Abstract
Chemotherapeutic drugs suffer from non-specific binding, undesired toxicity, and poor blood circulation which contribute to poor therapeutic efficacy. In this study, antibody–drug nanoparticles (ADNs) are engineered by synthesizing pure anti-cancer drug nanorods (NRs) in the core of nanoparticles with a therapeutic monoclonal antibody, Trastuzumab on the surface of NRs for specific targeting and synergistic treatments of human epidermal growth factor receptor 2 (HER2) positive breast cancer cells. ADNs were designed by first synthesizing ~ 95 nm diameter × ~ 500 nm long paclitaxel (PTX) NRs using the nanoprecipitation method. The surface of PTXNRs was functionalized at 2′ OH nucleophilic site using carbonyldiimidazole and conjugated to TTZ through the lysine residue interaction forming PTXNR-TTZ conjugates (ADNs). The size, shape, and surface charge of ADNs were characterized using scanning electron microscopy (SEM), SEM, and zeta potential, respectively. Using fluorophore labeling and response surface analysis, the percentage conjugation efficiency was found > 95% with a PTX to TTZ mass ratio of 4 (molar ratio ≈ 682). In vitro therapeutic efficiency of PTXNR-TTZ was evaluated in two HER2 positive breast cancer cell lines: BT-474 and SK-BR-3, and a HER2 negative MDA-MB-231 breast cancer cell using MTT assay. PTXNR-TTZ inhibited > 80% of BT-474 and SK-BR-3 cells at a higher efficiency than individual PTX and TTZ treatments alone after 72 h. A combination index analysis indicated a synergistic combination of PTXNR-TTZ compared with the doses of single-drug treatment. Relatively lower cytotoxicity was observed in MCF-10A human breast epithelial cell control. The molecular mechanisms of PTXNR-TTZ were investigated using cell cycle and Western blot analyses. The cell cycle analysis showed PTXNR-TTZ arrested > 80% of BT-474 breast cancer cells in the G2/M phase, while > 70% of untreated cells were found in the G0/G1 phase indicating that G2/M arrest induced apoptosis. A similar percentage of G2/M arrested cells was found to induce caspase-dependent apoptosis in PTXNR-TTZ treated BT-474 cells as revealed using Western blot analysis. PTXNR-TTZ treated BT-474 cells showed ~ 1.3, 1.4, and 1.6-fold higher expressions of cleaved caspase-9, cytochrome C, and cleaved caspase-3, respectively than untreated cells, indicating up-regulation of caspase-dependent activation of apoptotic pathways. The PTXNR-TTZ ADN represents a novel nanoparticle design that holds promise for targeted and efficient anti-cancer therapy by selective targeting and cancer cell death via apoptosis and mitotic cell cycle arrest.
Subject terms: Biotechnology, Biomaterials, Drug delivery
Introduction
Engineering nanoscale drug delivery systems have been studied extensively because of several advantages including high drug payloads1–5, improved drug release6,7, enhanced bioavailability8,9, and increased multivalence effects through receptor-ligand interactions10–13. The use of a ligand allows direct delivery of the cytotoxic agent to target cells, however, the clinical success of targeted antibody–drug conjugates is limited due to poor bioavailability and low therapeutic efficacy14–16. Improvements in drug design in combination with active targeting are shown to enhance cellular uptake, targeting, and higher therapeutic efficacy which have led to the design of antibody–drug nanoparticles (ADNs). ADNs are comprised of an anticancer drug core nanoparticle conjugated with a therapeutic monoclonal antibody on the nanoparticle surface for targeted delivery and enhanced therapeutic efficacy. Trastuzumab (TTZ) is used to treat early-stage human epidermal growth factor receptor 2 (HER2) positive breast cancer as the first line of treatment. However, the complete response of the treatment is not achieved in 20–60% cases of the HER2 positive breast cancer patients17. The TTZ treatment is associated with the risk of therapeutic resistance and tumor recurrence after a certain period of the treatment. A lower risk of tumor recurrence and a better outcome can be expected when TTZ is administered with another chemotherapeutic drug in combination18,19. Till 2013, four Phase III clinical trials involving more than 8000 patients showed that the risk of recurrence was decreased by 50% when TTZ was administered with or after chemotherapy in combination20–24. The American Society of Clinical Oncology (ASCO) guideline recommended anti-HER2 therapy in combination with taxane as the first line of treatment for metastatic HER2 positive breast cancer25.
Combination therapy using paclitaxel (PTX) and TTZ for adjuvant HER2 positive breast cancer showed promising long-term clinical outcomes in recent years. In 2015, after a median follow up period of 4 yr, adjuvant PTX and TTZ combination treatment trials reported that the 3 yr disease-free survival rate was 98.7% in HER2 positive breast cancer patients26. In 2019, after 7 yr follow up period, the disease-free survival rate was 93% with an overall survival rate (OS) of 95%27. While the combination therapy of PTX and TTZ are promising, the following challenges still have to overcome in terms of synergy between the drugs, toxicity, and therapeutic efficacy28. The current clinically used intratumoral concentration of PTX (1–9 μM) is shown to be insufficient for breast cancer treatment29. Moreover, PTX is poorly soluble in the aqueous phase causing a great limitation and trade-off in terms of toxicity in intravenous administration. The high concentration of TTZ used in clinical trials has always been associated with the risk of severe cardiac arrest26–28.
To overcome the limitations of toxicity and off-site targeting, nanoparticles have been engineered with advanced chemical composition14–16,30, surface functionalization31–35, and geometric modification35–38. Nanoparticles made of human serum albumin (Abraxane) have been used for the delivery of PTX to breast cancer cells39. Another example of a nanoparticle-based drug delivery system is liposome-based doxorubicin (Doxil) for the treatment of ovarian cancer40,41 and breast cancer42. However, the efficiency of these nanoparticles is controversial due to the lack of active targeting and controlled drug delivery. Currently, a large number of nanoparticles such as liposomes43, polymers44, micelles45, solid lipid nanoparticles46, and other organic and inorganic nanoparticles have been explored to improve the therapeutic efficiency by active targeting of cancer cells through the design of nanoparticle size, shape and surface charge.
Nanoparticle size in the range of 50–500 nm is desirable for enhanced blood circulation, tissue penetration, and cellular interaction36,47,48. Size less than 10 nm is cleared by renal excretion, while larger particles are cleared by phagocytosis11,12,49,50. Non-spherical nanoparticles have been reported to be less clearance by macrophages than spherical nanoparticles as dictated by the contact angle of nanoparticles with the macrophage membrane51. Nanoparticle also leads to a dramatic increase in surface area for surface functionalization with targeting ligands such as monoclonal antibodies, affibodies, peptides, and aptamers. For example, binding of rod-shaped nanoparticles can be high if the particle aligns with its major axis parallel to the cell membrane increasing the multivalence effects of receptor-ligand interactions10–13.
In this study, we synthesized PTX nanorods (PTXNRs) of (96.8 ± 33) × (503.4 ± 210) nm using the nanoprecipitation method. TTZ was used as a HER2 targeting antibody for specific PTX delivery to HER2 positive breast cancer cells (BT-474 and SK-BR-3). The surface of PTXNRs was conjugated via carbonyldiimidazole (CDI), a highly reactive crosslinker to form an active N-acyl imidazole group capable of coupling with amine-containing TTZ and formed a stable amide linkage. Flow cytometry analysis of a fluorophore-labeled PTXNR-TTZ and response surface analysis confirmed >95% TTZ conjugation efficiency. A combination approach of PTXNR-TTZ ADNs was explored in exploiting the therapeutic efficacy of ADNs towards HER2 positive breast cancer cells. It is hypothesized that a combination system using PTXNR-TTZ ADNs enhances therapeutic payloads and treatment efficiency by selective targeting of HER2 positive breast cancer cells.
Materials and methods
Synthesis of PTXNRs
PTXNRs were prepared using the nanoprecipitation method (SI Fig. 1)10,52,53. Briefly, PTX powder (> 99.5%, Alfa Aesar) was dissolved in 5 ml of ethanol at a concentration of 3 mg/ml. The solution was homogenized at 1000 rpm, sonicated, and injected into 20 ml deionized (DI) water at a flow rate of 2 ml/min to form PTXNRs. PTXNRs were washed three times using DI water by centrifuging at 18,000 rcf for 2 h, suspended and lyophilized (Labconco) overnight to obtain dried particles.
Characterization of PTXNR
The size and shape of PTXNRs were analyzed using a scanning electron microscope (Hitachi S4700 FESEM). The images were obtained at 10.0 kV accelerating voltage with 4.5 mm working distance and 40,000× magnification. The size distribution of NRs was analyzed using Fiji image processing software (Image J; Win 64 Java 1.8.0). The surface charge of PTXNRs was measured in DI water and phosphate-buffered saline (PBS) using zeta potential (Nano series Zetasizer, Malvern).
Activation of 2′ OH nucleophilic site of PTXNR
TTZ (Genentech) was conjugated with the 2′ OH nucleophilic site group of PTX using the CDI activation reaction. Briefly, 1 mg of dry PTXNRs was suspended in 1 ml of PBS. 5 mg CDI was added directly to the NR suspension with a continuous stirring speed of 600 rpm. The reaction was carried out at 4 °C for 20 min to form the amine-reactive PTX-carbamate (PTXNR-CDI). The PTXNRs-CDI were centrifuged at 16,000× g for 30 min and washed five times using DI water to remove the unreacted CDI or imidazole. The particles were lyophilized for subsequent TTZ conjugation reactions.
1H-NMR analysis of activated PTXNR
To confirm the linkage of the activated carbamate group at the 2′ OH site of PTXNR, the 1H-NMR experiment was carried out. In carrying out the experiment, 2 mg of each unmodified PTXNR and surface functionalized PTXNR particles were dissolved in 600 µl of chloroform-d solvent (Alfa Aesar). The solvent was used as the internal reference to determine chemical shifts (δ) in ppm. 1H-NMR spectra were then recorded using Bruker advanced III 400 MHz Liquid-State NMR instrument at R.T.
Conjugation of TTZ with activated PTXNR through the lysine residue interaction
The CDI activated PTXNRs were subsequently reacted with the ε-amino group of lysine residues of TTZ (pKa 10.53) for subsequent conjugation54. 100 µg of TTZ powder was dissolved in 100 µl carbonate buffer at pH 9.3–9.5 and added to the CDI activated 100 µl of 1 mg PTXNR-CDI particle suspension. The reaction was allowed to proceed for 48 h at room temperature (~ 22 °C). The resulting TTZ conjugated PTXNRs (PTXNR-TTZ) were centrifuged at 16,000× g for 25 min and washed using carbonate buffer at pH 9.3–9.5 (3 × 1 ml). The supernatant was collected after each washing for quantifying the unbound TTZ. The concentrate was collected by centrifuging the membrane filter at 1000× g for 2 min. The concentrated PTXNR-TTZ particles were finally re-suspended in 300 µl of PBS (pH 7.5). The amount of PTXNR in PTXNR-TTZ was quantified by measuring absorbance at 230 nm (BioTek Synergy 2; BioTek, Winooski, VT, USA) using a PTX calibration curve (SI Fig. 2). The amount of unbound antibody was quantified using a BCA protein Assay (Pierce Biotechnology, Rockford, IL, USA) and a TTZ calibration curve (SI Fig. 3). The size and shape of PTXNR-TTZ particles were investigated using SEM (10.0 kV; accelerating voltage with 5.6 mm working distance and 20,000× magnification). The surface charge of PTXNR-TTZ was measured in DI water and PBS using a Nano series Zetasizer (Malvern).
Fluorescence data analysis to confirm the conjugation of TTZ with PTXNR
To confirm the successful conjugation of TTZ with PTXNR, TTZ was tagged with Alexa 594 red fluorescent dye molecule (Invitrogen) according to the manufacturer’s protocol before conjugating with PTXNR. The fluorescence data of both unconjugated bare PTXNR and conjugated Alexa 594 tagged PTXNR-TTZ particles were obtained using a flow cytometer (BD Accuri C6 plus). The fluorescence signal of particles was acquired using a 585/40 bandpass filter with 488 nm laser excitation.
