Abstract
Purpose
To evaluate the performance, agreement, and efficiency of a fully convolutional network (FCN) for liver lesion detection and segmentation at CT examinations in patients with colorectal liver metastases (CLMs).
Materials and Methods
This retrospective study evaluated an automated method using an FCN that was trained, validated, and tested with 115, 15, and 26 contrast material–enhanced CT examinations containing 261, 22, and 105 lesions, respectively. Manual detection and segmentation by a radiologist was the reference standard. Performance of fully automated and user-corrected segmentations was compared with that of manual segmentations. The interuser agreement and interaction time of manual and user-corrected segmentations were assessed. Analyses included sensitivity and positive predictive value of detection, segmentation accuracy, Cohen κ, Bland-Altman analyses, and analysis of variance.
Results
In the test cohort, for lesion size smaller than 10 mm (n = 30), 10–20 mm (n = 35), and larger than 20 mm (n = 40), the detection sensitivity of the automated method was 10%, 71%, and 85%; positive predictive value was 25%, 83%, and 94%; Dice similarity coefficient was 0.14, 0.53, and 0.68; maximum symmetric surface distance was 5.2, 6.0, and 10.4 mm; and average symmetric surface distance was 2.7, 1.7, and 2.8 mm, respectively. For manual and user-corrected segmentation, κ values were 0.42 (95% confidence interval: 0.24, 0.63) and 0.52 (95% confidence interval: 0.36, 0.72); normalized interreader agreement for lesion volume was −0.10 ± 0.07 (95% confidence interval) and −0.10 ± 0.08; and mean interaction time was 7.7 minutes ± 2.4 (standard deviation) and 4.8 minutes ± 2.1 (P < .001), respectively.
Conclusion
Automated detection and segmentation of CLM by using deep learning with convolutional neural networks, when manually corrected, improved efficiency but did not substantially change agreement on volumetric measurements.
© RSNA, 2019
Summary
A deep learning method shows promise for facilitating detection and segmentation of colorectal liver metastases; user correction of three-dimensional automated segmentations can generally resolve deficiencies of fully automated segmentation for small metastases and is faster than manual three-dimensional segmentation.
Key Points
■ Per-lesion sensitivity for lesions smaller than 10 mm was very low with automated segmentation (0.10) but was higher for user-corrected segmentation (0.30–0.57) and manual segmentation (0.58–0.70).
■ The overall per-lesion Dice similarity coefficients were 0.64–0.82 for the manual, 0.62–0.78 for the user-corrected, and 0.14–0.68 for the automated segmentation methods.
■ Intermethod variation was higher than that of manual segmentation, as shown by significantly (P < .001) higher repeatability coefficients.
Introduction
Colorectal cancer is the third most commonly diagnosed cancer worldwide (1,2). More than half of patients with colorectal cancer develop liver metastases at the time of diagnosis or later during the progression of their disease (3). The 5-year survival decreases from 64.3% to 11.7% in the presence of colorectal liver metastases (CLMs), which constitute the leading cause of death in these patients (4). Surgical resection of CLMs, when possible, is the standard of care (5), achieving long-term survival (6) and possibility of cure (7). More efficient chemotherapy, evolution of surgical resectability criteria, and strategies to improve CLM resectability (5) have contributed to improvement in patient survival (8).
Preoperative images of the liver, often obtained with CT (9), can help determine the number and size of CLMs to inform prognosis (10) and the location of CLMs with respect to vascular structures to evaluate resectability (11). The current approach for assessment of tumor burden relies on uni- or bidimensional measurements of the tumor along the largest axes (12), applied to lesions measuring more than 1 cm (12) and for a total of five lesions with a maximum of two per organ according to Response Evaluation Criteria in Solid Tumors version 1.1 (12). However, uni- or bidimensional diameter measurement of liver metastases (13) may not accurately reflect tumor size and growth because tumors often have an irregular shape (14). Tumor segmentation could more accurately capture the tumor volume, growth, and shape. Yet, tumor segmentation is a tedious and time-consuming task susceptible to intra- and interoperator variability if performed manually by a human (15,16). Hence, there is an emerging opportunity for automated tumor segmentation to address all these shortcomings.
