Abstract
Background
To reveal the function of deubiquitylating enzyme USP41 in lung adenocarcinoma.
Methods
The relationship between USP41 and lung cancer was determined by analyzing data from The Cancer Genome Atlas (TCGA). A549 and H1299 cell lines were transfected with short hairpin RNA against USP41 (shUSP41 group) or negative control (shCon group). Western blotting was used to verify the transfection efficacy and marker expression. Cell proliferation and apoptosis were analyzed by EdU assay, MTT assay, and flow cytometry after USP41 knockdown. Transwell assay was used to determine the effect of USP41 downregulation on cell migration.
Results
Analysis of lung cancer data from TCGA database indicated a higher level of USP41 expression in lung cancer tumor tissue compared with that in noncancerous tissue, and USP41 overexpression was correlated with poor overall survival of lung cancer patients (p < 0.01). The outcomes of the EdU, MTT, and flow cytometry assays indicated decreased cell proliferation and enhanced apoptosis in shUSP41‐transfected cells. Transwell assay further demonstrated that USP41 knockdown increased the migration rate of A549 and H1299 cells.
Conclusions
In our study, USP41 was overexpressed in lung cancer tissue and associated with poor prognosis of lung cancer. USP41 knockdown inhibits cell proliferation and migration and induces cell apoptosis of lung cancer.
Keywords: deubiquitylating enzyme, invasion, migration, non‐small cell lung cancer, USP41
USP41 is overexpressed in lung cancer tissue and associated with poor prognosis of lung cancer. USP41 overexpression promotes cell proliferation and migration and inhibits apoptosis of lung cancer cells.

INTRODUCTION
Lung cancer has recently become the leading cause of cancer deaths worldwide, among which non‐small cell lung cancer (NSCLC) has shown a significant growth trend as its main pathological type. 1 , 2 Numerous studies have reported that the functions of the ubiquitin family (deubiquitylating enzymes, DUBs) are closely related to the occurrence and development of NSCLC. 3 The ubiquitin‐specific protease (USP) subfamily has the most members, and it is also the most widely studied family. 4 Members of the USP family are structurally diverse cysteine proteases that remove the ubiquitin chain from the target protein by hydrolysis, promote protein stability and function, and are involved in cellular DNA repair, cell cycle progression, and gene transcriptional regulation. 5 , 6 , 7 Because the number of studies targeting specifically activated members of the USP family for drug development is increasing, it is crucial to elucidate the function and mechanism of action of members of the USP family. The role of USP41, a member of the USP family, in NSCLC is unclear. Therefore, the aim of this study was to analyze the expression and function of USP41 in lung cancer to provide more evidence for its use as a target in the clinical treatment of lung cancer.
METHODS
Online database analysis
The GEPIA website (http://gepia2.cancer-pku.cn/#help) is an online database that includes information about cancer patients in The Cancer Genome Atlas (TCGA) database and gene expression data of normal human tissue in the Genotype‐Tissue Expression (GTEx) database. We used this website to collectively analyze data from 483 lung cancer patients in the TCGA database and the normal tissue data in the GTEx database. We compared the differences in USP41 expression in lung cancer and normal tissue and analyzed the overall survival rate of lung cancer patients using the Kaplan–Meier method.
Cell lines
We purchased NSCLC cell lines A549 and H1299 from the Shanghai Cell Bank of the Chinese Academy of Sciences (China). The two cell lines were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% fetal bovine serum and placed in a constant temperature incubator at 37°C with 5% CO2 and humidity saturation. The experiment was performed on cells in the logarithmic growth stage.
Antibodies and reagents
RPMI 1640 cell culture medium, fetal bovine serum, penicilli streptomycin mixture, trypsin, RIPA protein lysate, and 3‐(4,5‐dimethyl‐2‐thiazolyl)‐2,5‐diphenyltetrazolium bromide (MTT) reagent were purchased from Sigma‐Aldrich Corporation. USP41 antibody was purchased from Thermofisher Inc. and GAPDH antibody was purchased from Cell Signaling Technology Inc. Kits for bicinchoninic acid (BCA) protein quantitation, enhanced chemiluminescence (ECL), EdU staining, and apoptosis were purchased from Thermo Fisher. Transwell migration chambers were purchased from Corning Inc.
