Abstract
In recent years, there has been enormous interest in applying artificial intelligence (AI) to radiology. Although some of this interest may have been driven by exaggerated expectations that the technology can outperform radiologists in some tasks, there is a growing body of evidence that illustrates its limitations in medical imaging. The true potential of the technique probably lies somewhere in the middle, and AI will ultimately play a key role in medical imaging in the future. The limitless power of computers makes AI an ideal candidate to provide the standardization, consistency, and dependability needed to support radiologists in their mission to provide excellent patient care. However, important roadblocks currently limit the expansion of this field in medical imaging. This article reviews some of the challenges and potential solutions to advance the field forward, with focus on the experience gained by hosting image-based competitions.
Summary
The article emphasizes two main points that are extremely important to advancements in the field of artificial intelligence in medical imaging: (a) recognition of the current roadblocks and (b) description of ways to overcome these challenges focusing specifically on the role of image-based competitions such as the ones the Radiological Society of North America has been hosting for the past 2 years.
Key Points
■ Activities such as the competitions organized by the Radiological Society of North America may prove to be an important way to address current roadblocks in applying artificial intelligence to medical imaging and to increase the dialogue among radiologists and data scientists, which serves to guide and move the field forward.
■ Although competitions may help move research forward, the field should still rely on standard rigorous scientific methodology to ensure safe and clinically relevant outcomes.
Introduction
Artificial intelligence (AI) is not a new concept, but recent advancements in computing power, data availability, and algorithm performance have given rise to an “AI renaissance,” whereupon AI is being incorporated into nearly every business vertical with the hope to maximize productivity, efficiency, and accuracy. An extraordinary amount of interest in AI followed reports that deep learning algorithms could achieve human-level performance at several specific tasks. Some of the nonmedical applications enabled by artificial neural networks include recognition of cursive handwriting, autonomous vehicles, optical telescope focusing, and so forth (1). Medicine and, more specifically, medical imaging, are particularly suitable to machine learning applications and may see tremendous positive impact in the near future as a result of new research and approaches enabled by this technology. One particular advantage of machine learning is that many of the advancements in the field can be shared across all industry segments. For example, the software frameworks used to create these applications (eg, TensorFlow, Caffe, MXNet, PyTorch, Chainer, and Keras) are typically adaptable to multiple domains and are not specifically designed for medical imaging (2–7). In addition, AI models created for one specific use case such as recognizing objects on standard photographic images (eg, Inception V3 originally created to classify photographic images on the ImageNet database) can be successfully used in other domains such as medical imaging (8–10). Even the weights of some nodes in the network that may have been used to learn basic image features, such as edges and shapes, can be adapted and applied to other seemingly unrelated use cases by using methodologies such as transfer learning or fine-tuning (8,11,12). Despite the similarities in development of AI algorithms, there are many unique considerations when applying AI to medical imaging; before we start discussing ways to advance the field forward, it is important to understand challenges specific to this area. The first part of this article will focus on current challenges related to AI research in medical imaging. The second portion will review some of the potential solutions to the barriers with special attention to the importance of image-based competitions.
Data Heterogeneity and Complexity of Medical Images
Many of the early successes in computer vision were based on photographic images of common objects such as fruits, cars, houses, and so forth. One of the most famous competitions, the ImageNet Large Scale Visual Recognition Challenge, started in 2010 with a training set containing more than 1.2 million Joint Photographic Expert Group (JPEG) photographic color images distributed in 1000 classes. The average image resolution in this collection was relatively small at 482 × 415 pixels (13). Medical images, which are usually stored in Digital Imaging and Communications in Medicine (DICOM) format, are inherently different from the ImageNet dataset. The DICOM standard was created to enable numerous processes in medical imaging and is responsible for a variety of advancements in the field, especially the digitization of radiology. The standard can support varying resolutions and bit-depth allocation. For example, mammographic studies can have resolutions up to 3000 × 4000 pixels (14). While most medical imaging modalities produce gray-scale images, there are some that are displayed and stored as color images, such as PET/CT, Doppler US, and secondary capture objects (eg, advanced visualization images).