Optimization of TTZ conjugation using response surface analysis
The optimum conditions for maximum TTZ conjugation efficiency were investigated by the response surface analysis method using JMP statistical modeling software. The design of the experiment involved two factors, initial PTXNR, and initial TTZ concentration. Three levels were assigned to each of the two factors. For initial PTXNR concentration, the levels were 5, 10, and 15 mg/ml, and for initial TTZ concentrations, the levels were 0.5, 1.0, and 1.5 mg/ml, respectively. Each experimental design unit was replicated three times resulting in a total of 27 experimental units. For each experimental unit, a random number was assigned using JMP. The experiments were performed according to a complete randomized design in three days with nine randomly assigned experiments in a single day. Conjugation efficiency of TTZ was chosen to be the yield or response of the experimental design. TTZ conjugation efficiency was calculated using Eq. (1):
| 1 |
In vitro therapeutic efficacy
HER2 positive BT-474 and SK-BR-3 breast cancer cells, and HER2 negative MDA-MB-231 breast cancer cells were cultured in hybricare medium (ATCC), McCoys medium (ATCC), and RPMI-1640 (Gibco), respectively supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. MCF-10A normal breast epithelial cells were cultured in mammary epithelial cell basal medium (ATCC) supplemented with mammary epithelial cell growth kit (ATCC). All cells were cultured in a 37 °C and 5% CO2 cell culture incubator. Approximately, 10,000 cells per well were plated in 96-well plates and treated with different doses of PTXNR-TTZ, PTXNRs, TTZ alone, PTX solution alone, and PTX solution in combination with TTZ. After 72 h of incubation, cell viability was assessed using MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide, MW 414) assay. MTT reagent was added to each well to convert into an insoluble formazan from water-soluble MTT. After 4 h, sodium dodecyl sulfate (SDS) solution prepared in 0.01 N HCl was added to each well to solubilize the formazan. The percentage of live cells relative to untreated control wells was quantified by measuring the absorbance at 570 nm (BioTek Synergy 2). Cell viability was calculated as a means of six wells containing BT-474 and MDA-MB-231 cells by subtracting the mean background level of wells containing medium only. Nonspecific formation of formazan due to the presence of medium was determined from triplicate wells. The number of viable cells was calculated as follows [Eq. (2)]:
| 2 |
In vitro nuclear condensation assay
To further understand cell cytotoxicity of PTXNR-TTZ on breast cancer cells, Hoechst 33342 dye (361/486 nm ex/em) was used to stain the DNA of BT-474, SK-BR-3, and MDA-MB-231 breast cancer cell lines. Approximately, 20,000 cells were seeded in 8 well plates (Corning) followed by 10,000 nM of PTXNR-TTZ treatments for 72 h and incubation with Hoechst (5 µM). Live cells were imaged using a fluorescence microscope (Zeiss Apotome 2.0) equipped with a (450 ± 40) nm wavelength filter cube. The fluorescence images were captured to identify changes in cell nuclei by PTXNR-TTZ treatments.
Quantitative analysis of synergistic effects of PTXNR-TTZ
We investigated the quantitative effects of PTXNR-TTZ using combination index analysis (CI) by the Chou-Talalay method55,56. The CI of PTXNR alone, TTZ alone, and conjugated PTXNR-TTZ were calculated using Eq. (3) and plotted as CI versus the fraction of cells being affected.
| 3 |
Apoptosis quantification
The percentage of apoptosis and necrosis in BT-474, SK-BR-3, and MDA-MB-231 cell lines were quantified using the annexin V-FITC and propidium iodide (PI) detection kit. The assay was carried out according to the manufacturer’s instructions. Briefly, cells were seeded into 24-well culture plates (50,000 cells/well), incubated overnight at 37 °C under 5% CO2, and treated with PTXNR-TTX (10,000 nM) or culture medium (negative control) for 0, 24, 48 and 72 h. Next, cells were washed using PBS, suspended in 1× binding buffer, and incubated in the dark for 30 min with annexin V-FITC (1:20 dilution) and PI (1:40 dilution). The results of fluorescence-activated cell sorting (FACS) analysis, carried out in a flow cytometer (BD Accuri C6 plus), were analyzed using the BD Accuri C6 plus software. At least three independent tests were performed. The percentage of cell death was calculated from the fluorescence intensity (F.I.) of annexin V-PI. At least 12,000 cells from the gate were acquired and analyzed using the histogram. At least three independent experiments were performed.
Cell cycle analysis
The effect of the PTXNR-TTZ on the cell cycle of BT-474 cells was investigated after 24, 48, and 72 h of treatments. BT-474 cells without any treatments, PTXNR alone treatment, and TTZ alone treatment were used as controls. The cell cycle analysis protocol was developed by modifying the previously reported method57,58. For each sample of the analysis, cells were seeded in a T25 cell culture flask. After 12 h, PTXNR-TTZ, PTXNR alone, and TTZ alone were added and incubated for 24, 48, and 72 h. The untreated sample was incubated with 1 ml of PBS solution. Before harvesting the cells, 100 µl of 5′-Bromo-2′-deoxyuridine (BrdU) (Alfa Aesar) stock solution per 5 ml of media was added to an active concentration of 40 µM and incubated for 1 h at 37 °C. The culture media was removed completely, and the cells were collected in 15 ml centrifuge tubes. The cells were centrifuged, and the cell pellets were fixed by adding 1 ml of 100% ice-cold ethanol. The cells were stored at 4 °C at least for 30 min. Ethanol was removed after centrifugation at 500× g for 5 min and was resuspended in 1 ml of 0.5% Triton X-100 prepared in 2 N HCl. After 30 min of incubation at room temperature, cells were pelleted down to mix with 1 ml of 0.1 M Na2B4O7.10 H2O (pH 8.5) solution and was incubated for at least 30 min at room temperature. The cells were pelleted down again and washed with PBS containing 1 mg/ml Bovine Serum Albumin (BSA) and 0.1% Tween-20 (PBST) solution. After washing, cells were resuspended in 50 µl prediluted mouse anti-BrdU monoclonal antibody (Novus Biologicals, Cat. NBP248373) stock solution. After 30 min of incubation in dark, 0.5 ml PBST stock solution was added and washed. A 50 µl prediluted FITC conjugated rabbit anti-mouse secondary antibody (SouthernBiotech, Cat. 617002) stock solution was added to cell pellets and were incubated for 30 min at room temperature in dark. After washing again with PBST solution, cells were resuspended in 100 µl of 1 mg/ml RNAse A (Alfa Aesar) stock solution and incubated for at least 20 min. Finally, 300–500 µl of 35 µg/ml propidium iodide (PI) (Alfa Aesar) was added to cell pellets and incubated in the dark at 37 °C for 45 min. Before analyzing the cells in a flow cytometer (BD accuri C6 Plus), cells were passed through a 40-micron cell strainer (Thermo Scientific) to remove cell aggregates. For flow cytometry analysis, cells were gated to sort out the singlet cells, and only the singlets were gated for subsequent analysis. For acquiring the fluorescent data, 530/30 and 585/40 bandpass filters were used to acquire the BrdU positive and PI-positive cells, respectively using 488 nm of laser excitation.
Western blot analysis
BT-474 and MDA-MB-231 cells were treated with PTXNR-TTZ, PTXNR alone, and TTZ alone for 48 h at 10,000 nM each for Western blot analysis. The cells were harvested and lysed at 4 °C using 50 mM Tris–HCl, pH 8.0, 150 mM NaCl, and 0.1% Triton X-100, 0.5% sodium deoxycholate and 0.1% sodium dodecyl sulfate (SDS). The lysate protein was quantified using the BCA assay and the BSA calibration curve (SI Fig. 4). For each experiment, 40 µg of each sample was taken and added to an equal volume of 2X Laemmli sample buffer. The cell lysate in the sample buffer was heated at 86 °C for 5 min before running the SDS-PAGE electrophoresis process. Novex Tris–Glycine SDS gels (8–16% and 16%) (Invitrogen) were used for gel electrophoresis. The electrophoresis was performed for 45 min to 1 h at 200 V. The separated proteins were transferred from gel to nitrocellulose membrane using a Power Blotter (ThermoFisher Scientific) according to the manufacturer’s protocol. After the transfer process, the membrane was blocked for 5 h with moderate shaking using 3% BSA protein in TBST buffer [20 mM Tris (pH 7.5), 150 mM NaCl, and 0.1% Tween 20]. The membrane was incubated with a primary monoclonal antibody overnight at 4 °C. GAPDH (ThermoFisher, Cat. MA5-15738, 1: 5000) monoclonal antibody was used as a loading control. Actin (ThermoFisher, Cat. MA5-11869, 1: 2000), caspase -9 (ThermoFisher, Cat PA5-16358, 1:500), caspase-3 (ThermoFisher, Cat. 43–7800, 1:500), cleaved caspase-3 (ThermoFisher Cat. PA5- 23921, 1:1000), XIAP (ThermoFisher, Cat. PA1-84846, 1:1000) and cytochrome-C (ThermoFisher, Cat. MA5-11674, 1:400) primary monoclonal antibodies were used to observe the corresponding proteins of interest. After primary antibody incubation, the membrane was washed 3–5 times for 5 min each with TBST buffer. The membrane was incubated in horseradish peroxidase (HRP) enzyme-conjugated secondary anti-mouse (ThermoFisher Cat.A27025, 1:10000 or anti-rabbit monoclonal antibody (Cell Signaling Technology, Cat.7074P2, 1:10000) depending on the primary antibody host species for 1 h followed by washing with TBST washing buffer. The electro-chemiluminescent substrate (Super Signal West Dura, ThermoFisher, and Cat. 34075) was added to the membrane and incubated for 5 min. The electro-chemiluminescent signal was captured using a Bio-Rad gel imager. The signals were analyzed by image processing using Fiji (Image J) software.
Statistical analysis
Data were analyzed using JMP statistical software (version 15) and presented as mean ± standard deviation. To observe the statistically significant differences in PTXNR-TTZ treated cells compared to controls, we performed the analysis of variance (ANOVA) using a one-tailed Student’s t-test with a p value ≤ 0.05 and a confidence interval ≤ 95%. The TTZ conjugation efficiency analysis was performed using a complete randomized (CR) model. The final conjugation efficiency data was predicted using a response surface analysis using JMP statistical software.
Results
Synthesis of PTXNRs
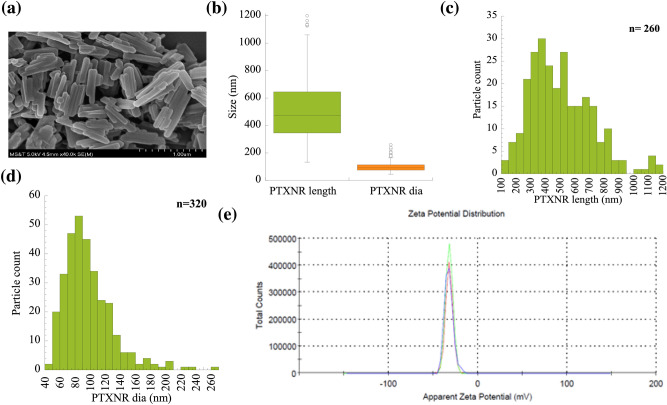
The SEM images of PTXNRs as shown in Fig. 1a confirms the elongated rod shape PTX drug NRs. The average diameter and length of PTXNRs were (96.87 ± 33.08) and (503.42 ± 210) nm, respectively (Fig. 1b). The size distribution plots (Fig. 1c, d) confirm the moderately narrow dispersity of the particle diameter and length. The polydispersity index (pDI) of the particles was measured as 0.20 ± 0.02. PTXNRs showed high colloidal stability in DI water having a zeta potential value of (− 32.60 ± 4.82) mV (Fig. 1e, SI Table 1). In 0.15 M PBS solution, the zeta potential value was measured (− 13.40 ± 2.81) mV, indicating the stability of nanoparticles even in high salt concentrations (SI Table 1).
Figure 1.
Characterization of PTXNRs: (a) Scanning electron microscopy (SEM) image of PTXNRs. (b) Particle size: Percentile plots of the PTXNR-TTZ particle length and diameter were analyzed using ImageJ/ Fiji software. (c) Size distribution of PTXNR particle length. (d) Size distribution of PTXNR particle diameter as calculated analyzing SEM images. The number ‘n’ denotes the particle population count for the analysis; and (e) Zeta potential (ξ) of PTXNR is -32.6 ± 4.8 mV in water. Three colors indicate three different experiments.
