Abstract
The photosynthetic pathway of plants is a fundamental trait that influences terrestrial environments from the local to global level. The distribution of different photosynthetic pathways in Australia is expected to undergo a substantial shift due to climate change and rising atmospheric CO2; however, tracking change is hindered by a lack of data on the pathways of species, as well as their distribution and relative cover within plant communities. Here we present the photosynthetic pathways for 2428 species recorded across 541 plots surveyed by Australia’s Terrestrial Ecosystem Research Network (TERN) between 2011 and 2017. This dataset was created to facilitate research exploring trends in vegetation change across Australia. Species were assigned a photosynthetic pathway using published literature and stable carbon isotope analysis of bulk tissue. The photosynthetic pathway of species can be extracted from the dataset individually, or used in conjunction with vegetation surveys to study the occurrence and abundance of pathways across the continent. This dataset will be updated as TERN’s plot network expands and new information becomes available.
Subject terms: Biodiversity, Stable isotope analysis, Photosynthesis
| Measurement(s) | oxidative photosynthetic carbon pathway |
| Technology Type(s) | digital curation • elemental analysis isotope ratio mass spectrometry |
| Factor Type(s) | date • geographic location |
| Sample Characteristic - Organism | Viridiplantae |
| Sample Characteristic - Environment | terrestrial natural environment |
| Sample Characteristic - Location | Australia |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.14134994
Background & Summary
The photosynthetic pathway of plants has a substantial impact on species productivity, abundance, and geographic distribution1–3. There are three primary photosynthetic pathways. C3 photosynthesis is the most common pathway. Plants that use this pathway include cool season grasses, most shrubs, and nearly all trees4,5. C4 plants include warm-season grasses, many sedges, and some forbs and shrubs6. Finally, Crassulacean acid metabolism (CAM) plants most commonly include epiphytes and succulents7. C3 plants have no special adaptations to prevent photorespiration, an energetically expensive process that occurs when the enzyme rubisco binds with oxygen to produce 2-phosphoglycolate8–10. The rate of photorespiration increases with increasing temperature11, restricting the photosynthetic capacity of C3 plants in warm environments. In contrast, C4 and CAM plants possess a series of biochemical, anatomical, and physiological adaptations that concentrate and isolate CO2 with rubisco, helping to eliminate photorespiration6,12. Consequently, C4 and CAM plants more easily live in hot or arid habitats3,13.
Global warming is expected to alter the competitive advantage of plants with different photosynthetic pathways14–16, changing species distributions and community composition, and leading to significant bottom-up effects on the structure, diversity and function of terrestrial communities17–19. Thus, the ecology and evolution of these different pathways has become a focus of recent botanical research20–22. Australia is an ecologically diverse continent that includes a wide variety of habitats and climatic zones23–25, making it an ideal environment to examine trends in C3, C4 and CAM distribution23,26. However, the photosynthetic pathway of numerous Australian species has not been assessed, and nationally systematic, compatible, and comparable vegetation surveys have not been historically available. The absence of these fundamental data severely limits national terrestrial research capacity.
Here we provide a dataset that lists the photosynthetic pathways of 2428 species found across Australia. These species were recorded at 541 vegetation survey plots established between 2011 and 2017 (inclusive; Fig. 1). These plots were established by the Terrestrial Ecosystem Research Network (TERN), Australia’s national terrestrial monitoring organisation. TERN is funded by the National Collaborative Research Infrastructure Strategy (NRCIS) and observes, records, and measures critical terrestrial ecosystem parameters and conditions for Australia over time. TERN Ecosystem Surveillance is one of three major branches within TERN, and is responsible for a nation-wide plot survey program. At each plot, TERN records vegetation composition and structural characteristics, and collects a range of soil and plant samples27,28. TERN data and resources are made freely accessible to scientists around the globe. The photosynthetic pathway dataset presented here was originally created by TERN to help facilitate research examining the distribution and abundance of C4 vegetation in Australia. This dataset will continue to be curated and updated as TERN increases its network of survey plots, and as new research investigates the photosynthetic pathways of terrestrial species.
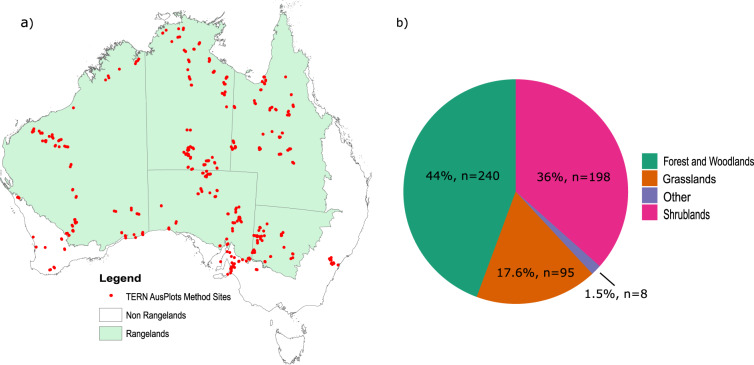
Fig. 1.
(a) Location of TERN Ecosystem Surveillance plots surveyed using the AusPlots Rangelands method from 2011–2017. Areas in green denote rangeland habitat (b) number (n) and proportion (%) of TERN Ecosystem Surveillance plots grouped by vegetation type.
Photosynthetic pathways were primarily assigned using peer-reviewed literature. We also measured the stable carbon isotope (δ13C) values of 540 species that had no recorded pathway. Tissue samples for δ13C analysis were acquired from plant specimens collected during TERN plot surveys. Using these techniques, we identified 2048 C3, 346 C4, 17 C3-CAM, and 7 C3-C4, 7 CAM, and 4 C4-CAM species across all plots. C4 species were found in 14 families and 84 genera. Most C4 species were Poaceae (228; 65.8%), followed by Cyperaceae (38; 10.9%) and Chenopodiaceae (25; 7.2%). CAM and CAM-facultative species were mainly found in Aizoaceae, Portulacaceae, and Crassulaceae. 14 genera included multiple photosynthetic pathways, specifically Tetragonia (Aizoaceae), Alternanthera (Amaranthaceae), Heliotropium (Boraginaceae), Polycarpaea (Caryophyllaceae), Tecticornia (Chenopodeceae), Cleome (Cleomaceae), Cyperus (Cyperaceae), Euphorbia (Euphorbiaceae), Aristida, Eragrostis, Neurachne, Panicum (Poaceae), and Tribulus (Zygophyllaceae). While data can be extracted for individual species, genera, or families, this dataset was designed to be used in conjunction with other TERN products. For example, photosynthetic pathway assignments can be directly combined with matching species records in TERN AusPlots vegetation surveys to obtain data on plant distribution, growth form, height and cover. These records can also be combined with other TERN plot data and products, including climate, soil, and landscape rasters. We expect this dataset will enable work examining patterns in plant occurrence, richness, and abundance, and ecosystem function at local to national scales.