The way medical images are acquired differs from photographic images. For example, some imaging studies in medicine (eg, radiographs) may require more than one view to determine three-dimensional position of structures within the body, whereas cross-sectional modalities such as CT and MRI acquire image data in a volumetric fashion. Also, multisequence modalities such as MRI contain multiple image types of the same body part to extract specific characteristics of the imaged tissue. The concept of image comparison, a vital feature in medical imaging used to determine alterations in patients’ health status over time, is not commonly explored in nonmedical applications. Moreover, medical imaging findings are often not specific for a single disease entity and can be identified in a variety of diseases. For example, a focal opacity on a chest radiograph can potentially represent infection, noninfectious inflammation, hemorrhage, scarring from prior trauma, or malignancy, and therefore requires correlation with other data such as comorbidities, symptoms, and laboratory test results. Proof of a diagnosis is often established with pathologic confirmation, yet an imaging finding is frequently acted on without pathologic proof in the instance of pneumonia or thromboembolic disease for example. To complicate evaluation further, there are also anatomic variants that do not have any pathologic implications to the patients but may simulate diseases at imaging.
Variety of Tasks and Endless Clinical Scenarios
Medical imaging is used to address a variety of clinical scenarios, ranging from disease detection to disease surveillance. Each scenario is composed of a series of specific tasks that may be suitable for machine learning, including disease detection (localization and classification), lesion segmentation, and classification. For example, when radiologists are reviewing a chest CT study, pulmonary nodules are detected, characterized, and classified as potentially benign or malignant (15). Other tasks requiring quantification such as the volumetric assessment of anatomic structures (eg, hippocampal volumetric assessment) require tools that can enable segmentation (16). In addition, there are instances in which the goal is calculation of a numerical value rather than to classify an object; an example is bone age assessment based on radiographic images of the hand (17,18). Image-based outcome prediction can also be performed as when trying to determine the likelihood of hematoma expansion on the basis of initial head CT presentation (19). The sheer number of clinical scenarios and the variety of tasks that each of these focused areas can contain is astronomical and clearly impossible to be tackled by one individual or a single organization with existing methodologies.
Challenges Associated with Medical Imaging Data Curation
ImageNet was transformational to computer vision research because it illustrated the importance of data curation in addition to feature and algorithm generation (20). One of the reasons ImageNet was so successful was that multiple individuals contributed to the effort, which resulted in a very large database of image classes (21,22). For the task of labeling standard photographic images containing relatively easily recognized objects, the general population was well-qualified for the task of classification or annotation, and ground-truth could be easily established. However, labeling of medical images requires the expertise of one or more trained radiologists, and labeling of images at a large scale requires reliance on multiple experienced and expensive domain experts, rather than relying on the general public to accomplish this task. Moreover, detection of subtle imaging findings, even by experts, can vary substantially by observer, which can have considerable impact on the final interpretation process. Inter- and intraobserver agreement, even among experts, can be very low for specific clinical conditions (23,24). Furthermore, the degree of specialization required for annotation limits the number of individuals who can contribute to the process, thereby imposing limitations on the ability to “crowdsource” this required information.
Concerns Regarding Patient Privacy
Historically, it has been notoriously difficult to create large public databases of medical images. This has been largely driven by fear of inadvertently exposing protected health information (PHI). This fear is further supported by the fact that DICOM images contain PHI hidden in unpredictable locations within the associated metadata (25,26). In addition, medical images can contain PHI embedded into the pixel data itself (burned-in annotations) or digitized films with handwritten PHI (25). Facial recognition software has been successfully used to reidentify patients from three-dimensional reconstructions of their facial structures, posing risks to confidentiality when these datasets are released to the public (27). In addition, necklaces, wristbands, and other accessories may contain patients’ names or be unique enough to allow patients to be recognized on volumetric images. Therefore, prior to making their datasets publicly available, many organizations manually curate each image for any potential identifiable information. This is both an expensive and labor-intensive process.
