Introduction
Any sufficiently advanced technology is indistinguishable from magic.
– Arthur C. Clarke
Deep learning is a popular form of artificial intelligence (AI) that has shown great effectiveness for many medical imaging tasks. In this article, we provide a set of instructions to access computing resources needed to perform deep learning on medical images. The article includes a simple deep learning example—categorizing medical images—to show how to use a notebook computing environment. This article will not provide a detailed description of the algorithm but will focus on helping people unfamiliar with deep learning computing to create an environment that will allow them to perform many deep learning tasks.
This article includes the “baby steps” that will help those with little or no AI programming experience or access to specialized hardware. The reader can use web-based resources to execute deep learning algorithms on medical images. It is important to have a computer with Internet access available while you read this article so you can execute the code; reading this article alone will not provide as much insight into deep learning.
Software Setup
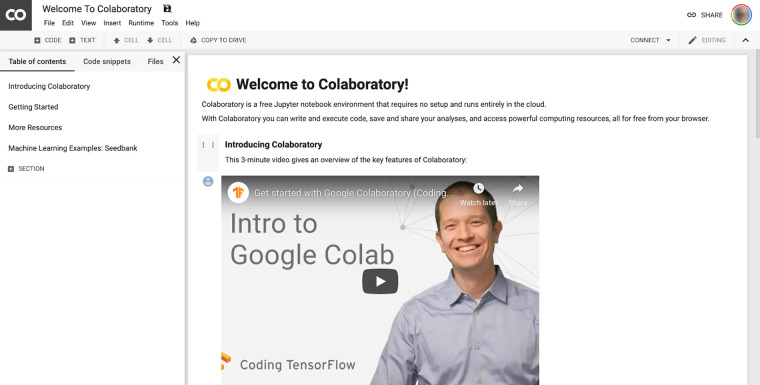
Cloud computing resources can enable any user to perform deep learning without an expensive computer. The easiest and cheapest option is to use Google’s Colab Notebooks, hereafter referred to as Colab (https://colab.research.google.com). The first recommendation is that when choosing Colab, use the Chrome browser. Once you have Chrome installed, open up the website https://colab.research.google.com to view an introduction to the Colab Notebook environment (Fig 1).
Figure 1:
The opening screen of Google’s Colab Notebook.
Interactive computing notebooks are web applications that allow creation and sharing of code, text, data, tools, and so forth, and are often used by software developers. Colab has many learning resources available, so those unfamiliar with Colab might take a few minutes to review those resources.
There are a few important concepts needed to use notebooks. Those who have written computer programs know we typically write the program using a text editor of some sort and then run it using either an interpreter or compiler. Notebooks are different: they have many “cells;” together, the cells will be a complete program. Any cell can be selected and run, without the requirement of executing prior or subsequent cells. You can also run a cell and then edit its contents and run it again until you get the cell to perform the desired function. This does require you have some visible result from that cell (which is displayed below the cell and is replaced with new output when the cell is rerun), and there is no debugger as in some conventional development environments where you can set stop points and view the values stored in variables without having to print them out. Once you are happy with the results of a particular cell, you can then create a new cell as follows: from the Insert > Code Cell menu, enter the code, and run and edit the code until the cell is correct. You continue adding cells of code and reviewing output until the entire program is written. Notebooks do not (currently) have a debugger, so you must print out values to see if the program is actually doing what you expected. The full list of Colab commands can be viewed by clicking Tools > Command Palette. (Please note the Colab menu labels used in this paragraph are as of this writing.)
Notebooks have several special commands that often prove useful. One can execute commands on the host computer by putting the “!” (exclamation point character) in front of the command. With Colab, the host computer uses a Unix operating system, so to see the files in the current directory, the user enters “!ls” (for list files) into a cell, and then executes that cell. We will use this capability to get data into the Colab host computer using a command called “git.” But first, you must get the actual notebook that has the code we have prepared for you.
