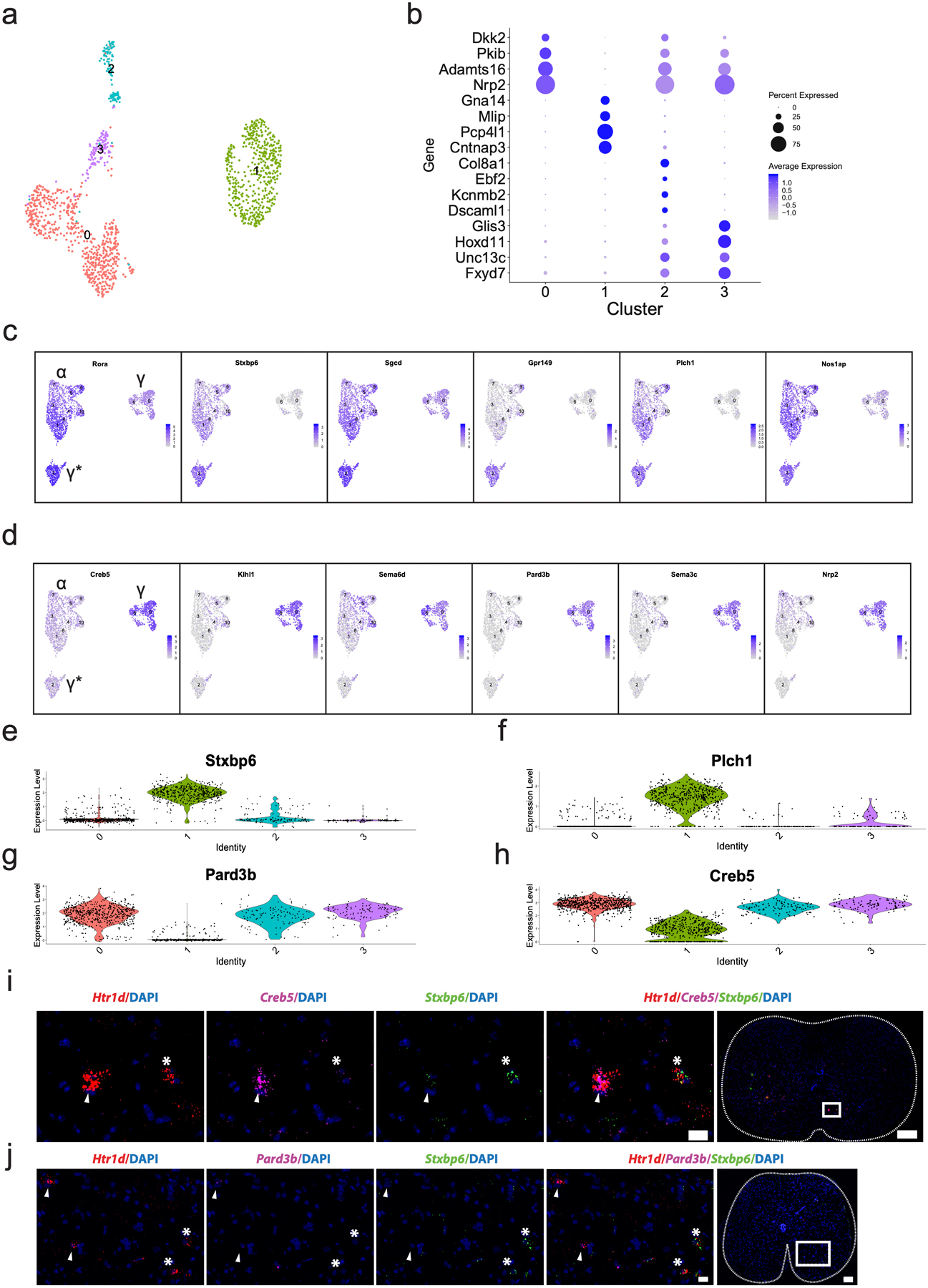
a, UMAP with 3 subclustered γ motor neurons populations. b, Novel marker gene expression across γ motor neuron subpopulations. Dot size is proportional to the percent of each cluster expressing the marker gene, while blue color intensity is correlated with expression level. c, Average log-normalized expression of genes enriched in γ* motor neurons over γ overlaid on UMAP. α, γ, and γ* populations are labeled. d, Average log-normalized expression of genes enriched in γ motor neurons over γ* overlaid on UMAP. α, γ, and γ* populations are labeled. e-h, Average expression of novel γ markers Stxbp6 (e) and Plch1 (f), as well as novel γ* markers Pard3b (g) and Creb5 (h) by cluster. i, Representative in situ hybridization against Htr1d/Creb5/Stxbp6 in transverse spinal cord shows mutual exclusion of novel markers Creb5 and Stxbp6 in Htr1d+ cells. n=4 biologically independent animals. Scale bar=20 µm (inset) and 200 µm (overview). j, Representative in situ hybridization against Htr1d/Pard3b/Stxbp6 in transverse spinal cord shows mutual exclusion of novel markers Pard3b and Stxbp6 in Htr1d+ cells. Arrowheads label canonical γ motor neurons and *labels γ*. n=5 biologically independent animals. Scale bar=20 µm (inset) and 100 µm (overview). Differentially expressed genes determined by Wilcoxon rank sum test implementation in Seurat and adjusted for multiple comparisons (Bonferroni method) (p_adj<0.01, log2-fold change >0.5).