Methods
The methods used to create this dataset are presented in the following order:
The TERN plot-based methodologies used to survey and identify plant species, and preserve plant specimens for stable isotope analysis
The procedures used to assign species a photosynthetic pathway using peer-reviewed literature
The procedures used to assign species a photosynthetic pathway using stable carbon isotope (δ13C) analysis.
TERN plot survey protocols, species identification, and sample collection
Plant species were identified at 541 one-hectare plots systemically surveyed by TERN between 2011 and 2017 (inclusive). Most TERN plots are located within the Australian rangelands (Fig. 1a). The Australian rangelands encompass 81% of the Australian landmass, and are characterised by vast spaces with highly weathered features, old and generally infertile soils29, highly variable rainfall, and diverse and variable plant and animal communities30. These areas have traditionally been underrepresented in Australian environmental monitoring programs, which typically focus on more mesic environments and areas closer to large population centres30. TERN’s AusPlots Rangelands method27,28 and location selection strategy was originally designed to address this underrepresentation by targeting these environments and developing and implementing survey methods that were consistent across the whole of the rangelands. Over time the network has expanded to include sampling in all the major terrestrial environments across the country, including alpine, heathland, and the subtropical systems of the east coast. The dominant vegetation types surveyed at the time of this work were woodlands and savannahs, tussock and hummock grasslands, and shrublands (including chenopod shrublands; Fig. 1b). Climate in TERN plots varies from monsoonal tropics in the north, arid deserts in the centre, to winter-dominant rainfall in the south.
The AusPlots Rangeland method27,28 consists of numerous survey modules designed to collect a wide suite of data on soil and vegetation attributes, as well as site contextual information (e.g. erosion, recent fires, etc.). These modules were conceived to provide the data level necessary to study plant community composition and structure, while also ensuring consistency in the collection of samples and data on vegetation, land, and soil characteristics. A complete description of TERN plot survey protocols is detailed in the TERN AusPlots Rangeland manual27,28. Only the protocols most relevant to plant surveys, identification, and specimen preservation are documented here.
TERN survey plots are 1 ha (100 × 100 m) permanently established sites located in a homogenous area of terrestrial vegetation (Fig. 2). Plots are usually surveyed only once, with an intention to revisit once per decade. Plots are surveyed as seasonal conditions permit, with the aim being to maximise the quality of the plant material collected and facilitate accurate herbarium identifications. Survey teams consist of between 2- and 6 people. A full complement of 6 people would include 1 to 2 people performing the vegetation survey modules, 1 to 2 people performing the soil survey modules, and the remaining team members undertaking other components of the Ausplots Rangelands method, such as recording site contextual information. The duration of each survey is variable and dependent on the density and diversity of the vegetation. Plot selection and orientation avoids major anthropogenic influences (such as roads, cattle yards, fences, bores, etc.). Ten transects (100 m long) are laid out within each plot in a grid pattern. Parallel transects running north to south are spaced 20 meters apart located at 10, 30, 50, 70, and 90 m both north and east from the SW corner (Fig. 2). Each plot is given a unique alphanumeric identifier that indicates the location of the plot, specifically its state (e.g. Western Australia, South Australia, Northern Territory, etc.) and Interim Biogeographic Regionalisation for Australia (IBRA) version 7 bioregion31, and a sequential number based on the number of plots in that bioregion. The date of the survey and GPS co-ordinates are also recorded for each plot.
Fig. 2.
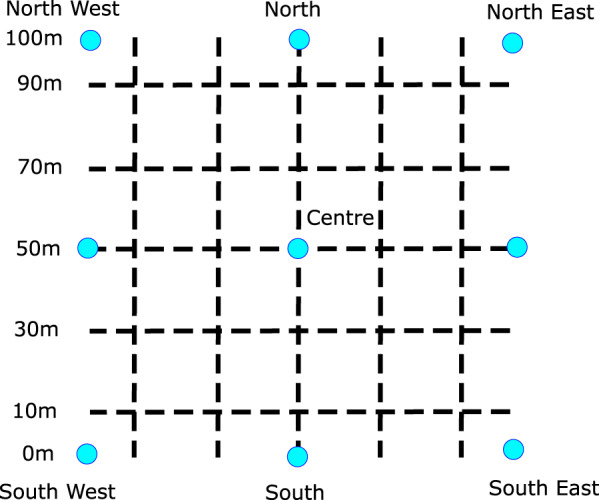
TERN Ecosystem Surveillance plot layout. The corners and centre of the plot (blue dots) are permanently marked with pickets and their locations recorded via GPS. Transects (dashed-lines, 100 m long) are laid in a grid pattern spaced 20 meters apart28.
Recording, collection, and identification of vascular flora is undertaken by specially trained members of the field survey team. One ground observer is tasked to perform line intercept transects. This ground observer records the species and substrate at each point (1 m) along each transect, resulting in survey data at 1010 points per plot. These point-intercept data are collected to calculate species cover (%) and other metrics. A second ground observer collects specimens of each vascular plant species in the plot, with enough material to fill an A3 size herbarium sheet (Fig. 3a,b). These members of the survey team work together to ensure the presence of each vascular plant species is recorded and enough specimens are collected. Each specimen ideally contains flowers or buds, leaves, fruit, and bark (for trees) to help enable identification. Each specimen is then tagged with a unique alphanumeric voucher barcode. All field and voucher data are recorded using a purpose-built app on a tablet to streamline data and sample collection32. The voucher specimen is ultimately delivered to a local herbarium for identification.
Fig. 3.
Collection procedures of vascular flora by TERN Ecosystem Surveillance team. (a) Collection of vascular flora by ground observers. (b) Voucher specimens are collected with enough material to fill an A3 size herbarium sheet, pressed, and ultimately sent to local herbaria for identification, (c) subsamples of each voucher specimen are collected from the main voucher sample to enable stable isotope analysis, the subsample is placed in a gauze “teabag” and (d) then sealed in a plastic container with 1 cm depth of silica granules (Photo Credit: TERN Ecosystem Surveillance program).
Subsamples of each voucher specimen are collected from the main voucher sample to enable stable isotope and molecular analysis (Fig. 3c). These subsamples are ideally free from disease, insect, or fungal contamination. The subsample is placed in a synthetic gauze ‘teabag’ and given its own unique alphanumeric barcode, referred to as the ‘primary genetic barcode’, which is linked to the date, plot, state, and voucher specimen from which it was collected. All teabags for a plot are then sealed in an air-tight, plastic container with 1 cm depth of silica granules (Fig. 3d). The container is stored in a cool location out of direct light for the duration of the survey. Upon return from the field, teabags are stored in dark conditions at room temperature at TERN facilities at the University of Adelaide (Adelaide, Australia). The silica granules are changed regularly until the samples are dehydrated and then replaced as necessary to keep the samples dry.