Construction of short hairpin RNA against USP41 (shUSP41) vector and cell transfection
The shUSP41 interference sequences were as follows: shUSP41‐1: sense, 5′‐CACCGCGTCTTGTTCAGGGCTCATCTCAAGAGGATGAGCCCTGAACAAGACGC‐3′ and antisense, 5′–AAAAGCGTCTTGTTCAGGGCTCATCCTCTTGAGATGAGCCCTGAACAAGACGC‐3′; and ShUSP41‐2: sense, 5′‐CACCGGGCTCATCAGTGTCAGTACGTCAAGAGCGTACTGACACTGATGAGCCC‐3′ and antisense, 5′‐AAAAGGGCTCATCAGTGTCAGTACGCTCTTGACGTACTGACACTGATGAGCCC‐3′. The interference sequence was connected with a pLKO vector to obtain pLKO‐shUSP41 core vector, which was then cotransfected with a pVSVG, pREV, and pGAG packaging system into HEK293T cells. After 24 h, we collected the supernatant containing the viral particles by centrifugation, filtered it through a 0.45 μ filter, and determined the viral titer by the dilution method to obtain the appropriate concentration of infected cells. Then, A549 and H1299 cells were inoculated into a 24‐well plate at a density of 3 × 104 cells/well, and virus and polybrene were added to each well. After 48 h, the medium containing puromycin was changed, and the plates were cultured for one week. The cell lines that were stably transfected with shUSP41 were screened out, and the successful knockout of USP41 was determined by western blot assay.
Western blotting
The transfected NSCLC cells were lysed, and the total protein was extracted with RIPA buffer and quantified using a BCA kit. Following denaturation in the sample buffer at 100°C for 5 min, 30 μg of each protein sample was subjected to 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis for 50 min. Sodium dodecyl sulfate‐polyacrylamide gel was placed on the nitrocellulose membrane, and protein transfer was performed at a constant current of 200 mA for 60 min. After blocking the nitrocellulose membrane in 5% skim milk at room temperature for 2 h, it was incubated at 4°C overnight in a proportional dilution of primary protein antibody. The next day, the membrane was washed three times with phosphate‐buffered saline (PBS) with Tween 20 (PBST) solution, placed in the secondary antibody, incubated at room temperature for 45 min, and washed with PBST. Finally, ECL solution was used to incubate and develop the color of the membrane to capture the target band.
EdU assay
The target cells were seeded in 96‐well plates and cultured for 48 h with 3–5 wells in each group. Then, 100 μl diluted EdU solution (final concentration, 50 M EdU) was added to each well and incubated for 2 h. The cells were fixed with 50 μl of 4% paraformaldehyde for 30 min, incubated with 100 μl permeation agent (containing 5% Triton X‐100 in PBS) for 15 min, and washed twice with PBS between each step. Thereafter, 100 μl diluted reaction solution was added to each well for staining and incubated on a shaker for 20 min before discarding the solution. Next, 100 μl permeation agent was added for 10 min. Finally, 200 μl 4′,6‐diamidino‐2‐phenylindole staining solution was added to each well and incubated in the dark for 5 min, followed by three washes with PBS. The number of positive cells was recorded, and the proliferation rate was calculated.
MTT assay
The USP41‐knockout A549 and H1299 cells and the control cells were digested and inoculated into 96‐well plates with 3000 cells per well and 100 μl fresh medium, which was changed every 24 h. At 24, 48, 72, and 96 h, 20 μl of MTT solution (5 mg/L) was added to each well, respectively. After 4 h, the medium was removed from the 96‐well plate, and 150 μl DMSO was added to each well. The absorbance value of each group at 490 nm was determined by a microplate reader and used for subsequent statistical analysis.
