Abstract
DJ-1/PARK7 mutations are linked with familial forms of early-onset Parkinson's disease (PD). We have studied the degradation of untagged DJ-1 wild type (WT) and missense mutants in mouse embryonic fibroblasts obtained from DJ-1-null mice, an approach closer to the situation in patients carrying homozygous mutations. The results showed that the mutants L10P, M26I, A107P, P158Δ, L166P, E163K, and L172Q are unstable proteins, while A39S, E64D, R98Q, A104T, D149A, A171S, K175E, and A179T are as stable as DJ-1 WT. Inhibition of proteasomal and autophagic-lysosomal pathways had little effect on their degradation. Immunofluorescence and biochemical fractionation studies indicated that M26I, A107P, P158Δ, L166P, E163K, and L172Q mutants associate with mitochondria. Silencing of mitochondrial matrix protease LonP1 produced a strong reduction of the degradation of the mitochondrial-associated DJ-1 mutants A107P, P158Δ, L166P, E163K, and L172Q but not of mutant L10P. These results demonstrated a mitochondrial pathway of degradation of those DJ-1 missense mutants implicated in PD pathogenesis.
Subject terms: Biochemistry, Cell biology, Molecular biology, Neuroscience, Pathogenesis
Introduction
Most of the Parkinson’s disease (PD) patients are sporadic cases; rare familiar forms of PD can account for 5–10% of all PD cases. Mutations in several genes are well established as a genetic cause in familial PD1. PARK7 /DJ-1 is one of those PD-linked genes whose pathogenic mutations show autosomal recessive inheritance and early-onset of the PD phenotype2. Those genetic mutations include CNVs, exonic deletions and truncations, splice-site mutations, homozygous (L10P, M26I, E64D, P158Δ, E163K, L166P and L172Q) and heterozygous (A39S, A104T and D149A) missense mutations, and rare polymorphisms (R98Q, A171S) in healthy individuals that are not associated with PD1,3.
Native DJ-1 protein is a dimer with a flavodoxin-like helix-strand-helix structure4–9. DJ-1, initially identified as a potential oncogene cooperating with Ha-Ras in cell transformation10, is implicated in several pathways, such as: transcriptional regulation11–15, RNA binding16,17, regulation of sumoylation18, protein folding as a chaperon19–21 or co-chaperon22 and cell death23,24. DJ-1 protein is cytoprotective being a sensor of oxidative stress and acting as antioxidant preventing apoptosis25–35.
Taking into account that some PARK7/DJ-1 gene mutations result in a loss of function of the gene (no protein produced), it is reasonable to hypothesize that some of the missense point mutants may produce a loss of function of the protein. In agreement with this hypothesis, accelerated protein degradation (increased protein instability in the cell) of the DJ-1 point mutants would produce a decrease in the steady-state levels of the protein mimicking, in part, the phenotype of loss of gene function (no protein produced). In fact, DJ-1 L166P mutant has reduced stability in the cell2,36–43. DJ-1 M26I mutant have increased degradation in the cell according to some reports40,41, while other groups found no effect of the M26I mutation on protein stability13,38,43,44. The mutants A104T and D149A have also increased rates of degradation41, but found stable by other groups13,38,43–45. More recently described missense mutants of DJ-1 L10P, P158Δ and L172Q are also unstable proteins46–49. Finally, the point mutant E163K retains similar properties to wild type DJ-1 (WT) protein respect to stability in cells50, but it reduces the thermal stability of DJ-1 in solution disrupting a salt bridge of E163 with R14551.
Several studies of the degradation of DJ-1 missense mutants use tagged versions of the mutants and transfection into recipient cells expressing their endogenous DJ-1 WT. From our point of view the use of tagged constructs is inadequate to study protein degradation. When a protein is tagged either in its N-terminus or its C-terminus it is assumed that the behaviour of this protein in the cell will be equivalent to that of the untagged endogenous protein, but this assumption is not necessarily true. As a proof of the previous statement, N-terminal tagged DJ-1 L166P has a reduced degradation rate compared to untagged DJ-1 L166P, while tagging L166P at the C-terminus has less effect52. Another caveat to correctly interpret the results reported is that the DJ-1 missense mutants variably heterodimerize with the endogenous DJ-1 WT of the recipient cells, and those interactions may also, in principle, modify the stability of the mutant protein. This situation will not occur in patient cells where only the missense mutant DJ-1 protein is expressed (homozygous mutation). A way to circumvent those caveats is the study of the stability of the untagged DJ-1 missense mutants in DJ-1-null cells. The use of DJ-1-null mouse embryonic fibroblasts (MEFs) to study DJ-1 missense mutants stability have been used in some reports47,48. Here we have systematically investigated the degradation of untagged human wild type DJ-1 and its missense point variants: L10P53, M26I54, A39S55, E64D56, R98Q54, A104T57, A107P58, D149A54, P158Δ58, E163K59, A171S57, L172Q49, K175E60 and A179T58,60 in MEFs from DJ-1-null mice. The results have shown that DJ-1 pathogenic point mutants L10P, M26I, A107P, P158Δ, E163K, L166P and L172Q showed a significant increase of the degradation rate respect to DJ-1 WT in MEFs from DJ-1-null mice, and no significant difference respect to WT was found for A39S, E64D, A104T, D149A, K175E, A179T missense mutants or the rare R98Q, A171S polymorphic variants.
Results
Degradation of human wild type DJ-1 and missense mutants
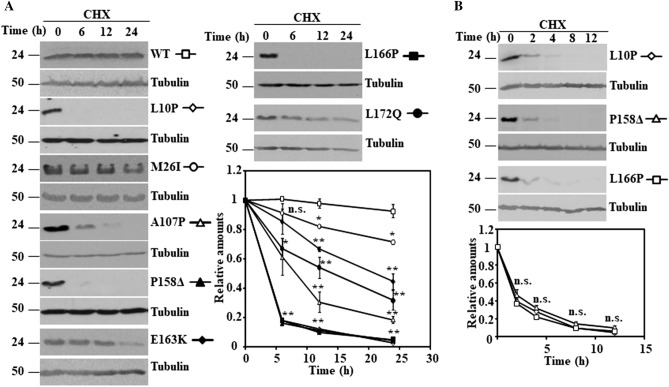
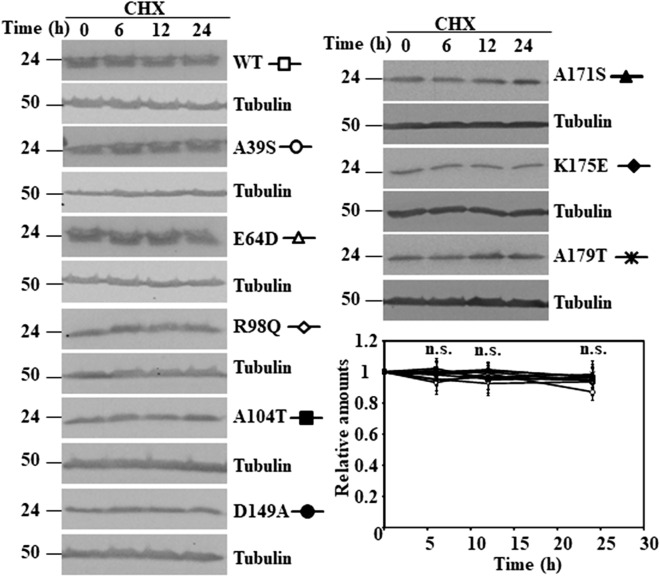
As previously stated in the introduction, degradation of ectopically expressed DJ-1 missense mutants could be affected by interactions with the cell endogenous DJ-1 and/or the use of tagged constructs. Accordingly, untagged wild type DJ-1 (WT) and the missense mutants were transfected into DJ-1-null mouse embryonic fibroblasts (MEFs). The mutants L10P, M26I, A107P, P158Δ, E163K, L166P and L172Q were unstable (Fig. 1A,B) after treatment of cells with cycloheximide (CHX) and showed different degradation rates. In contrast, DJ-1 WT and the missense mutants A39S, E64D, R98Q, A104T, D149A, A171S, K175E and A179T were not significantly degraded after treatment of the cells for 24 h with CHX (Fig. 2). The apparent degradation rate of DJ-1 missense mutants, evaluated by the corresponding half-life and ordered from shortest to longest half-life, was: L10P, P158Δ and L166P < A107P < L172Q < E163K < M26I (Supplementary Table I). We have previously shown that transfected human DJ-1 WT and point mutants M26I, R98Q, A104T, D149A are stable proteins in N2a mouse cells (expressing the endogenous mouse DJ-1) and L166P is unstable43. We extended those previous studies with similar assays to other missense DJ-1 mutants in the same cell line. In these experiments, the protein levels of the missense mutants A39S, A171S, K175E and A179T did not significantly change after treatment of transfected cells with CHX for 24 h (Supplementary Fig. 1A). In contrast, the point mutants L10P, A107P, P158Δ, E163K, L166P (shown again for comparison) and L172Q were degraded at different rates (Supplementary Fig. 1B).
Figure 1.
