Abstract
Purpose
To delineate image data curation needs and describe a locally designed graphical user interface (GUI) to aid radiologists in image annotation for artificial intelligence (AI) applications in medical imaging.
Materials and Methods
GUI components support image analysis toolboxes, picture archiving and communication system integration, third-party applications, processing of scripting languages, and integration of deep learning libraries. For clinical AI applications, GUI components included two-dimensional segmentation and classification; three-dimensional segmentation and quantification; and three-dimensional segmentation, quantification, and classification. To assess radiologist engagement and performance efficiency associated with GUI-related capabilities, image annotation rate (studies per day) and speed (minutes per case) were evaluated in two clinical scenarios of varying complexity: hip fracture detection and coronary atherosclerotic plaque demarcation and stenosis grading.
Results
For hip fracture, 1050 radiographs were annotated over 7 days (150 studies per day; median speed: 10 seconds per study [interquartile range, 3–21 seconds per study]). A total of 294 coronary CT angiographic studies with 1843 arteries and branches were annotated for atherosclerotic plaque over 23 days (15.2 studies [80.1 vessels] per day; median speed: 6.08 minutes per study [interquartile range, 2.8–10.6 minutes per study] and 73 seconds per vessel [interquartile range, 20.9–155 seconds per vessel]).
Conclusion
GUI-component compatibility with common image analysis tools facilitates radiologist engagement in image data curation, including image annotation, supporting AI application development and evolution for medical imaging. When complemented by other GUI elements, a continuous integrated workflow supporting formation of an agile deep neural network life cycle results.
Supplemental material is available for this article.
© RSNA, 2019
Summary
A graphical user interface promoted involvement of local-expert subspecialty radiologists in ongoing image data curation, including case-by-case annotation, potentially concurrent with artificial intelligence–enhanced routine image review in support of ongoing model learning.
Key Point
■ A custom-designed, clinically oriented graphical user interface provides radiologists with an environment to annotate medical images efficiently to reduce barriers in the development of artificial intelligence models.
Introduction
Deep neural network (DNN) formation for artificial intelligence (AI) in medical imaging necessitates curation of large amounts image data (1,2) . While natural language processing tools can curate from radiology reports, they do not provide the precise description of abnormalities essential to train specific models; while automated natural language processing may train other models, inherent noise within generated data makes it unsuitable for judging performance (3). Other strategies (eg, crowdsourcing [4]) also have limitations. Therefore, there is a role for expert image annotation, but little attention has been given to facilitating radiologist engagement in image data curation for AI algorithm development.
Image annotation, a major component of curation, has been a labor-intensive and imperfect process for ground-truth experts. Many image-directed toolboxes for annotation are application specific (5), unsuitable for clinical application (6), and/or nonconducive to radiologist utilization. On the other hand, more clinically focused toolboxes designed for radiologists fail to include annotation capabilities or integration platforms to process custom software programs (7).
This study delineated stepwise needs for image data curation and described locally designed components of a graphical user interface (GUI) promoting radiologist engagement to support AI algorithms.
Materials and Methods
This work was completed within the Laboratory for Augmented Intelligence in Imaging (LAI2) in collaboration (technical support only) with Siemens Healthineers (Erlangen, Germany and Malvern, Pa) and NVIDIA (Santa Clara, Calif). Institutional review board approval with waiver of informed consent applied to all image data use which was controlled by, and limited to, LAI2 faculty or researchers.
Software Platform
Components of the LAI2-designed thick-client GUI were created with Windows-based MeVisLab version 2.8 (MeVis Medical Solutions, Bremen, Germany) (8), providing analysis toolboxes for region detection, registration, and volume-based rendering supported by Digital Imaging and Communications in Medicine (DICOM) compatibility, picture archiving and communication system (PACS) integration, and third-party applications (eg, Qt [9]). It also serves as a platform to process scripting languages (eg, Python and C++) for building custom modules and allows integration of common deep learning libraries (eg, TensorFlow and Keras).