Migration assay
The cells were resuspended in serum‐free medium to form a single cell suspension, and 3 × 104 cells/well were seeded in the upper compartment of the Transwell chamber. A complete medium containing 10% fetal bovine serum was added to the lower compartment, and the chamber was placed in an 37°C incubator with 5% CO2 and humidity saturation. After 24 h, the cells that could not pass through the upper layer of the chamber were wiped off with a cotton swab, and the cells were stained with 0.1% crystal violet at room temperature for 15 min. We randomly selected five fields from each chamber for photography, and ImageJ software (https://imagej.nih.gov/ij/download.html) was used for quantitative analysis.
Flow cytometry assay
The cells were washed twice with PBS and centrifuged to remove the supernatant. Then, they were incubated in diluted annexin V staining solution for 30 min in each tube at room temperature, followed by the addition of 1 μl propidium iodide (PI) staining solution to each tube and incubation at room temperature for 5 min. Apoptosis was detected using a FACS Calibur flow cytometer (BD Biosciences, USA) for subsequent analysis and representation.
Statistical analysis
SPSS 21.0 (IBM Corp., USA) and GraphPad Prism 7.0 (GraphPad Software, USA) were used for statistical analysis and data mapping. The data with a normal distribution were expressed as means ± SD. Student's t‐test or one‐way ANOVA was used to compare the data from the two independent samples. A p‐value <0.05 was considered statistically significant.
RESULTS
Correlation analysis between USP41 and lung cancer prognosis
Using the GEPIA website, we first analyzed the expression of USP41 in lung cancer patients in TCGA database and in normal human tissue in the GTEx database. The level of deubiquitinase USP41 in lung cancer patients was significantly higher than that in normal tissue (Figure 1(a)), and a high expression level of USP41 predicted a poor overall survival rate in lung cancer patients (Figure 1(b)), indicating that USP41 promotes lung cancer progression and may be a prognostic indicator for lung cancer patients.
FIGURE 1.
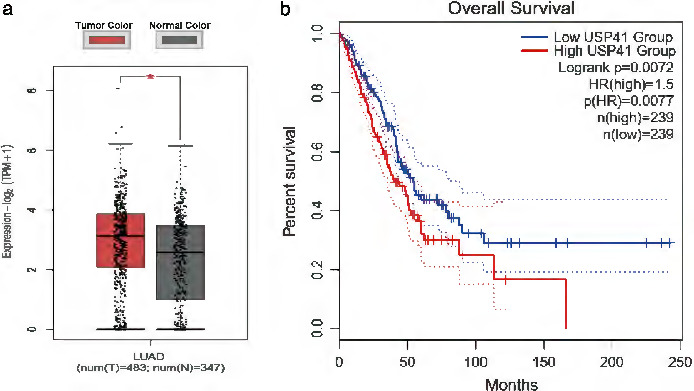
USP41 expression in lung cancer patients and its effect on prognosis. (a) Analysis of TCGA and GTEx databases showed that the level of deubiquitination enzyme USP41 in lung cancer patients was significantly higher than that in normal tissue. (b) The overall survival rate of lung cancer patients with a high level of USP41 expression was poor
USP41 affects the proliferation of lung cancer cells
We investigated whether USP41 affects the proliferation of lung cancer cells to further determine the oncogenic role of USP41 in lung cancer. USP41 knockdown was performed on A549 and H1299 lung cancer cell lines using shRNA, and the transfection efficiency was evaluated by western blot assay to determine the effectiveness of shUSP41 (Figure 2(a)). We analyzed the proliferation and survival of lung cancer cells using EdU and MTT assays. A549 and H1299 cells with USP41 knockdown were significantly less proliferative and the cell survival rate after 96 h of culture was significantly lower compared with those cells transfected with the control plasmid (Figure 2(b) and (c)).
FIGURE 2.