Time-course of the degradation of human DJ-1 unstable missense mutants transfected in DJ-1-null MEFs. DJ-1-null MEFs were transiently transfected with the indicated untagged human DJ-1 (hDJ-1) constructs and 48 h after transfection were treated with 25 µg/mL cycloheximide (CHX) for the times indicated. (A) Panels show representative immunoblots probed with anti-DJ-1 polyclonal antibodies of cells transfected with hDJ-1 WT and the missense mutants L10P, M26I, A107P, P158Δ, E163K, L166P and L172Q. (B) Panels show the results obtained with transfected DJ-1 missense mutants L10P, P158Δ and L166P in cells treated with CHX for shorter times, as indicated. Anti-tubulin antibodies were used as total protein loading control. Quantifications from three different experiments are shown in the graphs as mean ± s.e.m. Significant differences were found between hDJ-1 WT and hDJ-1 L10P at time points 6 h (**p = 8E−07), 12 h (**p = 1E−06) and 24 h (**p = 0.0008), between hDJ-1 WT and hDJ-1 M26I at time points 12 h (**p = 0.004) and 24 h (**p = 0.002), between hDJ-1 WT and hDJ-1 A107P at time points 6 h (*p = 0.03), 12 h (**p = 0.0007) and 24 h (**p = 2E−05), between hDJ-1 WT and hDJ-1 P158Δ at time points 6 h (**p = 4E−08), 12 h (**p = 1E−06) and 24 h (**p = 0.0008), between hDJ-1 WT and hDJ-1 E163K at time points 12 h (**p = 0.0001) and 24 h (**p = 0.001), between hDJ-1 WT and hDJ-1 L166P at time points 6 h (**p = 4E−08), 12 h (**p = 1E−06) and 24 h (**p = 5E−06) and between hDJ-1 WT and hDJ-1 L172Q at time points 6 h (*p = 0.04), 12 h (**p = 0.004) and 24 h (**p = 0.001). n.s., not significant.
Figure 2.
Degradation of human wild type DJ-1 and missense mutants in transfected DJ-1-null MEFs. DJ-1-null MEFs were transiently transfected with the indicated untagged human DJ-1 (hDJ-1) constructs and 48 h after transfection were treated with 25 µg/mL cycloheximide (CHX) for the times indicated. Panels show representative immunoblots developed with anti-DJ-1 polyclonal antibodies of human wild type DJ-1 (WT), A39S, E64D, R98Q, A104T, D149A, A171S, K175E and A179T. Anti-tubulin antibodies were used as total protein loading control. Below is shown the graph of quantification of the corresponding immunoblots. Data are mean ± s.e.m from three different experiments. n.s. not significant.
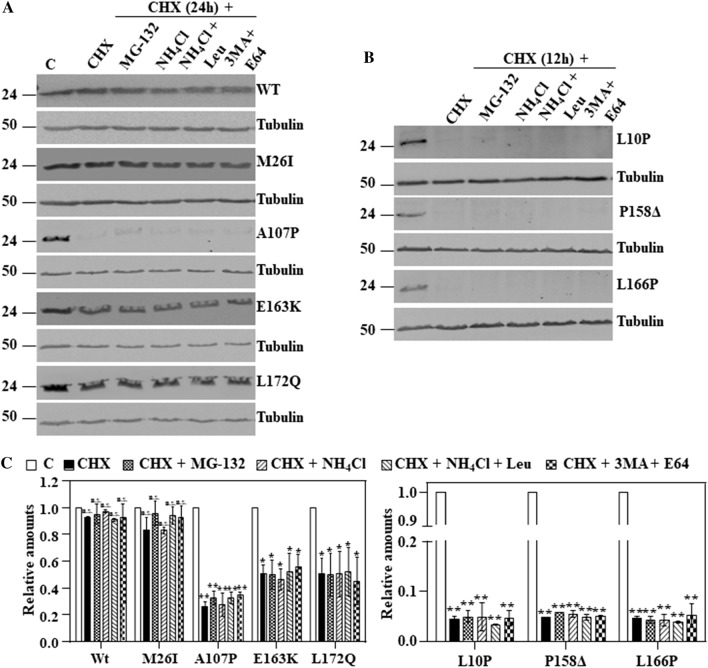
Although the proteasomal pathway is implicated in the degradation of DJ-L166P, its degradation is only partially prevented by proteasome inhibitors43. Therefore, we decided to re-evaluate those results and to test if inhibition of other proteolytic pathways can be more effective. To that end, transfected cells were treated with CHX for 24 h for those missense mutants that have shown longer half-lives: M26I, A107P, E163K and L172Q (Fig. 3A) and for 12 h for those missense mutants with shorter half-lives: L10P, P158Δ and L166P (Fig. 3B), in the absence or in the presence of several protease inhibitors. As shown in Fig. 3, addition of MG132 (proteasome inhibitor) or inhibitors of the autophagic-lyssosomal pathway (NH4Cl, NH4Cl in combination with leupeptin or 3-methyl adenine together with E64) had no significant effect on the degradation of the missense mutants or DJ-1 WT. Those results suggested that neither the proteasomal nor the autophagic-lysosomal pathways of protein degradation played a major role in the degradation of the unstable DJ-1 missense mutants.
Figure 3.
Effect of inhibitors of the proteasomal and autophagic-lysosomal pathways on the degradation of human wild type DJ-1 and missense mutants transfected in DJ-1-null MEFs. DJ-1-null MEFs were transiently transfected with the indicated untagged human DJ-1 (hDJ-1) constructs and 48 h after transfection were kept in complete medium (C) or treated with 25 µg/mL cycloheximide (CHX) in the absence (DMSO) or the presence of 10 µM MG-132, 20 mM NH4Cl, 20 mM NH4Cl plus 50 µM leupeptin (Leu) or 10 mM 3-methyl adenine (3MA) plus 5 µM E64 for 12 or 24 h, as indicated. Total cell lysates were analysed by Western and immunoblot with the corresponding specific antibodies. (A) Panels show representative immunoblots with anti-DJ-1 polyclonal antibody of human wild type DJ-1 (WT), M26I, A107P, E163K and L172Q. (B) Panels show the results obtained with transfected hDJ-1 L10P, P158Δ and L166P developed with anti-DJ-1 polyclonal antibody. Anti-tubulin antibodies were used as total protein loading control. (C) Graphs below each panel show the quantifications of the levels of DJ-1 protein respect to untreated cells as control. Values are expressed as mean ± s.e.m from three different experiments. Significant differences were found for hDJ-1 A107P between control cells and cells treated with CHX (**p = 0.002), CHX in combination with MG-132 (**p = 0.004), CHX in combination with NH4Cl (**p = 0.005), CHX in combination with NH4Cl plus leupeptin (**p = 0.003) and CHX in combination with 3MA plus E64 (**p = 0.001); for hDJ-1 E163K between control cells and cells treated with CHX (*p = 0.01), CHX in combination with MG-132 (*p = 0.03), CHX in combination with NH4Cl (*p = 0.02), CHX in combination with NH4Cl plus leupeptin (*p = 0.01) and CHX in combination with 3MA plus E64 (*p = 0.03); for hDJ-1 L172Q between control cells and cells treated with CHX (*p = 0.01), CHX in combination with MG-132 (*p = 0.03), CHX in combination with NH4Cl (*p = 0.04), CHX in combination with NH4Cl plus leupeptin (*p = 0.04) and CHX in combination with 3MA plus E64 (*p = 0.04); for hDJ-1 L10P between control cells and cells treated with CHX (**p = 0.0001), CHX in combination with MG-132 (**p = 6E−05), CHX in combination with NH4Cl (**p = 0.0003), CHX in combination with NH4Cl plus leupeptin (**p = 9E−05) and CHX in combination with 3MA plus E64 (**p = 0.0002); for hDJ-1 P158Δ between control cells and cells treated with CHX (**p = 5E−06), CHX in combination with MG-132 (**p = 9E−06), CHX in combination with NH4Cl (**p = 1E−05), CHX in combination with NH4Cl plus leupeptin (**p = 1E−05) and CHX in combination with 3MA plus E64 (**p = 4E−07) and for hDJ-1 L166P between control cells and cells treated with CHX (**p = 3E−06), CHX in combination with MG-132 (**p = 2E−05), CHX in combination with NH4Cl (**p = 5E−05), CHX in combination with NH4Cl plus leupeptin (**p = 2E−06) and CHX in combination with 3MA plus E64 (**p = 0.0002). n.s. not significant.