Photosynthetic pathway assignment
All TERN plant data were processed in the R statistical environment33 using the ausplotsR package34,35. The ausplotsR package was created by TERN to enable the live extraction, preparation, visualisation, and analysis of TERN Ecosystem Surveillance monitoring data. A list of all vascular plant species at each TERN plot was extracted using the get_ausplots function. This produced an initial list of 4002 unique records. Scientific names for each record are provided by herbaria and are the most commonly used names in the state where the voucher specimen was collected. However, scientific names sometimes vary between states due to jurisdictional differences in taxonomy and nomenclature. TERN Ecosystem Surveillance uses the scientific names as determined by the herbaria as the point of truth in all its analysis and data sets. State herbaria identify species to the lowest possible taxonomic level. Specimens that were only identified to the family or genus level were excluded from the photosynthetic pathway dataset. Hybrids were also excluded from the final species list. Varieties and subspecies were assumed to have the same photosynthetic pathway36, therefore photosynthetic pathways were assigned to the species (i.e. Genus species) rank. This process of elimination generated a final list of 2613 unique species.
To assign each species a photosynthetic pathway, scientific names were first cross-referenced against well-known plant trait databases including Kattge, et al.24, Osborne, et al.36, and Watson and Dallwitz37. We then conducted literature searches of the remaining unassigned species via Google Scholar with combinations of the key words “C3”, “C4”, “CAM”, “photosynthesis” and “photosynthetic pathway”. We used a total of 34 peer-reviewed sources to assign species photosynthetic pathways (Table 1). If species-specific information was not available, but the species belonged to a genus known to be exclusively C3, C4 or CAM it was assigned to that pathway (e.g. Acacia spp., Eucalyptus spp. are presumptive C3). Using these combined strategies, 1888 species were assigned a photosynthetic pathway. Discrepancies between sources were rare (total of 5). In cases where species were assigned different photosynthetic pathways by different sources, the photosynthetic pathway from the source that provided the best direct evidence to support the assignment was selected. If it was not possible to assign a photosynthetic pathway using published sources or presumptive reasoning, then that species was selected for stable carbon isotope analysis.
Table 1.
List of databases and peer-reviewed literature used to assign species in TERN plots a photosynthetic pathway.
| Source | Number of species assigned a photosynthetic pathway |
|---|---|
| Besnard, et al.60 | 1 |
| Bohley, et al.61 | 1 |
| Bruhl and Wilson62 | 8 |
| Caddy-Retalic63 | 154 |
| Carolin, et al.64 | 2 |
| Clayton, et al.65 | 3 |
| D’andrea, et al.66 | 1 |
| Ehleringer and Monson67 | 3 |
| Feodorova, et al.68 | 1 |
| Guillaume, et al.69 | 9 |
| Hancock, et al.45 | 7 |
| Herppich and Herppich70 | 1 |
| Holtum, et al.71 | 2 |
| Holtum, et al.72 | 1 |
| Horn, et al.73 | 10 |
| Kadereit, et al.74 | 1 |
| Kattge, et al.24 | 1013 |
| Koch and Kennedy75 | 1 |
| Madhusudana Rao, et al.76 | 1 |
| Metcalfe77 | 3 |
| Osborne, et al.36 | 6 |
| Pate, et al.78 | 8 |
| Sage5 | 657 |
| Sage, et al.25 | 27 |
| Sayed7 | 2 |
| Schmidt and Stewart79 | 1 |
| Taylor, et al.80. | 1 |
| Thiede and Eggli81 | 2 |
| Ting82 | 1 |
| Watson and Dallwitz37 | 637 |
| Watson and Dallwitz83 | 2 |
| Winter12 | 1 |
| Winter, et al.84 | 2 |
| Winter, et al.54 | 3 |
Sometimes multiple sources were used to justify the photosynthetic pathway assignment of a single species, as a result the total ‘Number of species assigned a photosynthetic pathway’ is greater than the number of unique species recorded in TERN plots.
The stable carbon isotope values of C3, C4, and CAM plants
The stable carbon isotope values of C3 plants range from −37‰ to −20‰ δ13C (mean = ~−27‰), while the values of C4 plants range from −12‰ to −16‰ δ13C (mean = ~−13‰)38,39. Therefore, for species where either a C3 or C4 pathway was possible (e.g. Poaceae), plants with δ13C values <−19‰ were designated C3, and plants with δ13C values >−19‰ were designated C426. Full CAM plants, or plants in which CAM is strongly expressed, have isotope values of >−20‰, and thus can be distinguished from C3 plants using δ13C39,40. However, CAM photosynthesis almost always co-exists with the C3 pathway (C3-CAM)12. The isotope values of C3-CAM plants are correlated with the proportion of carbon that is obtained during light and dark periods. As a result, C3-CAM δ13C values are highly variable (approximately −13‰ to −27‰) and are dependent upon the species, its developmental stage, and/or the time of day and conditions during which the plant was sampled40–42. For example, the CAM pathway is often upregulated during periods of stress, such as drought43,44. Therefore, although the δ13C of wild plant samples can be used to indicate CAM potential, stable isotope values are not a reliable way to distinguish CAM and C4, identify CAM when it is weakly expressed, or a definitive method to discriminate C3 and C3-CAM plants41,42. To confirm the presence of CAM, additional measures of other physiological and biochemical variables are usually required45. With this limitation in mind, for genera with previously confirmed C3-CAM potential, we followed past authors and tentatively denoted plants with a δ13C value >−20‰ as CAM, −21‰ to −24‰ as potentially C3 + CAM, and plants <−24‰ as C340,45,46.
Isotope analysis
540 species were selected for stable isotope analysis. The remaining 184 unassigned species were not included in δ13C analysis because no suitable tissue samples were available. TERN plant tissue samples were identified and selected using the ausplotsR package. Each species record is associated with a full list of the available silica-dried tissue samples. One sample was selected for stable isotope analysis based on overall condition and availability (i.e. the amount of sample available from a given plot).
A 2 g subsample of material was taken from each silica-dried tissue sample. Each subsample was placed in an Eppendorf tube with two small ball bearings and pulverised for approximately one minute at 30 htz using a Retsch Mixer Mill. If samples had not homogenised during this initial process, samples were transferred to a stainless-steel ball-mill grinder and were ground for a further one minute at 30 htz. Sample preparation procedures were performed at the Mawson Analytical Spectrometry Services (MASS) Facility, University of Adelaide. An initial group of 378 samples were analysed for stable isotopes at both MASS and the Stable Isotope Facility at the Waite Campus of CSIRO in 2019. A subsequent group of 162 plant samples were analysed in 2020 at MASS.