Recent studies have demonstrated successful application of deep learning techniques for automated detection, segmentation, and classification tasks (17). Tumor segmentation tasks may be performed by using semiautomated techniques that rely on a combination of interactive or contouring approaches or achieve fully automated segmentation followed by manual corrections, if needed (18,19). Among deep learning techniques, fully convolutional networks (FCNs), consisting of multilayer neural networks, have become the preferred approach for analysis of medical images (20,21). Deep FCNs are capable of learning from examples and building a hierarchical representation of the data (22). Barriers to the development and clinical adoption of fully automated methods include insufficient training datasets (23–27), lack of labeled data, and undefined performance metrics adapted to clinical needs. We hypothesize that recent advances in deep learning (17,22) may be applied for fully automated detection and segmentation of CLM. However, there is a need to systematically assess the performance, agreement, and efficiency of deep learning–based detection and segmentation of CLM.
Therefore, the purpose of this study was to evaluate the performance, agreement, and efficiency of an FCN architecture for lesion detection and segmentation on CT images in patients with CLM, with manual segmentation as the reference standard.
Materials and Methods
Study Design and Subjects
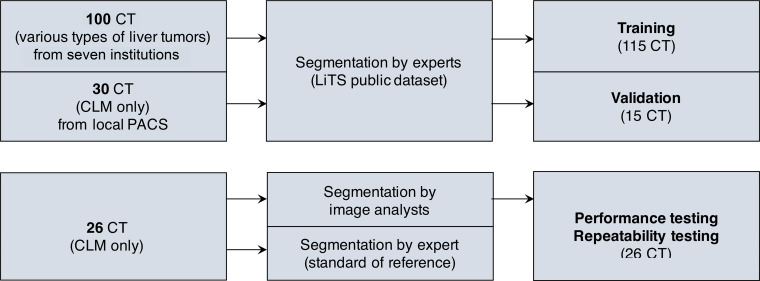
Our institutional review board at the Centre Hospitalier de l’Université de Montréal approved this retrospective cross-sectional study. Patient consent was waived for access to the training, validation, and test datasets. This study included two stages: a training and validation stage for various types of liver tumors on a public dataset, as well as a testing stage for specific CLM on an imaging dataset of patients seen at our institution. The study workflow is illustrated in Figure 1.
Figure 1:
Study workflow and datasets used in this study. CLM = colorectal liver metastasis, LiTS = Liver Tumor Segmentation, PACS = picture archiving and communication system.
Industry Support
Imagia Cybernetics (Montreal, Canada) provided financial support and graphics processing units to train models for this work. The authors had control of the data and the information submitted for publication. Authors who are not employees of Imagia determined data inclusion and performed the data analysis.
Training and Validation Dataset
Contrast material–enhanced CT examinations from the public Liver Tumor Segmentation (LiTS) challenge (25) dataset were used for training and validation. The LiTS dataset contains contrast-enhanced abdominal CT examinations in patients with primary or secondary liver tumors and was publicly shared with participants of the challenge. The training and validation sets included 115 CT examinations (261 liver lesions) and 15 CT examinations (22 liver lesions), respectively. Patients had various types of liver tumors, either primary (hepatocellular carcinoma) or metastatic (ie, colorectal, breast, and lung cancer). Cases from LiTS were provided by seven institutions and therefore obtained with different CT scanners with various protocols. Image resolution ranged from 0.56 to 1.0 mm in the axial plane, and the number of sections ranged from 42 to 1026. The cases included masks of liver metastases segmented at each clinical site by different radiologists and were reviewed by three experienced radiologists (25).
Testing Dataset
The testing dataset included 26 CT examinations (not included in the training dataset) in patients referred to our tertiary center for surgery. The cases were randomly selected from a biobank registered by the Canadian Tissue Repository Network (28). Inclusion criteria were: (a) patients with resected CLMs registered in the biobank and (b) who had undergone baseline (prechemotherapy and pretreatment) contrast-enhanced abdominal CT between October 2010 and December 2015. Patients from the testing set were 16 men and 10 women, with a mean age of 68 years ± 10 (standard deviation). The total number of lesions was 105, and the average number of lesions per patient was 4.0 ± 2.6. The lesions were stratified by size smaller than 10 mm (n = 30), 10–20 mm (n = 35), and larger than 20 mm in diameter (n = 40), with a mean size of 19.4 mm ± 15.0 (range, 1.10–89.9 mm). Preoperative CT studies were retrieved from our local picture archiving and communication system and were obtained at our institution (n = 10) and at other institutions (n = 16). Typical CT imaging technique parameters used in our center are reported in Table E1 (supplement).