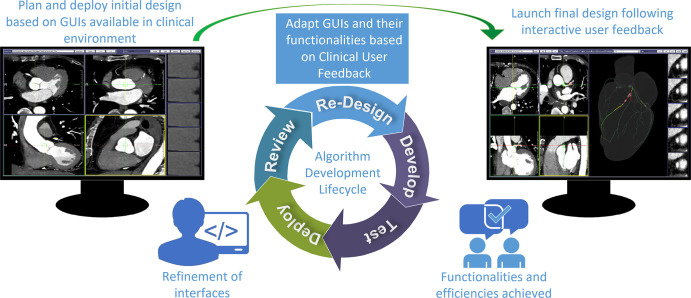
Essential image data curation steps were identified for comprehensive agile DNN formation for AI medical imaging applications (Fig 1). They facilitated design of use-case components of the locally developed GUI promoting curation efficiencies.
Figure 1:
Artificial intelligence algorithm development life cycle. User interfaces are built using iterative development processes under physician guidance. In this example, interface development for coronary stenosis labeling for deep neural network classification development is shown. After gathering user requirements, an interface (left) using existing clinical graphical user interfaces (GUIs) as a reference was initially built and deployed for labeling images. Later, following initial labeling of cases with end-user (expert cardiac imager) feedback, additional functionalities were developed and deployed. The optimized interface (right) features a volumetric navigational display, enabling vessel label tracking in three dimensions in sync with multiplanar reconstructions and arterial branching tree displays.
Image Data Importation
The first step involves selective manual image data importation from an institutional PACS. In this GUI-supported workflow, direct interaction with PACS (eg, IMPAX 6.6.1.3525: Agfa HealthCare, Mortsel, Belgium) is accomplished via an open-source software package, Orthanc (10), operating on a separate DICOM server (11). This interface relies on Representational State Transfer application program interface (RESTful API) technology (12,13) and Python script to automate DICOM query-retrieve user commands to a PACS, allowing a predefined case list to be automatically retrieved and placed into project-specific folders for future de-identification and annotation.
While Orthanc facilitates RESTful APIs via its own custom API (functionally resembling DICOMweb or a DICOMweb plug-in), the internal RESTful API was used for simplicity. Within MeVisLab it is possible to invoke Python scripts and make requests to RESTful APIs, allowing RESTful calls and management transfers from MeVisLab. During calls, patient identifiers (eg, medical record numbers) can be utilized as parameters. If institutional review board–specific de-identification is needed, DICOM-compliant files are produced while maintaining mappings for future referencing.
Patient De-Identification
Patient privacy must be considered throughout curation. During image data importation that is GUI functioning within the busy LAI2, personal and health care information on patients is protected using de-identification processes removing unwanted tags from DICOM header files (14). In this process, all private tags are removed from files, and all patient-identifiable tags (nonprivate) are either removed or hashed and/or de-identified in accordance with institutional review board approvals. All cases receive new unique identifiers at patient, study, and image levels, ensuring privacy and allowing for different aggregations during annotation or inference.
Image Annotation
Medical images must be viewed during manual annotation. When done using a platform familiar to radiologists, annotating can potentially be more efficient and accurate. Although annotation should ideally coincide with clinical image review, this integration is challenging because image-interpretation tools are Food and Drug Administration regulated, following complex development routes not conducive to an agile development life cycle needed in research. In addition, some tasks (eg, segmentation) are time-consuming and are not typically necessary for clinical interpretation.
The GUI combines the typical interactive viewing capabilities (eg, pan and zoom) of a commercial DICOM-based clinical image viewing system (eg, syngo.via; Siemens Healthineers, Erlangen, Germany) with advanced interactive delineation and annotation features (eg, lesion segmentation and labeling) in a thick-client application, thereby reducing duplicative image-manipulation workload and facilitating more confident output of manual annotations.
Formatting Image Annotation Output
The final step involves exportation of annotated images; this output must be delivered in a standardized format suitable for incorporation into a DNN for AI algorithm training, validation, and testing (15). Within the GUI, radiologist-dependent operations are recorded in a text file analyzed by a customized JavaScript object-notation parser, allowing users to resume or edit previous annotations. JavaScript Object Notation was selected as the annotation-recording format because of associated availability of open-source packages permitting easier integration with NoSQL databases (eg, MongoDB) for flexible storage of files reflecting key metrics (eg, region-of-interest [ROI] coordinates, classification label). In addition, it allows images to (a) be histogram equalized for normalization (16); (b) have custom size in format suitable for input into pretrained DNN models (17); and/or (c) be saved as either NumPy arrays or TensorFlow records for usage in DNN formation.