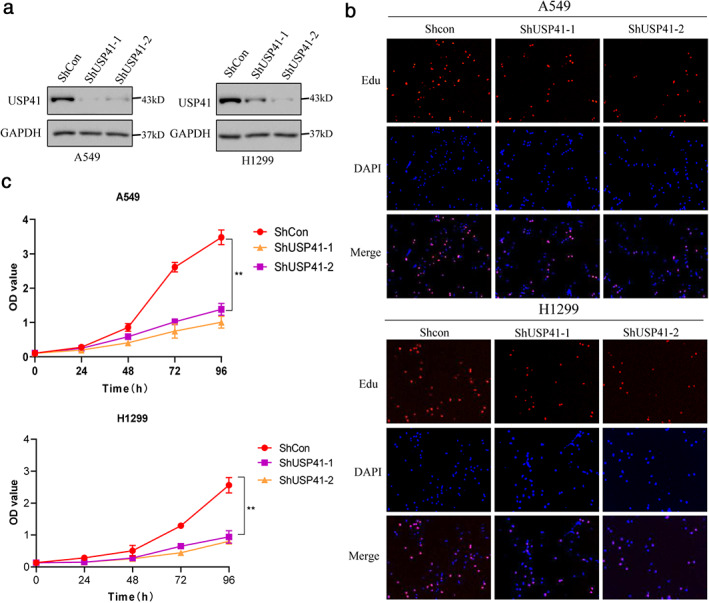
USP41 affects lung cancer cell proliferation. Three independent experiments were performed. **p < 0.01 (a) Western blot assay confirmed that USP41 knockdown was effective in A549 and H1299 lung cancer cell lines. (b) The EdU assay showed that the proliferation ability of A549 and H1299 cells was significantly decreased after USP41 knockdown compared with the control group. (c) MTT assay showed that the survival rate of A549 and H1299 cells cultured for 96 h after USP41 knockdown was significantly lower than that of the control group
USP41 knockdown promotes lung cancer cell apoptosis
We used annexin V/PI double‐staining to detect apoptosis in lung cancer cells. The apoptosis levels of A549 and H1299 lung cancer cells were significantly increased after USP41 knockdown. Compared with the control group, apoptosis of A549 cells increased from 2% to 8% (Figure 3(a)), and apoptosis of H1299 cells increased from 3% to approximately 10% (Figure 3(b)). Western blot results showed that USP41 knockdown significantly increased apoptotic marker proteins in A549 and H1299 cells (Figure 3(c)).
FIGURE 3.
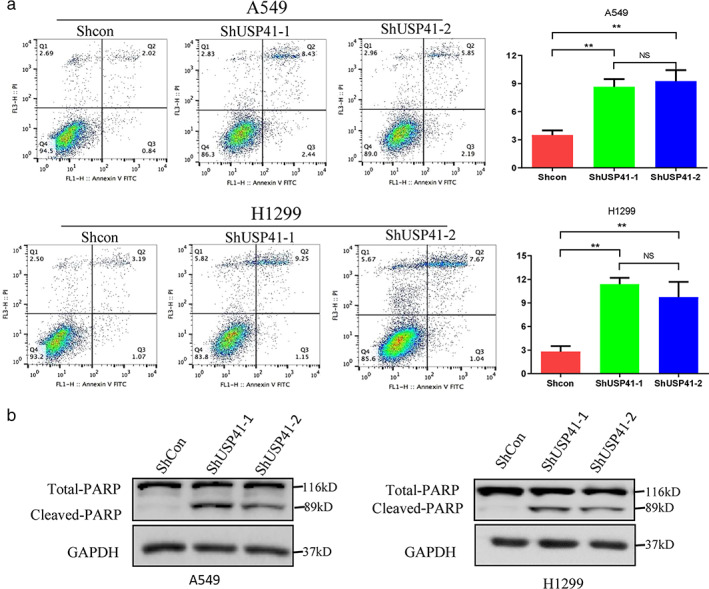
Effect of USP41 knockdown on lung cancer cell apoptosis. Three independent experiments were performed. **p < 0.01. (a, b) The amount of apoptotic A549 and H1299 cells increased when compared to the control group. (b) USP41 knockdown significantly increased cleaved PARP proteins in A549 and H1299 cells
Effect of USP41 knockdown on lung cancer cell migration
Transwell assay results showed that the number of migratory A549 and H1299 cells significantly decreased compared with the control group after USP41 knockdown (Figure 4(a)). Western blot results showed that after USP41 knockdown, the expression of N‐cadherin in A549 and H1299 cells was significantly decreased, while the expression of E‐cadherin, a migration inhibitor, was significantly increased (Figure 4(b)). These results indicated that USP41 promoted lung cancer cell migration. We knocked down E‐cadherin in lung cancer cell lines using siRNA (Figure 4(c)), and demonstrated that E‐Cadherin knockout offseted the effect of inhibition of USP41 on cell migration, and inhibition of USP41 may reverse EMT of lung cancer cells (Figure 4(d)).