Considerations for Algorithm Design and Measures of Performance in Medical Imaging
Although existing programmatic frameworks and libraries to create AI algorithms are shared across many domains, there are unique requirements to consider when creating tools for medical applications. When selecting the appropriate performance metrics for a medical AI algorithm, attention needs to be given to potential clinical implications, and the measures should be carefully chosen to ensure that performance reflects the clinical question. For example, an algorithm built to expedite decision making when using coronary CT angiography in patients suspected of having myocardial infarction may have disastrous consequences if the false-negative rate is not designed to be considerably low. Because positive cases are less common than normal cases in this setting, the unbalanced nature of the data makes accuracy and negative predictive value poor metrics to assess algorithm performance. Recall and area under the receiver operating characteristic curve would better reflect performance in this case. The scope of the algorithm is also an important part of design. A narrow algorithm designed for the single specific task of recognizing pneumonia on a chest radiograph cannot be used to independently interpret these examination findings despite high performance because it would fail to recognize other potentially equally important findings such as pneumoperitoneum with potentially tragic consequences.
Lack of Algorithm Transparency and Issues with Validation and Testing
Another issue with existing mechanisms of algorithm design relates to lack of transparency in the underlying methodologies employed to create them and the difficulties associated with clinical implementation. Testing reproducibility of a proprietary algorithm can already be very difficult in one single site; expanding the evaluation to other sites and different datasets can be extremely complex. This is especially true because machine learning applications do not always follow the same pipeline from data ingestion to output, and no standardization of the process exists. For example, algorithms with similar performance may have very different approaches to solve the same problem and may require unique preprocessing methodologies prior to inference. Therefore, each application may require its own server or virtual environment, making scalability difficult. One solution to implementation challenges is the creation of separate application containers in which multiple isolated applications or services can run on a single host and access the same operating system kernel. However, even in this scenario, the work required to integrate these applications with local clinical systems may be quite challenging if the right infrastructure is not in place.
Machine learning algorithms are created with the assumption that they will be applied to datasets with identical characteristics and probability distributions. This property is known as the generalizability of the algorithm. However, there is the practical reality that the patient population, the image acquisition devices, and image protocols can vary greatly between institutions, and therefore, the transferability (ability to transfer performance to data containing different probability distributions) of an algorithm may prove difficult even when the algorithm performance is excellent on data from a single source. Unfortunately, no statistical method exists to test transferability of an algorithm except for testing the algorithm on the new data at disparate locations.
Knowledge Dissemination Is Prolonged with Traditional Hypothesis-driven Research
It has long been recognized that there is a large temporal gap between the time that knowledge discoveries happen in the medical research setting relative to when they are clinically implemented. Prior work suggests that this gap may be as long as 17 years in the public health sector (28). One contributing factor to prolonged knowledge discovery and dissemination of the results is that traditional research methodologies follow a serial rather than a parallel path. The traditional research process is hypothesis-driven whereby a researcher or group of researchers in an organization formulate a research question that they set out to prove or disprove. Next, they request approval from their institutional review board to conduct the research in their own organization. Once approval is obtained, the research is conducted, and research findings are published in a journal not necessarily accessible to the entire community. Once a research group from another organization becomes aware of those findings, they will try to validate them and publish their own experience (Figure, A). The process can be repeated multiple times until some degree of confidence is reached that the initial findings are generalizable. While this activity is extremely important and has been the cornerstone of the scientific advancements in many decades, it could potentially benefit from a more decentralized approach of initial knowledge discovery, such as image analysis competitions. In this case, multiple individuals or groups can compete for the best solution to a specific problem and openly share their new findings, methods, and approaches, which can later be further tested with standard scientific rigor.
