Abstract
Introduction
Therapeutic acetaminophen (APAP) ingestion causes asymptomatic drug–induced liver injury in some patients. In most cases, elevations in alanine aminotransferase (ALT) are transient and return to the normal range, even with continued APAP ingestion, though ALT elevation persists in some patients unpredictably. The etiology of this liver injury or adaption is unclear. Our objective was to identify new pharmacogenomic variants associated with elevated ALT or elevated protein adduct concentrations in patients receiving therapeutic acetaminophen.
Methods
We performed genome-wide sequencing analysis on eight patients using leftover blood samples from an observational study that administered four grams of acetaminophen for up to 16 days to all patients. Two patients with ALT elevations > two times the upper limit of normal, two patients with no adduct formation, and four control patients were sequenced. The genomes were aligned with the GRCh38 reference sequence, and variants with predicted low, moderate, or high impact on the subsequent proteins were first manually curated for biologic plausibility, then organized and examined in the REACTOME pathway analysis program.
Results
We found 394 variants in 107 genes associated with elevated ALT. Variants associated with ALT elevation predominantly involved genes in the immune system (MHC class II complex genes), endoplasmic reticulum stress response (SEC23B and XBP1), oxidative phosphorylation (NDUFB9), and WNT/beta-catenin signaling (FZD5). Variants associated with elevated adducts were primarily in signal transduction (MUC20) and DNA repair mechanisms (P53).
Conclusions
While underpowered, genetic variants in immune system genes may be associated with drug-induced liver injury at therapeutic doses of acetaminophen.
Electronic supplementary material
The online version of this article (10.1007/s13181-020-00815-2) contains supplementary material, which is available to authorized users.
Keywords: Acetaminophen, ALT, Drug induced liver injury, Hepatotoxicity, Pharmacogenomics
Introduction
Acetaminophen is a commonly used analgesic that is available without a prescription in the USA. It is frequently taken daily for prolonged periods for treatment of osteoarthritis or other conditions that cause mild to moderate pain. While acetaminophen is well tolerated, between 31 and 44% patients taking 4 g of APAP daily in an inpatient study unit developed ALT elevation greater than 3 times the upper limit of normal [1]. Another study demonstrated this same phenomenon when 4 g acetaminophen was take for 16 days, though some patients ALT never returned to the normal range [2]. A systematic review of prospective studies in patients taking repeated dosing of therapeutic acetaminophen demonstrated that slight transaminase elevations may occur, but death or liver failure have not been reported [3]. These elevations resolve with continued administration suggesting hepatic adaptation [2]. Similar, self-limited changes have been reported for statins, isoniazid, and amiodarone [4]. However, the mechanism of these elevations and the subsequent resolution are not known.
Acetaminophen metabolism is well described. While the majority of acetaminophen undergoes glucuronidation and sulfation, a small amount is oxidized by CYP 2E1 to the radical N-acetyl-p-benzoquinone imine (NAPQI). With therapeutic doses, the vast majority of NAPQI is reduced by glutathione. This detoxification process is overwhelmed in overdose, allowing the NAPQI to bind to hepatic macromolecules forming acetaminophen-protein adducts. NAPQI binding is the accepted mechanism of injury that leads to cell death in acetaminophen poisoning [5], and high concentrations of adducts have been suggested as a marker of acetaminophen toxicity. However, small amounts of adducts are generally formed during therapeutic dosing as well. The concentrations of adducts with therapeutic dosing vary more than 10-fold, and understanding the genetics that determine adduct concentrations may provide insight into this variation and help refine the interpretation of adduct measurements [6]. The variation in adduct formation may provide insight into the low-level elevation in ALT reported during repeated therapeutic dosing.