Subcellular localization of human wild type DJ-1 and missense mutants
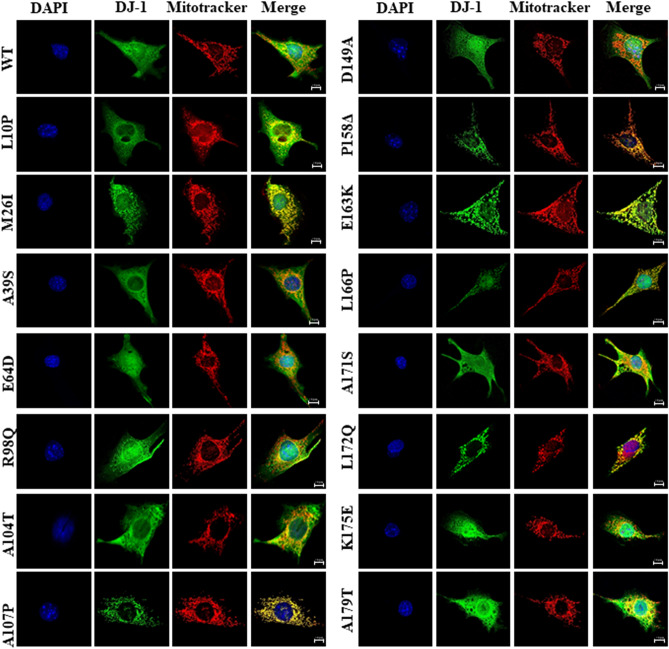
A possible clue to understand the pathway of degradation of DJ-1 missense mutants could be their subcellular localization. DJ-1 protein is described to be localized both in the cytoplasm and in the nucleus2,10,61 and to translocate to the mitochondria upon cell oxidative stress45,62. L166P missense mutant is reported to be localized in the cytoplasm and mitochondria2,63, E163K mutant was shown to be localized in the mitochondria63,64, as well as M26I63 and L172Q49 missense mutants. Therefore, the subcellular localization of the human DJ-1 WT and missense mutants was determined after transfection of DJ-1-null MEFs by indirect immunofluorescence. Figure 4 shows the results obtained and Supplementary Fig. 2 shows the analysis of the co-localization of DJ-1 and mitotracker fluorescence. Without doubt, it can be concluded that the immunofluorescence signal of M26I, A107P, P158Δ, E163K, L166P and L172Q co-localize with mitotracker fluorescence (Pearson coefficient ≥ 0.6). In contrast, it was observed a wide-spread pattern of fluorescence through the cell with some mitochondrial co-localization (naked-eye observation, Pearson coefficient > 0.2 and < 0.4) with mitotracker for E64D, R98Q, A104T, D149A and A171S missense mutants. Finally, a wide-spread non-mitochondrial distribution of fluorescence without significant co-localization with mitotracker fluorescence (Pearson coefficient ≤ 0.2) was found for DJ-1 WT, L10P, A39S, A171S and K175E. These results suggested that a good approach to study both, protein degradation and subcellular localization, would be to obtain fusion constructs of DJ-1 mutants with fluorescent proteins. To that end, constructs of M26I and L166P fused to the N-terminus of EGFP were produced. The results obtained after transfection in MEFs from DJ-1-null mice (see Supplementary Fig. 3) clearly indicated that the C-terminal tagging of the unstable L166P missense mutant greatly slows down its degradation rate compared to the untagged version (see Fig. 1 for comparison). Similarly, M26I fusion construct was not significantly degraded after 24 h of incubation in the presence of CHX. Furthermore, the fusion proteins did not reproduce the preferential mitochondrial localization observed by indirect immunofluorescence of the untagged M26I and L166P DJ-1 missense mutants (compare Supplementary Figs. 3, 4). In conclusion, the approach with fluorescent fusion constructs to study protein degradation and subcellular localization was discarded.
Figure 4.
Subcellular localization by indirect immunofluorescence of human wild type DJ-1 and missense mutants transfected in DJ-1-null MEFs. Confocal fluorescence images of the indicated untagged human DJ-1 constructs in transfected DJ-1-null MEFs grown under basal conditions (complete medium), stained with Mitotracker (red), processed for immunofluorescence with anti-DJ-1 polyclonal specific antibodies (green) and counterstained with DAPI for nuclei visualization (blue). Quantifications of the co-localization of red (mitochondria) and green (DJ-1 immunofluorescence) pixels are presented in Supplementary Fig. 2.
To get independent evidence of the subcellular localization, biochemical fractionation studies of DJ-1 mutants transfected in DJ-1-null MEFs were performed. Supplementary Fig. 4 shows the results obtained. Cell fractionation studies showed that DJ-1 point mutants M26I, A107P, P158Δ, L166P, E163K and L172Q showed significant association with the mitochondrial fraction, while they were also clearly present in the cytoplasmic fraction. This interpretation is further strengthened by comparison to similar fractionation studies with transfected DJ-1 WT and missense point mutants L10P, A39S, E64D, R98Q, A104T, D149A, A171S, K175E and A179T that showed a predominant cytoplasmic distribution and were absent in the mitochondrial fraction (Supplementary Fig. 4).
Pathway of degradation of DJ-1 unstable missense mutants
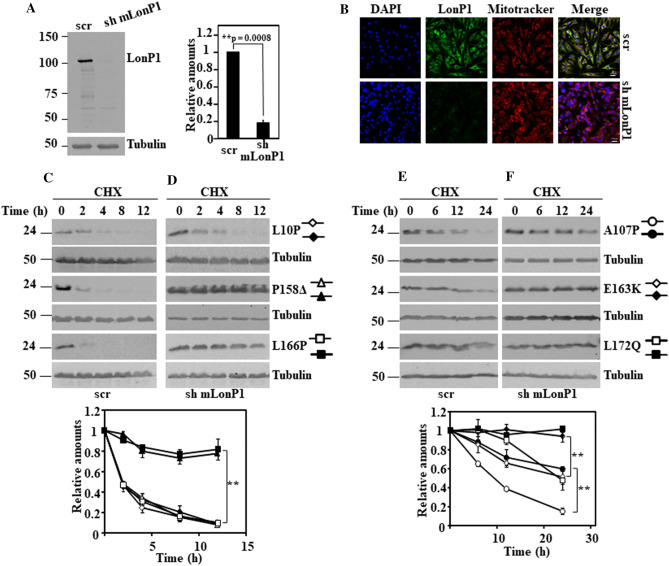
The above results seemed to indicate that a mitochondrial pathway could be responsible of the degradation of the unstable DJ-1 mutants. The mitochondrial localization of some unstable DJ-1 mutants and some previous reports describing that DJ-1 may translocate to the mitochondria matrix63,64 prompt us to study the possible involvement of mitochondrial matrix proteases in the degradation of DJ-1 missense mutants. The matrix mitochondrial LonP1 protease was examined as a possible candidate, based on it is reported implication in PINK1 degradation (see “Discussion”). LONP1 is an essential gene in mice65, as consequence we were unable to get cells from LonP1 null mice. Consequently, shRNA interference of mouse LonP1 (mLonP1) in DJ-1-null MEFS or N2a cells were used to test this hypothesis. As shown in Fig. 5, the down regulation of LonP1 expression in DJ-1-null MEFs (Fig. 5A immunoblot and Fig. 5B immunofluorescence) resulted in a significant decrease in the degradation rate of unstable DJ-1 missense mutants A107P, P158Δ, E163K, L166P and L172Q (Fig. 5C,D), but not of DJ-1 L0P. Similar results were obtained with mLonP1 silencing in N2a cells expressing those missense DJ-1 mutants (Supplementary Fig. 5).
Figure 5.
Effect of LonP1 silencing on the degradation of DJ-1 unstable mutants transfected in DJ-1-null MEFs. DJ-1-null MEFs were transduced with either scrambled shRNA (scr) or specific shRNA for mouse LonP1 (sh mLonP1), as described under the Material and Methods section. (A) Panel shows a representative immunoblot developed with specific anti-LonP1 antibodies of LonP1 shRNA-mediated knockdown in DJ-1-null MEFs. Quantification is shown in the right graph. (B) Confocal fluorescence images of non-target shRNA lentiviral transduced (scr) or LonP1 shRNA lentiviral transduced (sh mLonP1) DJ-1-null MEFs growing under basal conditions and stained with Mitotracker (red), anti-LonP1 specific antibodies (green) and counterstained with DAPI (blue) for nuclei visualization. Scr DJ-1-null MEFs, panels (C) and (E) or sh mLonP1 DJ-1-null MEFs, panels (D) and (F), were transiently transfected with hDJ-1 L10P, P158Δ and L166P (C,D) or hDJ-1 A107P, E163K and L172Q (E,F) and 48 h after transfection were treated with 25 µg/mL cycloheximide (CHX) for the times indicated. Panels show representative immunoblots with anti-DJ-1 polyclonal antibody of the indicated hDJ-1 missense mutants transfected MEFs. Anti-tubulin antibodies were used as total protein loading control. Quantifications are shown in the graphs below as mean ± s.e.m from three different experiments. Significant differences were found between scr and sh mLonP1 DJ-1-null MEFs transfected with P158Δ at the time points 2 h (**p = 0.0001), 4 h (**p = 0.003), 8 h (**p = 0.0008) and 12 h (**p = 0.009), between scr and sh mLonP1 DJ-1-null MEFs transfected with L166P at the time points 2 h (**p = 0.0002), 4 h (**p = 0.0001), 8 h (**p = 0.0008) and 12 h (**p = 0.002), between scr and sh mLonP1 DJ-1-null MEFs transfected with A107P at the time points 6 h (**p = 0.0008), 12 h (*p = 0.02) and 24 h (**p = 8E−05), between scr and sh mLonP1 DJ-1-null MEFs transfected with E163K at the time points 12 h (**p = 0.009) and 24 h (*p = 0.01) and between scr and sh mLonP1 DJ-1-null MEFs transfected with L172Q at the time point 24 h (**p = 0.001).