Figure:
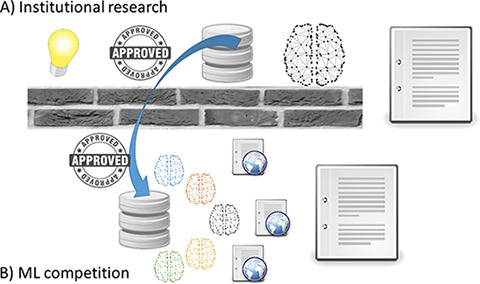
Diagram demonstrates the differences between, A, traditional institutional research and, B, the model of research promoted by image-based machine learning competitions. While traditional model follows a serial sequence of events (hypothesis formulation, institutional review board approval, data acquisition, knowledge discovery, and manuscript submission), image competitions promote a parallel approach to knowledge discovery and dissemination in which multiple participants of varying disciplines find solutions to the same problem and openly share their discoveries on the web or manuscripts in a short time period.
Potential Solutions to Current Challenges and the Role of Image Competitions in Fostering AI Research
AI holds extremely exciting opportunities for medical imaging, but several of the aforementioned challenges related to data complexity, data access and curation, concern for patient privacy, transferability of algorithms to the mass market, as well as integration of these tools into the clinical workflow, need to be addressed effectively to bring this technology to the forefront where it can augment patient care.
In the past 2 years, the Radiological Society of North America (RSNA) has hosted several public AI competitions to promote AI research in radiology. In 2017, the goal of the competition was prediction of the age of pediatric patients based on hand radiographs by using machine learning (29). In 2018, the competition focused on pneumonia detection (localization and classification) (30). In the following sections, we offer some of the lessons learned, which address some of the aforementioned idiosyncratic challenges when applying machine learning to medical imaging.
Addressing Data Complexity, Data Access, and a Variety of Clinical Scenarios While Ensuring Patient Privacy
The advent of deep learning has invigorated research in neural networks and imaging informatics in general. Recognizing that these techniques rely heavily on large datasets, several organizations started to focus on creating medical imaging databases to advance AI research for a variety of clinical scenarios. The fear of exposing patient health information has been at least partially mitigated by converting DICOM files into other formats such as JPEG, Neuroimaging Informatics Technology Initiative, or NIfTI, and Portable Network Graphics (PNG), ensuring the metadata will not be exposed. This has been the strategy of some of the publicly released medical imaging datasets. For example, the database used for the first RSNA Machine Learning Challenge converted radiographs of the hand in pediatric patients obtained from Stanford University and University of Colorado to JPEG format. The second competition for pneumonia detection leveraged an existing public dataset by creating new annotations for a subset of the 112 000 radiographs of the chest from the National Institutes of Health Clinical Center (NIHCC) (31). In this case, DICOM images were initially converted to PNG prior to release to the public for complete removal of the associated metadata by the NIHCC group. To allow participants of the competition to familiarize themselves with the medical imaging standard and to facilitate annotation, the PNG images were converted back to DICOM format. This process ensured complete removal of any patient identifiable information while maintaining the DICOM format for the competition.
Multiple additional databases of medical images have become available, including 30 000 CT imaging studies of multiple lesions in the chest, abdomen, and pelvis recently released by the NIHCC (32). The Cancer Imaging Archive (TCIA) has a large collection of anonymized DICOM images of various cancer types donated from numerous organizations and is cross correlated to the tissue and genomic bank of The Cancer Genome Atlas. The anonymization process employed by the TCIA is quite rigorous to ensure patient privacy (33). Recently, a dataset search engine has become available (34), listing datasets of multiple organized databases and prior competitions, potentially improving discoverability of existing data sources.
A newer strategy to protect patient privacy is to retain the datasets within each organization firewall and only move the algorithms or trained parameters to each site. The intent is to expedite model validation when applied to different settings or when a single institution dataset is small (35).