Prior investigators have examined genetic variation in patients with liver toxicity and acetaminophen exposure [7–11]. That work is limited by the failure to adequately establish or quantify acetaminophen exposure and use of candidate gene or microarray approaches that fails to acknowledge unrecognized mechanisms of hepatoxicity [9, 12]. Thus, previously identified variants associated with worse clinical outcomes may not have been associated with acetaminophen toxicity, but rather, other factors. New pathway analysis programs allow for organization of genome wide association results into well-curated biologic pathways. These peer-reviewed tools allow for distillation of huge amounts of complex bioinformatic data to provide insights into new biologic mechanisms of disease. It is unclear if APAP-induced ALT elevation is due to NAPQI production or another mechanism. APAP is a prototypical hepatotoxin therefore understanding the genetic mechanisms of this ALT elevation, and subsequent adaption, may provide insight to the mechanisms of other hepatotoxins. We believe that examining the genomics in patients with drug induced liver injury (DILI) due to acetaminophen in a well-controlled study can provide insight into the mechanisms of APAP induced liver injury, hepatic adaption, and possibly other hepatotoxins.
This report is a secondary analysis of samples collected during a clinical trial where subjects were administered 4 g/day of acetaminophen for a minimum of 16 days [2]. The subjects were followed for changes in serum transaminase activity, and serum adduct concentrations were also measured. The primary objective of this pilot study was to determine if there are genetic variants associated with ALT elevations and changes in serum acetaminophen-protein adduct concentrations in a subset of patients with discordant ALT and/or adduct responses from taking therapeutic doses of APAP.
Methods
We performed whole genome sequencing in 8 patients that ingested 4 g of acetaminophen for up to 16 days as part of a previously completed randomized controlled clinical trial (NCT00743093) [2]. For sequencing, we chose baseline blood samples from two patients that developed acetaminophen DILI as defined by an ALT > two times the upper limit of normal, two patients that never had elevations in their ALT and never had adducts formed, and four control patients that never had ALT elevations but had adduct elevation during the study. Baseline whole blood samples were stored at − 80 °C prior to sequencing. Both the parent study and this genomic analysis were approved by the Colorado Multiple Institutional Review Board.
Parent Study
The parent study was an outpatient study in which subjects were recruited from research list-serves, posters, a classified advertisement website (Craigslist), and media advertising. All subjects provided informed consent. We included male and female subjects who were age 18 years or older and who did not have any of the following exclusion criteria: history of acetaminophen ingestion on any of the 4 days preceding study enrollment or a measurable serum acetaminophen concentration at time of enrollment; laboratory testing suggesting active viral hepatitis A, B, or C infection; any of the following tests greater than the upper limit of normal at screening: serum aminotransferase or total bilirubin, international normalized ratio (INR). or alkaline phosphatase activity; platelet count less than 125,000/mL; positive pregnancy test; history of cholelithiasis (without cholecystectomy); history of heavy ethanol use defined as consuming more than an average of 3 alcohol containing drinks daily or 3 or more alcohol containing drinks on any given day over the preceding 2 weeks prior to study enrollment; new prescription medication started within the previous 30 days; taking isoniazid or warfarin; currently has anorexia nervosa or reports a fasting type diet; clinically intoxicated, psychiatrically impaired or unable to give informed consent for any reason; and known hypersensitivity or allergy to acetaminophen.
Intervention
Subjects were administered acetaminophen 4 g/day. Subjects were instructed to take 2 × 500 mg tablets every 4 h for 4 doses each day (the exact timing varied but subjects were asked to take the doses every 4 h after the first dose each day). The subjects noted the time of ingestion for each dose in a study diary. They also recorded other medications and alcohol consumption. Compliance was verified by study diary and pill counts at each study visit. ALT was drawn on study days 0 (baseline), 4, 7, 10, 13, and 16. The parent study also included a placebo arm, whose patients were not investigated as part of this study.
Genomics
DNA was extracted from whole blood using Qiagen® QuiAMP DNA blood kits per the manufacturer instructions. We performed whole genome sequencing in eight patients using an Illumina NovaSEQ6000 instrument with × 30 coverage per sample. We performed 150 bp paired-end read cycles and × 15 coverage per genome, with an average of 139 million reads. Reads trimmed shorter than 80 bases were removed (on average 18.5 million reads per sample). Runs with less than ×10 coverage of the reads were removed from the analysis. The genomes were aligned with the GRCh38 reference sequence.