GUI Component Descriptions
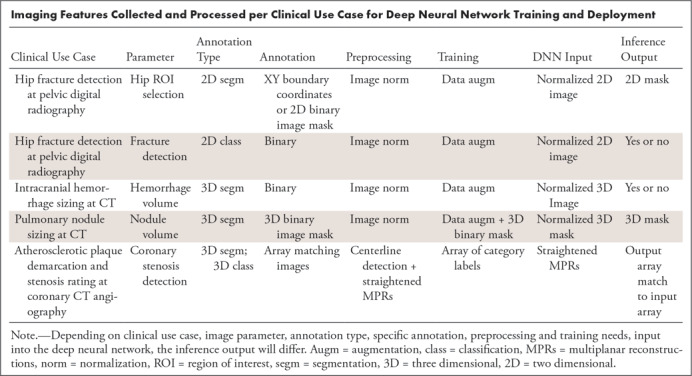
In response to aforementioned curation needs, specific GUI components were designed. For the distinct clinical use-case AI applications described later in this article, combined capabilities were incorporated into three components: two-dimensional segmentation and classification; three-dimensional (3D) segmentation and quantification; and 3D segmentation, quantification, and classification (Table).
Imaging Features Collected and Processed per Clinical Use Case for Deep Neural Network Training and Deployment
Results
During the evaluation period, local-expert subspecialty radiologists contributed directly to component design and were primarily responsible for image annotation.
Hip Segmentation and Proximal Femoral Fracture Classification
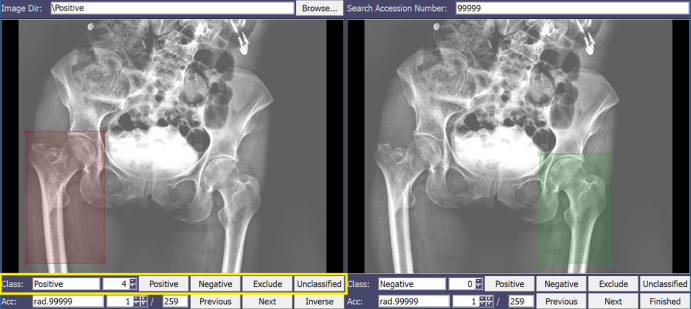
This component was developed for automated segmentation of hips on pelvic digital radiographs for classification of proximal femoral injury as fracture versus nonfracture (Fig 2).
Figure 2:
Graphical user interface (GUI) component for automated segmentation of hips on pelvic digital radiographs used for classification of proximal femoral injury as fracture versus nonfracture. The radiologist localizes the region of interest (ROI) with a bounding box and assigns a corresponding label (eg, fracture presence and severity) by selecting button options. A color code for each label (eg, red for fractured ROI, green for nonfractured ROI) provides visual feedback.
Its design and annotation efforts were led by an American Board of Radiology–certified subspecialty-trained radiologist with 28 years of postresidency experience in musculoskeletal imaging and special interest in acute trauma (J.S.Y).
To annotate images, the radiologist localizes the ROI and labels (fracture or nonfracture). The ROI allows the algorithm to focus on the area of interest and decrease dependence on high image volume. The view panel has visualization functionalities resembling a standard DICOM viewer; the simple GUI design allows the radiologist to quickly apply the ROI and localize it with a bounding box, and assign a corresponding label (eg, fracture and fracture severity, if applicable) by selecting button options. A color code for each label (eg, red for fractured ROI) provides visual feedback assisting the radiologist during annotation. The ROI coordinates, labels, and severity grades are saved in a text file for annotation revisiting. This process is depicted in Movie 1 (supplement).
Movie 1.
A video demonstration of 2D pelvis digital radiograph annotation with GUI. The video demonstrates image selection/review, image manipulation, ROI creation, and ROI labeling processes.
For hip fracture detection, 1050 radiographs were annotated over 7 days, indicating a rate of 150 studies per day and median speed of 10 seconds per study (interquartile range, 3–21 seconds per study).