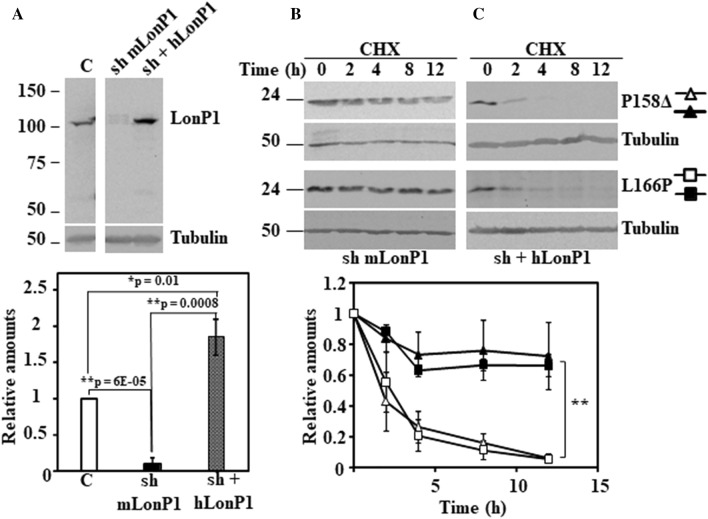
To check for off-target effects of mLonP1 shRNA, human LonP1 (hLonP1) cDNA was used to rescue the phenotype of mLonP1 silenced cells respect to degradation of two of the unstable DJ-1 mutants. As shown in Fig. 6, over expression of hLonP1 completely rescued the inhibition of degradation of the unstable DJ-1 P158Δ and L166P mutants produced by mLonP1 shRNAs. Taken together, those results clearly allowed the conclusion that LonP1 is the mitochondrial protease is clearly involved in the degradation of unstable DJ-1 mutants A107P, P158Δ, E163K, L166P and L172Q mutants but not in the degradation of DJ-1 L10P mutant.
Figure 6.
Rescue of the degradation of DJ-1 unstable mutants transfected in LonP1-silenced DJ-1-null MEFs by transfection of human LonP1. LonP1 shRNA-transduced DJ-1-null MEFs (sh mLonP1) were complemented with ectopically expressed human LonP1 (hLonP1). (A) Panel shows a representative immunoblot of LonP1 shRNA-mediated knockdown in DJ-1-null MEFs and the recovery of LonP1 protein levels after transfection with hLonP1. Immunoblot was developed with specific antibodies anti-LonP1. Quantification is shown in the right graph. LonP1-silenced DJ-1-null MEFs (sh mLonP1), panel (B) and rescued LonP1-silenced DJ-1-null MEFs (sh + hLonP1), panel (C), were transiently transfected with hDJ-1 P158Δ or L166P and 48 h after transfection were treated with 25 µg/mL cycloheximide (CHX) for the times indicated. Panels (B) and (C) show representative immunoblots developed with anti DJ-1 polyclonal antibody of the indicated constructs. Anti-tubulin antibodies were used as total protein loading control. Quantifications are shown in the graphs below as mean ± s.e.m from three different experiments. Significant differences were found between sh mLonP1 and sh + hLonP1 DJ-1-null MEFs transfected with P158Δ at the time points 2 h (*p = 0.01), 4 h (*p = 0.03), 8 h (*p = 0.03) and 12 h (*p = 0.04) and between sh mLonP1 and sh + hLonP1 DJ-1-null MEFs transfected with L166P at the time points 4 h (*p = 0.01), 8 h (**p = 0.001) and 12 h (**p = 0.001).
Discussion
DJ-1 WT is a stable protein when expressed by transfection in DJ-1-null MEFs, similar to what is found for endogenously expressed DJ-1 in other cells36,38,43,66–68. The PD missense mutations A39S, E64D, A104T, D149A, K175E and the polymorphic missense variants R98Q and A171S are as stable as DJ-1WT, both in MEFs from DJ-1-null mice (Fig. 1) and in N2a cells (Supplementary Fig. 1), expressing the endogenous DJ-1 WT. The DJ-1 point mutants L10P, A107P, P158Δ, E163K, L166P and L172Q were degraded with similar half-lives in MEFs from DJ-1-null mice and in N2a cells. The M26I DJ-1 mutant is slowly degraded in MEFs from DJ-1-null mice (see Fig. 1 and Supplementary Table 1), but degradation is not apparent in N2a cells after treatment of cells with CHX for 24 h, as reported previously43. These results indicated that the unstable (and untagged) DJ-1 mutants, qualitatively, are unstable even in the presence of the expression of endogenous DJ-1 WT.
Several laboratories (including ourselves) have reported that the missense DJ-1 L166P mutant is degraded by the ubiquitin–proteasome pathway2,36–43. Similar conclusions were reported for the degradation of L10P, P158Δ46,47 and L172Q49 point mutants. Nevertheless, several groups reported that proteasome inhibitors are not able to completely prevent the degradation of the unstable mutants L166P37,39,42,43,46, L10P and P158Δ46. Furthermore, as shown here, treatment with different inhibitors of the autophagic-lysosomal pathway (NH4Cl, leupeptin 3-methyl-adenine and E64) fail to prevent their degradation (Fig. 3). Similar results with those inhibitors have been reported previously for the degradation of DJ-1 L166P missense mutant37,39,41.
In the search for alternative pathways of degradation, experiments of subcellular localization of DJ-1 by immunofluorescence and biochemical cell fractionation studies were performed (Fig. 4 and Supplementary Fig. 4, respectively). Those data showed that the DJ-1 point mutants M26I, A107P, P158Δ, E163K, L166P and L172Q, but not L10P, are significantly associated with mitochondria. Those results are in agreement with previously published work of the localization of M26I, P158Δ, E163K, L166P and L172Q2,45,49,63,64,69; no data are available for A107P point mutant. The widely used experimental approach of using fusion constructs of the missense mutants with fluorescent proteins as reporters for both degradation and subcellular localization was discarded, as the DJ-1 L166P and M26I proteins fused to EGFP (Supplementary Fig. 3) did not meet the simple criteria of similar degradation rates and subcellular localization as their respective untagged DJ-1 missense mutants, another clear demonstration of the caveat of using tagged versions of proteins for studying DJ-1 point mutant behaviour. Similar results for the E63K mutant fused to GFP have been reported63.
To study the possible involvement of mitochondria in the degradation of missense DJ-1 mutants, we hypothesized that mitochondrial matrix LonP1 could be implicated. The experimental evidence presented (Fig. 5 and Supplementary Fig. 5) clearly indicated that mitochondrial LonP1 is implicated in the degradation of A107P, P158Δ, E163K, L166P and L172Q. We showed (Fig. 6) that the strong inhibition of their degradation by interrupting the expression of mouse LonP1 can be rescued by overexpression of human LonP1 (whose expression is not suppressed by the action of the specific mouse LonP1-targeting shRNAs), indicating the specificity of the observed inhibitory effects. Formally, it can be argued that rescue by a variant mLonp1 mutant construct insensitive to the shRNA may provide additional, but not essential, confirmation. Furthermore, while certainly hLonp1 is overexpressed in the rescue experiments, the results obtained suggest that those DJ-1 point mutants are also substrates for the hLonP1 protease, as expected. In contrast, there is no effect on L10P degradation. The elucidation of the degradation pathway of this missense mutant requires further investigation.
The localization of DJ-1 in the mitochondria and translocation to mitochondrial matrix have been studied63,69. The L10P mutation located in strand 1 prevents homodimer formation while the mutant interacts with DJ-1 WT46,48. The L10P mutation may hinder the mitochondrial localization, as that region of strand 1 of the 3D-protein structure4,5,7 has been implicated in mitochondrial localization. The DJ-1 E18A point mutant (a-helix 1) localizes to the mitochondria, but the substitution by alanine of the wild -type sequence at leucine (a.a. 7), valine (a.a. 8), isoleucine (a.a. 9) and leucine (a.a. 10, changed to Pro in the missense mutant) within strand 1 prevents mitochondrial localization69. The fact that the mutants ΔC15 and M26I (shown also here, Fig. 4) also localized to the mitochondria63, further supports the role of the aminoacid sequence of strand 1 and a-helix 1 (aminoacids 5–28) in the N-terminus as the sequence that either prevents the translocation of DJ-1 to mitochondria, or promotes its cytoplasmic retention. But those N-terminal structural elements are clearly insufficient to explain the location to mitochondria, as DJ-1 with point mutations at the C-terminus having an intact N-terminal sequence, also localize to the mitochondria. Clearly further experiments will allow elucidating the structural requirements for the mitochondrial localization and matrix translocation of DJ-1 missense mutants.