We sequenced two patients with APAP-induced ALT elevation, two patients with no ALT elevation and no APAP-cysteine adduct production, and four patients with no ALT elevation but detectable APAP-cysteine adducts (as expected with 4 g per day dosing). Determination of variant significance was performed using the ExAC and Genome Aggregation Databases (gnomAD), version 2.1 (http://gnomad.broadinstitute.org/) [13]. This program provides functional significance of the identified variants as modifiers, low, moderate, or high impact on protein function. Manual curation of the damaging variants of moderate or high impact on the resulting protein was performed using OMIM to determine biologic plausibility.
Pathway Analysis
We used the REACTOME pathway analysis software to map genes to biologic pathways. REACTOME is an open-source, open access, manually curated, and peer-reviewed pathway database [14]. Variants with functionally significant mutations, as defined as low, moderate, or high impact in gnomAD, were entered into the database to populate a linked analysis of biologic pathways associated with APAP induced ALT and adduct elevation. This program allows linkage between genetic variants, proteins, and other portions of the transcription/translation pathway to demonstrate interaction between genetic variants.
Results
The parent study enrolled 205 patients in the APAP intervention arm and, 47 patients in the placebo arm. Table 1 contains the demographics and clinical variables of the eight patients examined in this study. Acetaminophen was detected in blood at day 7 in all these patients, confirming exposure.
Table 1.
Patient demographics and clinical variables.
| Patient # | Age years (Gender) | Race/ethnicity | Alcohol drinks per week | Co-medications | Peak ALT IU, (study day) | Peak Adduct concentration CYS/mg protein (study day) |
|---|---|---|---|---|---|---|
|
1 ALT elevation |
20 (male) | Caucasian | < 1 | None | 191 (7) | 0.195 (16) |
|
2 ALT elevation |
55 (female) | Caucasian | < 1 | Multivitamin, levothyroxine, glucosamine, chondroitin, flax oil, bismuth subsalicylate, calcium, vitamin D, magnesium | 162 (10) | 0.509 (34) |
|
3 Control |
40 (female) | African American | 1–2 | Ibuprofen, naproxen | 24 (13) | 0.096 (16) |
|
4 Control |
49 (female) | Caucasian | 1–2 | Multivitamin, acidophillis, fish oil, niacin, fiber, lorazepam, furosemide, glucosamine, chondroitin | 29 (10) | 0.684 (7) |
|
5 Control |
48 (female) | Caucasian | < 1 | Ibuprofen, chlortrimeton, emergen C, fish oil, B complex, glucosamine, citalopram, vitamin E, Ester C vitamin, calcium with vitamin D, | 20 (10) | 0.132 (10) |
|
6 Control |
26 (male) | Caucasian | < 1 | None | 17 (13) | 0.131 (10) |
|
7 No adducts |
42 (male) | Caucasian | < 1 | None | 25 (13) | 0 (all study days) |
|
8 No adducts |
20 (male) | Hispanic | < 1 | Chondroitin, emergen C, glucosamine | 43 (10) | 0 (all study days) |
Comedications and Alcohol History
There were no associations between elevated ALT or APAP-adducts and either concomitant medications or alcohol use.
Genomic Results
We found 394 variants in 107 genes associated with elevated ALT. Most variants were deemed to be moderate impact on protein function (Table 2), by gnomAD designation. There were variants in 194 variants in 65 genes, also with a predominance of moderate impact. See Supplementary Appendix for all genes with damaging variants.
Table 2.