Stable carbon isotope analysis at CSIRO
2 to 2.5 mg of powdered plant samples were weighed into tin cups and analysed for δ13C using a continuous flow isotope ratio mass spectrometer (IRMS Delta V, ThermoBremen, Germany) equipped with an elemental analyser (Flash EA, Thermo, Bremen, Germany). Stable isotope ratios were expressed in δ notation as deviations from a standard in parts per mil (‰):
| 1 |
where Rsa is the ratio of abundances of 13C/12C in the sample, and Rref is this ratio in the reference gas47. δ13C was reported relative to the standard Vienna Pee Dee Belemnite (VPDB). See the “Technical Validation” section for normalisation methods and precision estimates.
Stable carbon isotope analysis at MASS, University of Adelaide
Like the procedures at CSIRO, 2 to 2.5 mg of powdered plant samples were weighed into tin cups and analysed for δ13C using a continuous flow isotope ratio mass spectrometer (Nu Horizon, Wrexham, UK) equipped with an elemental analyser (EA3000, EuroVector, Pavia, Italy). Stable isotope ratios were expressed in δ notation as deviations from a standard in parts per mil (‰) using Eq. 1. δ13C was reported relative to the standard Vienna Pee Dee Belemnite (VPDB). See the “Technical Validation” section for normalisation methods and precision estimates. Once all stable isotope analysis was complete, a final dataset was compiled that listed the photosynthetic pathway of 2429 plant species detected in TERN plots47.
Data Records
All data records are stored in the TERN Geospatial Catalogue repository and can be found via the TERN Data Discovery Portal47. Data has been released under a CC‐BY Creative Commons license (https://creativecommons.org/licenses/by/4.0/), which allows reuse with attribution. Any work or publications using these data should cite this descriptor and, if applicable, the original sources (Table 1). The data set is comprised of two data tables and one data descriptor file that defines the values in the two data tables (Table 2). All tables and files are in MS Excel (.xlsx). The first table contains a list of each species and its photosynthetic pathway. It specifies the method used to determine the photosynthetic pathway (i.e. peer-reviewed literature, inferred from lineage, or δ13C analysis), as well as the peer-reviewed source or δ13C value of the tested specimen, as applicable. The plot number, location, and date that specimens were collected, the facility where the stable isotope analysis was conducted, and any replicate δ13C values are also provided. Details on commonly used species name synonyms are also listed (see Usage Notes for details). Any discrepancies in photosynthetic pathway assignments between sources, or notes about the need for further testing to confirm tentative assignments, are also recorded for each species. The second table includes a list of all the peer-reviewed sources used to create this dataset. Updates to the dataset will be managed through the TERN Geospatial Catalogue by creating a new version of the dataset. As TERN continues to expand its plot network, we will aim to include new species on an annual basis. We will also re-evaluate species taxonomy and photosynthetic pathways as new information becomes available.
Table 2.
Description of database “The photosynthetic pathways of plant species surveyed in TERN Ecosystem Surveillance plots” with file locations.
| Source | Document Name | n. records | Data Description | Methods |
|---|---|---|---|---|
| Link | Plant Photosynthetic Pathway | 2428 | Photosynthetic Pathway of vascular plant species detected in TERN Ecosystem Surveillance plots | Literature search and stable isotope analysis |
| Link | List of Studies | 34 | Alphabetical list of references for species photosynthetic pathways | Literature Search |
| Link | Data Descriptor | 26 | Alphabetical list of descriptions for each data column in the “Plant Photosynthesis Pathway” data table | NA |
Technical Validation
TERN Ecosystem Surveillance plot surveys have been performed by different individuals and teams, which has the potential to introduce errors in plant identification in the field by ground observers. For this reason, all collections are given a temporary field name identification and assigned a permanent primary genetic barcode that is associated with a physical plant sample. Each data point and sample are tracked and recorded using the primary genetic barcode, which ensures each data point in the transect is correctly associated with a physical sample for later identification. TERN data is not published until the temporary field names are confirmed or corrected by expert local taxonomists at regional herbaria. Prior to publication of plot plant data, each species is cross-referenced against the Australian Plant Census (https://www.anbg.gov.au/chah/apc/) to confirm correct nomenclature. The whole database is also routinely compared to the Plant Census to detect changes in taxonomy over time.
Photosynthetic pathway assignments obtained from published sources have already been subject to scientific scrutiny and are well-validated. The assumption that all species within a given genus possess the same photosynthetic pathway is realistic in most circumstances3. However, our own work and the work of others has identified multiple exceptions. C4 and CAM photosynthesis have independently evolved multiple times across dozens of lineages48,49, which introduces the potential for misclassifications. To minimise this potential source of error, all species within a given family that are known to include C4 species were targeted for δ13C analysis. We targeted species in the families Aizoaceae, Asteraceae, Boraginaceae, Caryophyllaceae, Chenopodiaceae, Euphorbiaceae, Poaceae, Portulacaceae, and Zygophyllaceae. We recognize that Chenopodiaceae is now a subfamily of Amaranthaceae; however, chenopods have traditionally been examined as a separate family in past C4 analysis50–52. Therefore, to enable consistent comparisons with previous work and datasets we distinguished Chenopodiaceae independent of Amaranthaceae. As previously discussed, CAM or C3-CAM photosynthesis is particularly difficult to identify using δ13C, therefore any CAM or C3-CAM designations based on δ13C values should be considered tentative and warrant further investigation. Special mention should also be made of the genus Portucula (Portulacaceae). Traditionally considered a C4 genus, recent evidence has found some Portucula species have CAM potential53,54. Until species-specific information becomes available, most Portucula species in the dataset have been assigned to the C4 pathway, but the possibility of C4-CAM should be considered.
Stable isotope analysis was performed at two different laboratories over multiple years, therefore technical validation needs to be considered. Each laboratory measured plant δ13C using well-established analytical techniques. All samples where corrected for instrument drift and normalized according to reference values55 using a combination of certified and in-house calibrated standards (Table 3). For the stable isotope analysis conducted at CSIRO in 2019, all samples were normalized using a multipoint linear regression, where the slope and intercept are used to correct the isotope data on the δ13CVPDB scale56. Using the multipoint normalization procedure, measured δ values for the analysed standards are plotted on the x-axis, and the “true” accepted δ values expressed on the δ13CVPDB scale are plotting on the y-axis. These points create a regression line (Eq. 2) that covers the range of δ values:
| 2 |
Where a is the slope and b is the intercept. To normalize data, the measured δ value of the sample (δMSpl) is multiplied by the slope and the value of the intercept is added. Stable carbon isotope values had uncertainties of ≤0.77‰ δ13C based on repeat analysis of all the standards (n = 141). The mean and standard deviation of the absolute difference between replicate samples (10% of all samples) was 0.20 ± 0.34‰ δ13C.