Mutations in LONP1 gene produce the CODAS syndrome with Cerebral, Ocular, Dental, Auricular and Skeletal abnormalities70,71, more recently a bi-allelic mutation (c.2282 C > T, (p.Pro761Leu) in LONP1 results in neurodegeneration with deep hypotonia and muscle weakness, severe intellectual disability and progressive cerebellar atrophy72. These observations clearly indicate a relationship of LonP1 with central nervous system abnormalities and neurodegeneration. LonP1 is implicated in the degradation of matrix mitochondrial proteins like aconitase73, 5-aminolevulinic acid synthase74, TFAM75, StAR76, complex 1 of the OXPHOS77, SDH578 and SDHB79 of complex II of OXPHOS and unfolded proteins, like the matrix located OTC Delta used to promote an unfolding protein response (mitUPR) in mitochondria80,81. LonP1 also participates in the default degradation in mitochondrial matrix of the PD linked protein kinase encoded by PINK1/PARK6 gene. Under mitUPR conditions, PINK1 accumulates (increased accumulation was produced with silencing of LonP1) and recruits the PD-linked E3 ligase PARKIN/PARK2 promoting mitochondrial clearance by mitoautophagy80,82. Other groups have observed the accumulation of PINK1 upon silencing LonP1 without the need of concomitant induction of mitUPR response83,84. PINK1 is initially processed by the mitochondrial processing protease (MPP), other mitochondrial proteases like PARL, ClpPX and AFG3L2 also participate82. The processed PINK1, in contrast to the above observations of degradation by LonP1, would be rapidly degraded by the ubiquitin-proteasome pathway85,86 after polyubiquitylation87. In Drosophila, overexpression of DJ-1 rescues the altered phenotype caused by the loss of PINK1, but not of Parkin88. Similarly, DJ-1 overexpression also rescues the increased sensitivity of Substantia Nigra to MPTP in PINK1 null mice89. Furthermore, the mitochondrial fragmentation phenotype of DJ-1-null cells can be rescued by overexpression of PINK1 or PARKIN90,91. Taken together all these results indicate that DJ-1 and PINK1 (PARKIN) probably act in parallel pathways and LonP1 is involved in DJ-1 point mutants and PINK1 degradation, with variable degrees of involvement of the ubiquitin–proteasome pathway and other degradation pathways that may also be subjected to cell-specific determinants.
In conclusion, the PD phenotype presented by patients harbouring homozygous mutations M26I, A107P, P158Δ, E163K, L166P and L172Q can be explained by a "loss of function", similar to the effects produced by DJ-1 mutation that resulted in strong down-regulation or absence of DJ-1 mRNA (deletions, CNV and splicing mutations) because of its instability due to its mitochondrial targeting and degradation mainly by matrix mitochondrial LonP1 protease. A similar situation is the case of patients with homozygous DJ-1 L10P mutation, while its main pathway of degradation remains to be determined. In contrast, the other point mutants (A39S, E64D, A104T, D149A, K175E and A179T) and the rare polymorphisms (R98Q, A171S) might not be truly pathogenic (likely sure for the polymorphic variants R98Q, A171S). Another possibility is that their half-life could still be lower than DJ-1 WT, but escape to our detection limits (24 h), requiring the use of quantitative proteomic studies using SILAC pulse-chase experiments and MS for its determination (far away from the scope and budget of our work). By those methods the DJ-1 WT protein shows a very long half-life (t1/2), from t1/2 = 187 h in mouse NIH3T3 cells66 to t1/2 = 199 to 299 h as determine in vivo in mouse heart92. Accordingly, the pathogenetic mechanism for A39S, E64D, R98Q, A104T, D149A, A171S, K175E and A179T remains to be determined.
Materials and methods
Plasmid constructs
The vectors for expression of untagged human wild type DJ-1 (hDJ-1 WT) and missense mutants M26I, R98Q, A104T, D149A and L166P have been previously described43. The mutant L10P was obtained by PCR (4 min, 97 °C; 30 cycles of 45 s, 95 °C; 1 min, 61 °C and 1.5 min, 72 °C, and a final polymerization cycle of 15 min, 72 °C) with the appropriate oligonucleotides, forward BamH1-hDJ-1 L10P: 5′ GAGCGGATCCATGGCTTCCAAGAGGGCTCTGGTCATCCCGGCTAAAGGAGCAGAGG 3′ and reverse XhoI-hDJ-1 L10P: 5′ GCGCCTCGAGCTAGTCTTTAAGAACAAGTGGAGCC 3′ and amplified DNA was subcloned into pCMV-Sport6 vector for expression in eukaryotes, as described43.
The mutations A39S, E64D, A107P, P158Δ, E163K, A171S, K175E and A179T of hDJ-1 were introduced by PCR (initial 4 min, 97 °C; 30 cycles of 45 s, 95 °C; 1 min, 61 °C and 10 min, 72 °C, and a final polymerization cycle of 15 min, 72 °C) from hDJ-1 WT inserted in pCMV-Sport6 using the method of QuickChange Site-Directed Mutagenesis Kit (Statagene, La Jolla, California, USA) with the following oligonucleotides: forward hDJ-1 A39S: 5′CCGTTGCAGGCCTGTCTGGAAAAGACCCAGTAC 3′ and reverse hDJ-1 A39S 5′GTACTGGGTCTTTTCCAGACAGGCCTGCAACGG 3′; forward hDJ-1 E64D: 5′GCAAAAAAAGAGGGACCATTTGATGTGGTGGTTC 3′, and reverse hDJ-1 E64D: 5′GAACCACCACATCAAATGGTCCCTCTTTTTTTGC 3′; forward hDJ-1 A107P: 5′GATAGCCGCCATCTGTCCAGGTCCTACTGCTCTG 3′ and reverse hDJ-1 A107P: 5′CAGAGCAGTAGGACCTGGACAGATGGCGGCTATC 3′; forward hDJ-1 P158Δ: 5′CTGATTCTTACAAGCCGGGGTGGGACCAGCTTCGAG 3′ and reverse hDJ-1 P158Δ: 5′CTCGAAGCTGGTCCCACCCCGGCTTGTAAGAATCAG 3′; forward hDJ-1 E163K: 5′GGACCAGCTTTAAGTTTGCGCTTGC 3′ and reverse hDJ-1 E163K: 5′GCAAGCGCAAACTTAAAGCTGGTCC 3′; forward hDJ-1 A171S 5′GCAATTGTTGAATCCCTGAATGGCAAGGAGG 3′ and reverse hDJ-1 A171S: 5′CCTCCTTGCCATTCAGGGATTCAACAATTGC 3′; forward hDJ-1 K175E: 5′GCCCTGAATGGCGAGGAGGTGGCGGCTCAGG 3′ and reverse hDJ-1 K175E: 5′CTTGAGCCGCCACCTCCTCGCCATTCAGGGC 3′; forward hDJ-1 A179T: 5′GCAAGGAGGTGGCGACTCAAGTGAAGGC 3′ and reverse hDJ-1 A179T 5′GCCTTCACTTGAGTCGCCACCTCCTTGC 3′. The hDJ-1 L172Q mutant was synthesized by GenScript (Piscataway, New Jersey, USA).
The EGFP fusion protein constructs of hDJ-1 M26I and L166P were obtained by gene synthesis of hDJ-1 cDNA missense mutants including restriction sites 5′ for XhoI and 3′ for AgeI and subcloning into the XhoI-AgeI sites of the pEGFP-N1 mammalian expression vector (GenScript).
Mouse LonP1 shRNA (RMM4534-NM_028782) and the corresponding scramble control shRNA cloned in the lentiviral vector pLKO.1 puro65 were provided by Dr. Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Universidad de Oviedo, Spain. For rescue experiments in mouse LonP1 silenced cells, human LonP1 cDNA (clone MGC:1498 IMAGE:3350958) was obtained from the pOTB7 construct by digestion with EcoR1 and XhoI and subcloned into pcDNA 3.1 vector, previously digested with the same restriction enzymes. The DNA sequence of all constructs was verified by Sanger sequencing in an ABI Prism 3130XL (Applied Biosystems, Foster City, California, USA).
Cell culture, transfections and treatments
Mouse embryonic fibroblasts (MEFs) from DJ-1-null mice were provided by Dr. Jie Shen, Center for Neurologic Diseases, Brigham and Women's Hospital and Harvard Medical School, Boston, USA and grown in Dulbecco’s modified Eagle’s medium with low glucose (1 g/L) and supplemented with 15% fetal bovine serum and 50 µg/mL gentamicin. DJ-1-null MEFs (3 × 106) were transiently transfected with 6 µg of the indicated DJ-1 constructs by nucleofection (Nucleofector II program T-016, Amaxa Nucleofector Technology, Basel, Switzerland) by using an in-house made transfection buffer (150 mM KH2PO4, 24 mM NaHCO3, 4 mM D-glucose, 6 mM ATP and 12 mM MgCl2). Mouse neuroblastoma N2a cells were cultured in Dulbecco’s modified Eagle’s medium with 4.5 g/L glucose supplemented with 10% fetal bovine serum and 50 µg/mL gentamicin. N2a cells (3 × 105) were transiently transfected using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer's instructions.
