The advent of deep neural networks as a new artificial intelligence (AI) technique has engendered a large number of medical applications, particularly in medical imaging. Such applications of AI must remain grounded in the fundamental tenets of science and scientific publication (1). Scientific results must be reproducible, and a scientific publication must describe the authors’ work in sufficient detail to enable readers to determine the rigor, quality, and generalizability of the work, and potentially to reproduce the work’s results. A number of valuable manuscript checklists have come into widespread use, including the Standards for Reporting of Diagnostic Accuracy Studies (STARD) (2–5), Strengthening the Reporting of Observational studies in Epidemiology (STROBE) (6), and Consolidated Standards of Reporting Trials (CONSORT) (7,8). A radiomics quality score has been proposed to assess the quality of radiomics studies (9).
Peer-reviewed medical journals have an opportunity to connect innovations in AI to clinical practice through rigorous validation (10). Various guidelines for reporting evaluation of machine learning models have been proposed (11–14). We have sought to codify these into a checklist in a format concordant with the EQUATOR Network guidelines (15,16) that also incorporates general manuscript review criteria (17,18).
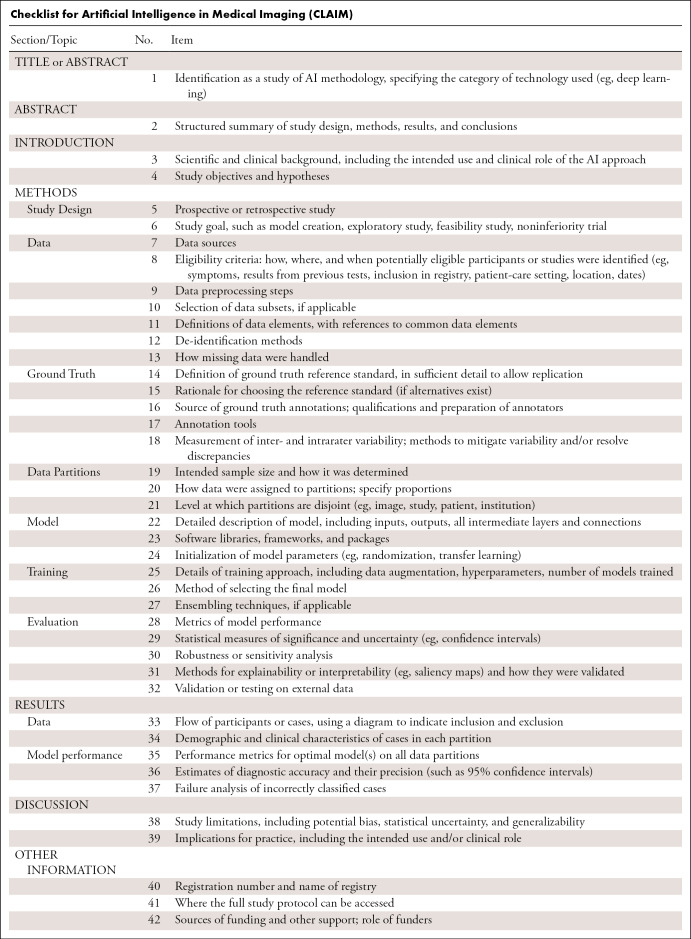
To aid authors and reviewers of AI manuscripts in medical imaging, we propose CLAIM, the Checklist for AI in Medical Imaging (see Table and downloadable Word document [supplement]). CLAIM is modeled after the STARD guideline and has been extended to address applications of AI in medical imaging that include classification, image reconstruction, text analysis, and workflow optimization. The elements described here should be viewed as a “best practice” to guide authors in presenting their research. The text below amplifies the checklist with greater detail.
Checklist for Artificial Intelligence in Medical Imaging (CLAIM)
Manuscript Title and Abstract
Item 1. Indicate the use of the AI techniques—such as “deep learning” or “random forests”—in the article’s title and/or abstract; use judgment regarding the level of specificity.
Item 2. The abstract should present a structured summary of the study’s design, methods, results, and conclusions; it should be understandable without reading the entire manuscript. Provide an overview of the study population (number of patients or examinations, number of images, age and sex distribution). Indicate if the study is prospective or retrospective, and summarize the statistical analysis that was performed. When presenting the results, be sure to include P values for any comparisons. Indicate whether the software, data, and/or resulting model are available publicly.
The Introduction
Item 3. Address an important clinical, scientific, or operational issue. Describe the study’s rationale, goals, and anticipated impact. Summarize related literature and highlight how the investigation builds upon and differs from that work. Guide readers to understand the context for the study, the underlying science, the assumptions underlying the methodology, and the nuances of the study.