Intracranial Hemorrhage and Pulmonary Nodule Segmentation and Quantification
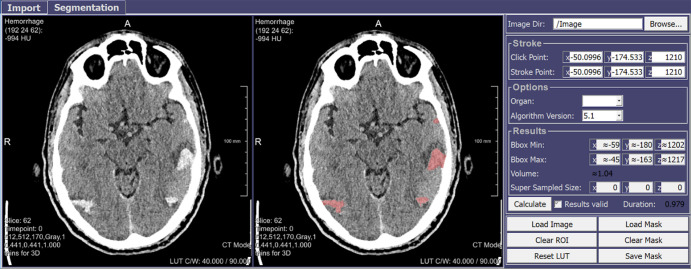
Automatic segmentation and quantification of either an intracranial hemorrhage (Fig 3) or a pulmonary nodule (Fig 4) on non-contrast material–enhanced CT images were incorporated into another component, with related design and annotation efforts supervised (input by subspecialty-trained thoracic imagers) by an American Board of Radiology–certified plus certificate of added qualification neuroradiologist with 14 years of postresidency experience (L.M.P.).
Figure 3:
Graphical user interface (GUI) component for automatic segmentation and quantification of intracranial hemorrhage on non–contrast-enhanced CT images. The radiologist clicks on the hemorrhagic region (left) to be segmented automatically and quantitated (right) by multiple possible parameters (eg, attenuation). Red represents hemorrhage.
Figure 4:
Graphical user interface (GUI) component for automatic segmentation and quantification of a lung nodule on non–contrast-enhanced CT images. A single click by the radiologist allows selection of the solid component of a nodule (red) for automatic segmentation; a line drawn across the nodule allowed the surrounding semisolid or ground-glass component (yellow) to be delineated.
It permits a radiologist to automatically segment an abnormal region using a single regional mouse action. To create a mask, a segmentation algorithm applies filters and detects regional intensity and texture features; masks are collected for DNN training.
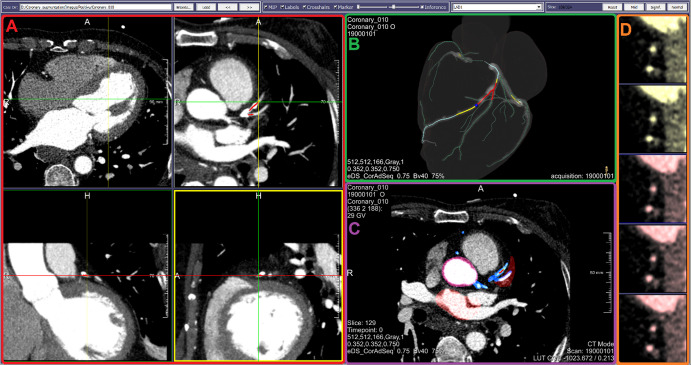
Coronary Atherosclerosis Segmentation and Stenosis Severity Classification
This component was developed to support a supervised DNN for (a) automatic detection versus exclusion of coronary artery atherosclerosis; (b) quantitation of maximum degree of associated luminal narrowing (percentage reduction) if plaque is detected; and (c) classification of quantitated stenosis as hemodynamically significant (eg, ≥50%) versus insignificant. An American Board of Radiology–certified subspecialty-trained radiologist with 35 years of postresidency experience in cardiovascular tomographic imaging (R.D.W.) led component design and annotation efforts.
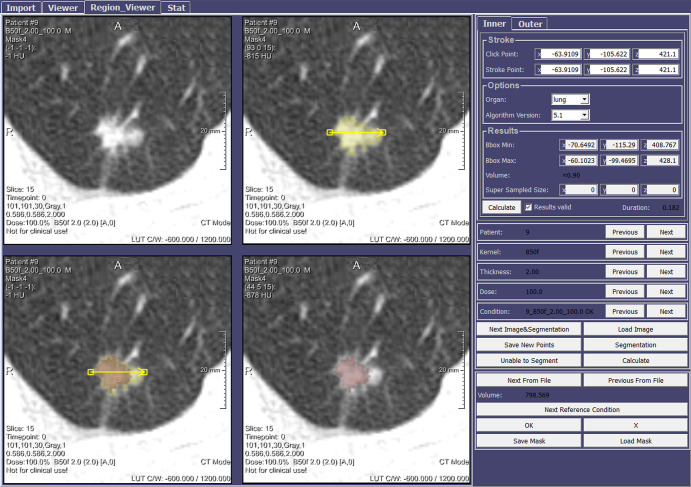
Due to the absence of an image-annotation capability in a commercial coronary CT angiography analysis tool (CT Cardiovascular Engine; Siemens Healthineers, Erlangen, Germany), it was essential to develop an image-annotation capability within this component. To that end, its image-viewing panel was made to resemble the clinical tool by including interactively synchronized modifiable views (eg, sliding orthogonal or multiplanar reconstruction of a 3D coronary CT angiography series (Fig 5). After cardiac-volume extraction from each coronary CT angiographic volume (18), coronary arteries and branches were automatically delineated utilizing work-in-progress proprietary centerline methodology (19).