Confirmation of linker conjugation with PTXNRs using 1H NMR analysis
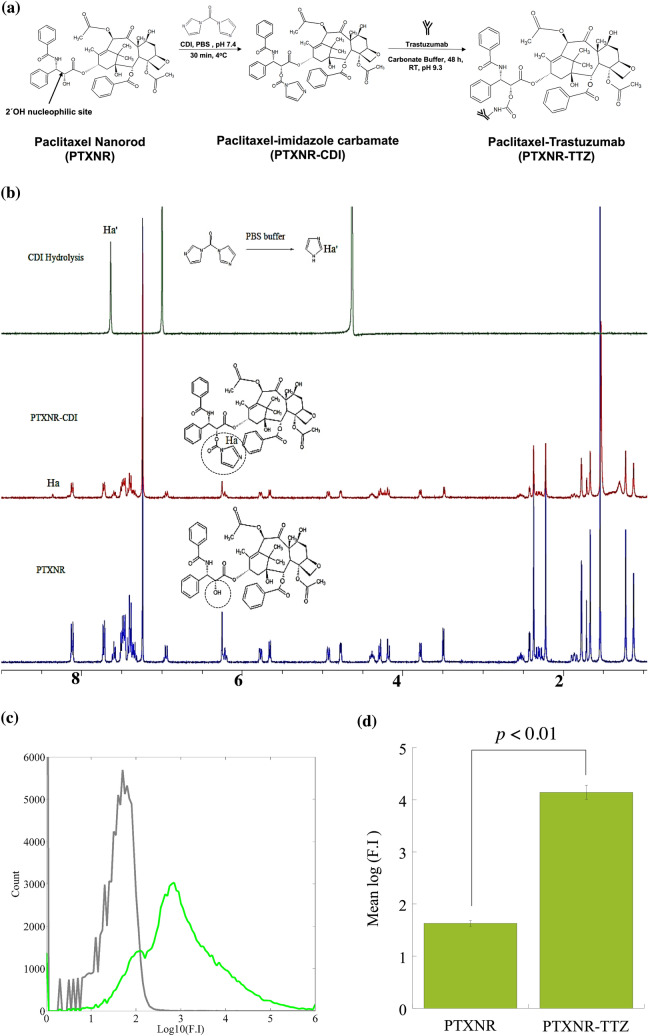
We activated the 2′ OH site of PTXNRs to form an imidazole carbamate intermediate for subsequent conjugation step with the ε- amino group in the lysine side chain of TTZ (Fig. 2a). In 1H NMR analysis, imidazole carbamate intermediate linkage was confirmed by the peak of the imidazole ring proton in ‘a’ position (Ha) at 8.35 ppm (Fig. 2b)59,60. In a parallel side reaction, CDI hydrolyzes in the aqueous phase of conjugation reaction with TTZ. Hydrolyzation of CDI produces a free imidazole ring and might interfere in the NMR analysis as a free surface-bound residuals. To further confirm the linkage of the imidazole carbamate with PTXNRs, we performed the NMR analysis of hydrolyzed CDI residuals. Residual imidazole group was identified by the peak of the imidazole proton in “a′” position (Ha′) at 7.6 ppm59. So, the shift of NMR peak at 8.35 ppm represents only the Ha proton, and hence, confirms the successful linkage of imidazole carbamate at 2′ OH site of PTXNR particles forming the PTX-imidazole carbamate intermediate (PTXNR-CDI) particles.
Figure 2.
1H-NMR analysis to characterize PTX-imidazole carbamate formation: (a) Schematic diagram of reactions involved for PTXNR functionalization and TTZ conjugation on the surface of PTXNRs. (b) 1H-NMR spectra of hydrolyzed carbonyldiimidazole (CDI), PTX-imidazole carbamate (PTXNR-CDI), and PTXNR. The NMR spectra were acquired at room temperature. For characterization of PTXNR and PTXNR-CDI deuterated chloroform (CDCl3, δ 7.24) and hydrolyzed CDI deuterated oxide (D2O, δ 4.65) were used as reference carrier solvents. (c) The fluorescence intensity of PTXNR-TTZ confirms the conjugation of TTZ on the surface of PTXNRs. The F.I. of bare PTXNRs (grey) and Alexa 594 fluorophore bound TTZ conjugated PTXNR (PTXNR-TTZ) (green) shows a significant increase in emission spectra. (d) The quantitative mean F.I. of PTXNR-TTZ increases by ~ 14,000 fold (2.54 logarithmic fold) compared to bare PTXNRs indicating TTZ is effectively conjugated on the surface of PTXNRs using a CDI linker.
Confirmation of TTZ conjugation with PTXNR-CDI
The ε-amino group in lysine residue of TTZ acts as an active electrophile at the reaction pH and attacks the CDI activated site of PTXNR through nucleophilic substitution reaction. Attack by the electrophile amine group releases the imidazole ring but leaves the carbonyl group resulting in a one-carbon spacer forming a stable carbamate linkage. To confirm the TTZ conjugation, Alexa 594 fluorescent molecule was conjugated with TTZ. The bare unconjugated PTXNR showed a mean autofluorescence intensity of 42.60 a.u. (arbitrary unit). For Alexa 594 fluorophore bound PTXNR-TTZ, the mean fluorescence intensity was measured 13,889.46 a.u. (Fig. 2c) that is 326 fold higher than the mean fluorescence intensity of bare PTXNR confirming a successful conjugation of TTZ with PTXNRs (Fig. 2d). PTXNR-TTZ showed good dispersibility in both water and PBS (SI Fig. 5).
Optimization of TTZ conjugation efficiency
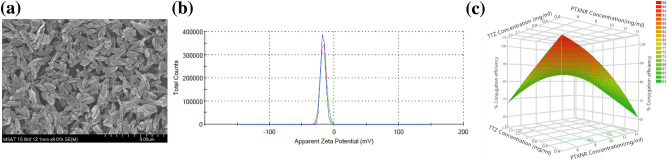
The SEM image of the PTXNR-TTZ particles confirms the consistency in the size and shape of particles after lyophilization (Fig. 3a). The zeta potential of the PTXNR-TTZ in DI water was measured − 17.1 ± 3.83 mV (Fig. 3b, SI Table 1). In PBS, the zeta potential was measured − 9.5 ± 0.02 mV (SI Table 1). The conjugated particles had a PDI of 0.21 ± 0.11, showing significant homogeneity in size and shape. Optimization of TTZ conjugation efficiency was performed using response surface analysis (Fig. 3c). The surface plot shows that the vicinity of lower concentrations of both initial PTXNR and TTZ result in 90–95% of conjugation efficiency, whereas the higher initial concentrations predict lower conjugation efficiency. The analysis suggests that the higher concentration of PTX results in precipitation of nanoparticles and less available surface for conjugation and several vacant reactive sites on the nanoparticle surface, whereas the higher concentration of TTZ produces excess unbound TTZ in the reaction medium. The maximum conjugation efficiency of TTZ to PTXNR was calculated as 95.24 ± 1.40% (SI Table 2), and hence, considered the optimum amount to use for subsequent experiments. The optimum condition for the maximum conjugation efficiency was 5 mg/ml and 0.5 mg/ml initial concentrations of PTXNR and TTZ, respectively. The drug to antibody ratio (DAR) of the conjugated particle was 4.0 ± 0.53 on a weight basis (molar ratio = 681.67 ± 90.23) (SI Table 2).
Figure 3.
Characterization of PTXNR-TTZ ADNs: (a) SEM image of PTXNR-TTZ ADNs. (b) The zeta potential (ξ) of PTXNR-TTZ was observed − 17.1 ± 3.83 mV in DI water. (c) Contour plot of response surface analysis showing % conjugation efficiency of TTZ on the surface of PTXNRs. In response surface analysis, two factors, PTXNR concentration and TTZ concentration were optimized for the highest conjugation efficiency. Each concentration factor had three levels. For PTXNR, the concentration levels were 5, 10, and 15 mg/ml, and for TTZ the levels were 0.5, 1.0, and 1.5 mg/ml, respectively. Each concentration was replicated at least three times resulting in a total of 27 experimental units. A maximum of ~ 95% conjugation efficiency was obtained using 5 mg/ml PTXNR and 0.5 mg/ml TTZ. The experiment was designed according to a complete randomized (CR) model using JMP statistical software.
In vitro anticancer efficacy
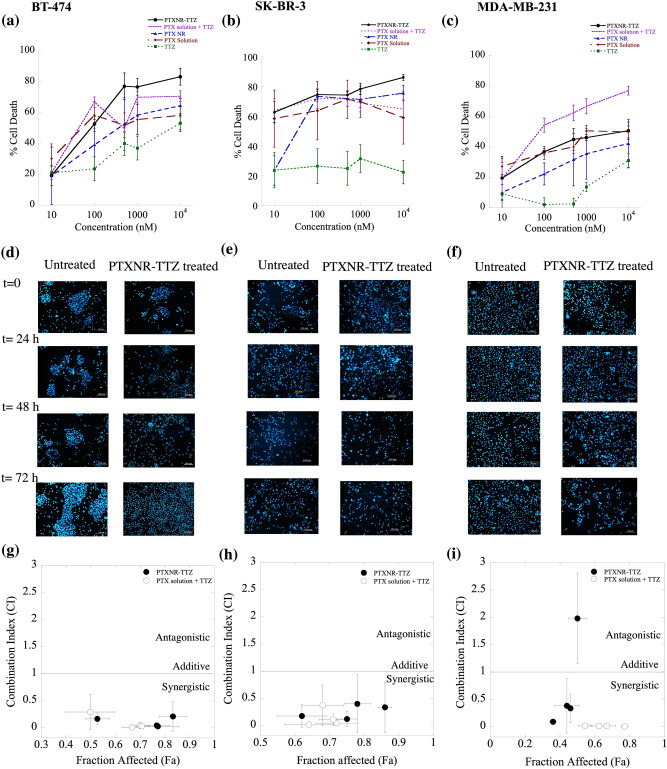
To determine the anti-cancer efficacy in vitro, the dose–response cytotoxicity was measured using PTXNR-TTZ, PTX NR alone, TTZ alone, PTX solution alone, and co-treatments of PTX and TTZ solutions in BT-474, SK-BR-3, and MDA-MB-231 cells (Fig. 4). The treatment of BT-474, MDA-MB-231, SK-BR-3, and MCF-10A with PTXNR-TTZ resulted in a concentration-dependent effect (Fig. 4). The cells were treated with PTXNR-TTZ and the following controls: cotreatment with PTX and TTZ solutions, PTX NR alone, PTX solution alone, and TTZ solution alone. The treatments using PTXNR-TTZ and controls showed dose-dependent response curves. The % cell death quantified using MTT assay is shown by the mean ± standard deviation. Differences between PTXNR-TTZ treatment and controls were analyzed using one way ANOVA followed by Student’s t-test method. *** projects significant differences with p < 0.05 and 95% confidence interval (SI Table 3). PTXNR-TTZ did not show significant differences in BT-474 and SK-BR-3 cells at concentrations ≤ 1000 nM compared with controls. A significant difference (p < 0.05) in the % of cell death is found at 10,000 nM of PTXNR-TTZ inhibiting > 83% of BT-474 cells (Fig. 4a). The cytotoxicity of PTX solution and TTZ was dose-dependent with the mean cell death of ~ 70% compared to that of PTXNR-TTZ. The mean cell death was increased from ~ 30% to 70% when the cells were treated with PTX NR or PTX solution alone. PTX solution and the co-treatment of PTX solution with TTZ showed slightly (~ 10%) higher mean cell death at concentrations ≤ 200 nM than that by the treatment using PTXNR-TTZ in BT-474 cells. PTXNR-TTZ may maintain a slow and sustained release of drugs owing to the dissolution from the nanorod conjugates. In our previous study, we showed similar in vitro drug release mechanisms of camptothecin from CPT-PCL-TTZ NRs following Fick’s diffusion model52. The low bioavailability of PTXNR-TTZ at concentrations ≤ 200 nM might cause lower cell death than PTX solution and the combination of PTX and TTZ solution. TTZ solution caused ~15-50% cell death .
Figure 4.