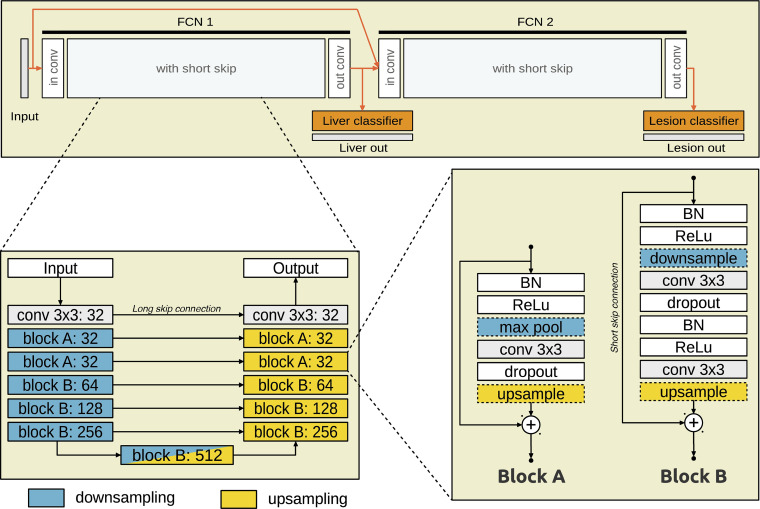
Model Architecture
The model is composed of two FCNs, one on top of the other (Fig 2). Both networks have a U-Net (29) type of architecture with short (between layers) and long (across the network) skip connections. FCN1 takes an axial CT section as input and outputs a probability of each pixel being within the liver. FCN2 takes as input the axial section concatenated with a stack of preclassifier features from FCN1 and outputs a probability of each pixel being a lesion. Each FCN is composed of a sequence of convolution blocks that extract features at the expense of spatial details along a contracting path (downsampling). The spatial details are then recovered along an expanding path. At each level along the expanding path, the corresponding feature representations are integrated to the spatial details through long skip connections. The technical details of the fully automated segmentation method of liver lesions are presented in Appendix E1 (supplement) and published in a previous work (30).
Figure 2:
Model structure of the convolutional neural network used in the study. The CT section is provided as input to FCN1, which outputs probability of each pixel being within the liver. FCN2 takes as input FCN1 output and the CT section and outputs probability for each liver pixel. BN = batch normalization, Conv = convolution kernel, FCN = fully convolutional network, ReLU = rectified linear unit.
Manual and User-corrected Segmentation
For each examination, CT images were imported into a free open-source image postprocessing software system (The Medical Imaging Interaction Toolkit [MITK]; Heidelberg, Germany) (31) as Digital Imaging and Communications in Medicine files. Binary lesion and nonlesion masks were created by manually segmenting the lesions on the CT images by using a cursor to contour the lesions. Lesion volumes were measured by counting the number of constituent voxels and considering the voxel size.
For user-corrected segmentations, CT examinations with binary lesion and nonlesion masks created by automated segmentation were imported into MITK, and each mask was manually corrected by analysts.
Reference Standard
Manual segmentation of liver lesions from 26 CT examinations of the testing set by a fellowship-trained abdominal radiologist (M.C., 8 years of experience) was used as the reference standard for lesion detection and segmentation tasks. Lesions were segmented during the portal venous phase as it accentuates the contrast between hypovascular metastatic lesions and normal liver parenchyma (32).
Agreement and Reliability
To assess the intra- and interobserver agreement and reliability, the testing set was independently segmented by two image analysts (Walid El Abyad and Assia Belblidia, 3 and 14 years of experience, respectively) who performed manual segmentations twice and corrections of automated segmentations twice. Each segmentation session was performed 1 week apart, and segmentations were performed in a randomized order to prevent recall bias.
Efficiency
User interaction time was recorded for manual segmentations and for manual corrections of automated segmentations. We evaluated efficiency as interaction time for manual segmentation compared with that of user-corrected segmentation.
Blinding
Image analysts were blinded to their previous segmentation results, those of the other analyst, and to the reference standard. The radiologist who performed the manual segmentation of the reference standard was blinded to the segmentation results of image analysts.
Statistical Analysis
Detection performance.—Detection performance was evaluated against the reference standard. Detection was established with intersection over union, while accounting for split and merge errors. Minimal thresholds were reported for overlap (intersection over union) greater than 0, greater than 0.25, and greater than 0.5 between the predicted lesion and the reference standard segmentation. While detection at intersection over union greater than 0.5 is biased toward better segmented lesions, it also favors positive predictive value (PPV) over sensitivity, downplaying false-positive findings. On the other hand, detection at intersection over union greater than 0 favors sensitivity over PPV.
Segmentation accuracy.—Accuracy was evaluated against the reference standard and assessed for overlap greater than 0, greater than 0.25, and greater than 0.5 by using Dice similarity coefficient and Jaccard index per detected lesion, maximum symmetric surface distance, and average symmetric surface distance (33). Measure definitions and formulas are given in Table E2 (supplement).