Figure 5:
Graphical user interface (GUI) component for sequential stepwise image annotation, including automatic detection versus exclusion of coronary artery atherosclerotic plaque; quantitation of maximum coronary luminal narrowing due to plaque; and classification of quantitated stenosis as hemodynamically significant (eg, ≥50%) versus insignificant. The display shown includes, A, multiple two-dimensional orthogonal or oblique multiplanar reconstruction (upper right) images, B, manually rotatable volume-rendered heart model including centerline-extracted three-dimensional branching coronary artery tree with artery enhancement (light blue) indicating manual selection of active vessel, and ball-marker (dark blue) and segment overlay (red) indicating specific level of manually demarcated plaque corresponding to cross-sectional slice in D. C, Axial view of CT scans with overlapped artery masks to monitor the quality segmentation. D, A stacked short-axis image series of active vessel, with manually applied tinting reflecting local presence of atherosclerosis causing significant stenosis (red) versus insignificant stenosis (yellow).
The resulting 3D heart model with maps of the branching coronary artery tree can be manually rotated, allowing a radiologist to examine each artery or branch for plaque with highlighting of the vessel of interest (“active vessel”), and position-indicator tracking along its course confirms the level being viewed or annotated for plaque. To support quantitation and labeling during annotation, the display also includes a cross-sectional series of the active vessel (resembling intravascular US [20]). This true-axial “fly through” view remains perpendicular to the centerline, thereby helping with annotation for plaque presence and assignment of a stenosis-significance grade; severity grading is aided further by simple button clicking prompting appropriate color tinting of images for ease of referencing during subsequent annotations. All operations on each coronary artery or branch (eg, annotation labels) are automatically saved in a text file, thereby enabling future revisiting of the annotation process per case. This process is depicted in Movie 2 (supplement).
Movie 2.
A video demonstration of 3D CCTA annotation with GUI. The video demonstrates available image views, vessel selection, image manipulation and annotation tools; labeling of images and review of previously labeled images is also shown.
A total of 294 coronary CT angiography studies with 1843 arteries and branches were annotated for atherosclerotic plaque over 23 days for a rate of 15.2 studies (80.1 vessels) per day with median speed of 6.08 minutes per study (interquartile range, 2.8–10.6 minutes per study) and 73 seconds per vessel (interquartile range, 20.9–155 seconds per vessel).
Discussion
The curation of sufficient image data for AI algorithm development and evolution supporting medical imaging is a recognized problem (21); it is laborious and time-consuming. Limitations in automated natural language processing and crowdsourcing (3,4) justify local curation involving ground-truth experts. To that end, involvement of radiologists in curation steps, but especially in image annotation, becomes extremely important. However, to date little attention has been given to improving curation and annotation processes to optimize radiologist engagement.
This work addressed progress in responding to image data curation needs, emphasizing specific combinations of capabilities within components of a locally developed GUI. It also focused on interfaces enabling ongoing contributions to AI models, such that once the initial algorithmic modeling is complete, the tools remain useable during clinical trials and/or within daily clinical workflows for ongoing processing and/or inference running. Hence, integration of these tools into routine activities, to foster regular image data curation for planned new DNN formation or refinement of implemented AI algorithms, is a high priority. Such process integration could help overcome issues of inter- and intraobserver variability due to subjectivity between otherwise detached activities (22,23).
For supporting AI use cases in medical imaging, capabilities are combined into three components of the GUI. Benefitting from compatibility with routine image review and analysis capabilities and endorsed by efficient annotation rates and speeds demonstrated in this work, they are currently being considered for clinical implementation.
Project limitations included lack of information on what annotation performance might be without these GUI tools, which were designed in response to important local clinical scenarios (24). For the coronary CT angiographic component, collection of data prior to GUI implementation is not possible as required capabilities are not routinely available on PACS. Moreover, annotation times were created based on time stamps provided by the application, and it is not possible to selectively distinguish between annotation duration and time spent on other activities. The potential negative effect on speed accuracy was minimized by reporting median and interquartile ranges to decrease outlier effects.