Extended Data Fig. 6. Retrograde CTB labeling of motor pools connects transcriptional subpopulations with motor pools.
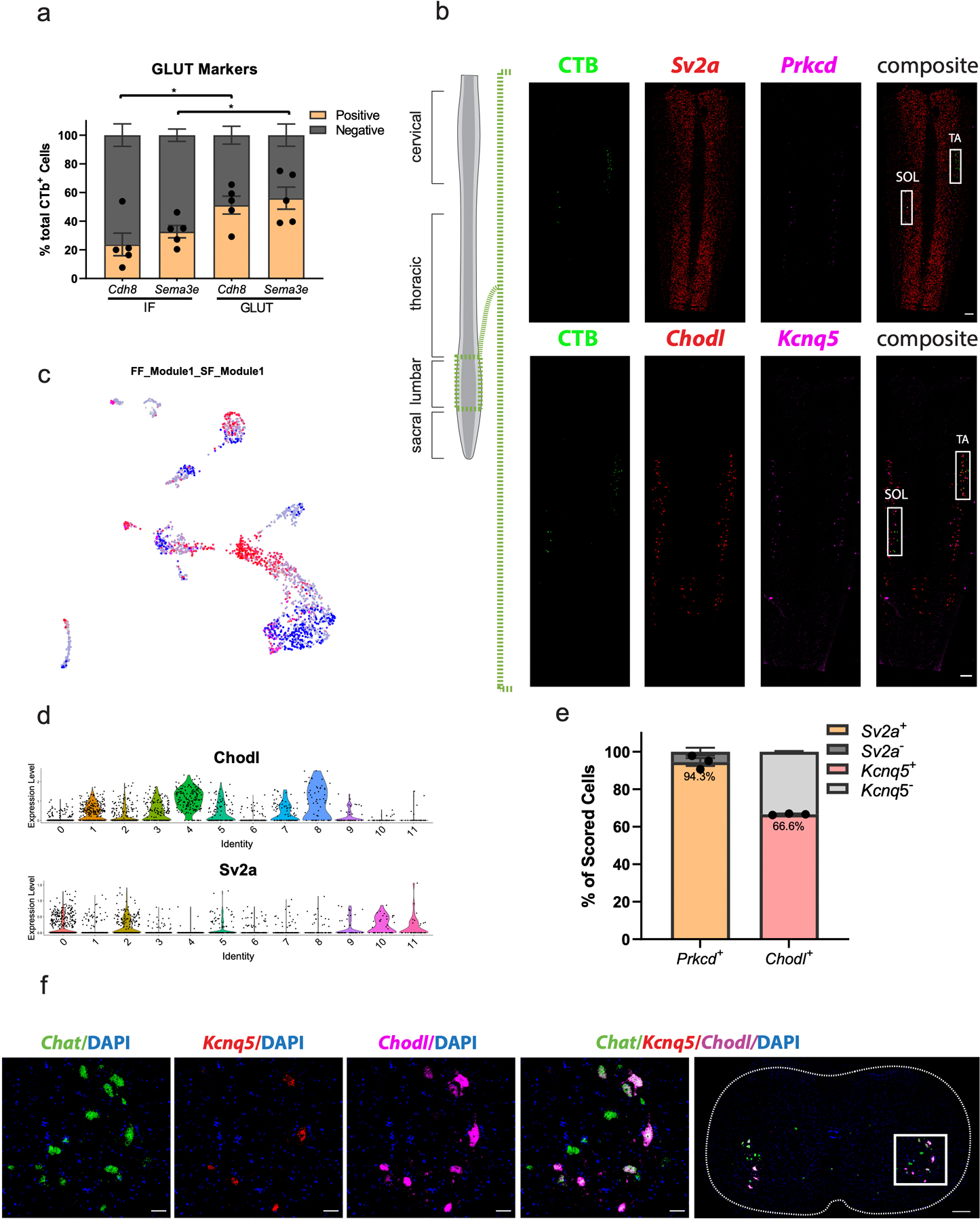
a, Proportion of CTB-labeled cells from GLUT and IF that are labeled with Cdh8 and Sema3e. The GLUT has a significantly larger proportion of Cdh8+ and Sema3e+ cells than the IF. n=5 biologically independent animals. b, Lower power view of in situ hybridization against Prkcd/Sv2a, and Kcnq5/Chodl (insets=Fig. 4h) in longitudinal sections demonstrates the specificity of CTB injections into the Soleus (SOL) and Tibialis anterior (TA). n=5 (TA) and 4 (SOL) biologically independent animals. One-way ANOVA with post-hoc Sidak multiple comparison test between same-gene conditions. Adjusted p-values=0.0212 (Cdh8) and 0.0499 (Sema3e). c, Expression of FF and SF gene modules overlaid on UMAP of all α motor neurons. d, Average log-normalized canonical marker expression of Chodl (fast-firing) and Sv2a (slow-firing). Scale bars = 250 µm. e, Proportion of Prkcd+ cells positive for Sv2a (left) and proportion of