Several additional competitions have already been conducted in the medical imaging domain, many of them presented at https://grand-challenge.org. These competitions cover a variety of clinical scenarios and multiple machine learning tasks. An extensive review of prior machine learning competitions in medical imaging has been provided by Maier-Hein et al with 150 competitions being reported up to 2016 (36). Most of the competitions focused on segmentation tasks (70%), and the most common imaging modality was MRI (62%). The reported number of training cases in these competitions has been relatively low, with a median of 15 cases and interquartile range varying between seven and 30 cases. The maximal number of cases reported was 32 468 (36). More recently, one of the competitions that received considerable attention was the 2017 Data Science Bowl challenge in which participants were asked to develop algorithms that attempted to predict cancerous lung lesions. The prize money for this competition was 1 million U.S. dollars, and as a result, it generated wide interest from individuals and organizations across the world, with 1972 teams joining the competition and multiple open-source models and modeling insights shared publicly on the web (37). While a cash prize can provide substantial financial incentive, many participants engage in the competitions for other reasons, including increased notoriety in the data science community, the excitement of collaboration and competition, and the long-term benefit of contributing to the public good. For example, the RSNA 2018 Machine Learning Challenge provided a conservative financial reward ($30 000), yet it had 360 teams join the competition just in the initial 4 days of its launch and 1399 teams during the total time of the competition.
Collaborative and Distributed Data Curation
The 2018 RSNA Machine Learning Challenge employed a collaborative and distributed process for data annotation. The curation process started with creating consensus definitions of what would be considered positive cases and a method for how they would be annotated. The more explicit the definitions are, the more consistent the labels become. The methodology employed in the data curation process is described in detail in Shih et al (38). Despite extensive effort to maintain consistency and uniformity of the labeling process, the annotated datasets showed a skewed distribution of annotation frequencies across users. Because the cases assigned to annotators were randomly selected, it is likely that some degree of interrater variability existed, further emphasizing the challenges related to data curation in medical imaging.
Automated tools such as natural language processing have also been used on radiology reports to curate large imaging datasets (31,32,39). Recent reports show promising results in these methodologies to label image datasets created for classification tasks and seem to indicate that imperfections of the natural language processing system can be counterbalanced by increasing the number of images in the training set (40). This is an important field of study because it would essentially minimize the need for expensive and time-consuming data curation in some scenarios; however, it would not replace the process of designating the location of an abnormality.
AI Research Fosters Collaboration and Community Building through Competition
One of the advantages of machine learning competitions over traditional hypothesis-driven research relates to the innate differences in the approach to problem solving. Data competitions by nature encourage multiple participants or groups to simultaneously address a specific problem independently and concurrently. This fosters rapid development of many unique solutions. While there is a competitive aspect in creating the best performing algorithm, there is often a simultaneous collaborative experience brokered through social media and sponsoring site forums. Visibility of individual algorithm performance is a principle prerequisite of any data competition. This is accomplished through posting algorithm performance of competitors on a public leaderboard such that each participant understands his or her relative performance. During the 2017 RSNA bone age competition, the participants gained additional knowledge to develop and enhance their algorithms through shared experiences from participants and the sponsors. The lessons learned from the competition organizers culminated in an article describing the process, as well as a detailed description of the top-five winning solutions (41). In the 2018 RSNA challenge, many of the solutions were openly described and posted on the discussion forums (30). The top 10 contributions will be shared with the community as open-source systems (training code), so people can learn from them and test performance of the algorithms with the standard competition dataset or by using their own local databases to test transferability. Another recent trend in machine learning publications has been the willingness of the authors to publish their work openly on forums, blogs, websites, or open manuscript platforms such as arXiv.org to share their work with the community and gather continuous feedback. The educational value of open-source codes, discussions forums, and open publications is immense for participants of the competition and other interested parties in the rapidly evolving field of machine learning.