FIGURE 4.
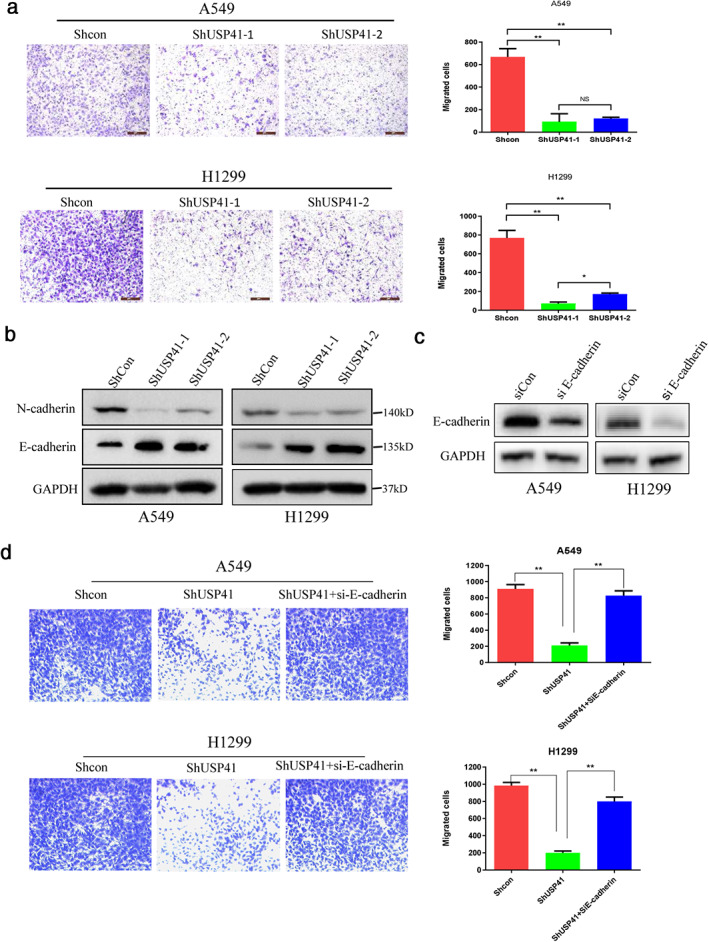
Effect of USP41 knockdown on lung cancer cell migration. Three independent experiments were performed. **p < 0.01. (a, b) The cell migration of A549 and H1299 cells were significantly reduced when compared to the control group. (c) Protein expression of N‐cadherin and E‐cadherin in the control and USP41‐knockdown group detected by western blot
DISCUSSION
Numerous clinical studies have shown that elevated tumor maker levels are a major feature of cancer occurrence. Ubiquitination and deubiquitination are important methods that regulate protein degradation and have attracted much attention in the field of cancer research. 8 The protein is linked to the ubiquitin chain under the action of a ubiquitin enzyme and then recognized by the proteasome and hydrolyzed. The DUBs are capable of regulating protein homeostasis and function by removing ubiquitin from proteins and participating in critical cellular physiological processes, such as cell signaling, DNA repair, cell cycle progression, and apoptosis. 1 , 9 When deubiquitinase is abnormally activated, the proteolytic process of corresponding oncoprotein substrates is impaired to varying degrees, 10 resulting in increased protein stability and the continuous function of oncoproteins, which promotes tumor formation and progression.