In conclusion, a radiologist-friendly GUI as described should facilitate active engagement in important initial and/or ongoing image data curation processes, but especially annotation, which are expected to support concurrent AI-enhanced clinical image interpretations. When blended with other GUI attributes, a continuous workflow integration supporting an agile AI algorithm development life cycle, beginning with image importation and finishing with annotated-image exportation to DNNs, was made possible.
Supported in part by a donation from the Edward J. DeBartolo, Jr Family, Master Research Agreement with Siemens Healthineers, and Master Research Agreement with NVIDIA.
Disclosures of Conflicts of Interest: M.D. Activities related to the present article: institution receives time-limited loaner access by OSU AI Lab to NVIDIA GPUs via Master Research Agreement between Ohio State University and NVIDIA; time-limited access by OSU AI lab to WIP postprocessing software via Master Research Agreement between Ohio State University and Siemens Healthineers. No money transferred; unrestrictive support of OSH AI lab from DeBartolo Family Funds. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. S.C. disclosed no relevant relationships. M.T.B. Activities related to the present article: institution receives time-limited loaner access by OSU AI Lab to NVIDIA GPUs via Master Research Agreement between Ohio State University and NVIDIA, no money transferred; time-limited access by OSU AI lab to WIP postprocessing software via Master Research Agreement between Ohio State University and Siemens Healthineers, no money transferred; unrestrictive support of OSH AI lab from DeBartolo Family Funds. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. S.M.Y. disclosed no relevant relationships. V.G. disclosed no relevant relationships. L.M.P. Activities related to the present article: institution receives time-limited loaner access by OSU AI Lab to NVIDIA GPUs via Master Research Agreement between Ohio State University and NVIDIA, no money transferred; time-limited access by OSU AI lab to WIP postprocessing software via Master Research Agreement between Ohio State University and Siemens Healthineers, no money transferred; unrestrictive support of OSH AI lab from DeBartolo Family Funds. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. R.D.W. Activities related to the present article: institution receives time-limited loaner access by OSU AI Lab to NVIDIA GPUs via Master Research Agreement between Ohio State University and NVIDIA, no money transferred; time-limited access by OSU AI lab to WIP postprocessing software via Master Research Agreement between Ohio State University and Siemens Healthineers, no money transferred; unrestrictive support of OSH AI lab from DeBartolo Family Funds. Activities not related to the present article: institution receives grant from NIH, Co-I on several unrelated R01 active or proposed grants. Other relationships: disclosed no relevant relationships. J.S.Y. disclosed no relevant relationships. R.G. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employed by Siemens Healthcare. Other relationships: disclosed no relevant relationships. M.W. disclosed no relevant relationships. A.W. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: disclosed no relevant relationships. Other relationships: employed by Siemens Healthineers. A.H.H. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employee of NVIDIA. Other relationships: disclosed no relevant relationships. A.I. disclosed no relevant relationships. T.P.O. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employee of Siemens Healthineers; has stock in Siemens Healthineers. Other relationships: disclosed no relevant relationships. B.S.E. Activities related to the present article: institution receives time-limited loaner access by OSU AI Lab to NVIDIA GPUs via Master Research Agreement between Ohio State University and NVIDIA, no money transferred; time-limited access by OSU AI lab to WIP postprocessing software via Master Research Agreement between Ohio State University and Siemens Healthineers, no money transferred; unrestrictive support of OSH AI lab from DeBartolo Family Funds. Activities not related to the present article: institution receives NIH grants; author is co-I on several unrelated NIH grants. Other relationships: disclosed no relevant relationships.