(a)–(c) In vitro anti-cancer efficiency of PTXNR-TTZ ADNs: The cytotoxic effects of PTXNR-TTZ (closed circle), PTXNR (triangle), PTX solution (diamond), and TTZ (square) on the growth of (a) BT-474, (b) SK-BR-3, and (c) MDA-MB-231 cells. Each experiment was replicated at least n = 6 times, and the average data is presented with mean ± standard deviation. The p values are given in SI Table 3. (d)–(f) The nuclear morphology of apoptotic cells stained with Hoechst 33342: Cells transfected with PTXNR-TTZ were stained with Hoechst 33342 72 h post-transfection and observed under a fluorescence microscope. The combined treatment with PTXNR-TTZ displayed the increased amount of nuclear fragmentation in (d) BT-474, (e) SK-BR-3, and (f) MDA-MB-231 cells. Untreated control cells remained uniformly stained with round and undamaged nuclei. The figures represent one of three independent experiments. (g)–(i) Combination index (C.I.) analysis of PTXNR-TTZ ADNs: The C.I. plot as a function of each fraction of cells affected by PTXNR-TTZ particles in (g) BT-474, (h) SK-BR-3, and (i) MDA-MB-231 cells. A C.I. value = 1; additive effect, < 1; synergistic effect, > 1; antagonistic effect.
TTZ alone exhibited very low toxicity in SK-BR-3 cells with cell death observed to be over 20% following exposure up to 10,000 nM concentration of the drug (Fig. 4b). The cytotoxic effect of PTNR-TTZ was more evident in SK-BR-3 than BT-474 cells, due to selective binding of the conjugate to cells that overexpress HER-2. These results are in good agreement with previous studies that showed that when maytansinoid was conjugated with TTZ (T-DM1), binding with HER-2 receptors was more specific both in vitro and in vivo than the free monoclonal antibody61. Other studies have confirmed the selective antiproliferative activity of PTX conjugated with TTZ in micelle drug carriers toward HER-2-positive SK-BR-3 cells compared to MDA-MB-231 cells62.
The addition of PTXNR-TTZ, PTX NR alone, PTX solution alone, and TTZ solution caused 20-50% cell death in HER2- MDA-MB-231 cells in the dose range of 1–10,000 nM (Fig. 4c). The mean cell death using PTXNR-TTZ did not differ significantly compared to single-agent treatments. Increasing cytotoxicity was observed by the treatment of PTX and TTZ solutions in a dose-dependent fashion. These data indicate non-specific therapeutic efficiency in HER2 negative MDA-MB-231 cells. The percentages of cell death in MCF-10A control cells varied between 5–40% at concentrations between 0.1–10,000 nM (SI Fig. 6) showing moderate toxicity in non-cancerous cells. A similar level of toxicities has been reported using paclitaxel63, taxane64, and nanoparticles65 in MCF-10A cells in vitro.
The half-maximal inhibitory concentrations (IC50), as defined as the concentration needed to kill 50% of cells, were found 34, 106, 609, and 9000 nM for co-treatment using PTX and TTZ, PTXNR-TTZ, PTX NR alone, and TTZ alone, respectively in BT-474 cells indicating a synergistic therapeutic efficiency using ADNs compared to the individual treatments (Fig. 4a). The IC50 values of PTX and TTZ solutions, PTXNR-TTZ, and PTX NR alone were 54, 31, and 51 nM, respectively in SK-BR-3 cells (Fig. 4b). The IC50s of PTX and TTZ co-treatments was slower than PTXNR-TTZ, which might be the consequence of slow drug release of PTX from PTXNR-TTZ due to its hydrophobicity. The IC50 of PTXNR-TTZ is higher in BT-474 than SK-BR-3 most likely due to almost tenfold HER2 higher concentrations in the cell membrane of BT-474 than that in SK-BR-3 cells66. MDA-MB-231 cells were resistant to PTXNR-TTZ exhibiting an IC50 > 3000 nM indicating intrinsic TTZ resistance in vitro (Fig. 4c). PTXNR alone and TTZ solution alone inhibited roughly 40% and 30% of the MDA-MB-231 cell growth, respectively. The cytotoxic effects of PTX solution were similar in both cell lines showing a dose-dependent cytotoxicity from 30 to 50%. PTXNR effectively killed more BT474 cells from 20 to 60% in dose-dependent manner than that in MDA-MB-231 cells from 10 to 30%. All these results suggest that PTXNR-TTZ exerts specific cytotoxicity in HER2 positive breast cancer cells.
Microscopic analyses on cell cytotoxicity
Chromatin condensation analysis by Hoechst staining was performed to study the cytotoxic effect of PTXNR-TTZ on BT-474 (Fig. 4d), SK-BR-3 (Fig. 4e), and MDA-MB-231 cells (Fig. 4f). During apoptosis, the chromatin becomes inert, highly condensed, undergoes fragmentation, and gets packaged into apoptotic bodies. The morphological changes induced by apoptosis can be detected by the blue-fluorescent Hoechst 33342 dye, which brightly stains the highly condensed, dense chromatin of apoptotic cells in comparison to the chromatin of untreated cells. After treatment with PTXNR-TTZ (10,000 nM) for 72 h, the number of condensed nuclei increased with time compared to untreated control cells exhibiting uncompromised cell nuclei after 72 h. The results indicate that PTXNR-TTZ is effective in inducing apoptosis in breast cancer cells.
The effects of PTXNR-TTZ is synergistic in HER2 positive breast cancer cells
The combination effects of PTXNR-TTZ and the cotreatments using PTX and TTZ solutions were evaluated in BT-474 (Fig. 4g), SK-BR-3 (Fig. 4h), and MDA-MB-231 cells (Fig. 4i) using the combination index (CI) analysis. The effects of the drugs were analyzed to determine whether a synergistic effect occurred by the treatment of PTXNR-TTZ in HER2 positive breast cancer cell lines. On the plot, any point showing CI < 1 indicates a synergistic effect, and the values showing CI > 1 are considered antagonistic. PTXNR-TTZ treatment in BT-474 cells resulted in CI values ranging from 0.16 ± 0.1 at 100 nM to 0.02 ± 0.02, 0.04, ± 0.04, and 0.2 ± 0.3 at 500, 1000, and 10,000 nM, respectively, indicating synergistic effects of drugs in the concentration range being tested (Fig. 4g). SK-BR-3 cells had relatively higher CI values than BT-474 with CI ranging from 0.12 ± 0.14, 0.60, ± 0.68, and 0.4 ± 0.6, and 0.34 ± 0.46 at 100, 500, 1000, and 10,000 nM, respectively (Fig. 4 h), indicating reduced synergy against breast cancer cells with lower HER2 overexpression on the cell membrane than BT-474 cells66. In contrast, very different CI vs. fraction affected line was observed in MDA-MB-231 cells showing increased CI values with increasing fraction affected cells up to 0.8 beyond which antagonistic effects were observed indicating a combination of PTX NR and TTZ is irrelevant in this cell line (Fig. 4 i). PTXNR-TTZ resulted in an antagonistic interaction (CI = 2 ± 0.8) at 10,000 nM in MDA-MB-231 cells, representing antagonism at high concentrations. Co-treatments of PTX with TTZ solutions showed synergistic antitumor effects in all three cell lines suggesting that the drug combination was effective for anticancer therapies.
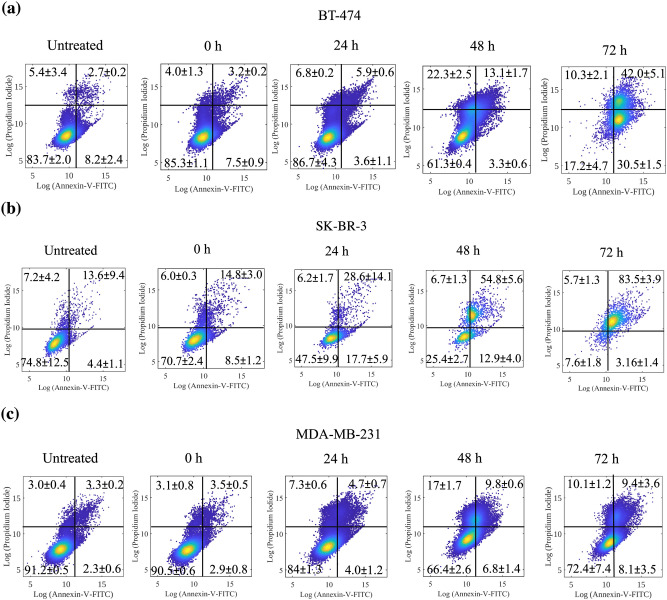
Measurement of apoptotic populations using Annexin V-FITC and PI
Annexin V and PI double-stained cells collected from the flow cytometry assay were observed for distinguishing live, early apoptosis, late apoptosis, and dead cell subtypes at the individual incubation time points of 24, 48, and 72 h. The Annexin V assay revealed that PTXNR-TTZ induced early and late stages of apoptosis in BT-474 cells (Fig. 5a). The cell distributions in live, early apoptosis, late apoptosis and dead cells are 83.7 ± 2.0%, 8.2 ± 2.4%, 2.7 ± 0.2%, and 5.4 ± 3.4%, respectively in untreated BT-474 cells. Induction of apoptosis by PTXNR-TTZ in BT-474 cells in a time-dependent manner was observed. The early apoptosis (Annexin V-FITC) plots show the BT-474 cell distribution of 7.5 ± 0.9%, 3.6 ± 1.1%, 3.3 ± 0.6%, and 30.5 ± 1.5% at t = 0, 24, 48, and 72 h, respectively following PTXNR-TTZ treatments. An increase in late apoptosis and dead cells was seen in all treated cells in comparison to the untreated cells. After 24 h of treatment, the percentage of late apoptotic cells increased from 3.2 ± 0.2% to 5.9 ± 0.6%. As the treatment duration reached 48 h, the percentage of apoptotic cells was increased further to 13.7 ± 1.7% and increased even more, to 42.0 ± 5.1%, after 72 h. The statistical differences are provided in SI Table 4. In untreated SK-BR-3 cells, the cell distribution in live, early apoptosis, late apoptosis, and dead cells are 74.8 ± 12.5%, 4.4 ± 1.1%, 13.6 ± 9.4%, and 7.2 ± 4.2%, respectively indicating a very low number of early, late apoptotic, and dead cells (Fig. 5b). However, the apoptotic cell distribution was increased after treatment with PTXNR-TTZ. The percentage of late apoptotic cells increased gradually with increasing treatment duration, being 28.6 ± 14.1%, 54.8 ± 5.6%, and 83.5 ± 3.9%, following 24, 48, and 72 h of incubation, respectively. The increase in cell distribution indicated a time-dependent increase in late apoptotic cells, parallel to that noted in BT-474 cells. Untreated MDA-MB-231 cells showed a distribution of 91.2 ± 0.5%, 2.3 ± 0.6%, 3.3 ± 0.2%, and 3.0 ± 0.4% in live, early apoptosis, late apoptosis, and dead cells, respectively (Fig. 5c). The percentage of late apoptosis increased to 3.5 ± 0.5%, 4.7 ± 0.7%, 9.8 ± 0.6%, and 9.4 ± 3.6% for 0, 24, 48 and 72 h of PTXNR-TTZ treatment, respectively. The % of apoptotic MDA-MB-231 cells displayed only a slight increase in cell distribution as a result of treatment with PTXNR-TTZ. Thus, the overall cell population shift indicated that PTXNR-TTZ caused a significant increase in apoptosis in HER2 positive breast cancer cells.
Figure 5.
Evaluation of apoptosis induction. Annexin V-FITC analysis of (a) BT-474, (b) SK-BR-3, and (c) MDA-MB-231 cells untreated and treated with PTXNR-TTZ for 0, 24, 48, and 72 h. Results represent one of three independent experiments. Viable cells are represented in the lower left quadrant. Early apoptosis (Annexin + / PI −) is shown in the lower right quadrant for each panel, while late apoptosis (Annexin + / PI +) is shown in the upper right quadrant. Necrosis (Annexin − / PI +) is shown in the upper left quadrant. Values are presented as means ± SD of triplicate experiments. Detailed statistical analysis (p < 0.05) of the data are shown in SI Table 4.