Detection and segmentation agreement.—Interreader, intrareader, and intermethod reliability of detection status evaluated against the reference standard were estimated by using the pooled Cohen κ coefficient (34).
Repeatability and reproducibility.—Per-lesion intrareader repeatability and interreader and intermethod reproducibility of lesion volume estimates were assessed with Bland-Altman analyses. Both replicates of measurements by each image analyst were used in the evaluations. Volume measurements were considered only for lesions in the reference standard that were detected in at least one replicate of either measurement being compared. To approximate homoscedasticity by removing proportional bias, the difference in volume measurements was normalized by the mean of volume measurements, yielding a proportional difference in volume measurements. To account for variable replicates, variance and limits of agreement were estimated according to equation 2, and 95% confidence intervals were estimated according to equation 4, by using the delta method presented by Zou (35). The significance of systematic bias was evaluated with a paired t test on proportional difference in volume measurements. A P value less than .001 indicated a statistically significant difference.
Bootstrapping.—For all metrics other than those related to Bland-Altman analyses, 95% confidence intervals were estimated by bootstrapping using random sampling. The approach would estimate the intervals by repeating the resampling process from the distribution of lesion measurements.
Efficiency.—We compared the user interaction time for manual segmentation only with user correction of automated segmentations by using two-factor repeated analysis of variance.
Results
Detection Performance
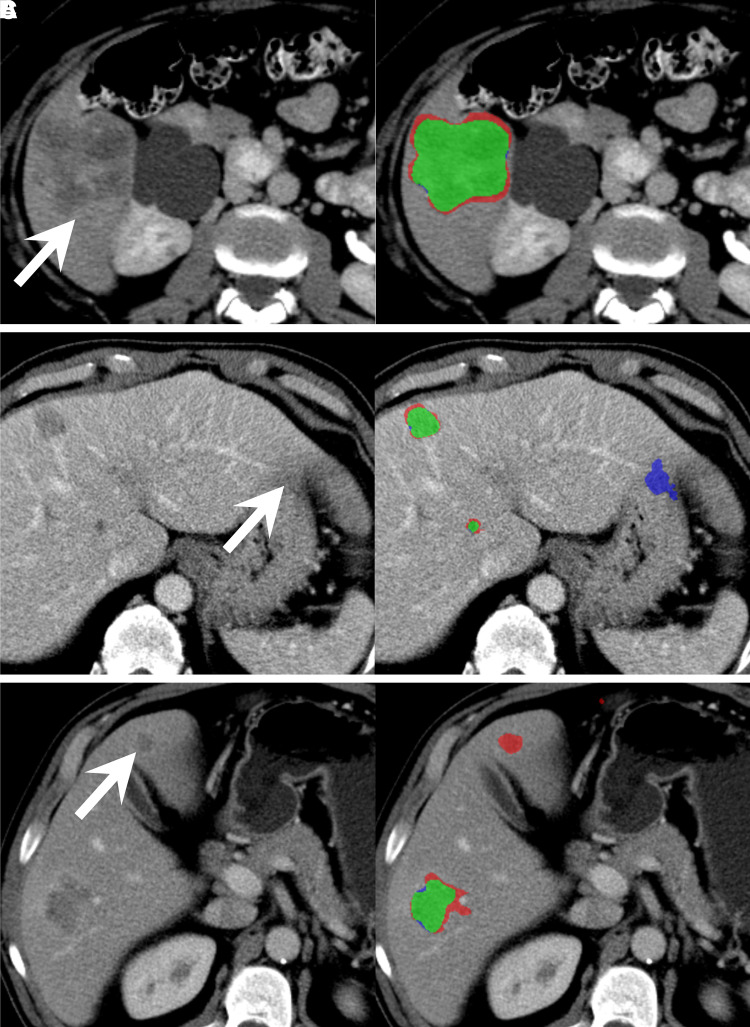
Examples of concordant and discordant detection are shown in Figure 3. Per-patient and per-lesion detection sensitivity and PPV for manual, user-corrected, and automated segmentation methods at greater than 0, greater than 0.25, and greater than 0.5 overlap are summarized in Table 1, Table E3 (supplement), and Table E4 (supplement), respectively. For an overlap greater than 0, manual segmentation had per-patient sensitivity of 0.82–0.83 and PPV of 0.86–0.91, user-corrected automatic segmentation had a sensitivity of 0.76–0.83 and PPV of 0.94–0.95, and automated segmentation had a sensitivity of 0.59 and PPV of 0.80.