Item 4. Define clearly the clinical or scientific question to be answered; avoid vague statements or descriptions of a process. Limit the chance of post hoc data dredging by specifying the study’s hypothesis a priori. Identify a compelling problem to address. The study’s objectives and hypothesis will guide sample size calculations and whether the hypothesis will be supported or not.
The Methods Section
Describe the study’s methodology in a sufficiently clear and complete manner to enable readers to reproduce the steps described. If a thorough description exceeds the journal’s word limits, summarize the work in the Methods section and provide full details in a supplement.
Study Design
Item 5. Indicate if the study is retrospective or prospective. Evaluate predictive models in a prospective setting, if possible.
Item 6. Define the study’s goal, such as model creation, exploratory study, feasibility study, or noninferiority trial. For classification systems, state the intended use, such as diagnosis, screening, staging, monitoring, surveillance, prediction, or prognosis (2). Indicate the proposed role of the AI algorithm relative to other approaches, such as triage, replacement, or add-on (2). Describe the type of predictive modeling to be performed, the target of predictions, and how it will solve the clinical or scientific question.
Data
Item 7. State the source of data and indicate how well the data match the intended use of the model. Describe the targeted application of the predictive model to allow readers to interpret the implications of reported accuracy estimates. Reference any previous studies that used the same dataset and specify how the current study differs. Adhere to ethical guidelines to assure that the study is conducted appropriately; describe the ethics review and informed consent (19). Provide links to data sources and/or images, if available. Authors are strongly encouraged to deposit data and/or software used for modeling or data analysis in a publicly accessible repository.
Item 8. Define how, where, and when potentially eligible participants or studies were identified. Specify inclusion and exclusion criteria such as location, dates, patient-care setting, symptoms, results from previous tests, or registry inclusion. Indicate whether a consecutive, random, or convenience series was selected. Specify the number of patients, studies, reports, and/or images.
Item 9. Preprocessing converts raw data from various sources into a well-defined, machine-readable format for analysis (20,21). Describe preprocessing steps fully and in sufficient detail so that other investigators could reproduce them. Specify the use of normalization, resampling of image size, change in bit depth, and/or adjustment of window/level settings. State whether or not the data have been rescaled, threshold-limited (“binarized”), and/or standardized. Specify how the following issues were handled: regional format, manual input, inconsistent data, missing data, wrong data types, file manipulations, and missing anonymization. Define any criteria to remove outliers (11). Specify the libraries, software (including manufacturer name and location), and version numbers, and all option and configuration settings employed.
Item 10. In some studies, investigators select subsets of the raw extracted data as a preprocessing step, for instance, selecting a subset of the images, cropping down to a portion of an image, or extracting a portion of a report. If this process is automated, describe the tools and parameters used; if done manually, specify the training of the personnel and the criteria they used. Justify how this manual step would be accommodated in the context of the clinical or scientific problem to be solved.
Item 11. Define the predictor and outcome variables. Map them to common data elements, if applicable, such as those maintained by the radiology community (22–24) or the U.S. National Institutes of Health (25,26).
Item 12. Describe the methods by which data have been de-identified and how protected health information has been removed to meet U.S. (HIPAA), European (GDPR), or other relevant laws. Because facial profiles can allow identification, specify the means by which such information has been removed or made unidentifiable (20).
Item 13. State clearly how missing data were handled, such as replacing them with approximate or predicted values. Describe the biases that the imputed data might introduce.
Ground Truth
Item 14. Include detailed, specific definitions of the ground truth annotations, ideally referencing common data elements. Avoid vague descriptions such as “size of liver lesion;” use more precise definitions, such as “greatest linear measurement in millimeters passing entirely through the lesion as measured on axial contrast-enhanced CT images of 2.5-mm thickness.” Provide an atlas of examples to annotators to illustrate subjective grading schemes (eg, mild/moderate/severe), and make that information available for review.
Item 15. Describe the rationale for the choice of the reference standard and the potential errors, biases, and limitations of that reference standard.
Item 16. Specify the number of human annotators and their qualifications. Describe the instructions and training given to annotators; include training materials as a supplement, if possible. Describe whether annotations were done independently and how any discrepancies among annotators were resolved.
Item 17. Specify the software used for manual, semiautomated, or automated annotation, including the version number. Describe if and how imaging labels were extracted from free-text imaging reports or electronic health records using natural language processing or recurrent neural networks (20,27,28).
Item 18. Describe the methods to measure inter- and intrarater variability, and any steps taken to reduce or mitigate this variability and/or resolve discrepancies.