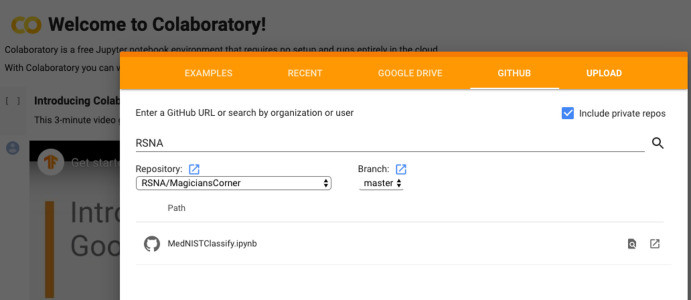
To access the notebook, click on the File > Open Notebook menu option. When you click that, you will be presented with a dialog box with the option “GitHub.” GitHub (Microsoft, Redmond, Wash) is a website that allows developers to save programs and associated data that can be accessed by the program called git to track program changes and version control. Enter “RSNA” into the search field and hit the search icon, and then select the repository “RSNA/MagiciansCorner” using the drop-down list arrows. Below that, you will see an entry called “MedNISTClassify.ipynb” (Fig 2). Select that and after a few seconds you will see the notebook appear. Congratulations—you are ready to start programming deep learning algorithms!
Figure 2:
Dialog to select the starting notebook for this article.
There are a few things to note: there is a slight color difference (light gray) in the background between cells and the output of cells. Clicking on a cell will also change the color of the left side and an arrowhead/triangle will appear. Select the first cell (it has “# Cell 1” as the first line) and click the arrowhead on the left, and you are now executing your first notebook commands. These commands install a library (fastai) that does the actual machine learning. You may get a note about executing a non-Google notebook and also about restarting a runtime; click “Run Anyway” to accept those alerts. Once that cell has finished executing, click the button that says “Restart Runtime.” Select the Runtime > Change Runtime Type from the Runtime menu, choose the options “Python3” and “GPU” and select “SAVE.” Note that these instructions are also shown below the output from executing the first cell. These instructions (and the title at the top of the notebook) are text cells, which are used for documentation or explanation, but do not contain code that is to be executed.
The next cell starts with:
# Cell 2
# clean out any old data just to be sure, such as if rerunning cells
!rm -rf MagiciansCorner
Click the arrowhead to execute this cell. Any previously downloaded data will be removed (by the Unix command “rm”), and then the data we will be using will be downloaded (by the command “git clone https://github.com/...”). The data are then unzipped and ready to be used. You might be asking yourself “What previously downloaded data?” and that is where the ability to rerun cells comes in. Let’s suppose you ran the whole notebook and then wanted to download a different dataset or wanted to change something in the notebook and run the whole thing (which the author did several times as a part of debugging). The “rm -rf MagiciansCorner” command is included because if the cell has already been run, the directory and old data would exist, and the command to unzip data into that existing directory would fail. If you are adventurous (and I hope you are), go ahead and click the arrowhead again! You will see it execute the cell of code again, and now it will delete the “MagiciansCorner” directory.
This ability to interactively rerun portions of a program is one example of how notebook coding is different from traditional program development methods. Note also that when the cell code is completed, the number in the brackets (where the arrowhead was) has incremented. This number reflects the last time that cell was run and can help to show the sequence of execution of cells. You do not have to execute each cell in sequence: you can skip cells and go back and execute them again (changed or unchanged). Note also that below each code cell, there is text showing the output from when the code was originally created and then run by me, before being stored on github. When you run a cell, the output from the original run is erased and the new results are shown. If you download the notebook from GitHub again, your results will be erased, and the notebook will be just like it was when you first downloaded (including showing original output, since that is what was saved on GitHub).
Nearly all deep learning libraries for imaging tasks are designed for photographic-type images, that is, images in the Joint Photographic Experts Group (JPEG) or Portable Network Graphics (PNG) format. We will use one such library in this article. Because we want to work with DICOM images, we must either alter the libraries to handle the unique aspects of medical images (namely, 16 bits of gray rather than three channels of 8 bits each of red, green, and blue intensity) or alter medical images to appear like photographic images (convert 16 bits to 8 bits and replicate into all three color channels). A basic understanding of how images are stored in computers is needed. A pixel on a screen gets its intensity from the numerical value stored. We use a two-dimensional array of such values to store a complete image. And, if we want color, each pixel is actually stored with three values: one each for red, green, and blue (RGB). If the RGB values are all the same, the pixel will be a gray color somewhere between black and white, depending on the value for the RGB triplet. For instance, black would be a triplet of (0,0,0) and white would be (255,255,255) because nearly all JPEG images use values from 0 to 255. As mentioned, the images you download have already been converted from DICOM to 8-bit “color” images with the red, green, and blue channels set to be equal.