Table 3.
List of standards (and their verified values) used to correct for instrument drift and normalize the δ13C of plant samples analyzed at the Stable Isotope Facility at the Waite Campus of CSIRO and the Mawson Analytical Spectrometry Services (MASS) Facility, University of Adelaide.
| Standard | Verified δ13C Value (‰) | Facility |
|---|---|---|
| USGS-40 | −26.39 | CSIRO/MASS |
| High Organic Sediment Standard OAS | −28.85 | CSIRO |
| Wheat Flour Standard OAS | −26.43 | CSIRO |
| Sorghum Flour Standard OAS | −13.78 | CSIRO |
| Glycine | −31.20 | MASS |
| Glutamic Acid | −16.72 | MASS |
| Triphenylamine | −29.20 | MASS |
| USGS 41 | −37.63 | MASS |
USGS-40 is a certified standard, all others were calibrated in-house by each facility.
MASS standards were calibrated using a two-point correction57:
| 3 |
where δsa,c is the corrected value of the measurement, δstd1,m and δstd2,m are the measured values of the standards, and δstd1 and δstd are the known values of the standards. For the isotope analysis conducted at MASS in 2019, isotope values had uncertainties of ≤0.31‰ δ13C based on repeat analysis of all the standards (n = 30). For the isotope analysis conducted at MASS in 2020, isotope values had uncertainties of ≤0.09‰ δ13C based on repeat analysis of all the standards (n = 75). The mean and standard deviation of the absolute difference between replicate samples (10% of all samples) in 2020 was 0.24 ± 0.48‰ δ13C. Given the broad but unique range of isotope values exhibited by C3 and C4 species, small deviations in values between laboratories are not likely to affect photosynthetic pathway assignment.
Usage Notes
All photosynthetic pathways assignments in this dataset are available in the public plant trait database ‘Austraits’, which aggregates trait values for Australian plants. Site descriptions and complete species and specimen lists can be freely accessed for all TERN plots via the TERN ausplotsR package (available via CRAN and with the latest development version and patches at https://github.com/ternaustralia/ausplotsR)34,35, or the TERN Data Discovery Portal (https://portal.tern.org.au/). As previously described, ausplotsR allows users to directly access all TERN plot-based data on vegetation and soils across Australia34,35. It also provides functions that calculate and visualise species presence, richness and cover (%) at all TERN plots. The photosynthetic pathway dataset presented here was designed to be easily combined with TERN ausplotsR species distribution data to investigate national distribution patterns of different photosynthetic pathways. As an example, we have provided sample code for the R statistical environment to demonstrate how the TERN photosynthetic pathway dataset presented here and % species cover calculated at TERN plots can be combined to calculate C4 plant cover (relative to C3) across Australia, and relate relative C4 cover values to changes in climate and local factors. As detailed in Supplementary File 1, simple functions in ausplotsR can quickly calculate % species cover at each TERN plot, and then each species in each plot can be assigned its correct photosynthetic pathway using the TERN photosynthetic pathway dataset. This enables the calculation of relative C4 plant cover at each plot. Relative C4 cover can then be regressed against climate and local parameters by using TERN plot coordinates to extract site-specific environmental data from other national climate58 and soil59 rasters.
Additional TERN data infrastructure can be found via the TERN Data Discovery Portal. For more information and tutorials on how to access TERN data, visit www.tern.org.
As previously discussed, scientific names for species in the TERN database are provided by state herbaria and are the most commonly used names in a given state. However, valid scientific names may vary between states due to differences in nomenclature (although this is rare). TERN Ecosystem Surveillance uses the scientific names as provided by the local herbaria as the point of truth in all its analysis and datasets. To enable the integration of this dataset with other data records, where there are known nomenclature issues between jurisdictions, we have notated alternative synonyms in the species name comments field of Table 1 in the dataset. When using this dataset, users should take care to select the most relevant synonym for their work.
Supplementary information
Acknowledgements
We acknowledge the TERN Ecosystem Surveillance field team for their work collecting the voucher specimens used in the δ13C analysis. We also acknowledge the support of TERN by the Australian government through the National Collaborative Research Infrastructure Strategy. Additional financial support for this project was provided by the AMP Foundation and the AMP Tomorrow Fund awarded to S.M., the Australian Research Council Future Fellowship (FT110100100793) awarded to F.A.M., and Australian Government Research Training Program and University of Adelaide Faculty of Sciences Divisional scholarships awarded to J.W.A.
Author contributions
S.M., F.A.M. and J.A. originally formulated the idea, S.M., F.A.M., J.A., E.L., G.G. and B.S. designed the study and developed the methodology. S.M., N.W., F.A.M., J.A., E.L., T.H., S.S., S.C.R. and R.A. collected plant samples, performed the experiments, and analysed the data. S.M. wrote the manuscript; all other authors provided editorial advice.
Code availability
No custom code was used in this analysis. Examples of how to combine this photosynthetic pathway dataset with other TERN data infrastructure in the R statistical environment has been provided in the supplementary material (Supplementary File 1).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-021-00877-z.
References
- 1.Collatz GJ, Berry JA, Clark JS. Effects of climate and atmospheric CO2 partial pressure on the global distribution of C4 grasses: present, past, and future. Oecologia. 1998;114:441–454. doi: 10.1007/s004420050468. [DOI] [PubMed] [Google Scholar]
- 2.von Fischer JC, Tieszen LL, Schimel DS. Climate controls on C3 vs. C4 productivity in North American grasslands from carbon isotope composition of soil organic matter. Glob. Chang. Biol. 2008;14:1141–1155. doi: 10.1111/j.1365-2486.2008.01552.x. [DOI] [Google Scholar]
- 3.Sage, R. F., Wedin, D. A. & Li, M. The biogeography of C4 photosynthesis: patterns and controlling factors. in C4plant biology (eds Rowan F. Sage & Russel K. Monson) 313–373 (Academic Press, 1999).
- 4.Kellogg EA. Evolutionary history of the grasses. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sage RF. A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and hall of fame. J Exp. Bot. 2016;68:11–28. doi: 10.1093/jxb/erw156. [DOI] [PubMed] [Google Scholar]
- 6.Sage RF, Sage TL, Kocacinar F. Photorespiration and the evolution of C4 photosynthesis. Ann. Rev. Plant. Biol. 2012;63:19–47. doi: 10.1146/annurev-arplant-042811-105511. [DOI] [PubMed] [Google Scholar]
- 7.Sayed OH. Crassulacean Acid Metabolism 1975–2000, a Check List. Photosynthetica. 2001;39:339–352. doi: 10.1023/A:1020292623960. [DOI] [Google Scholar]
- 8.Andrews, J. T. & Lorimer, G. H. Rubisco: structure, mechanisms, and prospects for improvement. in The Biochemistry of Plants: A Comprehensive Treatise Vol. 10 (eds MD Haleh & NK Boardman) 132–207 (Academic Press, 1987).