Expediting Knowledge Discovery and Dissemination
While traditional hypothesis-driven research follows a serial path employing a single investigator or group, competitions promote a parallel process of iterative knowledge discovery and dissemination. There is also cross-pollination among different specialties and domains because of the open nature of the research. Once the competition opens and a dataset is made publicly available, multiple individuals from varied backgrounds and disciplines work toward solving the problem. In a relatively short time, numerous solutions to the same problem are presented and discussed openly. The 2017 RSNA Bone Age Challenge illustrated how a parallel approach can accelerate discovery; in less than 3 months, more than 10 solutions surpassed performance of previously published state-of-the-art algorithms. Issues identified with the datasets including the methodologies employed to create them were also openly discussed to determine their limitations. Solutions to the problems were presented and shared back with the community in the form of web posts or manuscripts (Figure, B). Researchers around the world continually benefit from the public dissemination of the datasets used in former and current image-based competitions.
Caveats and Limitations of Machine Learning Competitions Compared with Traditional Approaches
Although competitions may be an excellent catalyzer to foster collaborative research in machine learning for medical imaging, it does not replace standard hypothesis-driven peer-reviewed publications with rigorous scientific scrutiny to ensure validity, generalizability, and transferability of the results. Recent publications have exposed several weaknesses of competitions, including how inconsistencies in their methodologies may affect reproduction, interpretation, and cross-comparison of the results (36). In addition, the rank order of the winning algorithms is sensitive to several variables related to design choices, including how the test set was created and the methodology employed to assess algorithm performance (36). Moreover, competitions with flawed design may allow competitors to “game the system” and achieve higher performance by submitting only selective or “easy cases” if the performance metric is not designed to penalize missing values (42). These problems emphasize the need for competitions to follow a standard or at least adhere to best practices as promoted by groups with extensive expertise in the subject (36,42).
In conclusion, medical imaging machine learning research is a new paradigm for traditional radiology researchers. There are new processes, limitations, and challenges that need to be learned and managed when performing state-of-art data science analysis. Readily available, well-curated, and labeled data of high quality is paramount to performing effective research in this area. The radiology community, as stewards of this imaging data, needs to remain cognizant of our patients’ privacy concerns tempered by the need for large volumes of high-quality data. Activities such as the competitions organized by RSNA may prove to be an important activity to collaboratively address these problems by facilitating dialogue between radiologists and data scientists, which serves to help guide and move the field forward while relying on standard rigorous scientific methodology to ensure safe and clinically relevant outcomes.
Disclosures of Conflicts of Interest: L.M.P. disclosed no relevant relationships. S.S.H. disclosed no relevant relationships. G.S. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: board member and shareholder of MD.ai. Other relationships: disclosed no relevant relationships. C.C.W. disclosed no relevant relationships. M.D.K. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: received travel reimbursement for board meetings from SIIM; consultant for Medical Sciences Corporation, consultant for National Library of Medicine; received ACR Innovation grant for development of LI-RADS integration with dictation products; gave one paid lecture at Gilead campus to staff regarding big data, machine learning, and imaging; received funds for travel for committee meetings from RSNA and SIIM. Other relationships: disclosed no relevant relationships. F.H.C. disclosed no relevant relationships. B.J.E. disclosed no relevant relationships. J.K. Activities related to the present article: supported by the following NIH grants U01CA154601 (NIH/NCI), U24CA180927 (NIH/NCI), U24CA180918 (NIH/NCI); supported by contract from Leidos. Activities not related to the present article: consultant for INFOTECH, Soft. Other relationships: disclosed no relevant relationships. K.P.A. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: consultant for McKinsey & Company; on RSNA Radiology Informatics Committee and Machine Learning and Standards Subcommittees; Senior Scientist for education at ACR Data Science Institute; Director of Research Strategy and Operations at the MGH and BWH Center for Clinical Data Science, which is funded in part by monies and resources from NVIDIA, GE, and Nuance; associate editor for Radiology: Artificial Intelligence and Journal of Medical Imaging. Other relationships: disclosed no relevant relationships. A.E.F. disclosed no relevant relationships.