PTXNR-TTZ arrests HER2 positive breast cancer cells in the G2/M phase
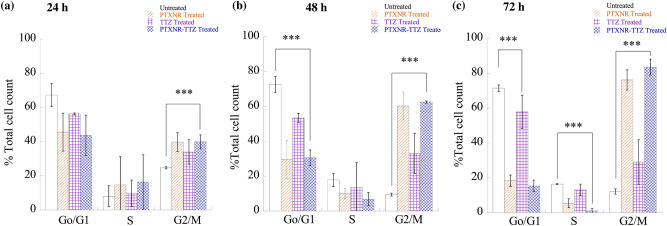
We quantified the population of BT-474 cells arrested in G0/G1, S, and G2/M cell cycle phases after treatment with PTXNR-TTZ, PTXNR alone, and TTZ alone (Fig. 6, SI Figs. 7–9 and SI Table 5). PTXNR-TTZ arrested 39.90, 62.56, and 83.55% of BT-474 cells in the G2/M phase after 24, 48, and 72 h treatments, respectively. Conversely, it decreased the proportion of the G0/G1 phase from 43.70 to 30.6 to 15.39% at 24, 48, and 72 h, respectively. PTXNR alone caused 39.76, 60.31, 76.18% arrest in the G2/M phase after 24, 48, and 72 h, respectively. TTZ alone treatment arrested 56.25, 53.39, 57.83% of BT-474 cells in the G1 phase after 24, 48, and 72 h, respectively. The cell cycle data confirms that PTXNR modulates the mitotic cell cycle arrest, or cell growth inhibition, and eventually apoptosis in BT-474 breast cancer cells.
Figure 6.
Cell cycle analysis: BT-474 cells were treated with PTXNR-TTZ, PTXNR alone, and TTZ alone. Cell cycle analysis was performed by flow cytometry after (a) 24 h, (b) 48 h, and (c) 72 h of treatment. Each experiment was replicated at least n = 2 times, and the average cell count data are presented as mean ± standard deviation. Statistically significant differences are shown using *** (p < 0.05; detailed p value calculations are shown in SI Table 5).
PTXNR-TTZ downregulates anti-apoptotic protein and induces apoptosis in a caspase-dependent intrinsic pathway
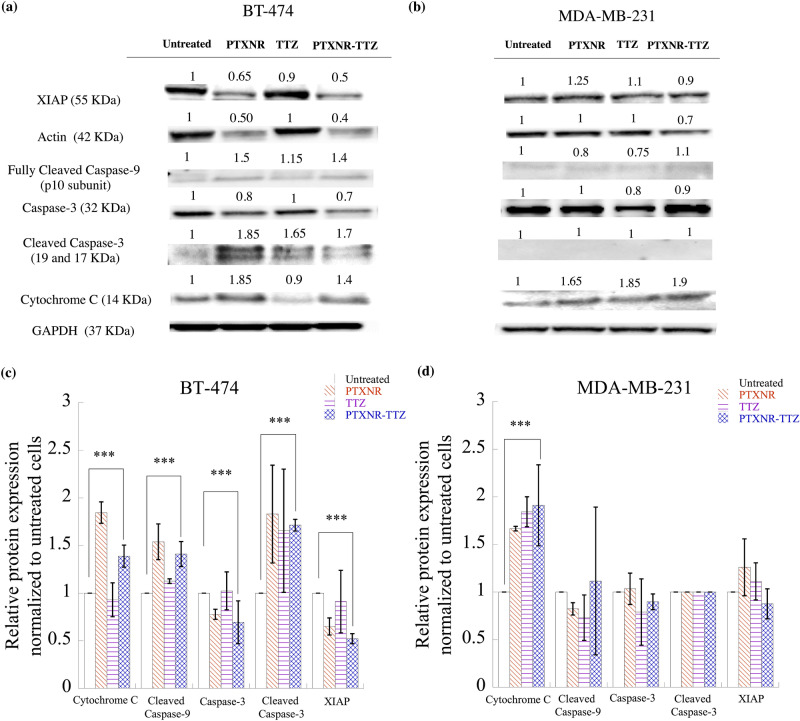
We found that > 80% of BT-474 cells were inhibited and arrested in the G2/M phase by PTXNR-TTZ ADN treatments. We investigated if the prolonged cell cycle arrest in the G2/M phase induced programmed cell death or apoptosis in cancer cells and the mechanisms of apoptotic induction by PTXNR-TTZ ADNs. PTXNR-TTZ induced apoptosis in HER2 positive BT-474 cells in an intrinsic cytochrome-C mediated pathway (Fig. 7a, c and SI Fig. 10a). In BT-474 cells, after 48 h of treatment using PTXNR-TTZ and PTX NR, we observed a significant release of cytochrome-C than that of untreated cells. We further observed a significantly higher expression of initiator apoptotic protein fully cleaved caspase-9. The lower expression of procaspase-3 was correlated with the upregulation of cleaved or active form of caspase-3. The observation is in agreement with the time-dependent expression study of caspase-3 on taxane treated HER2 positive SK-BR-3 cell line67. In BT-474 cells, the anti-apoptotic protein XIAP was downregulated by PTXNR-TTZ and PTXNR treatments in comparison to untreated and TTZ treated control cells. XIAP is a natural inhibitor of caspase-3 and caspase-947,68–70. High XIAP activity may cause the inactivation of early apoptotic protein caspase-9 and inactivation of active caspase-3 at the later phase of apoptosis. The significant downregulation of XIAP by PTXNR-TTZ treatment facilitates the late apoptosis by overexpressing the effector caspase-3 apoptotic protein that attributes to the synergistic cytotoxic effect in BT-474 cells. The high concentration of PTXNR and PTXNR-TTZ significantly downregulated the actin production in BT-474 cells compared to MDA-MB-231 cells. In MDA-MB-231 cells, although the cytochrome-C release was observed for both PTXNR-TTZ and PTXNR treatments, there was no significant change in the expression of active caspase-9 suggesting that the apoptosome was not activated in the cytosolic area (Fig. 7b, d and SI Fig. 10b). The procaspase-3 expression in PTXNR-TTZ treated cells was not statistically different from that of untreated cells. Eventually, the cleaved form of the caspase-3 signal was not observed at all with the individual drug treatment and PTXNR-TTZ treatment. XIAP activity was observed in individually treated cells as well as in PTXNR-TTZ treated cells. XIAP activity in the drug-treated cells was found to be as high as that in untreated MDA-MB-231 cancer cells. XIAP is a potent anti-apoptotic protein and abnormal expression of XIAP can block the cell death pathways47.
Figure 7.
Western blot analysis: The cell signaling protein expressions of (a) BT-474 and (b) MDA-MB-231 cells using PBS, PTX alone, TTZ alone, and PTXNR-TTX treatments. The relative intensity of XIAP, actin, cleaved caspase-9, caspase 3, cleaved caspase 3, and cytochrome C is analyzed after 48 h of treatment in (c) BT-474 and (d) MDA-MB-231 cells using ImageJ/ Fiji. For each protein expression the experiment was replicated at least n = 2–4 times and the average data is presented as mean ± standard deviation. The relative protein expressions are normalized to the untreated control cells and to the housekeeping GAPDH protein expression (hypothesized expression mean, µ = 1). The p values for overexpression or downregulation of cytochrome-C, cleaved caspase-3, and XIAP in BT-474 are 0.039, 0.003, and 0.000049, respectively. The p value for the overexpression of cytochrome-C in MDA-MB-231 is 0.047. ***Represents p < 0.05 (for detailed p value calculations, please see SI Table 6).
Discussion
Many chemotherapeutic drugs including PTX is a water-insoluble drug that results in low circulation time, poor bioavailability in the tumor tissue, and higher administrative dosage71–73. Nano-sized drug carriers, liposomes, polymeric nanoparticles, and micelles have been designed over the past decades to improve water dispersibility, stability, and blood circulation half-life of chemotherapeutic drugs74–77. However, most nanoparticles suffer from poor specificity targeting only 1.6% of the administered dose of the linked small molecule drug to be delivered in the targeted tumor vicinity78,79. In this study, we designed ADNs by conjugating TTZ monoclonal antibodies on the surface of PTXNRs through a single carbonyl group spacing with > 95% conjugation efficiency. PTXNR-TTZ was synthesized in two steps. First, PTX NRs were synthesized by phase separation of PTX drug solution from ethanol into water. This method is simple and scalable. Secondly, PTXNRs were reacted with CDI linker in water to form intermediate PTXNR-carbamate followed by coupling with the amine groups of TTZ using carbodiimide chemistry that forms PTXNR-TTZ ADN80,81. The reaction is chemically defined, efficient, and scalable with an overall yield of > 80% and a high PTX: TTZ w/w ratio ≈ 4 (molar ratio ≈ 682). The cytotoxic payload in ADNs is 170-fold more than conventional antibody–drug conjugates (molar ratio ≈ 4) which improves the ability to deliver more active drugs at the target site and greater in vitro potency. The PTXNR-TTZ ADN showed greatly enhanced water dispersibility (> 1 mg/ml) compared to PTX aqueous solution, indicating little aggregation in water52.
PTXNR-TTZ ADNs are designed to target HER2 receptors overexpressed in breast cancer cells so that the ADN binds to target cells, are internalized via endocytosis, and exerts apoptosis mediated therapeutic effects10,48,50,51,82–95. The NRs, when coated with TTZ, exhibit high binding toward HER2-overexpressing breast cancer cells53,96. The adhesion strength of NRs has been shown high both experimentally and by theoretical modeling92,97–100. After targeting and internalization, PTXNR-TTZ ADNs activated intrinsic apoptosis pathways and inhibited > 80% of HER2 positive BT-474 and SK-BR-3 cells which was 43% more than the cytotoxicity in HER2 negative MDA-MB-231 cells. No difference in cytotoxicity was observed for PTXNR-TTZ, unconjugated PTXNR, PTX solution alone, and TTZ alone treatments in MDA-MB-231 cells, indicating non-specific cell death. The inhibitory effect of PTX and TTZ co-treatments is concentration-dependent inducing non-specific cell death in both HER2 positive and HER2 negative breast cancer cells after 72 h of treatment. The IC50 values of PTXNR-TTZ in BT-474 cells and SK-BR-3 cells are lower than single drug treatments which are consistent with the previously published literature. The IC50s of PTX vary between 2–200 nM in breast cancer cells due to differential basal level expressions of β-tubulin subunits of microtubules101–103, sensitivity of PTX toward binding with β-tubulin101, the expression of tau proteins involved in microtubule polymerization104, and HER2 expression levels105. PTXNR-TTZ inhibits tubulin polymerization and microtubule formation, blocks the cell cycle in G2/M and induces apoptosis.
The CI analysis showed that PXNR-TTZ augments apoptosis in BT-474 cells synergistically indicating an induction in apoptosis through multiple pathways including intrinsic caspase-dependent pathway and cell cycle arrest. PTX and TTZ arrest cells in G2/M and G0/G1 phase, respectively by disrupting the microtubule dynamics and binding with the HER2 extracellular domain106–110. Actin polymerization is a universal mechanism to drive cell cycle in dividing cells including the accelerated outgrowth of breast cancer cells1–3. PTX, as microtubule-binding drugs, stabilizes microtubules4,5, blocks their dynamics6,7, and induces apoptosis in cancer cells8,9. PTX has been widely used to treat a variety of cancers, including breast cancer, head and neck cancer, gastroesophageal cancer, Kaposi's sarcoma, leukemia, lymphoma, non-small cell lung cancer, and ovarian cancer10. Nevertheless, the natural or acquired resistance to PTX limits its therapeutic use. This fact, together with the key role of microtubule dynamics in cancer growth, led to the concept that PTXNR-TTZ directly inhibits actin polymerization causing apoptosis in cancer cells. Using the densitometry of Western blot results, we show that HER2 + breast cancer cells treated with PTXNR-TTZ contained a reduction in actin polymerization by ∼60% compared with untreated cells. In this study, individual treatment using PTXNR alone and TTZ alone arrested 76% and 57% of BT-474 cells in G2/M and G0/G1, respectively after 72 h of treatment. In contrast, PTXNR-TTZ arrested 83.5% of BT-474 cells in G2/M after 72 h of treatment indicating dominant effects of PTX in sensitizing and synchronizing cells in the G2/M phase which correlates with the percentage of apoptotic dead cells after PTXNR-TTZ treatment111–113. The cleaved caspase-3 and cytochrome C were significantly increased, while anti-apoptotic XIAP was decreased in BT-474 cells after 72 h of PTXNR and PTXNR-TTZ treatments compared with untreated control. Cytochrome-C binds to apoptosis protease-activating factor-1 (APAF1), inducing its oligomerization to form apoptosome that recruits and activates an apoptosis initiator protein, caspase-9, cleaves caspase-9 into its active small subunits and initiates the late apoptotic caspase cascade reactions47,70,114,115. The cleaved caspase-9 eventually activates the late apoptotic protein caspase-3116–118. The overexpression of cleaved or active form of caspase-3 ultimately confirms the late phase of apoptosis in BT-474 cells using PTXNR-TTZ treatment116,119–121. In the late phase, apoptosis is facilitated by two-fold downregulation of anti-apoptotic XIAP protein after PTXNR-TTZ treatment compared to that in the untreated control. In contrast, MDA-MB-231 cells did not show any sign of mid- or late phase apoptosis although having overexpression of the cytochrome-C protein. No changes in XIAP expression was observed in MDA-MB-231 cells treated with PTXNR-TTZ. XIAP, a key member of the inhibitor of apoptosis protein (IAP) family122, has been shown to be a direct inhibitor of caspase-3 and to interfere with the cytochrome-C pathway by inhibiting caspase-9 in lung cancer cells123–125, and leukemia cells126–129. In this study, the downregulation of XIAP and activation of caspase cascades in BT-474 cells by PTXNR-TTZ has proven to be an effective approach for the treatment of HER2 positive breast cancer cells.