- 9.Ogren WL. Photorespiration: pathways, regulation, and modification. Annu. Rev. Plant. Physiol. 1984;35:415–442. doi: 10.1146/annurev.pp.35.060184.002215. [DOI] [Google Scholar]
- 10.Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR. The costs of photorespiration to food production now and in the future. Annu. Rev. Plant. Biol. 2016;67:107–129. doi: 10.1146/annurev-arplant-043015-111709. [DOI] [PubMed] [Google Scholar]
- 11.Dusenge ME, Duarte AG, Way DA. Plant carbon metabolism and climate change: elevated CO2 and temperature impacts on photosynthesis, photorespiration and respiration. New Phytol. 2019;221:32–49. doi: 10.1111/nph.15283. [DOI] [PubMed] [Google Scholar]
- 12.Winter K. Ecophysiology of constitutive and facultative CAM photosynthesis. J Exp. Bot. 2019;70:6495–6508. doi: 10.1093/jxb/erz002. [DOI] [PubMed] [Google Scholar]
- 13.Edwards EJ, Still CJ. Climate, phylogeny and the ecological distribution of C4 grasses. Ecol. Lett. 2008;11:266–276. doi: 10.1111/j.1461-0248.2007.01144.x. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa S, et al. Elevated CO2 concentrations reduce C4 cover and decrease diversity of understorey plant community in a Eucalyptus woodland. J Ecol. 2018;106:1483–1494. doi: 10.1111/1365-2745.12943. [DOI] [Google Scholar]
- 15.Wittmer MHOM, Auerswald K, Bai Y, Schaufele R, Schnyder H. Changes in the abundance of C3/C4 species of Inner Mongolia grassland: evidence from isotopic composition of soil and vegetation. Glob. Chang. Biol. 2010;16:605–616. doi: 10.1111/j.1365-2486.2009.02033.x. [DOI] [Google Scholar]
- 16.Winslow JC, Hunt ER, Jr, Piper SC. The influence of seasonal water availability on global C3 versus C4 grassland biomass and its implications for climate change research. Ecol. Model. 2003;163:153–173. doi: 10.1016/S0304-3800(02)00415-5. [DOI] [Google Scholar]
- 17.Haveles AW, Fox DL, Fox-Dobbs K. Carbon isoscapes of rodent diets in the Great Plains USA deviate from regional gradients in C4 grass abundance due to a preference for C3 plant resources. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2019;527:53–66. doi: 10.1016/j.palaeo.2019.04.003. [DOI] [Google Scholar]
- 18.Haddad NM, et al. Plant species loss decreases arthropod diversity and shifts trophic structure. Ecol. Lett. 2009;12:1029–1039. doi: 10.1111/j.1461-0248.2009.01356.x. [DOI] [PubMed] [Google Scholar]
- 19.Warne RW, Pershall AD, Wolf BO. Linking precipitation and C3–C4 plant production to resource dynamics in higher‐trophic‐level consumers. Ecology. 2010;91:1628–1638. doi: 10.1890/08-1471.1. [DOI] [PubMed] [Google Scholar]
- 20.Griffith DM, et al. Biogeographically distinct controls on C3 and C4 grass distributions: merging community and physiological ecology. Glob. Ecol. Biogeogr. 2015;24:304–313. doi: 10.1111/geb.12265. [DOI] [Google Scholar]
- 21.Still CJ, Cotton JM, Griffith DM. Assessing earth system model predictions of C4 grass cover in North America: From the glacial era to the end of this century. Glob. Ecol. Biogeogr. 2019;28:145–157. doi: 10.1111/geb.12830. [DOI] [Google Scholar]
- 22.Griffith DM, Cotton JM, Powell RL, Sheldon ND, Still CJ. Multi-century stasis in C3 and C4 grass distributions across the contiguous United States since the industrial revolution. J Biogeogr. 2017;44:2564–2574. doi: 10.1111/jbi.13061. [DOI] [Google Scholar]
- 23.Hattersley P. The distribution of C3 and C4 grasses in Australia in relation to climate. Oecologia. 1983;57:113–128. doi: 10.1007/BF00379569. [DOI] [PubMed] [Google Scholar]
- 24.Kattge J, et al. TRY plant trait database – enhanced coverage and open access. Glob. Chang. Biol. 2020;26:119–188. doi: 10.1111/gcb.14904. [DOI] [PubMed] [Google Scholar]
- 25.Sage RF, Sage TL, Pearcy RW, Borsch T. The taxonomic distribution of C4 photosynthesis in Amaranthaceae sensu stricto. Am J Bot. 2007;94:1992–2003. doi: 10.3732/ajb.94.12.1992. [DOI] [PubMed] [Google Scholar]
- 26.Murphy BP, Bowman DM. Seasonal water availability predicts the relative abundance of C3 and C4 grasses in Australia. Glob. Ecol. Biogeogr. 2007;16:160–169. doi: 10.1111/j.1466-8238.2006.00285.x. [DOI] [Google Scholar]
- 27.White, A. et al. AUSPLOTS rangelands survey protocols manual. (The University of Adelaide Press, 2012).
- 28.Sparrow, B. D. et al. A vegetation and soil survey method for surveillance monitoring of rangeland environments. Front. Ecol. Evol. 8 (2020).
- 29.Orians GH, Milewski AV. Ecology of Australia: the effects of nutrient‐poor soils and intense fires. Biol. Rev. 2007;82:393–423. doi: 10.1111/j.1469-185X.2007.00017.x. [DOI] [PubMed] [Google Scholar]
- 30.Sparrow, B. et al. Our capacity to tell an Australian ecological story. in Biodiversity and Environmental Change: Monitoring, Challenges and Direction 51–84 (CSIRO Publishing Collingwood, Victoria, 2014).
- 31.Thackway, R. & Cresswell, I. An Interim Biogeographic Regionalisation for Australia: a framework for establishing the national system of reserves, Version 4.0. (Australian Nature Conservation Agency, Canberra, 1995).
- 32.Tokmakoff A, Sparrow B, Turner D, Lowe A. AusPlots Rangelands field data collection and publication: Infrastructure for ecological monitoring. Future Gener. Comp. Sy. 2016;56:537–549. doi: 10.1016/j.future.2015.08.016. [DOI] [Google Scholar]
- 33.R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2019).
- 34.Guerin, G. et al. ausplotsR: TERN AusPlots analysis package. https://cran.r-project.org/web/packages/ausplotsR/index.html (2020).