Chodl+ cells that are positive for Kcnq5 (right). n=3 biologically independent animals. f, Representative in situ hybridization showing Kcnq5 is expressed in a subset of Chodl+ cells. n=4 biologically independent animals. Scale bar=50 µm inset and 200 µm overview. *=p value<0.05, **=p value <0.01, ***=p value<0.001, ****=p value<0.0001. Error bars are SEM.
Extended Data Fig. 7. Retrograde CTB labeling of motor pools enables the identification of transcriptionally distinct classes of fast and slow-firing motor neurons in the adult spinal cord.
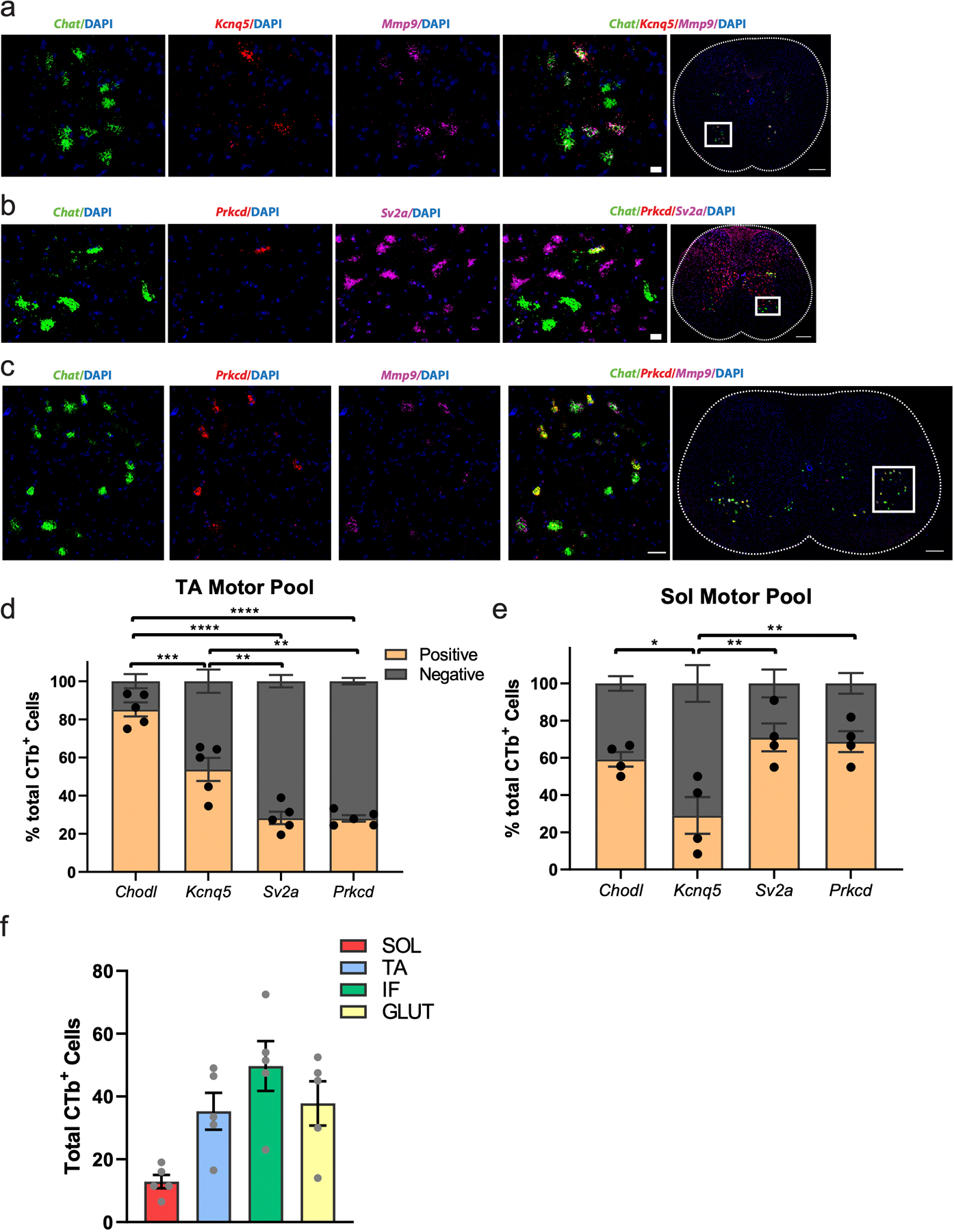
a, Representative in situ hybridization against Chat/Mmp9/Kcnq5 in transverse spinal cord shows that Kcnq5 is expressed in a subset of Mmp9+ fast-firing motor neurons. n=4 biologically independent animals. Scale bar=20 µm inset and 200 µm overview. b, Representative in situ hybridization against Chat/Sv2a/Prkcd in transverse spinal cord shows that Prkcd is expressed in almost every Chat+/Sv2a+ slow-firing motor neuron. n=2 biologically independent animals. Scale bar=20 µm inset and 200 µm overview. c, Representative in situ hybridization against Chat/Mmp9/Prkcd in transverse spinal cord shows that Prkcd is excluded from almost every Chat+/Mmp9+ fast-firing motor neuron. n=4 biologically independent animals. Scale bar=30 µm inset and 200 µm overview. d-e, Proportion of cells expressing fast and slow-firing markers in the CTB-labeled TA (d) and SOL (e) motor pools. There is a significantly higher proportion of cells expressing both known and novel fast-firing markers in TA than SOL (d), and a higher proportion of cells expressing both known and novel slow-firing markers in SOL than TA. Adjusted p-value=0.0456 (Chodl+>Kcnq5+). f, Total number of CTB-positive cells labeled across biologically independent animals. One-way ANOVA with post-hoc Tukey multiple comparison test among all conditions. n=4–5 biologically independent animals (d-f). *=p value<0.05, **=p value <0.01, ***=p value<0.001, ****=p value<0.0001. Error bars are SEM.
Extended Data Fig. 8. Cross-replicate variability in single-nucleus transcriptomic experiments.