Conclusion
The first synthesis of PTXNR-TTZ ADNs and its therapeutic efficiency in vitro are presented which induces higher apoptosis responses than single drug treatments in HER2 positive breast cancer cells. The use of PTXNR of ~ 95 nm in diameter and 500 nm in length remove the compromising need for toxic organic carrier solvent for intravenous administration of PTX alone. The particles are stable in the aqueous phase that makes it suitable for intravenous administration without any need for any organic solvent. The surface of PTXNR was functionalized by CDI activation reaction in the aqueous phase and successfully conjugated TTZ without altering the size and shape of the nanoparticles. PTXNR-TTZ synergistically inhibited HER2 positive breast cancer cells. The IC50 values of PTXNR-TTZ are 31 and 106 nM in SK-BR-3 and BT-474 cancer cells, respectively which facilitates lower dose, and hence, fewer side effects than individual treatments. PTXNR-TTZ arrested > 83% of the proliferating cells in the G2/M phase of the cell cycle after 72 h of the treatment, which was well corroborated with cell cycle analysis. The higher number of cells (> 80%) arrested in the G2/M phase after PTXNR-TTZ treatment than individual treatments using PTXNR and TTZ (arrests in the G0/G1 phase) confirms the synergistic crosstalk between PTX and TTZ. The Western blot analysis demonstrates that PTXNR-TTZ activates the effector caspase proteins in HER2 positive breast cancer cells, and downregulates the anti-apoptotic XIAP facilitating caspase-dependent apoptosis followed by the cell cycle arrest in the G2/M phase. Taken together, it is envisaged that the results from this study will facilitate the rational design of combination therapy through a novel design of ADNs.
Supplementary Information
Acknowledgements
This work was supported by the Center for Biomedical Research (CBR) at Missouri S&T and by using Missouri soybean farmers’ checkoff dollars provided by the Missouri Soybean Merchandising Council (MSMC; Project No. 20-447-21).
Author contributions
M.R.A. and S.B. conceived of the presented idea. M.R.A. designed and performed experiments, and analyzed the data. K.P., R.A., and D.B. contributed to the cytotoxicity and Western blot analyses, and to the statistical analysis of the results. S.B. verified the analytical methods and supervised the findings of this work. All authors discussed the results and contributed to the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86762-6.
References
- 1.Vega-Vásquez P, Mosier NS, Irudayaraj J. Nanoscale drug delivery systems: From medicine to agriculture. Front. Bioeng. Biotechnol. 2020;8:79. doi: 10.3389/fbioe.2020.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patra JK, Das G, Fraceto LF, Campos EVR, Rodriguez-Torres MDP, Acosta-Torres LS, Diaz-Torres LA, Grillo R, Swamy MK, Sharma S, Habtemariam S, Shin H-S. Nano based drug delivery systems: recent developments and future prospects. J. Nanobiotechnol. 2018;16:71–71. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scicluna MC, Vella-Zarb L. Evolution of nanocarrier drug-delivery systems and recent advancements in covalent organic framework-drug systems. ACS Appl. Nano Mater. 2020;3:3097–3115. doi: 10.1021/acsanm.9b02603. [DOI] [Google Scholar]
- 4.Hossen S, Hossain MK, Basher MK, Mia MNH, Rahman MT, Uddin MJ. Smart nanocarrier-based drug delivery systems for cancer therapy and toxicity studies: A review. J. Adv. Res. 2019;15:1–18. doi: 10.1016/j.jare.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liechty WB, Kryscio DR, Slaughter BV, Peppas NA. Polymers for drug delivery systems. Annu. Rev. Chem. Biomol. Eng. 2010;1:149–173. doi: 10.1146/annurev-chembioeng-073009-100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldorera-Moore M, Guimard N, Shi L, Roy K. Designer nanoparticles: Incorporating size, shape and triggered release into nanoscale drug carriers. Expert Opin. Drug Deliv. 2010;7:479–495. doi: 10.1517/17425240903579971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malam Y, Loizidou M, Seifalian AM. Liposomes and nanoparticles: Nanosized vehicles for drug delivery in cancer. Trends Pharmacol. Sci. 2009;30:592–599. doi: 10.1016/j.tips.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Lombardo D, Kiselev MA, Caccamo MT. Smart nanoparticles for drug delivery application: Development of versatile nanocarrier platforms in biotechnology and nanomedicine. J. Nanomater. 2019;2019:3702518. doi: 10.1155/2019/3702518. [DOI] [Google Scholar]
- 9.Qin S-Y, Zhang A-Q, Cheng S-X, Rong L, Zhang X-Z. Drug self-delivery systems for cancer therapy. Biomaterials. 2017;112:234–247. doi: 10.1016/j.biomaterials.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 10.Barua S, Yoo J-W, Kolhar P, Wakankar A, Gokarn YR, Mitragotri S. Particle shape enhances specificity of antibody-displaying nanoparticles. Proc. Natl. Acad. Sci. 2013;110:3270–3275. doi: 10.1073/pnas.1216893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson N, Ghandehari H. Polymeric conjugates for drug delivery. Chem. Mater. 2012;24:840–853. doi: 10.1021/cm2031569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasarao M, Galliford CV, Low PS. Principles in the design of ligand-targeted cancer therapeutics and imaging agents. Nat. Rev. Drug Discov. 2015;14:203–219. doi: 10.1038/nrd4519. [DOI] [PubMed] [Google Scholar]
- 14.Chari RVJ. Targeted delivery of chemotherapeutics: Tumor-activated prodrug therapy. Adv. Drug Deliv. Rev. 1998;31:89–104. doi: 10.1016/S0169-409X(97)00095-1. [DOI] [PubMed] [Google Scholar]
- 15.Lin K, Tibbitts J. Pharmacokinetic considerations for antibody drug conjugates. Pharm. Res. 2012;29:2354–2366. doi: 10.1007/s11095-012-0800-y. [DOI] [PubMed] [Google Scholar]
- 16.Mack F, Ritchie M, Sapra P. The next generation of antibody drug conjugates. Semin. Oncol. 2014;41:637–652. doi: 10.1053/j.seminoncol.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Brasó-Maristany F, Griguolo G, Pascual T, Paré L, Nuciforo P, Llombart-Cussac A, Bermejo B, Oliveira M, Morales S, Martínez N, Vidal M, Adamo B, Martínez O, Pernas S, López R, Muñoz M, Chic N, Galván P, Garau I, Manso L, Alarcón J, Martínez E, Gregorio S, Gomis RR, Villagrasa P, Cortés J, Ciruelos E, Prat A. Phenotypic changes of HER2-positive breast cancer during and after dual HER2 blockade. Nat. Commun. 2020;11:385. doi: 10.1038/s41467-019-14111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marty M, Cognetti F, Maraninchi D, Snyder R, Mauriac L, Tubiana-Hulin M, Chan S, Grimes D, Antón A, Lluch A. Randomized phase II trial of the efficacy and safety of trastuzumab combined with docetaxel in patients with human epidermal growth factor receptor 2–positive metastatic breast cancer administered as first-line treatment: The M77001 study group. J. Clin. Oncol. 2005;23:4265–4274. doi: 10.1200/JCO.2005.04.173. [DOI] [PubMed] [Google Scholar]
- 19.Von Minckwitz G, Procter M, De Azambuja E, Zardavas D, Benyunes M, Viale G, Suter T, Arahmani A, Rouchet N, Clark E. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer. N. Engl. J. Med. 2017;377:122–131. doi: 10.1056/NEJMoa1703643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldhirsch A, Gelber RD, Piccart-Gebhart MJ, De Azambuja E, Procter M, Suter TM, Jackisch C, Cameron D, Weber HA, Heinzmann D. 2 years versus 1 year of adjuvant trastuzumab for HER2-positive breast cancer (HERA): An open-label, randomised controlled trial. The Lancet. 2013;382:1021–1028. doi: 10.1016/S0140-6736(13)61094-6. [DOI] [PubMed] [Google Scholar]
- 21.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 22.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 23.Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, Mackey J, Glaspy J, Chan A, Pawlicki M. Adjuvant trastuzumab in HER2-positive breast cancer. N. Engl. J. Med. 2011;365:1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez E, Romond E, Suman V. Trastuzumab plus adjuvant chemotherapy for HER2-positive breast cancer: Final planned joint analysis of overall survival (OS) from NSABP B-31 and NCCTG N9831. J. Clin. Oncol. 2014;32:3744–3752. doi: 10.1200/JCO.2014.55.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giordano SH, Temin S, Chandarlapaty S, Crews JR, Esteva FJ, Kirshner JJ, Krop IE, Levinson J, Lin NU, Modi S, Patt DA, Perlmutter J, Ramakrishna N, Winer EP, Davidson NE. Systemic therapy for patients with advanced human epidermal growth factor receptor 2-positive breast cancer: ASCO clinical practice guideline update. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018;36:2736–2740. doi: 10.1200/JCO.2018.79.2697. [DOI] [PubMed] [Google Scholar]
- 26.Tolaney SM, Barry WT, Dang CT, Yardley DA, Moy B, Marcom PK, Albain KS, Rugo HS, Ellis M, Shapira I, Wolff AC, Carey LA, Overmoyer BA, Partridge AH, Guo H, Hudis CA, Krop IE, Burstein HJ, Winer EP. Adjuvant paclitaxel and trastuzumab for node-negative, HER2-positive breast cancer. N. Engl. J. Med. 2015;372:134–141. doi: 10.1056/NEJMoa1406281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolaney SM, Guo H, Pernas S, Barry WT, Dillon DA, Ritterhouse L, Schneider BP, Shen F, Fuhrman K, Baltay M, Dang CT, Yardley DA, Moy B, Marcom PK, Albain KS, Rugo HS, Ellis MJ, Shapira I, Wolff AC, Carey LA, Overmoyer B, Partridge AH, Hudis CA, Krop IE, Burstein HJ, Winer EP. Seven-year follow-up analysis of adjuvant paclitaxel and trastuzumab trial for node-negative, human epidermal growth factor receptor 2-positive breast cancer. J. Clin. Oncol. 2019;37:1868–1875. doi: 10.1200/JCO.19.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bullock K, Blackwell K. Clinical efficacy of taxane–trastuzumab combination regimens for HER-2–positive metastatic breast cancer. Oncologist. 2008;13:515–525. doi: 10.1634/theoncologist.2007-0204. [DOI] [PubMed] [Google Scholar]
- 29.Weaver BA. How taxol/paclitaxel kills cancer cells. Mol. Biol. Cell. 2014;25:2677–2681. doi: 10.1091/mbc.e14-04-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller RH, Keck CM. Challenges and solutions for the delivery of biotech drugs—A review of drug nanocrystal technology and lipid nanoparticles. J. Biotechnol. 2004;113:151–170. doi: 10.1016/j.jbiotec.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Schrama D, Reisfeld RA, Becker JC. Antibody targeted drugs as cancer therapeutics. Nat. Rev. Drug Discov. 2006;5:147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 32.Beck A, Goetsch L, Dumontet C, Corvaïa N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat. Rev. Drug Discovery. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- 33.Panowski S, Bhakta S, Raab H, Polakis P, Junutula JR. Site-specific antibody drug conjugates for cancer therapy. MAbs. 2014;6:34–45. doi: 10.4161/mabs.27022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchikama K, An Z. Antibody-drug conjugates: Recent advances in conjugation and linker chemistries. Protein Cell. 2018;9:33–46. doi: 10.1007/s13238-016-0323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flygare JA, Pillow TH, Aristoff P. Antibody–drug conjugates for the treatment of cancer. Chem. Biol. Drug Des. 2013;81:113–121. doi: 10.1111/cbdd.12085. [DOI] [PubMed] [Google Scholar]