- 35.Munroe, S. et al. ausplotsR: An R package for rapid extraction and analysis of vegetation and soil data collected by Australia’s Terrestrial Ecosystem Research Network. Preprint at https://ecoevorxiv.org/25phx/ (2020).
- 36.Osborne CP, et al. A global database of C4 photosynthesis in grasses. New Phytol. 2014;204:441–446. doi: 10.1111/nph.12942. [DOI] [PubMed] [Google Scholar]
- 37.Watson, L., & Dallwitz, M. J. The Families of Flowering Plants: Descriptions, Illustrations, Identification, and Information Retrieval. http://www1.biologie.uni-hamburg.de/b-online/delta/angio/index.htm (1992).
- 38.Kohn MJ. Carbon isotope compositions of terrestrial C3 plants as indicators of (paleo)ecology and (paleo)climate. PNAS. 2010;107:19691–19695. doi: 10.1073/pnas.1004933107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Leary MH. Carbon isotopes in photosynthesis. Bioscience. 1988;38:328–336. doi: 10.2307/1310735. [DOI] [Google Scholar]
- 40.Winter K, Holtum JAM, Smith JAC. Crassulacean acid metabolism: a continuous or discrete trait? New Phytol. 2015;208:73–78. doi: 10.1111/nph.13446. [DOI] [PubMed] [Google Scholar]
- 41.Winter K, Holtum JA. How closely do the δ13C values of crassulacean acid metabolism plants reflect the proportion of CO2 fixed during day and night? Plant Physiol. 2002;129:1843–1851. doi: 10.1104/pp.002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cernusak LA, et al. Environmental and physiological determinants of carbon isotope discrimination in terrestrial plants. New Phytol. 2013;200:950–965. doi: 10.1111/nph.12423. [DOI] [PubMed] [Google Scholar]
- 43.Winter K, Holtum JAM. Facultative crassulacean acid metabolism (CAM) plants: powerful tools for unravelling the functional elements of CAM photosynthesis. J Exp. Bot. 2014;65:3425–3441. doi: 10.1093/jxb/eru063. [DOI] [PubMed] [Google Scholar]
- 44.Bloom AJ, Troughton JH. High productivity and photosynthetic flexibility in a CAM plant. Oecologia. 1979;38:35–43. doi: 10.1007/BF00347822. [DOI] [PubMed] [Google Scholar]
- 45.Hancock LP, Holtum JAM, Edwards EJ. The evolution of CAM photosynthesis in Australian Calandrinia reveals lability in C3+ CAM phenotypes and a possible constraint to the evolution of strong CAM. Integr. Comp. Biol. 2019;59:517–534. doi: 10.1093/icb/icz089. [DOI] [PubMed] [Google Scholar]
- 46.Guralnick LJ, Cline A, Smith M, Sage RF. Evolutionary physiology: the extent of C4 and CAM photosynthesis in the genera Anacampseros and Grahamia of the Portulacaceae. J Exp. Bot. 2008;59:1735–1742. doi: 10.1093/jxb/ern081. [DOI] [PubMed] [Google Scholar]
- 47.Munroe S, 2020. The Photosynthetic Pathways of Plant Species surveyed in TERN Ecosystem Surveillance Plots. Terrestrial Ecosystem Research Network (TERN) [DOI]
- 48.Sage RF. The evolution of C4 photosynthesis. New Phytol. 2004;161:341–370. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- 49.Keeley JE, Rundel PW. Evolution of CAM and C4 carbon-concentrating mechanisms. Int. J Plant Sci. 2003;164:S55–S77. doi: 10.1086/374192. [DOI] [Google Scholar]
- 50.Wang R, Ma L. Climate-driven C4 plant distributions in China: divergence in C4 taxa. Sci. Rep. 2016;6:27977. doi: 10.1038/srep27977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stowe LG, Teeri JA. The geographic distribution of C4 species of the Dicotyledonae in relation to climate. Am. Nat. 1978;112:609–623. doi: 10.1086/283301. [DOI] [Google Scholar]
- 52.Pyankov, V. I., Gunin, P. D., Tsoog, S. & Black, C. C. C4 plants in the vegetation of Mongolia: their natural occurrence and geographical distribution in relation to climate. 123, 15-31 (2000). [DOI] [PubMed]
- 53.Guralnick LJ, Edwards G, Ku MS, Hockema B, Franceschi V. Photosynthetic and anatomical characteristics in the C4–crassulacean acid metabolism-cycling plant Portulaca grandiflora. Funct. Plant Biol. 2002;29:763–773. doi: 10.1071/PP01176. [DOI] [PubMed] [Google Scholar]
- 54.Winter K, Sage RF, Edwards EJ, Virgo A, Holtum JAM. Facultative crassulacean acid metabolism in a C3–C4 intermediate. J Exp. Bot. 2019;70:6571–6579. doi: 10.1093/jxb/erz085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coplen TB, et al. New guidelines for δ13C measurements. Anal. Chem. 2006;78:2439–2441. doi: 10.1021/ac052027c. [DOI] [PubMed] [Google Scholar]
- 56.Skrzypek G. Normalization procedures and reference material selection in stable HCNOS isotope analyses: an overview. Anal. Bioanal. Chem. 2013;405:2815–2823. doi: 10.1007/s00216-012-6517-2. [DOI] [PubMed] [Google Scholar]
- 57.Ke L, Lin Z, Guoxing Z. Study of normalization method of isotopic compositions to isotope reference scales. J Chem. Pharmaceut. Res. 2014;6:1. [Google Scholar]
- 58.Harwood T, 2016. 9s climatology for continental Australia 1976–2005: Summary variables with elevation and radiative adjustment, version 3. Commonwealth Scientific and Industrial Research Organisation (CSIRO) [DOI]
- 59.Viscarra Rossel R, 2014. Soil and Landscape Grid National Soil Attribute Maps - pH - CaCl2 (3” resolution), version 3. Commonwealth Scientific and Industrial Research Organisation (CSIRO) [DOI]
- 60.Besnard G, et al. Phylogenomics of C4 photosynthesis in sedges (Cyperaceae): multiple appearances and genetic convergence. Mol. Biol. Evol. 2009;26:1909–1919. doi: 10.1093/molbev/msp103. [DOI] [PubMed] [Google Scholar]
- 61.Bohley K, et al. Phylogeny of Sesuvioideae (Aizoaceae)–Biogeography, leaf anatomy and the evolution of C4 photosynthesis. Perspect. Plant Ecol. Evol. Syst. 2015;17:116–130. doi: 10.1016/j.ppees.2014.12.003. [DOI] [Google Scholar]
- 62.Bruhl JJ, Wilson KL. Towards a comprehensive survey of C3 and C4 photosynthetic pathways in Cyperaceae. Aliso. 2007;23:99–148. doi: 10.5642/aliso.20072301.11. [DOI] [Google Scholar]
- 63.Caddy-Retalic, S. Quantifying responses of ecological communities to bioclimatic gradients PhD thesis, University of Adelaide, School of Biological Sciences (2017).