Abbreviations:
- AI
- artificial intelligence
- DICOM
- Digital Imaging and Communications in Medicine
- JPEG
- Joint Photographic Expert Group
- NIHCC
- National Institutes of Health Clinical Center
- PHI
- protected health information
- PNG
- Portable Network Graphics
- RSNA
- Radiological Society of North America
- TCIA
- The Cancer Imaging Archive
References
- 1.Widrow B, Rumelhart DE, Lehr MA. Neural networks: applications in industry, business and science. Commun ACM 1994;37(3):93–105. [Google Scholar]
- 2.TensorFlow . TensorFlow. https://www.tensorflow.org/. Accessed August 26, 2018.
- 3.Caffe Deep Learning Framework . http://caffe.berkeleyvision.org/. Accessed August 26, 2018.
- 4.MXNet . https://mxnet.apache.org/. Accessed August 26, 2018.
- 5.PyTorch . https://pytorch.org/. Accessed August 26, 2018.
- 6.Chainer: A flexible framework for neural networks . https://chainer.org/. Accessed December 4, 2018.
- 7.Home: Keras documentation . https://keras.io/. Accessed December 4, 2018.
- 8.Kim DH, MacKinnon T. Artificial intelligence in fracture detection: transfer learning from deep convolutional neural networks. Clin Radiol 2018;73(5):439–445. [DOI] [PubMed] [Google Scholar]
- 9.Cheng PM, Tejura TK, Tran KN, Whang G. Detection of high-grade small bowel obstruction on conventional radiography with convolutional neural networks. Abdom Radiol (NY) 2018;43(5):1120–1127. [DOI] [PubMed] [Google Scholar]
- 10.Huh M, Agrawal P, Efros AA. What makes ImageNet good for transfer learning? arXiv:160808614 [cs]. 2016.[preprint] http://arxiv.org/abs/1608.08614. Posted August 30, 2016. Revised December 10, 2016. Accessed August 26, 2018.
- 11.Ribeiro E, Uhl A, Wimmer G, Häfner M. Exploring deep learning and transfer learning for colonic polyp classification. Comput Math Methods Med 2016;2016:6584725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tajbakhsh N, Shin JY, Gurudu SR, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 2016;35(5):1299–1312. [DOI] [PubMed] [Google Scholar]
- 13.ImageNet Large Scale Visual Recognition Competition 2014 (ILSVRC2014) . http://www.image-net.org/challenges/LSVRC/2014/. Accessed December 4, 2018.
- 14.Huda W, Abrahams RB. X-ray-based medical imaging and resolution. AJR Am J Roentgenol 2015;204(4):W393–W397. [DOI] [PubMed] [Google Scholar]
- 15.Dhara AK, Mukhopadhyay S, Dutta A, Garg M, Khandelwal N. Content-based image retrieval system for pulmonary nodules: assisting radiologists in self-learning and diagnosis of lung cancer. J Digit Imaging 2017;30(1):63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim HK, Hong SC, Jung WS, et al. Automated segmentation of hippocampal subfields in drug-naïve patients with Alzheimer disease. AJNR Am J Neuroradiol 2013;34(4):747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology 2018;287(1):313–322. [DOI] [PubMed] [Google Scholar]
- 18.Ren X, Li T, Yang X, et al. Regression convolutional neural network for automated pediatric bone age assessment from hand radiograph. IEEE J Biomed Health Inform . doi:10.1109/JBHI.2018.2876916. Published online October 19, 2018 . [DOI] [PubMed] [Google Scholar]
- 19.Shen Q, Shan Y, Hu Z, et al. Quantitative parameters of CT texture analysis as potential markersfor early prediction of spontaneous intracranial hemorrhage enlargement. Eur Radiol 2018;28(10):4389–4396. [DOI] [PubMed] [Google Scholar]
- 20.Gershgorn D. The data that transformed AI research—and possibly the world. Quartz. https://qz.com/1034972/the-data-that-changed-the-direction-of-ai-research-and-possibly-the-world/. Accessed August 31, 2018.