Transfected cells were grown for 48 h and then protein degradation was studied by treatment of cells with 25 µg/mL cycloheximide (CHX) for the times indicated. At the dose of CHX indicated, cell viability (MEFs and N2a, trypan blue exclusion) was > 95% after 24 h of incubation. For some experiments, transiently transfected DJ-1-null MEFs were kept in complete medium or treated with 25 µg/mL CHX in the absence or the presence of 10 µM MG-132, 20 mM NH4Cl, 20 mM NH4Cl plus 50 µM leupeptin (Leu) or 10 mM 3-methyl adenine (3MA) plus 5 µM E64 for 12 or 24 h, as indicated. Cells were then washed three times with cold phosphate-buffered saline (PBS), pelleted by centrifugation and used for the obtention of total cell extracts.
Lentiviral production
For lentiviral production, 3.5 × 106 HEK293T cells (cultured in Dulbecco’s modified Eagle’s medium with 4.5 g/L glucose supplemented with 10% fetal bovine serum and 50 µg/mL gentamicin) were seeded in 20 mm × 100 mm culture dishes and the next day transfected with 3.2 µg of the packaging plasmid psPAX2, 1.6 µg of the envelope plasmid pM2.G and 5 µg of the corresponding lentiviral transfer plasmid, scramble control shRNA or mouse LonP1 shRNA in pLKO.1 puro plasmid65, by using Lipofectamine 2000 reagent according to the manufacturer's protocol (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The culture medium was removed 24 h after transfection and complete fresh medium was added. Lentiviral particles were harvested 48 h after transfection, again fresh culture medium was added and 72 h after transfection a second harvest of lentiviral particles was collected. The collected culture media were centrifuged at 200 × g for 5 min, filtered-sterilized through 0.45 µm filters (Millipore, Darmstadt, Germany), aliquoted and stored frozen at − 80 °C until used for further experiments.
Cell transduction for LonP1 silencing and rescue experiments
DJ-1-null MEFs (1.3 × 105 per well) or N2a cells (3 × 105 per well) were seeded in 6-well culture plates and incubated with 700 µL of filtered lentivirus-containing medium (scramble control shRNA or mouse LonP1 shRNA) in the presence of 8 µg/mL polybrene to a final volume of 1.4 mL per well. Culture media was replaced 24 h after infection, 48 h after infection transduced cells were selected by addition of 8 µg/mL puromycin to the culture media for four days. Puromycin-resistant cells were transiently transfected with the indicated human DJ-1 constructs and treated with CHX for studying protein degradation, as described above.
For rescue experiments in mouse LonP1 silenced MEFs, puromycin-resistant MEFs were transiently transfected with human LonP1, selected with culture medium containing 250 µg/mL zeocin for four days, transiently transfected with the indicated human DJ-1 constructs and treated with CHX for studying protein degradation, as described above.
Cell fractionation studies
For biochemical subcellular fractionation experiments, DJ-1-null MEFs were transfected with the indicated untagged human DJ-1 constructs and 48 h after transfection cells were washed with cold PBS for three times, pelleted by centrifugation at 110 × g for 5 min at 4 °C, suspended in lysis buffer (20 mM HEPES pH 7.4, 250 mM sucrose, 5 mM MgCl2, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 25 mM NaF, 1 mM orthovanadate, 10 µM leupeptin, 1 µg/mL pepstatin and 1 mM PMSF) and incubated on ice for 10 min, with thoroughly pipetting up and down the suspension each 2 min. Next, the cell suspension was incubated for 5 min at room temperature with lysis buffer containing digitonin (to a final concentration of 50 µg/mL), cell lysis was verified by Trypan blue staining. Cell suspensions were then transfered to ice and centrifuged at 1000 × g for 5 min at 4 °C to remove nuclei, debris and non lysed cells. The supernatant was used as the total fraction (input) and was further centrifuged at 15,000 × g for 15 min at 4 °C. The supernatant was used as the cytosolic fraction whereas the pellet, containing mitochondria, was washed twice with 200 µL of lysis buffer without digitonin and centrifuged at 15,000 × g for 15 min at 4 °C. After the last wash, the pellet was resuspended in a final volume identical to the volume of the cytosolic fraction with a buffer containing 10 mM HEPES pH 7.4, 10 mM KCl, 1 mM DTT, 0.6% NP-40, 1 mM EDTA, 1 mM EGTA, 25 mM NaF, 1 mM orthovanadate, 10 µM leupeptin, 1 µg/mL pepstatin and 1 mM PMSF, vortexed for 10 s and centrifuged for 30 s at 15,000 × g at 4 °C. The supernatant of this centrifugation was used as the mitochondrial fraction. Samples from the total input, and mitochondrial and cytoplasmic fractions were loaded onto SDS-PAGE and analyzed by Western and immunoblot, as described below.
Western immunoblotting
After the treatments, cells were directly lysed in SDS-buffer (62.5 mM Tris–HCl pH 6.8, 2% SDS, 20% glycerol, 10 µM leupeptin, 1 µg/mL pepstatin and 1 mM PMSF). Cell extracts were sonicated for 10 min on ice, centrifuged at 15,000 × g for 10 min and supernatants used to measure total protein concentration with BCA protein assay kit (Thermo Scientific-Pierce, Waltham, Massachusetts, USA). Equal amounts of total protein were loaded onto 10% or 14% SDS-PAGE for Western transfer to PVDF membranes and immunoblotting.
Immunoblots were probed with mouse anti-DJ-1 monoclonal antibody (1:1000, MBL, Woburn, Massachusetts, USA, Clone 3E8); rabbit anti-DJ-1 polyclonal antibody (1:1000, Abcam, Cambridge, UK, ab18257); rabbit anti-LonP1 polyclonal antibody (1:1000, Abcam ab103809) and rabbit anti-Tim23 polyclonal antibody (1:1000, Abcam ab230253). Mouse α-Tubulin (1:10,000, DM1A, Sigma, Darmstadt, Germany) monoclonal antibody was used as loading control. The blots were developed with a peroxidase-labeled goat anti-mouse or anti-rabbit secondary antibody (1:5000, Biorad, Hercules, California, USA) and chemiluminiscence detection MF-ChemiBIS 3.2 (DNR Bio-Imaging Systems, Neve Yamin, Israel). Blots were analyzed by quantitative densitometry using Totallab TL100 software (version 1.0, TotalLab Ltd., Newcastle upon Tyne, UK) and protein levels were normalized respect to tubulin.
Immunofluorescence, confocal microscopy and image analysis
Cells grown on coverslips in 24-well plates were stained for 45 min with 250 nM MitoTracker Red (Invitrogen). The coverslips were then washed with cold PBS three times, fixed with 4% paraformaldehyde in PBS for 20 min at room temperature, permeabilized in PBS with 0.1% Triton X-100 and blocked with PBS and 3% BSA for 1 h at room temperature. The coverslips were processed for indirect immunofluorescence by incubation with rabbit anti-DJ-1 polyclonal antibody (1:200, Abcam) or rabbit anti-LonP1 polyclonal antibody (1:100, Abcam) for 3 h at room temperature, washed with PBS three times for 10 min followed by incubation with Alexa-488 or Alexa-647 fluorescent-labelled secondary antibodies (1:1000) in blocking buffer for 1 h. Next, coverslips were washed with PBS three times for 10 min and 1 μg/mL DAPI (4′,6-diamidino-2-phenylindole) was included for nuclear counterstaining in the second of the washing steps. Coverslips with transfected cells with EGFP fusion protein constructs of DJ-1 were processed for direct fluorescence visualization by incubation with 1 μg/mL DAPI for 5 min. Coverslips were mounted with Prolong Gold antifade reagent (Invitrogen) for confocal microscopy observation in a laser scanning microscope (Leica TCS SP5, Wetzlar, Germany).
To quantify the co-localization between MitoTracker fluorescence (red channel) and the immunofluorescence of transfected human DJ-1 constructs (green channel), single planes along the z-axis of the confocal fluorescence images obtained were analysed with the Image Correlation Analysis (ICA) plugin of ImageJ software93. After background subtraction, a single cell was defined as a region of interest (ROI) and quantitative co-localization analysis of the pixels for both channels (red and green) was measured using Pearson’s correlation coefficient the hDJ-1/MitoTracker. Data are presented as mean ± s.e.m from at least 20 individual cells from two different experiments.
Statistical analysis
Quantitative data are reported as means ± s.e.m from three different experiments. When indicated, values are expressed as mean ± upper and lower values from two different experiments. Statistical significance between groups was calculated using a two-tailored Student’s t-test.
Supplementary Information
Acknowledgements
This work was supported by Grants from MICIU SAF2017-85199, and by Grant S2017/BMD-3700 (NEUROMETAB-CM) from Comunidad de Madrid co-financed with the Structural Funds of the European Union.to J.G.C.. We thank Dr. Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Universidad de Oviedo, Spain for providing us the shRNA constructs for LonP1 and Dr. Jie Shen, Center for Neurologic Diseases, Brigham and Women's Hospital and Harvard Medical School, Boston, USA for the DJ-1 null MEFs used in this work.