The USP family is the largest deubiquitination enzyme subfamily, comprising over 50 members. USP structure is highly conserved, and USPs participates in many physiological processes, such as DNA repair, cell cycle progression, and transcriptional regulation. 7 USP family members play different functions in cancer, and most of them, such as USP1, USP2, USP7, USP14, and USP17, contribute to cancer promotion. USP2 expression levels in breast cancer, cervical cancer, glioma, and prostate cancer tissue are significantly higher than those in the corresponding paracancer tissue. 11 , 12 , 13 , 14 USP7 is highly expressed in lung cancer and multiple myeloma 15 , 16 and is associated with poor prognosis of colon cancer, glioma, liver cancer, and other cancers. 17 , 18 , 19 A few USPs, such as CYLD, are characterized as tumor suppressor factors that inhibit the occurrence or metastasis of liver, skin, and colon cancers. 20 , 21 , 22 Also, several USPs play both carcinogenic and anticancer roles in different cancers. For example, USP9X inhibits tumor formation in colon cancer, while it is highly expressed in lung cancer and various blood cancers. 23 , 24 , 25
NSCLC studies have shown that high expression levels of USP5 and USP7 in lung cancer tissue promote lung cancer cell proliferation by stabilizing beta‐catenin. 15 , 26 , 27 USP14 expression by miR‐124 promotes stem cell characteristics of NSCLC cells and gemcitabine resistance, 28 and USP17 expression is associated with NSCLC metastasis and poor prognosis. 29 , 30 Furthermore, CYLD is a tumor suppressor, and its defects lead to lung cancer metastasis. 31
At present, the function and regulatory mechanism of many USP family members, including USP41, have not been studied. In this study, the analysis of lung cancer cases in TCGA database showed that USP41 was highly expressed in lung cancer tumor tissue and predicted poor overall survival rate of lung cancer patients, indicating that USP41 functions as an oncogene in lung cancer. MTT, EdU, flow cytometry, and transwell assays further determined that USP41 enhanced the proliferation and migration of lung cancer cells. Based on the mechanisms of involvement of the USP family in many solid tumors, 14 , 32 , 33 we can speculate that USP41 acts through regulating MDM2, Cyclin D1, FAS, and TGFBR1, which promote tumor cell survival and proliferation, inhibit cell apoptosis, promote cell migration, and may be associated with the beta‐catenin signaling pathway. We intend to continue our exploration of USP41‐specific regulatory mechanisms in NSCLC in subsequent experiments. Due to the important role of the USP family in cancer, many USP inhibitors have been developed. For example, small‐molecule inhibitor P5091 inhibits USP7 in colon cancer and chronic leukemia, 16 , 17 , 34 and small‐molecule inhibitor b‐AP15 plays a significant role in inhibiting the cell growth of leukemia and esophageal squamous cell carcinoma. 35 , 36 Further studies on USP41 in NSCLC may provide more evidence for its use as a target in the clinical treatment of lung cancer.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ACKNOWLEDGMENTS
This study was supported by the grants from the National Natural Science Foundation of China (to Ying Li, No.81601996).
Ji J, Yang S, Zu L, Li Y, Li Y. Deubiquitinating enzyme USP41 promotes lung cancer cell proliferation and migration. Thorac Cancer. 2021;12:1041–1047. 10.1111/1759-7714.13843
Jiaqi Ji and Shuping Yang contributed equally to this work
Contributor Information
Yongwen Li, Email: leeywen@163.com.