Variant impact and counts associated with ALT and adduct elevation.
| Variant impact | ALT variant count (probability) | Adduct variant Count (probability) |
|---|---|---|
| High | 6 (0.02) | 9 (0.05) |
| Moderate | 221 (0.56) | 105 (0.54) |
| Low | 10 (0.03) | 8 (0.04) |
| Modifier | 157 (0.40) | 72 (0.37) |
Pathway Analysis
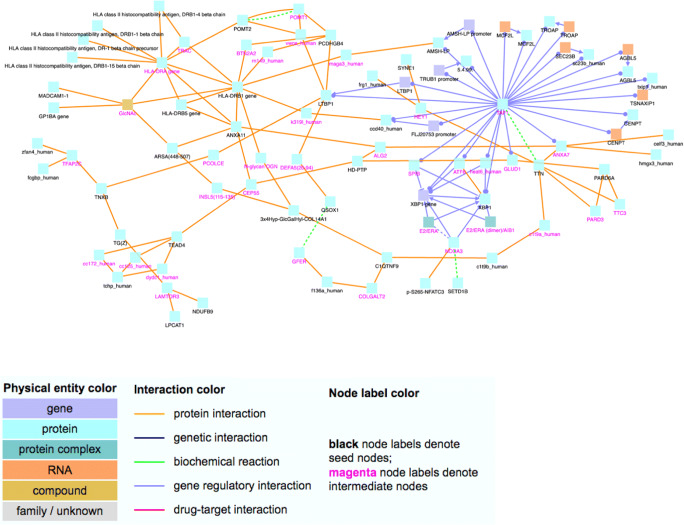
Among the variants associated with ALT elevation, many were found to be within genes involved in immune function, based on REACTOME analysis (Figs. 1 and 2). There were 25 variants in immune system genes and 14 variants in MHC class II antigen presenting genes. Additionally, we found variants in interferon signaling genes and other inflammatory cascades. At therapeutic acetaminophen doses, DILI is likely to be due to immune induced hepatocyte damage, lending credence to these findings.
Fig. 1.
a REACTOME pathway analysis. a Damaging variants associated with ALT elevation. Heat map indicating the number of genes in each pathway. Orange line denotes selected pathways with predominance of moderate impact variants. b Damaging variants associated with adduct elevation. Heat map indicating the number of genes in each pathway. Orange line denotes selected pathways with predominance of moderate impact variants.
Fig. 2.
Network analysis of genes with moderate or high impact variants associated with ALT elevation, performed in REACTOME analysis. Nodes with black labels are from the input gene list (seed nodes) and nodes with a purple label are intermediate nodes that are not in the input list but connect seed nodes and have significantly many links in the network module. Each edge represents an interaction (physical, biochemical, regulatory or drug–target interaction). Gene list available in Supplement.
Variants associated with ALT elevation involved genes in the endoplasmic reticulum stress response (SEC23B and XBP1) [15, 16], oxidative phosphorylation (NDUFB9) [17], and WNT/beta-catenin signaling (FZD5) [18, 19]. All of these pathways have known roles in liver toxicity and repair. Interestingly, WNT/ beta-catenin signaling has been implicated in expression pathways of CYP2E1, among other aspects of hepatic regeneration [20].
Patients with elevated adducts were more likely to have damaging variants in genes involved in signal transduction (Fig. 1b). A variant in the Rho GTPase activating protein 11A was identified; in response to DNA damage, the encoded protein interacts with the p53 tumor suppressor protein and stimulates its tetramerization, which results in cell-cycle arrest and apoptosis. There were variants in the gene that encodes mucin 20, a cell surface associated protein (MUC20), which is a member of the mucin protein family. Mucins are high molecular weight glycoproteins secreted by many epithelial tissues to form an insoluble mucous barrier. The C-terminus of this family member associates with the multifunctional docking site of the MET proto-oncogene and suppresses activation of some downstream MET signaling cascades. MET is a hepatocyte growth factor receptor. Thus, damage to MUC20 may lead to failure of hepatocytes to recover from APAP-induced oxidative stress. Polymorphism in this gene is not known to be associated with a clinical trait; thus, the implications of a damaging variant in this gene are unclear.
Discussion
In this preliminary study examining patients receiving 4 g of acetaminophen per day, variants associated with ALT elevation were found in genes associated with the immune system function. While underpowered to confirm these findings, this study provides novel insight in a unique population. The subjects and exposure were well characterized. The predominant biologic pathways identified have been examined by other investigators; both cultured hepatocyte and animal models have [21] demonstrated importance of the identified genes or proteins outlined in our results.