Author contributions
J.G.C. did the formulation and evolution of the project, R.S.L. contribute to the discussion and planning of the experiments to reach the objectives. Both authors, J.G.C. and R.S.L., are responsible for data management. R.S.L. is responsible for primary data acquisition and mathematical calculations that were revised by J.G.C. J.G.C. is responsible for funding acquisition to perform the work presented. R.S.L. performed the experiments and collected the evidence. Both authors, R.S.L. and J.G.C., designed and discuss the methodology used to perform the experiments. Both authors, J.G.C. and R.S.L., acted in the management and coordination responsibility for the research activity planning and execution. J.G.C. was responsible of the provision of materials, reagents etc. and contact other investigators that provide materials used in the development of the work. R.S.L. and J.G.C. validated replication/reproducibility of results/experiments presented. R.S.L. did the preparation and creation of specifically visualization/data presentation. J.G.C. made the original draft with contributions by R.S.L. J.G.C. wrote the revised version and RSL provided comments and prepared the materials to be presented.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86847-2.
References
- 1.Hernandez DG, Reed X, Singleton AB. Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J. Neurochem. 2016;139(Suppl 1):59–74. doi: 10.1111/jnc.13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifati V, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 3.Nuytemans K, Theuns J, Cruts M, van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: A mutation update. Hum. Mutat. 2010;31:763–780. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao X, Tong L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson's disease. J. Biol. Chem. 2003;278:31372–31379. doi: 10.1074/jbc.M304221200. [DOI] [PubMed] [Google Scholar]
- 5.Honbou K, et al. The crystal structure of DJ-1, a protein related to male fertility and Parkinson's disease. J. Biol. Chem. 2003;278:31380–31384. doi: 10.1074/jbc.M305878200. [DOI] [PubMed] [Google Scholar]
- 6.Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA. The 11-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huai Q, et al. Crystal structure of DJ-1/RS and implication on familial Parkinson's disease. FEBS Lett. 2003;549:171–175. doi: 10.1016/S0014-5793(03)00764-6. [DOI] [PubMed] [Google Scholar]
- 8.Lee SJ, et al. Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J. Biol. Chem. 2003;278:44552–44559. doi: 10.1074/jbc.M304517200. [DOI] [PubMed] [Google Scholar]
- 9.Malgieri G, Eliezer D. Structural effects of Parkinson's disease linked DJ-1 mutations. Protein Sci. 2008;17:855–868. doi: 10.1110/ps.073411608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagakubo D, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, et al. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J. Biol. Chem. 2001;276:37556–37563. doi: 10.1074/jbc.M101730200. [DOI] [PubMed] [Google Scholar]
- 12.Taira T, Iguchi-Ariga SM, Ariga H. Co-localization with DJ-1 is essential for the androgen receptor to exert its transcription activity that has been impaired by androgen antagonists. Biol. Pharm. Bull. 2004;27:574–577. doi: 10.1248/bpb.27.574. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, et al. The Parkinson's disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum. Mol. Genet. 2005;14:1231–1241. doi: 10.1093/hmg/ddi134. [DOI] [PubMed] [Google Scholar]
- 14.Shinbo Y, Taira T, Niki T, Iguchi-Ariga SM, Ariga H. DJ-1 restores p53 transcription activity inhibited by Topors/p53BP3. Int. J. Oncol. 2005;26:641–648. [PubMed] [Google Scholar]
- 15.Zhong N, et al. DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor. J. Biol. Chem. 2006;281:20940–20948. doi: 10.1074/jbc.M601935200. [DOI] [PubMed] [Google Scholar]
- 16.Hod Y, Pentyala SN, Whyard TC, El Maghrabi MR. Identification and characterization of a novel protein that regulates RNA-protein interaction. J. Cell Biochem. 1999;72:435–444. doi: 10.1002/(SICI)1097-4644(19990301)72:3<435::AID-JCB12>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 17.van der Brug MP, et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinbo Y, et al. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 2006;13:96–108. doi: 10.1038/sj.cdd.4401704. [DOI] [PubMed] [Google Scholar]
- 19.Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004;2:e362. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou W, Zhu M, Wilson MA, Petsko GA, Fink AL. The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J. Mol. Biol. 2006;356:1036–1048. doi: 10.1016/j.jmb.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 21.Deeg S, et al. BAG1 restores formation of functional DJ-1 L166P dimers and DJ-1 chaperone activity. J. Cell Biol. 2010;188:505–513. doi: 10.1083/jcb.200904103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee DH, et al. PARK7 modulates autophagic proteolysis through binding to the N-terminally arginylated form of the molecular chaperone HSPA5. Autophagy. 2018;14:1870–1885. doi: 10.1080/15548627.2018.1491212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Junn E, et al. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9691–9696. doi: 10.1073/pnas.0409635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai-Nagara I, Matsuoka S, Ariga H, Suda T. Mortalin and DJ-1 coordinately regulate hematopoietic stem cell function through the control of oxidative stress. Blood. 2014;123:41–50. doi: 10.1182/blood-2013-06-508333. [DOI] [PubMed] [Google Scholar]
- 25.Meulener MC, Xu K, Thomson L, Ischiropoulos H, Bonini NM. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12517–12522. doi: 10.1073/pnas.0601891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canet-Aviles RM, et al. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taira T, et al. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou W, Freed CR. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J. Biol. Chem. 2005;280:43150–43158. doi: 10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- 29.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aleyasin H, et al. The Parkinson's disease gene DJ-1 is also a key regulator of stroke-induced damage. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18748–18753. doi: 10.1073/pnas.0709379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F, Nguyen JL, Hulleman JD, Li L, Rochet JC. Mechanisms of DJ-1 neuroprotection in a cellular model of Parkinson's disease. J. Neurochem. 2008;105:2435–2453. doi: 10.1111/j.1471-4159.2008.05333.x. [DOI] [PubMed] [Google Scholar]
- 32.Blackinton J, et al. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J. Biol. Chem. 2009;284:6476–6485. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aleyasin H, et al. DJ-1 protects the nigrostriatal axis from the neurotoxin MPTP by modulation of the AKT pathway. Proc. Natl. Acad. Sci. U. S. A. 2010;197:3186–3191. doi: 10.1073/pnas.0914876107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Billia F, et al. Parkinson-susceptibility gene DJ-1/PARK7 protects the murine heart from oxidative damage in vivo. Proc. Natl. Acad. Sci. U. S. A. 2013;110:6085–6090. doi: 10.1073/pnas.1303444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi MS, et al. Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in Parkinson's disease models. J. Neurosci. 2014;34:15123–15131. doi: 10.1523/JNEUROSCI.4751-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macedo MG, et al. The DJ-1L166P mutant protein associated with early onset Parkinson's disease is unstable and forms higher-order protein complexes. Hum. Mol. Genet. 2003;12:2807–2816. doi: 10.1093/hmg/ddg304. [DOI] [PubMed] [Google Scholar]
- 37.Miller DW, et al. L166P mutant DJ-1, causative for recessive Parkinson's disease, is degraded through the ubiquitin-proteasome system. J. Biol. Chem. 2003;278:36588–36595. doi: 10.1074/jbc.M304272200. [DOI] [PubMed] [Google Scholar]
- 38.Moore DJ, Zhang L, Dawson TM, Dawson VL. A missense mutation (L166P) in DJ-1, linked to familial Parkinson's disease, confers reduced protein stability and impairs homo-oligomerization. J. Neurochem. 2003;87:1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- 39.Olzmann JA, et al. Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 2004;279:8506–8515. doi: 10.1074/jbc.M311017200. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi-Niki K, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson's disease patients. Biochem. Biophys. Res. Commun. 2004;320:389–397. doi: 10.1016/j.bbrc.2004.05.187. [DOI] [PubMed] [Google Scholar]
- 41.Blackinton J, et al. Effects of DJ-1 mutations and polymorphisms on protein stability and subcellular localization. Mol. Brain Res. 2005;134:76–83. doi: 10.1016/j.molbrainres.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Gorner K, et al. Structural determinants of the C-terminal helix-kink-helix motif essential for protein stability and survival promoting activity of DJ-1. J. Biol. Chem. 2007;282:13680–13691. doi: 10.1074/jbc.M609821200. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez-Castelao B, et al. Reduced protein stability of human DJ-1/PARK7 L166P, linked to autosomal recessive Parkinson disease, is due to direct endoproteolytic cleavage by the proteasome. Biochim. Biophys. Acta. 2012;1823:524–533. doi: 10.1016/j.bbamcr.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 44.Baulac S, LaVoie MJ, Strahle J, Schlossmacher MG, Xia W. Dimerization of Parkinson's disease-causing DJ-1 and formation of high molecular weight complexes in human brain. Mol. Cell Neurosci. 2004;27:236–246. doi: 10.1016/j.mcn.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, et al. Mitochondrial localization of the Parkinson's disease related protein DJ-1: Implications for pathogenesis. Hum. Mol. Genet. 2005;14:2063–2073. doi: 10.1093/hmg/ddi211. [DOI] [PubMed] [Google Scholar]