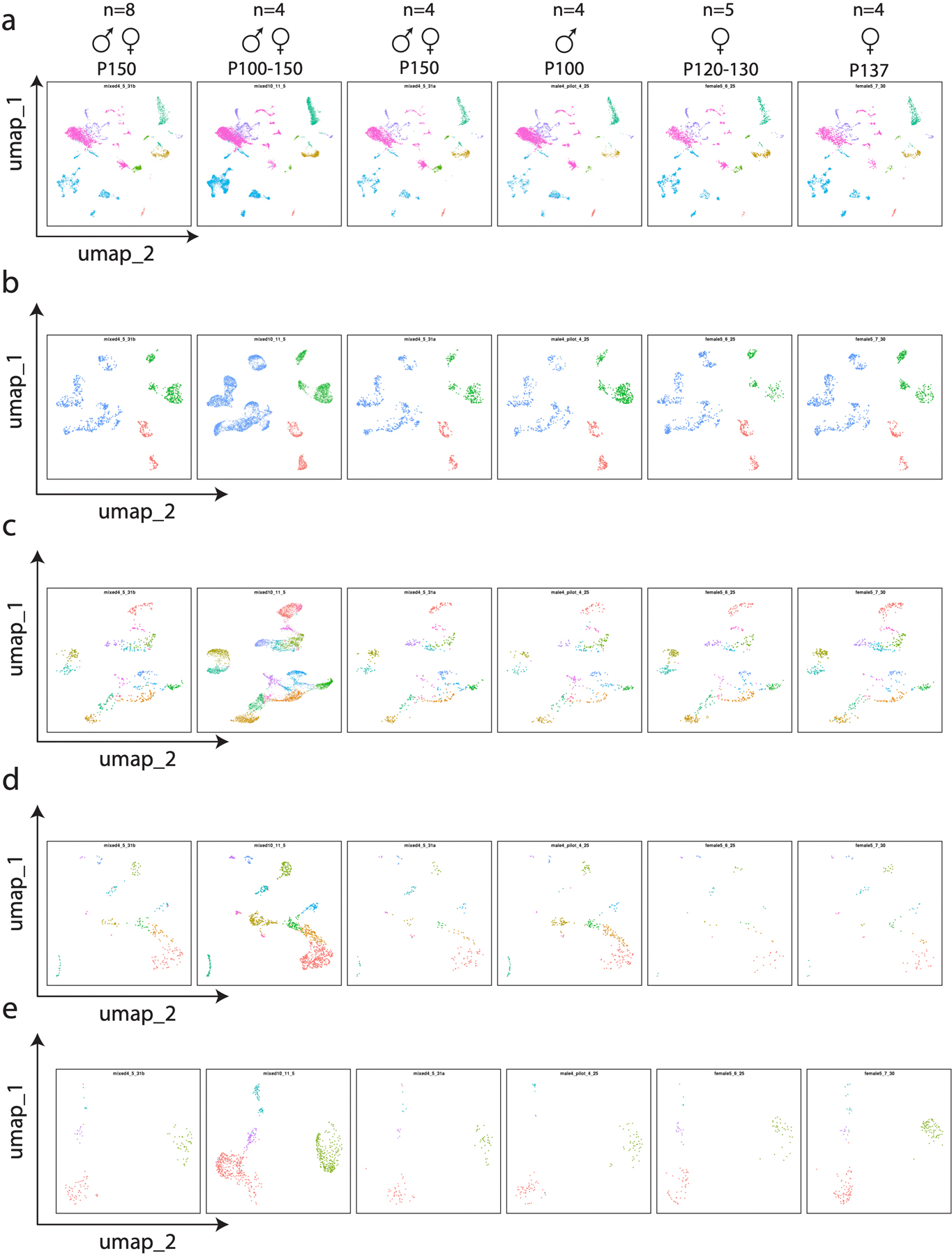
a–e, Each spinal cord sequencing replicate, plotted side-by-side and visualized by UMAP. Note that we observe minimal batch-to-batch variability along sex, age, or replicate number axes in terms of cluster identification and overall shape of the dimensionality reduced data. This does not preclude sex or age-related transcriptional changes but demonstrates that they do not fundamentally alter the transcriptomic classes that we focus on in the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R35NS097263 (A.D.G.) and R01NS083998 (J.A.K.), the Robert Packard Center for ALS Research at Johns Hopkins (A.D.G.), the Blavatnik Family Foundation (J.A.B.), and the Brain Rejuvenation Project of the Wu Tsai Neurosciences Institute. Sorting was performed on an instrument in the Shared FACS Facility obtained using NIH S10 Shared Instrument Grant S10RR025518–01. Portions of Fig. 1d and Fig. 2b were generated using objects from BioRender.com, and figures were generated using Adobe Illustrator.
Footnotes
Code availability
All custom code used to analyze data and generate figures is available in the form of a Jupyter Notebook at https://github.com/neurojacob/blum_et_al_2020.
Accession codes
All raw and processed sequencing data has been deposited in the Gene Expression Omnibus (GEO), accession number: GSE161621.
Competing Interests
A.D.G. has served as a consultant for Aquinnah Pharmaceuticals, Prevail Therapeutics and Third Rock Ventures and is a scientific founder of Maze Therapeutics. W.J.G. has affiliations with 10x Genomics (consultant), Guardant Health (consultant) and Protillion Biosciences (co-founder and consultant). J.A.B. has served as a consultant for Maze Therapeutics.