We demonstrate that some genes involved in pathways facilitating and rescuing cellular damage are functionally altered in individuals with elevated ALT due to therapeutic doses of APAP, but not in patients without ALT elevation. The clinical implications of these ALT elevations are unclear, since these patients did not develop clinical symptoms associated with this laboratory abnormality. We suggest that these patients may not have the same capacity to adapt to this hepatic insult, and this may represent a risk for DILI. We hypothesize that these mutations create an environment that is less able to handle the APAP load—even at therapeutic doses—resulting in acute liver injury. Pathway analysis suggests that necrotic hepatocytes activate the innate immune system, which initially increases the liver injury. Eventually, these pathways promote liver regeneration and recovery, which is consistent with the natural course of ALT resolution in these patients. However, some individuals may have altered hepatocyte regeneration pathways leading to worse clinical outcomes. Mutations in genes related to the innate immune response, platelet activation, and liver regeneration were present in individuals with elevated ALT in our cohort. This hypothesis is supported by work that demonstrated increased transcripts reflective of upregulation of T-cells, mast cells, eosinophils, mononuclear cells, and lymphocytes in patients with increased ALT following 7 days of therapeutic APAP [22].
HLA variants have been implicated in DILI from other drugs [23, 24], and similar variants may be involved in APAP induced DILI. HLA genotyping is not commonly performed during targeted analyses due to the highly polymorphic nature of these genes [25], the need for phasing to provide accurate genotype calls, sequence similarity between the genes, and a large degree of linkage disequilibrium in the locus. We also found variants in genes in the MHC II complex genes, which were upregulated in patients with ALT elevation in a small group of patients receiving therapeutic APAP dosing [22]. Thus, it is likely that this mechanism of APAP-induced DILI has been underappreciated by prior investigators. Future work in this field should focus on the mechanisms of immune mediated hepatotoxicity and hepatic adaption when faced with hepatotoxins. Understanding the genomics of these patients, paired with longitudinal biomarker assessments, can provide insight into which patients are at risk of DILI and the mechanisms of hepatic adaption are. Performing whole genome sequencing allows for a more complete understanding of how genetic variants and genomic architecture interact in this complex polygenic condition.
Limitations
This pilot study has important limitations. Performing whole genome sequencing in only eight patients is likely to reveal type 1 errors. While genetic polymorphism may be observed simply because the subjects are different, we have included twice as many controls than cases and we have attempted to report variants with biologic plausibility. However, confirmation will require thousands of patients with heterogenous racial and ethnic backgrounds. Our manual curation of variants with moderate to high impact on protein function may minimize our appreciation of the lower impact polymorphisms on this condition. We felt it important to present the most impactful variants as part of this pilot report, for fear of examining clinically insignificant variants in such a small cohort. If we are able to perform GWAS in all these patients, we will not curate the analysis in this way to minimize the impact of that introduced bias. This study is clearly underpowered for this complex clinical outcome, and we recognize that this work is primarily proof of concept. We acknowledge this and plan to use these data as hypothesis generating data for future pharmacogenomic studies. The pathways identified in this study will allow more focused targeted genotyping studies in the remainder of the cohort and other cohorts in the future. These variants may not be associated with more severe outcomes in overdose patients, since outcomes in overdose may also involve variants associated with treatment, in addition to the pre-treatment hepatotoxic phenotype. This will alter the statistical significance of variants identified in overdose patients, since hepatotoxicity is a culmination of both drug-induced toxicity and treatment effectiveness. Larger studies with more diverse racial and ethnic populations are needed to validate this pilot study. Certain variants are more common in populations from different ancestry backgrounds. These variants, which may not be well represented in this small cohort, such as CYP2E1 ultrarapid metabolizers, may have functional implications on ALT elevation. The complete lack of adducts in two subjects suggests some patients may not generate adducts at therapeutic dose. Both subjects recorded compliance with dosing in their daily diaries and one subject had detectable acetaminophen during one of his visits further supporting compliance.