Data Partitions
Item 19. Describe the sample size and how it was determined. Use traditional power calculation methods, if applicable, to estimate the required sample size to allow for generalizability in a larger population and how many cases are needed to show an effect (29).
Item 20. Specify how the data were assigned into training, validation (“tuning”), and testing partitions; indicate the proportion of data in each partition and justify that selection. Indicate if there are any systematic differences between the data in each partition, and if so, why.
Item 21. Describe the level at which the partitions are disjoint. Sets of medical images generally should be disjoint at the patient level or higher so that images of the same patient do not appear in each partition.
Model
Item 22. Provide a complete and detailed structure of the model, including inputs, outputs, and all intermediate layers, in sufficient detail that another investigator could exactly reconstruct the network. For neural network models, include all details of pooling, normalization, regularization, and activation in the layer descriptions. Model inputs must match the form of the preprocessed data. Model outputs must correspond to the requirements of the stated clinical problem, and for supervised learning should match the form of the ground truth annotations. If a previously published model architecture is employed, cite a reference that meets the preceding standards and fully describe every modification made to the model. In some cases, it may be more convenient to provide the structure of the model in code as supplemental data.
Item 23. Specify the names and version numbers of all software libraries, frameworks, and packages. Avoid detailed description of hardware unless benchmarking computational performance is a focus of the work.
Item 24. Indicate how the parameters of the model were initialized. Describe the distribution from which random values were drawn for randomly initialized parameters. Specify the source of the starting weights if transfer learning is employed to initialize parameters. When there is a combination of random initialization and transfer learning, make it clear which portions of the model were initialized with which strategies.
Training
Item 25. Completely describe all of the training procedures and hyperparameters in sufficient detail that another investigator could exactly duplicate the training process. Typically, to fully document training, a manuscript would: Describe how training data were augmented (eg, for images the types and ranges of transformations). State how convergence of training of each model was monitored and what the criteria for stopping training were. Indicate the values that were used for every hyperparameter, which of these were varied between models, over what range, and using what search strategy. For neural networks, descriptions of hyperparameters should include at least learning rate schedule, optimization algorithm, minibatch size, dropout rates (if any), and regularization parameters (if any). Discuss what objective function was employed, why it was selected, and to what extent it matches the performance required for the clinical or scientific use case. Define criteria used to select the best-performing model. If some model parameters are frozen or restricted from modification, as is often the case in transfer learning, clearly indicate which parameters are involved, the method by which they are restricted, and the portion of the training for which the restriction applies. It may be more concise to describe these details in code in the form of a succinct training script, particularly for neural network models when using a standard framework.
Item 26. Describe the method and performance parameters used to select the best-performing model among all the models trained for evaluation against the held-out test set. If more than one model is selected, justify why this is appropriate.
Item 27. If the final algorithm involves an ensemble of models, describe each model comprising the ensemble in complete detail in accordance with the preceding recommendations. Indicate how the outputs of the component models are weighted and/or combined.
Evaluation
Item 28. Describe the metric(s) used to measure the model’s performance and indicate how they address the performance characteristics most important to the clinical or scientific problem. Compare the presented model to previously published models.
Item 29. Indicate the uncertainty of the performance metrics’ values, such as with standard deviation and/or confidence intervals. Compute appropriate tests of statistical significance to compare metrics. Specify the statistical software.
Item 30. Analyze the robustness or sensitivity of the model to various assumptions or initial conditions.
Item 31. If applied, describe the methods that allow one to explain or interpret the model’s results and provide the parameters used to generate them (14). Describe how any such methods were validated in the current study.
Item 32. Describe the data used to evaluate performance of the completed algorithm. When these data are not drawn from a different data source than the training data, note and justify this limitation. If there are differences in structure of annotations or data between the training set and evaluation set, explain the differences, and describe and justify the approach taken to accommodate the differences.
The Results Section
Present the outcomes of the experiment in sufficient detail. If the description of the results would exceed the word count or other journal requirements, the data can be offered in a supplement to the manuscript.
Data
Item 33. Specify the criteria to include and exclude patients or examinations or pieces of information and document the numbers of cases that met each criterion. We strongly recommend including a flowchart/diagram in your results to show initial patient population and those excluded for any reason. Describe the summary of the technical characteristics of the dataset. For example, for images: modality vendors/models, acquisition parameters, reformat parameters; for reports: practice setting, number and training of report authors, extent of structured reporting.
Item 34. Demographic and clinical characteristics of cases in each partition should be specified. State the performance metrics on all data partitions.