Abbreviations:
- AI
- artificial intelligence
- DICOM
- Digital Imaging and Communications in Medicine
- DNN
- deep neural network
- GUI
- graphical user interface
- PACS
- picture archiving and communication system
- ROI
- region of interest
- 3D
- three dimensional
References
- 1.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 2.Ravi D, Wong C, Deligianni F, et al. Deep learning for health informatics. IEEE J Biomed Health Inform 2017;21(1):4–21. [DOI] [PubMed] [Google Scholar]
- 3.Pons E, Braun LM, Hunink MG, Kors JA. Natural language processing in radiology: A systematic review. Radiology 2016;279(2):329–343. [DOI] [PubMed] [Google Scholar]
- 4.Yank V, Agarwal S, Loftus P, Asch S, Rehkopf D. Crowdsourced health data: Comparability to a US national survey, 2013–2015. Am J Public Health 2017;107(8):1283–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iakovidis D, Smailis C. A semantic model for multimodal data mining in healthcare information systems. Stud Health Technol Inform 2012;180:574–578. [PubMed] [Google Scholar]
- 6.Russell BC, Torralba A, Murphy KP, Freeman WT. LabelMe: A database and web-based tool for image annotation. Int J Comput Vis 2008;77(1-3):157–173. [Google Scholar]
- 7.Syngo.via. Siemens Healthineers site . https://usa.healthcare.siemens.com/medical-imaging-it/advanced-visualization-solutions/syngovia. Accessed December 16, 2018.
- 8.Ritter F, Boskamp T, Homeyer A, et al. Medical image analysis. IEEE Pulse 2011;2(6):60–70. [DOI] [PubMed] [Google Scholar]
- 9.Qt. Qt site. https://www.qt.io. Accessed December 16, 2018.
- 10.Orthanc. Orthanc server site. https://www.orthanc-server.com/. Accessed May 20, 2019.
- 11.Jodogne S. The Orthanc ecosystem for medical imaging. J Digit Imaging 2018;31(3):341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.REST API Tutorial . https://restfulapi.net/. Accessed December 16, 2018.
- 13.Fielding RT. Architectural styles and the design of network-based software architectures. [PhD thesis] University of California, Irvine: 2000. https://www.ics.uci.edu/∼fielding/pubs/dissertation/top.htm. Accessed December 16, 2018. [Google Scholar]
- 14.HHS.gov site . Guidance regarding methods for de-identification of protected health information in accordance with the health insurance portability and accountability act privacy rule. https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html. Accessed December 16, 2018.
- 15.Erickson BJ, Korfiatis P, Kline TL, Akkus Z, Philbrick K, Weston AD. Deep learning in Radiology: Does one size fit all? J Am Coll Radiol 2018;15(3 Pt B):521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hummel RA. Image enhancement by histogram transformation. Comput Graph Image Process 1977;6(2):184–195. [Google Scholar]
- 17.Yosinski Y, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? Appears in Advances in neural information processing systems 27 (NIPS 2014) 2014:3320-3328. arXiv:1411.1792. [preprint] https://arxiv.org/abs/1411.1792. Posted November 6, 2014. Accessed December 16, 2018. [Google Scholar]
- 18.Seo JD. Towards Data Science site. Generating pixelated images from segmentation masks using conditional adversarial networks with interactive code. https://towardsdatascience.com/generating-pixelated-images-from-segmentation-masks-using-conditional-adversarial-networks-with-6479c304ea5f. Accessed December 16, 2018. [Google Scholar]
- 19.Zheng Y, Tek H, Funka-Lea G. Robust and accurate coronary artery centerline extraction in CTA by combining model-driven and data-driven approaches. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. MICCAI 2013. Lecture Notes in Computer Science, Vol 8151. Berlin, Germany: Springer, 2013; 74–81. [DOI] [PubMed] [Google Scholar]
- 20.Schoenhagen P, Nissen S. Understanding coronary artery disease: tomographic imaging with intravascular ultrasound. Heart 2002;88(1):91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang A, Tam R, Cadrin-Chênevert A, et al. Canadian Association of Radiologists white paper on artificial intelligence in Radiology. Can Assoc Radiol J 2018;69(2):120–135. [DOI] [PubMed] [Google Scholar]
- 22.Udupa JK, Leblanc VR, Zhuge Y, et al. A framework for evaluating image segmentation algorithms. Comput Med Imaging Graph 2006;30(2):75–87. [DOI] [PubMed] [Google Scholar]
- 23.Karargyris A, Siegelman J, Tzortzis D, et al. Combination of texture and shape features to detect pulmonary abnormalities in digital chest X-rays. Int J CARS 2016;11(1):99–106. [DOI] [PubMed] [Google Scholar]
- 24.Prevedello LM, Erdal BS, Ryu JL, et al. Automated critical test findings identification and online notification system using artificial intelligence in imaging. Radiology 2017;285(3):923–931. [DOI] [PubMed] [Google Scholar]