- 36.Simone EA, Dziubla TD, Muzykantov VR. Polymeric carriers: Role of geometry in drug delivery. Expert Opin. Drug Deliv. 2008;5:1283–1300. doi: 10.1517/17425240802567846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitragotri S, Burke PA, Langer R. Overcoming the challenges in administering biopharmaceuticals: Formulation and delivery strategies. Nat. Rev. Drug Discov. 2014;13:655–672. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vicent MJ, Duncan R. Polymer conjugates: Nanosized medicines for treating cancer. Trends Biotechnol. 2006;24:39–47. doi: 10.1016/j.tibtech.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Gradishar WJ, Tjulandin S, Davidson N, Shaw H, Desai N, Bhar P, Hawkins M, O'Shaughnessy J. Phase III trial of nanoparticle albumin-bound paclitaxel compared with polyethylated castor oil-based paclitaxel in women with breast cancer. J. Clin. Oncol. 2005;23:7794–7803. doi: 10.1200/JCO.2005.04.937. [DOI] [PubMed] [Google Scholar]
- 40.McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N. Engl. J. Med. 1996;334:1–6. doi: 10.1056/NEJM199601043340101. [DOI] [PubMed] [Google Scholar]
- 41.Campos SM, Penson RT, Mays AR, Berkowitz RS, Fuller AF, Goodman A, Matulonis UA, Muzikansky A, Seiden MV. The clinical utility of liposomal doxorubicin in recurrent ovarian cancer. Gynecol. Oncol. 2001;81:206–212. doi: 10.1006/gyno.2000.5980. [DOI] [PubMed] [Google Scholar]
- 42.Ranson MR, Carmichael J, O'Byrne K, Stewart S, Smith D, Howell A. Treatment of advanced breast cancer with sterically stabilized liposomal doxorubicin: Results of a multicenter phase II trial. J. Clin. Oncol. 1997;15:3185–3191. doi: 10.1200/JCO.1997.15.10.3185. [DOI] [PubMed] [Google Scholar]
- 43.Abu Lila A, Ishida T, Kiwada H. Targeting anticancer drugs to tumor vasculature using cationic liposomes. Pharm. Res. 2010;27:1171–1183. doi: 10.1007/s11095-010-0110-1. [DOI] [PubMed] [Google Scholar]
- 44.Muro S, Garnacho C, Champion JA, Leferovich J, Gajewski C, Schuchman EH, Mitragotri S, Muzykantov VR. Control of endothelial targeting and intracellular delivery of therapeutic enzymes by modulating the size and shape of ICAM-1-targeted carriers. Mol. Ther. 2008;16:1450–1458. doi: 10.1038/mt.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Yang T, Wang X, Wang J, Zhang X, Zhang Q. Targeted polymeric micelle system for delivery of combretastatin A4 to tumor vasculature in vitro. Pharm. Res. 2010;27:1861–1868. doi: 10.1007/s11095-010-0184-9. [DOI] [PubMed] [Google Scholar]
- 46.Zih-rou H, Shu-chiou H, Yueh-lung Y, Jia-you F. Development and evaluation of lipid nanoparticles for camptothecin delivery: A comparison of solid lipid nanoparticles, nanostructured lipid carriers, and lipid emulsion. Acta Pharmacol. Sin. 2008;29:1094–1102. doi: 10.1111/j.1745-7254.2008.00829.x. [DOI] [PubMed] [Google Scholar]
- 47.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 48.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janat-Amsbury MM, Ray A, Peterson CM, Ghandehari H. Geometry and surface characteristics of gold nanoparticles influence their biodistribution and uptake by macrophages. Eur. J Pharm. Biopharm. Off. J. Arbeitsgemeinschaft Pharm. Verfahrenstech. 2011;77:417–423. doi: 10.1016/j.ejpb.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Champion JA, Mitragotri S. Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4930–4934. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laemthong T, Kim HH, Dunlap K, Brocker C, Barua D, Forciniti D, Huang Y-W, Barua S. Bioresponsive polymer coated drug nanorods for breast cancer treatment. Nanotechnology. 2016;28:045601. doi: 10.1088/1361-6528/28/4/045601. [DOI] [PubMed] [Google Scholar]
- 53.Barua S, Mitragotri S. Synergistic targeting of cell membrane, cytoplasm, and nucleus of cancer cells using rod-shaped nanoparticles. ACS Nano. 2013;7:9558–9570. doi: 10.1021/nn403913k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dennler P, Fischer E, Schibli R. Antibody conjugates: from heterogeneous populations to defined reagents. Antibodies. 2015;4:197–224. doi: 10.3390/antib4030197. [DOI] [Google Scholar]
- 55.Chou TC. Drug combination studies and their synergy quantification using the Chou–Talalay method. Can. Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 56.Chou T-C, Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 57.Cecchini, M. J., Amiri, M., Dick, F. A. Analysis of cell cycle position in mammalian cells. JoVE (Journal of Visualized Experiments) e3491 (2012). [DOI] [PMC free article] [PubMed]
- 58.Sarsour EH, Agarwal M, Pandita TK, Oberley LW, Goswami PC. Manganese superoxide dismutase protects the proliferative capacity of confluent normal human fibroblasts. J. Biol. Chem. 2005;280:18033–18041. doi: 10.1074/jbc.M501939200. [DOI] [PubMed] [Google Scholar]
- 59.Olsson JV, Hult D, Cai Y, García-Gallego S, Malkoch M. Reactive imidazole intermediates: Simplified synthetic approach to functional aliphatic cyclic carbonates. Polym. Chem. 2014;5:6651–6655. doi: 10.1039/C4PY00911H. [DOI] [Google Scholar]
- 60.Hornig S, Liebert T, Heinze T. Structure design of multifunctional furoate and pyroglutamate esters of dextran by polymer-analogous reactions. Macromol. Biosci. 2007;7:297–306. doi: 10.1002/mabi.200600189. [DOI] [PubMed] [Google Scholar]
- 61.Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, Blättler WA, Lambert JM, Chari RVJ, Lutz RJ, Wong WLT, Jacobson FS, Koeppen H, Schwall RH, Kenkare-Mitra SR, Spencer SD, Sliwkowski MX. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Can. Res. 2008;68:9280–9290. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- 62.Peng J, Chen J, Xie F, Bao W, Xu H, Wang H, Xu Y, Du Z. Herceptin-conjugated paclitaxel loaded PCL-PEG worm-like nanocrystal micelles for the combinatorial treatment of HER2-positive breast cancer. Biomaterials. 2019;222:119420. doi: 10.1016/j.biomaterials.2019.119420. [DOI] [PubMed] [Google Scholar]
- 63.Wang J, Li Y, Li S, Yao W, Liu X, Zhu Y, Li W, Sun L, Jin N, Li X. Anti-tumor synergistic effect of a dual cancer-specific recombinant adenovirus and paclitaxel on breast cancer. Front. Oncol. 2020;10:244–244. doi: 10.3389/fonc.2020.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ciardiello F, Caputo R, Pomatico G, De Laurentiis M, De Placido S, Bianco AR, Tortora G. Resistance to taxanes is induced by c-erbB-2 overexpression in human MCF-10A mammary epithelial cells and is blocked by combined treatment with an antisense oligonucleotide targeting type I protein kinase A. Int. J. Cancer. 2000;85:710–715. doi: 10.1002/(SICI)1097-0215(20000301)85:5<710::AID-IJC18>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 65.Xue HY, Narvikar M, Zhao J-B, Wong HL. Lipid encapsulation of cationic polymers in hybrid nanocarriers reduces their non-specific toxicity to breast epithelial cells. Pharm. Res. 2013;30:572–583. doi: 10.1007/s11095-012-0902-6. [DOI] [PubMed] [Google Scholar]
- 66.McCluskey AJ, Olive AJ, Starnbach MN, Collier RJ. Targeting HER2-positive cancer cells with receptor-redirected anthrax protective antigen. Mol. Oncol. 2013;7:440–451. doi: 10.1016/j.molonc.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jelínek M, Balušíková K, Schmiedlová M, Němcová-Fürstová V, Šrámek J, Stančíková J, Zanardi I, Ojima I, Kovář J. The role of individual caspases in cell death induction by taxanes in breast cancer cells. Cancer Cell Int. 2015;15:8. doi: 10.1186/s12935-015-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Allan LA, Clarke PR. Apoptosis and autophagy: Regulation of caspase-9 by phosphorylation. FEBS J. 2009;276:6063–6073. doi: 10.1111/j.1742-4658.2009.07330.x. [DOI] [PubMed] [Google Scholar]
- 69.Galbán S, Duckett CS. XIAP as a ubiquitin ligase in cellular signaling. Cell Death Differ. 2010;17:54. doi: 10.1038/cdd.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Würstle ML, Laussmann MA, Rehm M. The central role of initiator caspase-9 in apoptosis signal transduction and the regulation of its activation and activity on the apoptosome. Exp. Cell Res. 2012;318:1213–1220. doi: 10.1016/j.yexcr.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 71.Kipp J. The role of solid nanoparticle technology in the parenteral delivery of poorly water-soluble drugs. Int. J. Pharm. 2004;284:109–122. doi: 10.1016/j.ijpharm.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 72.Lukyanov AN, Torchilin VP. Micelles from lipid derivatives of water-soluble polymers as delivery systems for poorly soluble drugs. Adv. Drug Deliv. Rev. 2004;56:1273–1289. doi: 10.1016/j.addr.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Z, Mei L, Feng S-S. Paclitaxel drug delivery systems. Expert Opin. Drug Deliv. 2013;10:325–340. doi: 10.1517/17425247.2013.752354. [DOI] [PubMed] [Google Scholar]
- 74.Senapati S, Mahanta AK, Kumar S, Maiti P. Controlled drug delivery vehicles for cancer treatment and their performance. Signal Transduct. Target. Ther. 2018;3:7. doi: 10.1038/s41392-017-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tong R, Cheng J. Anticancer polymeric nanomedicines. Polym. Rev. 2007;47:345–381. doi: 10.1080/15583720701455079. [DOI] [Google Scholar]
- 76.Mishra B, Patel BB, Tiwari S. Colloidal nanocarriers: A review on formulation technology, types and applications toward targeted drug delivery. Nanomed. Nanotechnol. Biol. Med. 2010;6:9–24. doi: 10.1016/j.nano.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 77.Naksuriya O, Okonogi S, Schiffelers RM, Hennink WE. Curcumin nanoformulations: A review of pharmaceutical properties and preclinical studies and clinical data related to cancer treatment. Biomaterials. 2014;35:3365–3383. doi: 10.1016/j.biomaterials.2013.12.090. [DOI] [PubMed] [Google Scholar]
- 78.Teicher BA, Chari RVJ. Antibody conjugate therapeutics: Challenges and potential. Clin. Cancer Res. 2011;17:6389. doi: 10.1158/1078-0432.CCR-11-1417. [DOI] [PubMed] [Google Scholar]
- 79.Nejadmoghaddam M-R, Minai-Tehrani A, Ghahremanzadeh R, Mahmoudi M, Dinarvand R, Zarnani A-H. Antibody-drug conjugates: Possibilities and challenges. Avicenna J Med Biotechnol. 2019;11:3–23. [PMC free article] [PubMed] [Google Scholar]
- 80.Ghosh AK, Brindisi M. Organic carbamates in drug design and medicinal chemistry. J. Med. Chem. 2015;58:2895–2940. doi: 10.1021/jm501371s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quiles S, Raisch KP, Sanford LL, Bonner JA, Safavy A. Synthesis and preliminary biological evaluation of high-drug-load paclitaxel-antibody conjugates for tumor-targeted chemotherapy. J. Med. Chem. 2010;53:586–594. doi: 10.1021/jm900899g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fahmy TM, Samstein RM, Harness CC, Saltzman WM. Surface modification of biodegradable polyesters with fatty acid conjugates for improved drug targeting. Biomaterials. 2005;26:5727–5736. doi: 10.1016/j.biomaterials.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 83.Steenblock ER, Fadel T, Labowsky M, Pober JS, Fahmy TM. An artificial antigen-presenting cell with paracrine delivery of IL-2 impacts the magnitude and direction of the T cell response. J. Biol. Chem. 2011;286:34883–34892. doi: 10.1074/jbc.M111.276329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Steenblock ER, Fahmy TM. A comprehensive platform for ex vivo T-cell expansion based on biodegradable polymeric artificial antigen-presenting cells. Mol. Ther. 2008;16:765–772. doi: 10.1038/mt.2008.11. [DOI] [PubMed] [Google Scholar]