Data availability
All sequencing data is available in raw and processed form from NCBI-GEO (accession: GSE161621). An interactive web portal for exploring the dataset is available at http://www.spinalcordatlas.org.
References
- 1.Haase G et al. GDNF Acts through PEA3 to Regulate Cell Body Positioning and Muscle Innervation of Specific Motor Neuron Pools. Neuron 35, 893–905 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Song M-R & Pfaff SL Hox Genes: The Instructors Working at Motor Pools. Cell 123, 363–365 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Arber S Motor Circuits in Action: Specification, Connectivity, and Function. Neuron 74, 975–989 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Dasen JS, Tice BC, Brenner-Morton S & Jessell TM A Hox Regulatory Network Establishes Motor Neuron Pool Identity and Target-Muscle Connectivity. Cell 123, 477–491 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Kaplan A et al. Neuronal matrix Metalloproteinase-9 is a determinant of selective Neurodegeneration. Neuron 81, 333–348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maniatis S et al. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science 364, 89–93 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Sathyamurthy A et al. Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior. Cell Rep 22, 2216–2225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweeney LB et al. Origin and Segmental Diversity of Spinal Inhibitory Interneurons. Neuron 97, 341–355.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prigge JR et al. Nuclear double-fluorescent reporter for in vivo and ex vivo analyses of biological transitions in mouse nuclei. Mamm. Genome (2013) 10.1007/s00335-013-9469-8. [DOI] [PMC free article] [PubMed]
- 10.Sunkin SM et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res 41, D996–D1008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burrill JD, Moran L, Goulding MD & Saueressig H PAX2 is expressed in multiple spinal cord interneurons, including a population of EN1+ interneurons that require PAX6 for their development. Development 124, 4493 LP – 4503 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Espinosa-Medina I et al. The sacral autonomic outflow is sympathetic. Science 354, 893–897 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg AB et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thaler JP et al. A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron 41, 337–350 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Dasen JS, De Camilli A, Wang B, Tucker PW & Jessell TM Hox Repertoires for Motor Neuron Diversity and Connectivity Gated by a Single Accessory Factor, FoxP1. Cell 134, 304–316 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Gilbey MP & Stein RD Characteristics of sympathetic preganglionic neurones in the lumbar spinal cord of the cat. J. Physiol 432, 427–443 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gladwell SJ & Coote JH Inhibitory and indirect excitatory effects of dopamine on sympathetic preganglionic neurones in the neonatal rat spinal cord in vitro. Brain Res 818, 397–407 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Yoshimura M, Polosa C & Nishi S Slow EPSP and the depolarizing action of noradrenaline on sympathetic preganglionic neurons. Brain Res 414, 138–142 (1987). [DOI] [PubMed] [Google Scholar]
- 19.de Groat WC et al. The role of neuropeptides in the sacral autonomic reflex pathways of the cat. J. Auton. Nerv. Syst (1983) 10.1016/0165-1838(83)90087-5. [DOI] [PubMed]
- 20.Parry LJ, McGUANE JT, Gehring HM, Kostic IGT & Siebel AL Mechanisms of relaxin action in the reproductive tract: Studies in the relaxin-deficient (Rlx−/−) mouse. Ann. N. Y. Acad. Sci 1041, 91–103 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Ivell R, Kotula-Balak M, Glynn D, Heng K & Anand-Ivell R Relaxin family peptides in the male reproductive system—a critical appraisal. MHR Basic Sci. Reprod. Med 17, 71–84 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Zogovic B & Pilowsky PM Intrathecal neurotensin is hypotensive, sympathoinhibitory and enhances the baroreflex in anaesthetized rat. Br. J. Pharmacol 166, 378–389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franz DN, Madsen PW, Peterson RG & Sangdee C Functional roles of monoaminergic pathways to sympathetic preganglionic neurons. Clin. Exp. Hypertens. A 4, 543–562 (1982). [DOI] [PubMed] [Google Scholar]