Conclusion
Acetaminophen toxicity is the most commonly seen condition by medical toxicologists, the mechanism of toxicity is well understood, and therefore, it represents a perfect disease model to examine the role of pharmacogenomic mediators of adverse drug events. In summary, we present preliminary data that immune system variants are associated with APAP induced DILI in a small number of patients with excellent exposure histories. Larger studies may confirm which genes are highly associated with this clinical condition and may provide hypotheses for pharmacogenomic guided treatment of overdose patients in the future.
Electronic supplementary material
(XLSX 30 kb)
(XLSX 45 kb)
Funding
The parent study was funded by Johnson and Johnson. A.A.M. received support from NIH 1R35GM124939 and NIH CTSI UL1 TR001082 for this work.
Compliance with Ethical Standards
Conflict of Interest
None
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Watkins PB, Kaplowitz N, Slattery JT, Colonese CR, Colucci SV, Stewart PW, Harris SC. Aminotransferase elevations in healthy adults receiving 4 grams of acetaminophen daily: a randomized controlled trial. JAMA. 2006;296(1):87–93. doi: 10.1001/jama.296.1.87. [DOI] [PubMed] [Google Scholar]
- 2.Heard K, Green JL, Anderson V, Bucher-Bartelson B, Dart RC. A randomized, placebo-controlled trial to determine the course of aminotransferase elevation during prolonged acetaminophen administration. BMC Pharmacol Toxicol. 2014;15:39. doi: 10.1186/2050-6511-15-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dart RC, Bailey E. Does therapeutic use of acetaminophen cause acute liver failure? Pharmacotherapy. 2007;27(9):1219–1230. doi: 10.1592/phco.27.9.1219. [DOI] [PubMed] [Google Scholar]
- 4.Fontana RJ. Pathogenesis of idiosyncratic drug-induced liver injury and clinical perspectives. Gastroenterology. 2014;146(4):914–928. doi: 10.1053/j.gastro.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heard KJ. Acetylcysteine for acetaminophen poisoning. N Engl J Med. 2008;359(3):285–292. doi: 10.1056/NEJMct0708278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davern TJ, 2nd, James LP, Hinson JA, et al. Measurement of serum acetaminophen-protein adducts in patients with acute liver failure. Gastroenterology. 2006;130(3):687–694. doi: 10.1053/j.gastro.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 7.Court MH, Freytsis M, Wang X, et al. The UDP-glucuronosyltransferase (UGT) 1A polymorphism c.2042C>G (rs8330) is associated with increased human liver acetaminophen glucuronidation, increased UGT1A exon 5a/5b splice variant mRNA ratio, and decreased risk of unintentional acetaminophen-induced acute liver failure. J Pharmacol Exp Ther. 2013;345(2):297–307. doi: 10.1124/jpet.112.202010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Court MH, Peter I, Hazarika S, Vasiadi M, Greenblatt DJ, Lee WM, The Acute Liver Failure Study Group Candidate gene polymorphisms in patients with acetaminophen-induced acute liver failure. Drug Metab Dispos. 2014;42(1):28–32. doi: 10.1124/dmd.113.053546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heruth DP, Shortt K, Zhang N, Li DY, Zhang LQ, Qing Ye S. Genetic Association of Single nucleotide polymorphisms with acetaminophen-induced hepatotoxicity. J Pharmacol Exp Ther. 2018;367(1):95–100. doi: 10.1124/jpet.118.248583. [DOI] [PubMed] [Google Scholar]
- 10.Navarro SL, Chen Y, Li L, Li SS, Chang JL, Schwarz Y, King IB, Potter JD, Bigler J, Lampe JW. UGT1A6 and UGT2B15 polymorphisms and acetaminophen conjugation in response to a randomized, controlled diet of select fruits and vegetables. Drug Metab Dispos. 2011;39(9):1650–1657. doi: 10.1124/dmd.111.039149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueshima Y, Tsutsumi M, Takase S, Matsuda Y, Kawahara H. Acetaminophen metabolism in patients with different cytochrome P-4502E1 genotypes. Alcohol Clin Exp Res. 1996;20(1 Suppl):25A–28A. doi: 10.1111/j.1530-0277.1996.tb01722.x. [DOI] [PubMed] [Google Scholar]