- 85.Ben-Akiva E, Meyer RA, Wilson DR, Green JJ. Surface engineering for lymphocyte programming. Adv. Drug Deliv. Rev. 2017;114:102–115. doi: 10.1016/j.addr.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Munisvaradass R, Kumar S, Govindasamy C, Alnumair KS, Mok PL. Human CD3+ T-cells with the anti-ERBB2 chimeric antigen receptor exhibit efficient targeting and induce apoptosis in ERBB2 overexpressing breast cancer cells. Int. J. Mol. Sci. 2017;18:1797. doi: 10.3390/ijms18091797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sunshine JC, Perica K, Schneck JP, Green JJ. Particle shape dependence of CD8+ T cell activation by artificial antigen presenting cells. Biomaterials. 2014;35:269–277. doi: 10.1016/j.biomaterials.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meyer RA, Sunshine JC, Perica K, Kosmides AK, Aje K, Schneck JP, Green JJ. Biodegradable nanoellipsoidal artificial antigen presenting cells for antigen specific T-cell activation. Small. 2015;11:1519–1525. doi: 10.1002/smll.201402369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schütz C, Fleck M, Mackensen A, Zoso A, Halbritter D, Schneck JP, Oelke M. Killer artificial antigen-presenting cells: A novel strategy to delete specific T cells. Blood. 2008;111:3546–3552. doi: 10.1182/blood-2007-09-113522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perica K, Tu A, Richter A, Bieler JG, Edidin M, Schneck JP. Magnetic field-induced t cell receptor clustering by nanoparticles enhances t cell activation and stimulates antitumor activity. ACS Nano. 2014;8:2252–2260. doi: 10.1021/nn405520d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Champion J, Mitragotri S. Shape induced inhibition of phagocytosis of polymer particles. Pharm. Res. 2009;26:244–249. doi: 10.1007/s11095-008-9626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Decuzzi P, Ferrari M. The adhesive strength of non-spherical particles mediated by specific interactions. Biomaterials. 2006;27:5307–5314. doi: 10.1016/j.biomaterials.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 93.Decuzzi P, Ferrari M. The receptor-mediated endocytosis of nonspherical particles. Biophys. J . 2008;94:3790–3797. doi: 10.1529/biophysj.107.120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yan G, Paul D, Shenshen C, Richard T, Manorama T, Tamara M, Dennis ED. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang X, Teng X, Chen D, Tang F, He J. The effect of the shape of mesoporous silica nanoparticles on cellular uptake and cell function. Biomaterials. 2010;31:438–448. doi: 10.1016/j.biomaterials.2009.09.060. [DOI] [PubMed] [Google Scholar]
- 96.Barua, S., Yoo, J. -W., Kolhar, P., Wakankar, A., Gokarn, Y. R., Mitragotri, S. Particle shape enhances specificity of antibody-displaying nanoparticles. Proc. Natl. Acad. Sci. U.S.A. (2013). [DOI] [PMC free article] [PubMed]
- 97.Gentile F, Chiappini C, Fine D, Bhavane RC, Peluccio MS, Cheng MM-C, Liu X, Ferrari M, Decuzzi P. The effect of shape on the margination dynamics of non-neutrally buoyant particles in two-dimensional shear flows. J. Biomech. 2008;41:2312–2318. doi: 10.1016/j.jbiomech.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 98.Decuzzi P, Godin B, Tanaka T, Lee SY, Chiappini C, Liu X, Ferrari M. Size and shape effects in the biodistribution of intravascularly injected particles. J. Control. Release. 2010;141:320–327. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 99.Adriani G, de Tullio MD, Ferrari M, Hussain F, Pascazio G, Liu X, Decuzzi P. The preferential targeting of the diseased microvasculature by disk-like particles. Biomaterials. 2012;33:5504–5513. doi: 10.1016/j.biomaterials.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Godin B, Chiappini C, Srinivasan S, Alexander JF, Yokoi K, Ferrari M, Decuzzi P, Liu X. Drug delivery: Discoidal porous silicon particles: Fabrication and biodistribution in breast cancer bearing mice. Adv. Funct. Mater. 2012;22:4186–4186. doi: 10.1002/adfm.201290121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tommasi S, Mangia A, Lacalamita R, Bellizzi A, Fedele V, Chiriatti A, Thomssen C, Kendzierski N, Latorre A, Lorusso V, Schittulli F, Zito F, Kavallaris M, Paradiso A. Cytoskeleton and paclitaxel sensitivity in breast cancer: The role of β-tubulins. Int. J. Cancer. 2007;120:2078–2085. doi: 10.1002/ijc.22557. [DOI] [PubMed] [Google Scholar]
- 102.Marc, S. G., Ahmed, T. A., Asok, B., Cassandra, D. M. C., Philip, W., Mariusz, K., Carol E., C., Richard, F. L., Jack A. T., Sambasivarao, D. IC50 Values for Paclitaxel and Analogs in Cytotoxicity Assays with Breast Cancer Cell Lines (2015).
- 103.McCloskey DE, Kaufmann SH, Prestigiacomo LJ, Davidson NE. Paclitaxel induces programmed cell death in MDA-MB-468 human breast cancer cells. Clin. Cancer Res. 1996;2:847–854. [PubMed] [Google Scholar]
- 104.Rouzier R, Rajan R, Wagner P, Hess KR, Gold DL, Stec J, Ayers M, Ross JS, Zhang P, Buchholz TA, Kuerer H, Green M, Arun B, Hortobagyi GN, Symmans WF, Pusztai L. Microtubule-associated protein tau: a marker of paclitaxel sensitivity in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8315–8320. doi: 10.1073/pnas.0408974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Haghnavaz N, Asghari F, Elieh Ali Komi D, Shanehbandi D, Baradaran B, Kazemi T. HER2 positivity may confer resistance to therapy with paclitaxel in breast cancer cell lines. Artif. Cells Nanomed. Biotechnol. 2018;46:518–523. doi: 10.1080/21691401.2017.1326927. [DOI] [PubMed] [Google Scholar]
- 106.Fisi V, Kátai E, Bogner P, Miseta A, Nagy T. Timed, sequential administration of paclitaxel improves its cytotoxic effectiveness in a cell culture model. Cell Cycle. 2016;15:1227–1233. doi: 10.1080/15384101.2016.1158361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sui M, Dziadyk JM, Zhu X, Fan W. Cell cycle-dependent antagonistic interactions between paclitaxel and γ-radiation in combination therapy. Clin. Cancer Res. 2004;10:4848–4857. doi: 10.1158/1078-0432.CCR-03-0707. [DOI] [PubMed] [Google Scholar]
- 108.Weissenstein U, Kunz M, Urech K, Regueiro U, Baumgartner S. Interaction of a standardized mistletoe (Viscum album) preparation with antitumor effects of Trastuzumab in vitro. BMC Complem. Altern. Med. 2016;16:271–271. doi: 10.1186/s12906-016-1246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rodríguez CE, Reidel SI, De Kier Joffé EDB, Jasnis MA, Fiszman GL. Autophagy protects from trastuzumab-induced cytotoxicity in HER2 overexpressing breast tumor spheroids. PLoS ONE. 2015;10:e0137920. doi: 10.1371/journal.pone.0137920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang C-X, Koay DC, Edwards A, Lu Z, Mor G, Ocal IT, DiGiovanna MP. In vitro and in vivo effects of combination of trastuzumab (herceptin) and tamoxifen in breast cancer. Breast Cancer Res. Treat. 2005;92:251–263. doi: 10.1007/s10549-005-3375-z. [DOI] [PubMed] [Google Scholar]
- 111.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013;14:25. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 2005;5:773. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 113.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr. Biol. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 114.Portt L, Norman G, Clapp C, Greenwood M, Greenwood MT. Anti-apoptosis and cell survival: A review. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2011;1813:238–259. doi: 10.1016/j.bbamcr.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 115.Twiddy D, Cain K. Caspase-9 cleavage, do you need it? Biochem. J. 2007;405:e1–e2. doi: 10.1042/BJ20070617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gupta S, Kass GEN, Szegezdi E, Joseph B. The mitochondrial death pathway: A promising therapeutic target in diseases. J. Cell Mol. Med. 2009;13:1004–1033. doi: 10.1111/j.1582-4934.2009.00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brunelle JK, Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Galluzzi L, Morselli E, Kepp O, Vitale I, Rigoni A, Vacchelli E, Michaud M, Zischka H, Castedo M, Kroemer G. Mitochondrial gateways to cancer. Mol. Asp. Med. 2010;31:1–20. doi: 10.1016/j.mam.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 119.Allan LA, Clarke PR. Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol. Cell. 2007;26:301–310. doi: 10.1016/j.molcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 120.Andersen JL, Johnson CE, Freel CD, Parrish AB, Day JL, Buchakjian MR, Nutt LK, Thompson JW, Moseley MA, Kornbluth S. Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase-2. EMBO J. 2009;28:3216–3227. doi: 10.1038/emboj.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ola MS, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol. Cell. Biochem. 2011;351:41–58. doi: 10.1007/s11010-010-0709-x. [DOI] [PubMed] [Google Scholar]
- 122.Deveraux QL, Reed JC. IAP family proteins—Suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 123.Pardo OE, Lesay A, Arcaro A, Lopes R, Ng BL, Warne PH, McNeish IA, Tetley TD, Lemoine NR, Mehmet H, Seckl MJ, Downward J. Fibroblast growth factor 2-mediated translational control of IAPs blocks mitochondrial release of Smac/DIABLO and apoptosis in small cell lung cancer cells. Mol. Cell Biol. 2003;23:7600–7610. doi: 10.1128/MCB.23.21.7600-7610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ferreira CG, van der Valk P, Span SW, Jonker JM, Postmus PE, Kruyt FAE, Giaccone G. Assessment of IAP (inhibitor of apoptosis) proteins as predictors of response to chemotherapy in advanced non-small-cell lung cancer patients. Ann. Oncol. 2001;12:799–805. doi: 10.1023/A:1011167113067. [DOI] [PubMed] [Google Scholar]
- 125.Dong F, Guo W, Zhang L, Wu S, Teraishi F, Davis JJ, Fang B. Downregulation of XIAP and induction of apoptosis by the synthetic cyclin-dependent kinase inhibitor GW8510 in non-small cell lung cancer cells. Cancer Biol. Ther. 2006;5:165–170. doi: 10.4161/cbt.5.2.2316. [DOI] [PubMed] [Google Scholar]
- 126.Asselin E, Mills GB, Tsang BK. XIAP regulates Akt activity and caspase-3-dependent cleavage during cisplatin-induced apoptosis in human ovarian epithelial cancer cells. Can. Res. 2001;61:1862–1868. [PubMed] [Google Scholar]
- 127.Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Almenara J, Rosato R, Grant S. Synergistic induction of mitochondrial damage and apoptosis in human leukemia cells by flavopiridol and the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) Leukemia. 2002;16:1331–1343. doi: 10.1038/sj.leu.2402535. [DOI] [PubMed] [Google Scholar]
- 129.Dai Y, Chen S, Wang L, Pei X-Y, Kramer LB, Dent P, Grant S. Bortezomib interacts synergistically with belinostat in human acute myeloid leukaemia and acute lymphoblastic leukaemia cells in association with perturbations in NF-κB and Bim. Br. J. Haematol. 2011;153:222–235. doi: 10.1111/j.1365-2141.2011.08591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.