- 64.Carolin R, Jacobs S, Vesk M. The chlorenchyma of some members of the Salicornieae (Chenopodiaceae) Aust. J. Bot. 1982;30:387–392. doi: 10.1071/BT9820387. [DOI] [Google Scholar]
- 65.Clayton, W. D., Vorontsova, M. S., Harman, K. T. & Williamson, H. World Grass Species: Synonymy. http://www.kew.org/data/grasses-syn.html (2002).
- 66.D’andrea RM, Andreo CS, Lara MV. Deciphering the mechanisms involved in Portulaca oleracea (C4) response to drought: metabolic changes including crassulacean acid‐like metabolism induction and reversal upon re‐watering. Physiol. Plant. 2014;152:414–430. doi: 10.1111/ppl.12194. [DOI] [PubMed] [Google Scholar]
- 67.Ehleringer JR, Monson RK. Evolutionary and ecological aspects of photosynthetic pathway variation. Annu. Rev. Ecol. Evol. Syst. 1993;24:411–439. doi: 10.1146/annurev.es.24.110193.002211. [DOI] [Google Scholar]
- 68.Feodorova TA, Voznesenskaya EV, Edwards GE, Roalson EH. Biogeographic patterns of diversification and the origins of C4 in Cleome (Cleomaceae) Syst. Bot. 2010;35:811–826. doi: 10.1600/036364410X539880. [DOI] [Google Scholar]
- 69.Guillaume K, Huard M, Gignoux J, Mariotti A, Abbadie L. Does the timing of litter inputs determine natural abundance of 13C in soil organic matter? Insights from an African tiger bush ecosystem. Oecologia. 2001;127:295–304. doi: 10.1007/s004420000598. [DOI] [PubMed] [Google Scholar]
- 70.Herppich WB, Herppich M. Ecophysiological investigations on plants of the genus Plectranthus (Fam. Lamiaceae) native to Yemen and southern Africa. Flora. 1996;191:401–408. doi: 10.1016/S0367-2530(17)33117-1. [DOI] [Google Scholar]
- 71.Holtum JA, et al. Australia lacks stem succulents but is it depauperate in plants with crassulacean acid metabolism (CAM)? Curr. Opin. Plant Biol. 2016;31:109–117. doi: 10.1016/j.pbi.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 72.Holtum JA, Hancock LP, Edwards EJ, Winter K. Facultative CAM photosynthesis (crassulacean acid metabolism) in four species of Calandrinia, ephemeral succulents of arid Australia. Photosynth. Res. 2017;134:17–25. doi: 10.1007/s11120-017-0359-x. [DOI] [PubMed] [Google Scholar]
- 73.Horn JW, et al. Evolutionary bursts in Euphorbia (Euphorbiaceae) are linked with photosynthetic pathway. Evolution. 2014;68:3485–3504. doi: 10.1111/evo.12534. [DOI] [PubMed] [Google Scholar]
- 74.Kadereit G, Borsch T, Weising K, Freitag H. Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. Int. J. Plant Sci. 2003;164:959–986. doi: 10.1086/378649. [DOI] [Google Scholar]
- 75.Koch KE, Kennedy RA. Crassulacean acid metabolism in the succulent C4 dicot, Portulaca oleracea L under natural environmental conditions. Plant. Physiol. 1982;69:757–761. doi: 10.1104/pp.69.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Madhusudana Rao I, Swamy PM, Das VSR. Some characteristics of crassulacean acid metabolism in five nonsucculent scrub species under natural semiarid conditions. Zeitschrift für Pflanzenphysiologie. 1979;94:201–210. doi: 10.1016/S0044-328X(79)80159-2. [DOI] [Google Scholar]
- 77.Metcalfe, C. R. Anatomy of the monocotyledons. 1. Gramineae. (Clarendon Press, 1960).
- 78.Pate JS, Unkovich MJ, Erskine PD, Stewart GR. Australian mulga ecosystems –13C and 15N natural abundances of biota components and their ecophysiological significance. Plant Cell Environ. 1998;21:1231–1242. doi: 10.1046/j.1365-3040.1998.00359.x. [DOI] [Google Scholar]
- 79.Schmidt S, Stewart G. δ15N values of tropical savanna and monsoon forest species reflect root specialisations and soil nitrogen status. Oecologia. 2003;134:569–577. doi: 10.1007/s00442-002-1150-y. [DOI] [PubMed] [Google Scholar]
- 80.Taylor SH, et al. Ecophysiological traits in C3 and C4 grasses: a phylogenetically controlled screening experiment. New Phytol. 2010;185:780–791. doi: 10.1111/j.1469-8137.2009.03102.x. [DOI] [PubMed] [Google Scholar]
- 81.Thiede, J. & Eggli, U. Crassulaceae. in Flowering Plants· Eudicots 83–118 (Springer, 2007).
- 82.Ting IP. Photosynthesis of arid and subtropical succulent plants. Aliso. 1989;12:387–406. doi: 10.5642/aliso.19891202.14. [DOI] [Google Scholar]
- 83.Watson, L., & Dallwitz, M. J. The grass genera of the world: descriptions, illustrations, identification, and information retrieval; including synonyms, morphology, anatomy, physiology, phytochemistry, cytology, classification, pathogens, world and local distribution, and references. https://www.delta-intkey.com/grass/intro.htm (1992).
- 84.Winter K, Garcia M, Virgo A, Holtum JA. Operating at the very low end of the crassulacean acid metabolism spectrum: Sesuvium portulacastrum (Aizoaceae) J. Exp. Bot. 2019;70:6561–6570. doi: 10.1093/jxb/ery431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Munroe S, 2020. The Photosynthetic Pathways of Plant Species surveyed in TERN Ecosystem Surveillance Plots. Terrestrial Ecosystem Research Network (TERN) [DOI]
- Harwood T, 2016. 9s climatology for continental Australia 1976–2005: Summary variables with elevation and radiative adjustment, version 3. Commonwealth Scientific and Industrial Research Organisation (CSIRO) [DOI]
- Viscarra Rossel R, 2014. Soil and Landscape Grid National Soil Attribute Maps - pH - CaCl2 (3” resolution), version 3. Commonwealth Scientific and Industrial Research Organisation (CSIRO) [DOI]
Supplementary Materials
Data Availability Statement
No custom code was used in this analysis. Examples of how to combine this photosynthetic pathway dataset with other TERN data infrastructure in the R statistical environment has been provided in the supplementary material (Supplementary File 1).