- 12.Bushel PR, Fannin RD, Gerrish K, Watkins PB, Paules RS. Blood gene expression profiling of an early acetaminophen response. Pharm J. 2017;17(3):230–236. doi: 10.1038/tpj.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sidiropoulos K, Viteri G, Sevilla C, Jupe S, Webber M, Orlic-Milacic M, Jassal B, May B, Shamovsky V, Duenas C, Rothfels K, Matthews L, Song H, Stein L, Haw R, D’Eustachio P, Ping P, Hermjakob H, Fabregat A. Reactome enhanced pathway visualization. Bioinformatics. 2017;33(21):3461–3467. doi: 10.1093/bioinformatics/btx441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusama H, Kon K, Ikejima K, Arai K, Aoyama T, Uchiyama A, Yamashina S, Watanabe S. Sodium 4-phenylbutyric acid prevents murine acetaminophen hepatotoxicity by minimizing endoplasmic reticulum stress. J Gastroenterol. 2017;52(5):611–622. doi: 10.1007/s00535-016-1256-3. [DOI] [PubMed] [Google Scholar]
- 16.Yehia L, Niazi F, Ni Y, Ngeow J, Sankunny M, Liu Z, Wei W, Mester JL, Keri RA, Zhang B, Eng C. Germline heterozygous variants in SEC23B are associated with Cowden syndrome and enriched in apparently sporadic thyroid cancer. Am J Hum Genet. 2015;97(5):661–676. doi: 10.1016/j.ajhg.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Briede JJ, Jennen DG, et al. Increased mitochondrial ROS formation by acetaminophen in human hepatic cells is associated with gene expression changes suggesting disruption of the mitochondrial electron transport chain. Toxicol Lett. 2015;234(2):139–150. doi: 10.1016/j.toxlet.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Wright SC, Canizal MCA, Benkel T, et al. FZD5 is a Galphaq-coupled receptor that exhibits the functional hallmarks of prototypical GPCRs. Sci Signal. 2018;11(559). 10.1126/scisignal.aar5536. [DOI] [PubMed]
- 19.Dadhania VP, Bhushan B, Apte U, Mehendale HM. Wnt/beta-catenin signaling drives thioacetamide-mediated heteroprotection against acetaminophen-induced lethal liver injury. Dose-Response. 2017;15(1):1559325817690287. doi: 10.1177/1559325817690287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerbal-Chaloin S, Dume AS, Briolotti P, et al. The WNT/beta-catenin pathway is a transcriptional regulator of CYP2E1, CYP1A2, and aryl hydrocarbon receptor gene expression in primary human hepatocytes. Mol Pharmacol. 2014;86(6):624–634. doi: 10.1124/mol.114.094797. [DOI] [PubMed] [Google Scholar]
- 21.Cullen M, Perfetto SP, Klitz W, Nelson G, Carrington M. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am J Hum Genet. 2002;71(4):759–776. doi: 10.1086/342973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fannin RD, Gerrish K, Sieber SO, Bushel PR, Watkins PB, Paules RS. Blood transcript immune signatures distinguish a subset of people with elevated serum ALT from others given acetaminophen. Clin Pharmacol Ther. 2016;99(4):432–441. doi: 10.1002/cpt.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hautekeete ML, Horsmans Y, Van Waeyenberge C, et al. HLA association of amoxicillin-clavulanate-induced hepatitis. Gastroenterology. 1999;117(5):1181–1186. doi: 10.1016/S0016-5085(99)70404-X. [DOI] [PubMed] [Google Scholar]
- 24.O'Donohue J, Oien KA, Donaldson P, et al. Co-amoxiclav jaundice: clinical and histological features and HLA class II association. Gut. 2000;47(5):717–720. doi: 10.1136/gut.47.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SGE. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43(Database issue):D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 30 kb)
(XLSX 45 kb)