Model Performance
Item 35. Report the final model’s performance on the test partition. Benchmark the performance of the AI model against current standards, such as histopathologic identification of disease or a panel of medical experts with an explicit method to resolve disagreements.
Item 36. For classification tasks, include estimates of diagnostic accuracy and their precision, such as 95% confidence intervals (2). Apply appropriate methodology such as receiver operating characteristic analysis and/or calibration curves. When the direct calculation of confidence intervals is not possible, report nonparametric estimates from bootstrap samples (11). State which variables were shown to be predictive of the response variable. Identify the subpopulation(s) for which the prediction model worked most and least effectively (11).
Item 37. Provide information to help understand incorrect results. If the task entails classification into two or more categories, provide a confusion matrix that shows tallies for predicted versus actual categories. Consider presenting examples of incorrectly classified cases to help readers better understand the strengths and limitations of the algorithm.
The Discussion Section
This section provides four pieces of information: summary, limitations, implications, and future directions.
Item 38. Summarize the results succinctly and place them into context; explain how the current work advances our knowledge and the state of the art. Identify the study’s limitations, including those involving the study’s methods, materials, biases, statistical uncertainty, unexpected results, and generalizability.
Item 39. Describe the implications for practice, including the intended use and possible clinical role of the AI model. Describe the key impact the work may have on the field. Envision the next steps that one might take to build upon the results. Discuss any issues that would impede successful translation of the model into practice.
Other Information
Item 40. Comply with the clinical trial registration statement from the International Committee of Medical Journal Editors (ICMJE). ICMJE recommends that all medical journal editors require registration of clinical trials in a public trials registry at or before the time of first patient enrollment as a condition of consideration for publication (30). Registration of the study protocol in a clinical trial registry, such as ClinicalTrials.gov or WHO Primary Registries, helps avoid overlapping or redundant studies and allows interested parties to contact the study coordinators (5).
Item 41. State where readers can access the full study protocol if it exceeds the journal’s word limit; this information can help readers evaluate the validity of the study and can help researchers who want to replicate the study (5). Describe the algorithms and software in sufficient detail to allow replication of the study. Authors should deposit all computer code used for modeling and/or data analysis into a publicly accessible repository.
Item 42. Specify the sources of funding and other support and the exact role of the funders in performing the study. Indicate whether the authors had independence in each phase of the study (5).
Conclusion
The CLAIM guideline provides a road map for authors and reviewers; its goal is to promote clear, transparent, and reproducible scientific communication about the application of AI to medical imaging. We recognize that not every manuscript will be able to address every CLAIM criterion, and some criteria may not apply to all works. Nevertheless, CLAIM provides a framework that addresses the key concerns to assure high-quality scientific communication. An analysis of published manuscripts is underway to provide empirical data about the use and applicability of the guideline.
This journal adopts the CLAIM guideline herewith as part of our standard for reviewing manuscripts, and we welcome comments from authors and reviewers.
Acknowledgments
Acknowledgments
The authors thank David Bluemke, MD, PhD, Adeline N. Boettcher, PhD, William Hsu, PhD, Jayashree Kalpathy-Cramer, PhD, Despina Kontos, PhD, Curtis P. Langlotz, MD, PhD, and Ronnie A. Sebro, MD, PhD, for their insightful comments.
Footnotes
Disclosures of Conflicts of Interest: J.M. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: author is consultant for Siemens; institution receives grants from GE, Nuance, and Enlitic. Other relationships: disclosed no relevant relationships. L.M. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: author is consultant for Lunit Insight and iCAD; institution receives grant from Siemens; travel accommodations from the Chinese Congress of Radiology and the Society of Breast Imaging. Other relationships: disclosed no relevant relationships. C.E.K. Activities related to the present article: institution receives salary support as Editor of Radiology: Artificial Intelligence. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships.