We will teach our deep learning model to categorize images into one of six classes: CT abdomen (CTAbd), CT chest (CTChest), CT head (CTHead), MR breast (MRBreast), MR brain (MRBrain), and chest radiograph (CXR). We have separated the images into a folder for each type. The next cell (cell 3—go ahead and execute it by clicking the arrowhead) lists the folders (or directories) so you can see the six classes of images.
For this lesson, we will be using the FastAI deep learning library (http://fast.ai) loaded in the first cell. This library was chosen because it produces very good results with little training and with little code modification. The next cell points the FastAI library to the directories with the data (classes_dir). There is also a randomizer that FastAI uses for deciding the subset of cases that will be used for training versus testing. We can set the fraction of cases used for testing using “valid_pct = 0.2” or 20% in this case, leaving 80% of the dataset for training. Please run cell 4.
It is important to know that in most deep learning tasks applied to radiology, we do “supervised learning,” which means that we give example model training images with known answers (also known as “labels”) to the algorithm. Of course, if we give all examples to the algorithm, we probably train it the best (more examples are better), but doing this makes assessing its performance inaccurate—these algorithms are very good at learning and may learn the specifics of the examples and not the general principles such that showing it an unknown would be much less likely a correct prediction. Therefore, most people hold out a set of cases to test the algorithm after it is trained to estimate the performance. Sometimes this is called the “testing” set, and sometimes the “validation” set. The reason it is important to pay attention to this term is because the algorithm training process usually also has a “hold out” set that it uses for assessing performance, and this set will be called “validation” if “test” is the holdout, and “test” if “validation” is used internally. Please run cell 5.
Now that our data are set up for training, it is wise to visually confirm that things look right, and that is the purpose of the next cell (cell 6). Running that cell (go ahead and run it) will display a few images from the training set and their associated label (class). This is a random sample, so you may not see an example image from each class.
Training the Model
Now that the data are prepared, we are ready to start the task of training the model. “Model” is the term used for the structure of the machine learning network and varies in the number of layers, the type of layers, number of nodes in a layer, and so forth. The user must define each of these, but we will use FastAI, which has good starting points. One popular model architecture is the ResNet34 model. This gets its name because it uses residual networks in its model and has 34 layers. (ResNet18 and ResNet50 are other popular models with 18 and 50 layers, respectively.) ResNets use skip connections to jump over layers, simplifying the network, speeding learning and reducing the impact of vanishing gradients (a topic for later). Error_rate is the metric that is displayed, though actually the system optimizes for a loss function, and that defaults to cross_entropy—more on cross entropy later.
You are ready to train the model! Go ahead, click the arrow to run cell 7.
Now the moment we have all been waiting for: click the arrowhead to run the cell “learn.fit_one_cycle(1).” As one might deduce, this line of code takes the model we created (called “learn”) and tells the computer to fit the data in the training set to the labels for that training set. The “1” in the parentheses tells the computer to do just one cycle—that is, all the examples in the training set are sampled just once. (Go ahead and run this cell, if you haven’t already.)
In practice we will do more training than one cycle, but it is quite impressive that with such a small amount of training, we are achieving better than 96% accuracy. Factors that contribute to this include the training algorithm, the large training set size, and relatively distinctive images in each class. But can we do better? Most likely yes, if we run it for more cycles. This is where notebooks shine: replace the “1” in the line of code “learn.fit_one_cycle (1)” with the number 5. Your line in cell 7 should now look like this: “learn.fit_one_cycle (5).” Then run cell 7 again by clicking the arrowhead. You will see it start to compute again, but because the model has not been discarded, the training resumes where it left off, and adds five more cycles of training, and the loss values, accuracy, and times are printed out. You are free to run this cell again as often as you like, but at some point, the loss/error will cease to improve. Have you hit the limit of what the computer can learn? Perhaps. This is a common problem faced by all machine learning experts: when to quit.
And here we draw this lesson to a close. Notebook environments, such as Google Colab, provide a simple way to use cloud computing resources to solve machine learning problems. Subsequent articles will use other libraries that will show the many decisions that must be made to build a deep learning network.
Footnotes
Disclosures of Conflicts of Interest: B.J.E. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: disclosed no relevant relationships. Other relationships: claimed a copyright on the processed datasets primarily so others cannot “take” them. No plans for patent or commercialization. (These are on public GitHub site to make it easy for readers to download).