- 24.Miles GB, Hartley R, Todd AJ & Brownstone RM Spinal cholinergic interneurons regulate the excitability of motoneurons during locomotion. Proc. Natl. Acad. Sci 104, 2448 LP – 2453 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zagoraiou L et al. A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron 64, 645–662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enjin A et al. Identification of novel spinal cholinergic genetic subtypes disclose chodl and Pitx2 as markers for fast motor neurons and partition cells. J. Comp. Neurol 518, 2284–2304 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Maluenda J et al. Mutations in GLDN, Encoding Gliomedin, a Critical Component of the Nodes of Ranvier, Are Responsible for Lethal Arthrogryposis. Am. J. Hum. Genet 99, 928–933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stepien AE, Tripodi M & Arber S Monosynaptic rabies virus reveals premotor network organization and synaptic specificity of cholinergic partition cells. Neuron 68, 456–472 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Demireva EY, Shapiro LS, Jessell TM & Zampieri N Motor neuron position and topographic order imposed by beta- and gamma-catenin activities. Cell 147, 641–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price SR & Briscoe J The generation and diversification of spinal motor neurons: signals and responses. Mech. Dev 121, 1103–1115 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Kaplan A et al. Neuronal matrix Metalloproteinase-9 is a determinant of selective Neurodegeneration. Neuron 81, 333–348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller D et al. Dlk1 promotes a fast motor neuron biophysical signature required for peak force execution. Science 343, 1264–1266 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Friese A et al. Gamma and alpha motor neurons distinguished by expression of transcription factor Err3. Proc. Natl. Acad. Sci. U. S. A 106, 13588–93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stifani N Motor neurons and the generation of spinal motor neuron diversity. Front. Cell. Neurosci 8, 1–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enjin A et al. Sensorimotor function is modulated by the serotonin receptor 1d, a novel marker for gamma motor neurons. Mol. Cell. Neurosci 49, 322–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morisaki Y et al. Selective Expression of Osteopontin in ALS-resistant Motor Neurons is a Critical Determinant of Late Phase Neurodegeneration Mediated by Matrix Metalloproteinase-9. Sci. Rep 6, 27354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barker D, Emonet-Dénand F, Harker DW, Jami L & Laporte Y Types of intra- and extrafusal muscle fibre innervated by dynamic skeleto-fusimotor axons in cat peroneus brevis and tenuissimus muscles, as determined by the glycogen-depletion method. J. Physiol 266, 713–726 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendelsohn AI, Dasen JS & Jessell TM Divergent Hox Coding and Evasion of Retinoid Signaling Specifies Motor Neurons Innervating Digit Muscles. Neuron 93, 792–805.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machado TA, Pnevmatikakis E, Paninski L, Jessell TM & Miri A Primacy of Flexor Locomotor Pattern Revealed by Ancestral Reversion of Motor Neuron Identity. Cell 162, 338–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landmesser LT The Generation of Neuromuscular Specificity. Annu. Rev. Neurosci 3, 279–302 (1980). [DOI] [PubMed] [Google Scholar]
- 41.Fukuhara K et al. Specificity of monosynaptic sensory-motor connections imposed by repellent Sema3E-PlexinD1 signaling. Cell Rep 5, 748–758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pecho-Vrieseling E, Sigrist M, Yoshida Y, Jessell TM & Arber S Specificity of sensory-motor connections encoded by Sema3e-Plxnd1 recognition. Nature 459, 842–846 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sürmeli G, Akay T, Ippolito GC, Tucker PW & Jessell TM Patterns of spinal sensory-motor connectivity prescribed by a dorsoventral positional template. Cell 147, 653–665 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin L et al. The Sleep Disorder Canine Narcolepsy Is Caused by a Mutation in the Hypocretin (Orexin) Receptor 2 Gene. Cell 98, 365–376 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Li H et al. Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 171, 1206–1220.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nijssen J, Comley LH & Hedlund E Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathol 133, 863–885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakkalakal JV, Nishimune H, Ruas JL, Spiegelman BM & Sanes JR Retrograde influence of muscle fibers on their innervation revealed by a novel marker for slow motoneurons. Development 137, 3489–3499 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kammoun M, Cassar-Malek I, Meunier B & Picard B A simplified immunohistochemical classification of skeletal muscle fibres in mouse. Eur. J. Histochem 58, 2254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor JP, Brown RHJ & Cleveland DW Decoding ALS: from genes to mechanism. Nature 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data is available in raw and processed form from NCBI-GEO (accession: GSE161621). An interactive web portal for exploring the dataset is available at http://www.spinalcordatlas.org.