Ying Li, Email: ly980910@163.com.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 3. Cheng J, Guo J, North BJ, Wang B, Cui C‐P, Li H, et al. Functional analysis of deubiquitylating enzymes in tumorigenesis and development. Biochim Biophys Acta Rev Cancer. 2019;1872(2):188312. [DOI] [PubMed] [Google Scholar]
- 4. Mevissen TET, Komander D. Mechanisms of Deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–92. [DOI] [PubMed] [Google Scholar]
- 5. Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–63. [DOI] [PubMed] [Google Scholar]
- 6. Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez‐Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31(19):2373–88. [DOI] [PubMed] [Google Scholar]
- 7. McClurg UL, Robson CN. Deubiquitinating enzymes as oncotargets. Oncotarget. 2015;6(12):9657–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumari N, Jaynes PW, Saei A, Iyengar PV, Richard JLC, Eichhorn PJA. The roles of ubiquitin modifying enzymes in neoplastic disease. Biochim Biophys Acta Rev Cancer. 2017;1868(2):456–83. [DOI] [PubMed] [Google Scholar]
- 9. Nijman SMB, Luna‐Vargas MPA, Velds A, Brummelkamp TR, AMG D, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–86. [DOI] [PubMed] [Google Scholar]
- 10. Singh N, Singh AB. Deubiquitinases and cancer: a snapshot. Crit Rev Oncol Hematol. 2016;103:22–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qu Q, Mao Y, Xiao G, Fei X, Wang J, Zhang Y, et al. USP2 promotes cell migration and invasion in triple negative breast cancer cell lines. Tumour Biol. 2015;36(7):5415–23. [DOI] [PubMed] [Google Scholar]
- 12. Yang Y, Hou JQ, Qu LY, Wang GQ, Ju HW, Zhao ZW, et al. Differential expression of USP2, USP14 and UBE4A between ovarian serous cystadenocarcinoma and adjacent normal tissues. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2007;23(6):504–6. [PubMed] [Google Scholar]
- 13. Boustani MR, Khoshnood RJ, Nikpasand F, Taleshi Z, Ahmadi K, Yahaghi E, et al. Overexpression of ubiquitin‐specific protease 2a (USP2a) and nuclear factor erythroid 2‐related factor 2 (Nrf2) in human gliomas. J Neurol Sci. 2016;363:249–52. [DOI] [PubMed] [Google Scholar]
- 14. Graner E, Tang D, Rossi S, Baron A, Migita T, Weinstein LJ, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5(3):253–61. [DOI] [PubMed] [Google Scholar]
- 15. Zhang C, Lu J, Zhang QW, Zhao W, Guo JH, Liu SL, et al. USP7 promotes cell proliferation through the stabilization of Ki‐67 protein in non‐small cell lung cancer cells. Int J Biochem Cell Biol. 2016;79:209–21. [DOI] [PubMed] [Google Scholar]
- 16. Chauhan D, Tian Z, Nicholson B, Kumar KGS, Zhou B, Carrasco R, et al. A small molecule inhibitor of ubiquitin‐specific protease‐7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. An T, Gong Y, Li X, Kong L, Ma P, Gong L, et al. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem Pharmacol. 2017;131:29–39. [DOI] [PubMed] [Google Scholar]
- 18. Yi L, Cui Y, Xu Q, Jiang Y. Stabilization of LSD1 by deubiquitinating enzyme USP7 promotes glioblastoma cell tumorigenesis and metastasis through suppression of the p53 signaling pathway. Oncol Rep. 2016;36(5):2935–45. [DOI] [PubMed] [Google Scholar]
- 19. Cai JB, Shi GM, Dong ZR, Ke AW, Ma HH, Gao Q, et al. Ubiquitin‐specific protease 7 accelerates p14(ARF) degradation by deubiquitinating thyroid hormone receptor‐interacting protein 12 and promotes hepatocellular carcinoma progression. Hepatology. 2015;61(5):1603–14. [DOI] [PubMed] [Google Scholar]