- 46.Ramsey CP, Giasson BI. L10p and P158DEL DJ-1 mutations cause protein instability, aggregation, and dimerization impairments. J. Neurosci. Res. 2010;88:3111–3124. doi: 10.1002/jnr.22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rannikko EH, et al. Loss of DJ-1 protein stability and cytoprotective function by Parkinson's disease-associated proline-158 deletion. J. Neurochem. 2012;125:314–327. doi: 10.1111/jnc.12126. [DOI] [PubMed] [Google Scholar]
- 48.Repici M, et al. Parkinson's disease-associated mutations in DJ-1 modulate its dimerization in living cells. J. Mol. Med. (Berl) 2013;91:599–611. doi: 10.1007/s00109-012-0976-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taipa R, et al. DJ-1 linked parkinsonism (PARK7) is associated with Lewy body pathology. Brain. 2016;139:1680–1687. doi: 10.1093/brain/aww080. [DOI] [PubMed] [Google Scholar]
- 50.Ramsey CP, Giasson BI. The E163K DJ-1 mutant shows specific antioxidant deficiency. Brain Res. 2008;1239:1–11. doi: 10.1016/j.brainres.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakshminarasimhan M, Maldonado MT, Zhou W, Fink AL, Wilson MA. Structural impact of three Parkinsonism-associated missense mutations on human DJ-1. Biochemistry. 2008;47:1381–1392. doi: 10.1021/bi701189c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alvarez-Castelao B, Ruiz-Rivas C, Castano JG. A critical appraisal of quantitative studies of protein degradation in the framework of cellular proteostasis. Biochem. Res. Int. 2012;2012:823597. doi: 10.1155/2012/823597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo JF, et al. Mutation analysis of Parkin, PINK1, DJ-1 and ATP13A2 genes in Chinese patients with autosomal recessive early-onset Parkinsonism. Mov. Disord. 2008;23:2074–2079. doi: 10.1002/mds.22156. [DOI] [PubMed] [Google Scholar]
- 54.Abou-Sleiman PM, Healy DG, Quinn N, Lees AJ, Wood NW. The role of pathogenic DJ-1 mutations in Parkinson's disease. Ann. Neurol. 2003;54:283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 55.Tang B, et al. Association of PINK1 and DJ-1 confers digenic inheritance of early-onset Parkinson's disease. Hum. Mol. Genet. 2006;15:1816–1825. doi: 10.1093/hmg/ddl104. [DOI] [PubMed] [Google Scholar]
- 56.Hering R, et al. Novel homozygous p.E64D mutation in DJ1 in early onset Parkinson disease (PARK7) Hum. Mutat. 2004;24:321–329. doi: 10.1002/humu.20089. [DOI] [PubMed] [Google Scholar]
- 57.Clark LN, et al. Analysis of an early-onset Parkinson's disease cohort for DJ-1 mutations. Mov. Disord. 2004;19:796–800. doi: 10.1002/mds.20131. [DOI] [PubMed] [Google Scholar]
- 58.Macedo MG, et al. Genotypic and phenotypic characteristics of Dutch patients with early onset Parkinson's disease. Mov. Disord. 2009;24:196–203. doi: 10.1002/mds.22287. [DOI] [PubMed] [Google Scholar]
- 59.Annesi G, et al. DJ-1 mutations and parkinsonism-dementia-amyotrophic lateral sclerosis complex. Ann. Neurol. 2005;58:803–807. doi: 10.1002/ana.20666. [DOI] [PubMed] [Google Scholar]
- 60.Nuytemans K, et al. Relative contribution of simple mutations vs. copy number variations in five Parkinson disease genes in the Belgian population. Hum. Mutat. 2009;30:1054–1061. doi: 10.1002/humu.21007. [DOI] [PubMed] [Google Scholar]
- 61.Bandyopadhyay S, Cookson MR. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol. Biol. 2004;4:6. doi: 10.1186/1471-2148-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li HM, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Association of DJ-1 with chaperones and enhanced association and colocalization with mitochondrial Hsp70 by oxidative stress. Free Radic. Res. 2005;39:1091–1099. doi: 10.1080/10715760500260348. [DOI] [PubMed] [Google Scholar]
- 63.Kojima W, et al. Unexpected mitochondrial matrix localization of Parkinson's disease-related DJ-1 mutants but not wild -type DJ-1. Genes Cells. 2016;21:772–788. doi: 10.1111/gtc.12382. [DOI] [PubMed] [Google Scholar]
- 64.Bjorkblom B, et al. Reactive oxygen species-mediated DJ-1 monomerization modulates intracellular trafficking involving karyopherin beta2. Mol. Cell Biol. 2014;34:3024–3040. doi: 10.1128/MCB.00286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quiros PM, et al. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8:542–556. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 66.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 67.Song IK, et al. Degradation of redox-sensitive proteins including peroxiredoxins and DJ-1 is promoted by oxidation-induced conformational changes and ubiquitination. Sci. Rep. 2016;6:34432. doi: 10.1038/srep34432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanchez-Lanzas R, Castano JG. Inhibitors of lysosomal function or serum starvation in control or LAMP2 deficient cells do not modify the cellular levels of Parkinson disease-associated DJ-1/PARK 7 protein. PLoS ONE. 2018;13:e0201152. doi: 10.1371/journal.pone.0201152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maita C, Maita H, Iguchi-Ariga SM, Ariga H. Monomer DJ-1 and its N-terminal sequence are necessary for mitochondrial localization of DJ-1 mutants. PLoS ONE. 2013;8:e54087. doi: 10.1371/journal.pone.0054087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dikoglu E, et al. Mutations in LONP1, a mitochondrial matrix protease, cause CODAS syndrome. Am. J. Med. Genet. A. 2015;167:1501–1509. doi: 10.1002/ajmg.a.37029. [DOI] [PubMed] [Google Scholar]
- 71.Strauss KA, et al. CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ Lon protease. Am. J. Hum. Genet. 2015;96:121–135. doi: 10.1016/j.ajhg.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nimmo GAM, et al. Bi-allelic mutations of LONP1 encoding the mitochondrial LonP1 protease cause pyruvate dehydrogenase deficiency and profound neurodegeneration with progressive cerebellar atrophy. Hum. Mol. Genet. 2019;28:290–306. doi: 10.1093/hmg/ddy351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 74.Tian Q, et al. Lon peptidase 1 (LONP1)-dependent breakdown of mitochondrial 5-aminolevulinic acid synthase protein by heme in human liver cells. J. Biol. Chem. 2011;286:26424–26430. doi: 10.1074/jbc.M110.215772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu B, et al. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell. 2013;49:121–132. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Granot Z, et al. Turnover of mitochondrial steroidogenic acute regulatory (StAR) protein by Lon protease: The unexpected effect of proteasome inhibitors. Mol. Endocrinol. 2007;21:2164–2177. doi: 10.1210/me.2005-0458. [DOI] [PubMed] [Google Scholar]
- 77.Pryde KR, Taanman JW, Schapira AH. A LON-ClpP proteolytic axis degrades complex i to extinguish ros production in depolarized mitochondria. Cell Rep. 2016;17:2522–2531. doi: 10.1016/j.celrep.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bezawork-Geleta A, Saiyed T, Dougan DA, Truscott KN. Mitochondrial matrix proteostasis is linked to hereditary paraganglioma: LON-mediated turnover of the human flavinylation factor SDH5 is regulated by its interaction with SDHA. FASEB J. 2014;28:1794–1804. doi: 10.1096/fj.13-242420. [DOI] [PubMed] [Google Scholar]
- 79.Maio N, et al. Disease-causing SDHAF1 mutations impair transfer of Fe-S clusters to SDHB. Cell Metab. 2016;23:292–302. doi: 10.1016/j.cmet.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bezawork-Geleta A, Brodie EJ, Dougan DA, Truscott KN. LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci. Rep. 2015;5:17397. doi: 10.1038/srep17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Greene AW, et al. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012;13:378–385. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomas RE, Andrews LA, Burman JL, Lin WY, Pallanck LJ. PINK1-Parkin pathway activity is regulated by degradation of PINK1 in the mitochondrial matrix. PLoS. Genet. 2014;10:e1004279. doi: 10.1371/journal.pgen.1004279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zurita, R. O. & Shoubridge, E. A., LONP1 is required for maturation of a subset of mitochondrial proteins, and its loss elicits an integrated stress response, Mol. Cell Biol.38 (2018). [DOI] [PMC free article] [PubMed]
- 85.Lin W, Kang UJ. Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem. 2008;106:464–474. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS. Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, et al. The ubiquitination of PINK1 is restricted to its mature 52-kDa form. Cell Rep. 2017;20:30–39. doi: 10.1016/j.celrep.2017.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9747–9752. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haque ME, et al. Inactivation of Pink1 gene in vivo sensitizes dopamine-producing neurons to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and can be rescued by autosomal recessive Parkinson disease genes, Parkin or DJ-1. J. Biol. Chem. 2012;287:23162–23170. doi: 10.1074/jbc.M112.346437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thomas KJ, et al. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 2010;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Irrcher I, et al. Loss of the Parkinson's Disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- 92.Lau E, et al. A large dataset of protein dynamics in the mammalian heart proteome. Sci. Data. 2016;3:160015. doi: 10.1038/sdata.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.