References
- 1.Kahn CE Jr. Artificial intelligence, real radiology. Radiol Artif Intell 2019;1(1):e184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Radiology 2015;277(3):826–832. [DOI] [PubMed] [Google Scholar]
- 3.Bossuyt PM, Reitsma JB, Bruns DE, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: The STARD Initiative. Radiology 2003;226(1):24–28. [DOI] [PubMed] [Google Scholar]
- 4.Bossuyt PM, Reitsma JB; Standards for Reporting of Diagnostic Accuracy. The STARD initiative. Lancet 2003;361(9351):71. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2016;6(11):e012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandenbroucke JP, von Elm E, Altman DG, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. PLoS Med 2007;4(10):e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz KF, Altman DG, Moher D; CONSORT Group . CONSORT 2010 statement: updated guidelines for reporting parallel group randomised trials. BMJ 2010;340:c332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begg C, Cho M, Eastwood S, et al. Improving the quality of reporting of randomized controlled trials. The CONSORT statement. JAMA 1996;276(8):637–639. [DOI] [PubMed] [Google Scholar]
- 9.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14(12):749–762. [DOI] [PubMed] [Google Scholar]
- 10.Park SH, Kressel HY. Connecting technological innovation in artificial intelligence to real-world medical practice through rigorous clinical validation: What peer-reviewed medical journals could do. J Korean Med Sci 2018;33(22):e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo W, Phung D, Tran T, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: A multidisciplinary view. J Med Internet Res 2016;18(12):e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handelman GS, Kok HK, Chandra RV, et al. Peering into the black box of artificial intelligence: Evaluation metrics of machine learning methods. AJR Am J Roentgenol 2019;212(1):38–43. [DOI] [PubMed] [Google Scholar]
- 13.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 2018;286(3):800–809. [DOI] [PubMed] [Google Scholar]
- 14.Bluemke DA, Moy L, Bredella MA, et al. Assessing Radiology Research on Artificial Intelligence: A Brief Guide for Authors, Reviewers, and Readers-From the Radiology Editorial Board. Radiology 2020;294(3):487–489. [DOI] [PubMed] [Google Scholar]
- 15.Altman DG, Simera I, Hoey J, Moher D, Schulz K. EQUATOR: reporting guidelines for health research. Lancet 2008;371(9619):1149–1150. [DOI] [PubMed] [Google Scholar]
- 16.Simera I, Altman DG. Reporting medical research. Int J Clin Pract 2013;67(8):710–716. [DOI] [PubMed] [Google Scholar]
- 17.Provenzale JM, Stanley RJ. A systematic guide to reviewing a manuscript. AJR Am J Roentgenol 2005;185(4):848–854. [DOI] [PubMed] [Google Scholar]
- 18.Budovec JJ, Kahn CE Jr. Evidence-based radiology: a primer in reading scientific articles. AJR Am J Roentgenol 2010;195(1):W1–W4. [DOI] [PubMed] [Google Scholar]
- 19.Geis JR, Brady AP, Wu CC, et al. Ethics of artificial intelligence in radiology: Summary of the joint European and North American multisociety statement. Radiology 2019;293(2):436–440. [DOI] [PubMed] [Google Scholar]
- 20.Willemink MJ, Koszek WA, Hardell C, et al. Preparing medical imaging data for machine learning. Radiology 2020 Feb 18:192224 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvey H, Glocker B. A standardised approach for preparing imaging data for machine learning tasks in radiology. In: Ranschaert ER, Morozov S, Algra PR, eds. Artificial Intelligence in Medical Imaging: Opportunities, Applications and Risks. New York, NY: Springer International, 2019. [Google Scholar]
- 22.Rubin DL, Kahn CE Jr. Common data elements in radiology. Radiology 2017;283(3):837–844. [DOI] [PubMed] [Google Scholar]
- 23.Kohli M, Alkasab T, Wang K, et al. Bending the artificial intelligence curve for radiology: Informatics tools from ACR and RSNA. J Am Coll Radiol 2019;16(10):1464–1470. [DOI] [PubMed] [Google Scholar]
- 24.Radiological Society of North America, American College of Radiology . RadElement: Common Data Elements. https://RadElement.org. Accessed February 1, 2020.
- 25.Sheehan J, Hirschfeld S, Foster E, et al. Improving the value of clinical research through the use of Common Data Elements. Clin Trials 2016;13(6):671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Institutes of Health . NIH Common Data Elements (CDE) Repository. Bethesda, MD: National Library of Medicine. https://cde.nlm.nih.gov/. Accessed February 20, 2020. [Google Scholar]
- 27.Lakhani P, Kim W, Langlotz CP. Automated extraction of critical test values and communications from unstructured radiology reports: an analysis of 9.3 million reports from 1990 to 2011. Radiology 2012;265(3):809–818. [DOI] [PubMed] [Google Scholar]
- 28.Lipton ZC, Berkowitz J, Elkan C. A critical review of recurrent neural networks for sequence learning. ArXiv 2015:1506.00019v4. [preprint] https://arxiv.org/abs/1506.00019v4. Posted October 17, 2015. Accessed January 27, 2020.
- 29.Eng J. Sample size estimation: how many individuals should be studied? Radiology 2003;227(2):309–313. [DOI] [PubMed] [Google Scholar]
- 30.International Committee of Medical Journal Editors . Clinical Trials. http://www.icmje.org/recommendations/browse/publishing-and-editorial-issues/clinical-trial-registration.html. Accessed February 20, 2020.