- 20. Pannem RR, Dorn C, Ahlqvist K, Bosserhoff AK, Hellerbrand C, Massoumi R. CYLD controls c‐MYC expression through the JNK‐dependent signaling pathway in hepatocellular carcinoma. Carcinogenesis. 2014;35(2):461–8. [DOI] [PubMed] [Google Scholar]
- 21. Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl‐3‐dependent NF‐kappaB signaling. Cell. 2006;125(4):665–77. [DOI] [PubMed] [Google Scholar]
- 22. Zhang J, Stirling B, Temmerman ST, Ma CA, Fuss IJ, Derry JMJ, et al. Impaired regulation of NF‐kappaB and increased susceptibility to colitis‐associated tumorigenesis in CYLD‐deficient mice. J Clin Invest. 2006;116(11):3042–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khan OM, Carvalho J, Spencer‐Dene B, Mitter R, Frith D, Snijders AP, et al. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J Clin Invest. 2018;128(4):1326–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen X, Yu C, Gao J, Zhu H, Cui B, Zhang T, et al. A novel USP9X substrate TTK contributes to tumorigenesis in non‐small‐cell lung cancer. Theranostics. 2018;8(9):2348–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463(7277):103–7. [DOI] [PubMed] [Google Scholar]
- 26. Wu N, Liu C, Bai C, Han YP, Cho WC, Li Q. Over‐expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of beta‐catenin. Int J Mol Sci. 2013;14(6):10749–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma X, Qi W, Pan H, Yang F, Deng J. Overexpression of USP5 contributes to tumorigenesis in non‐small cell lung cancer via the stabilization of beta‐catenin protein. Am J Cancer Res. 2018;8(11):2284–95. [PMC free article] [PubMed] [Google Scholar]
- 28. Yu F, Liu JB, Wu ZJ, Xie WT, Zhong XJ, Hou LK, et al. Tumor suppressive microRNA‐124a inhibits stemness and enhances gefitinib sensitivity of non‐small cell lung cancer cells by targeting ubiquitin‐specific protease 14. Cancer Lett. 2018;427:74–84. [DOI] [PubMed] [Google Scholar]
- 29. Zhang S, Yuan J, Zheng R. Suppression of ubiquitin‐specific peptidase 17 (USP17) inhibits tumorigenesis and invasion in non‐small cell lung cancer cells. Oncol Res. 2016;24(4):263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McFarlane C, McFarlane S, Paul I, Arthur K, Scheaff M, Kerr K, et al. The deubiquitinating enzyme USP17 is associated with non‐small cell lung cancer (NSCLC) recurrence and metastasis. Oncotarget. 2013;4(10):1836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahmed N, Zeng M, Sinha I, Polin L, Wei WZ, Rathinam C, et al. The E3 ligase itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat Immunol. 2011;12(12):1176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benassi B, Flavin R, Marchionni L, Zanata S, Pan Y, Chowdhury D, et al. MYC is activated by USP2a‐mediated modulation of microRNAs in prostate cancer. Cancer Discov. 2012;2(3):236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell. 2009;36(3):469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fu C, Zhu X, Xu P, Li Y. Pharmacological inhibition of USP7 promotes antitumor immunity and contributes to colon cancer therapy. Onco Targets Ther. 2019;12:609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang L, Sun Y, Wang J, He Q, Chen X, Lan X, et al. Proteasomal cysteine deubiquitinase inhibitor b‐AP15 suppresses migration and induces apoptosis in diffuse large B cell lymphoma. J Exp Clin Cancer Res. 2019;38(1):453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sha B, Chen X, Wu H, Li M, Shi J, Wang L, et al. Deubiquitylatinase inhibitor b‐AP15 induces c‐Myc‐Noxa‐mediated apoptosis in esophageal squamous cell carcinoma. Apoptosis. 2019;24(9–10):826–36. [DOI] [PubMed] [Google Scholar]