- 21.Deng J, Dong W, Socher R, Li LJ, Li K, Li FF. ImageNet: a large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition, 2009; 248–255. [Google Scholar]
- 22.Russakovsky O, Deng J, Su H, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis 2015;115(3):211–252. [Google Scholar]
- 23.Winklhofer S, Held U, Burgstaller JM, et al. Degenerative lumbar spinal canal stenosis: intra- and inter-reader agreement for magnetic resonance imaging parameters. Eur Spine J 2017;26(2):353–361. [DOI] [PubMed] [Google Scholar]
- 24.Hoang JK, Middleton WD, Farjat AE, et al. Interobserver variability of sonographic features used in the American College of Radiology Thyroid Imaging Reporting and Data System. AJR Am J Roentgenol 2018;211(1):162–167. [DOI] [PubMed] [Google Scholar]
- 25.Monteiro E, Costa C, Oliveira JLA. A de-identification pipeline for ultrasound medical images in DICOM format. J Med Syst 2017;41(5):89. [DOI] [PubMed] [Google Scholar]
- 26.Freymann JB, Kirby JS, Perry JH, Clunie DA, Jaffe CC. Image data sharing for biomedical research--meeting HIPAA requirements for De-identification. J Digit Imaging 2012;25(1):14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazura JC, Juluru K, Chen JJ, Morgan TA, John M, Siegel EL. Facial recognition software success rates for the identification of 3D surface reconstructed facial images: implications for patient privacy and security. J Digit Imaging 2012;25(3):347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brownson RC, Kreuter MW, Arrington BA, True WR. Translating scientific discoveries into public health action: how can schools of public health move us forward? Public Health Rep 2006;121(1):97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.RSNA Pediatric Bone Age Challenge . http://rsnachallenges.cloudapp.net/competitions/4. Accessed August 27, 2018.
- 30.RSNA Pneumonia Detection Challenge . https://www.kaggle.com/c/rsna-pneumonia-detection-challenge. Accessed December 6, 2018.
- 31.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. arXiv:170502315 [cs]. 2017.[preprint] http://arxiv.org/abs/1705.02315. Posted May 5, 2017. Revised December 14, 2017. Accessed August 31, 2018.
- 32.Yan K, Wang X, Lu L, Summers RM. DeepLesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J Med Imaging (Bellingham) 2018;5(3):036501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dataset search . https://toolbox.google.com/datasetsearch. Accessed September 7, 2018.
- 35.Chang K, Balachandar N, Lam C, et al. Distributed deep learning networks among institutions for medical imaging. J Am Med Inform Assoc 2018;25(8):945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier-Hein L, Eisenmann M, Reinke A, et al. Is the winner really the best? a critical analysis of common research practice in biomedical image analysis competitions. arXiv:180602051 [cs]. 2018. [preprint] http://arxiv.org/abs/1806.02051. Posted June 6, 2018. Accessed December 4, 2018.
- 37.Data Science Bowl . 2017. https://www.kaggle.com/c/data-science-bowl-2017. Accessed September 1, 2018.
- 38.Shih G, Wu CC, Halabi SS, et al. Augmenting the National Institutes of Health chest radiograph dataset with expert annotations of possible pneumonia. Radiol Artif Intell 2019; 1(1): e180041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajpurkar P, Irvin J, Ball RL, et al. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 2018;15(11):e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rolnick D, Veit A, Belongie S, Shavit N. Deep learning is robust to massive label noise. arXiv:170510694 [cs]. 2017. [preprint] http://arxiv.org/abs/1705.10694. Posted May 30, 2017. Revised February 26, 2018. Accessed December 4, 2018. [Google Scholar]
- 41.Halabi SS, Prevedello LM, Kalpathy-Cramer J, et al. The RSNA Pediatric Bone Age Machine Learning Challenge. Radiology 10.1148/radiol.2018180736. Published online November 27, 2018. [DOI] [PMC free article] [PubMed]
- 42.Reinke A, Eisenmann M, Onogur S, et al. How to exploit weaknesses in biomedical challenge design and organization. In: Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C, Fichtinger G, eds. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Cham, Switzerland: Springer International Publishing, 2018; 388–395. Accessed December 4, 2018. [Google Scholar]