Figure 3:
Contrast-enhanced axial CT images in three different patients with colorectal liver metastases demonstrate representative cases of, A, good agreement (green) of the automated segmentation mask with the ground-truth segmentation mask of a metastasis in segment VI (arrow), B, false-positive pixels (blue) of partial volume in segment II (arrow) from the automated segmentation mask, and, C, false-negative pixels (red) of a metastasis missed by automated segmentation in segment IVb (arrow).
Table 1:
Detection Performance at Minimum Overlap Greater than 0
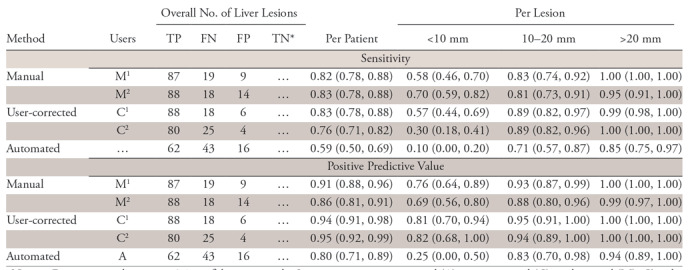
Note.—Data in parentheses are 95% confidence intervals. Segmentations are automated (A), user-corrected (C), and manual (M). Ci and Mi are both replicates of user-corrected or manual segmentation by reader i ϵ {1, 2}. FN = false negative, FP = false positive, TN = true negative, TP = true positive.
*True-negative findings are not counted because a true-negative finding cannot be defined per lesion.
Per-lesion sensitivity for small lesions less than 10 mm was very low with automated segmentation (0.10) but higher for user-corrected segmentation (0.30–0.57) and manual segmentation (0.58–0.70). Per-lesion sensitivity for lesions 10–20 mm was moderate with automated segmentation (0.71) but higher for user-corrected segmentation (0.89) and manual segmentation (0.81–0.83).
Segmentation Accuracy
Figure 4 shows an example lesion surface, produced by the automated method, with a color error map. Metrics of segmentation accuracy for manual, user-corrected, and automated segmentation at greater than 0, greater than 0.25, and greater than 0.5 overlap are summarized in Table 2, Table E5 (supplement), and Table E6 (supplement), respectively. The overall per-lesion Dice similarity coefficients were 0.64–0.82 for manual, 0.62–0.78 for user-corrected, and 0.14–0.68 for automated segmentation method. For the same voxel sizes and image resolution, the per-voxel effect of voxel misclassification (lesion vs not lesion) on volumetric metrics such as Dice coefficient increases as the lesion size decreases. This explains why the Dice score drops with lesion size. The reference standard was produced by manual detection and segmentation by a radiologist and was used for all metrics. The overall per-lesion Jaccard index for overlap greater than 0, greater than 0.25, and greater than 0.5 is summarized in Table E7 (supplement). The overall maximum symmetric surface distances were 3.07–6.44 mm for manual, 2.98–7.13 mm for user-corrected, and 5.15–10.42 mm for automated segmentation methods. The overall average symmetric surface distances were 0.68–0.89 mm for manual, 0.65–1.20 mm for user-corrected, and 1.65–2.82 mm for automated segmentation methods.
Figure 4:
Color error map shows an example of a liver lesion segmented with the automated method, shown from two views. The surface is color mapped according to a signed distance (in millimeters, represented by vertical scales to the right of the liver maps) to the reference surface. Green indicates small errors; blue, positive errors; and red, negative errors.
Table 2:
Segmentation Performance Measures at Minimum Overlap Greater than 0
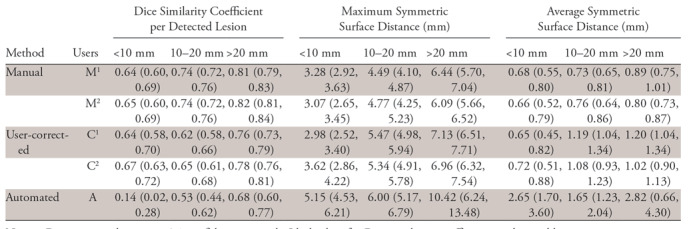
Note.—Data in parentheses are 95% confidence intervals. Ideal values for Dice similarity coefficient per detected lesion, maximum symmetric surface distance, and average symmetric surface distance are 1, 0 mm, and 0 mm, respectively. Segmentations are automated (A), user-corrected (C), and manual (M). Ci and Mi are both replicates of user-corrected or manual segmentation by reader i ϵ {1, 2}.
Detection Reliability
Intrareader and interreader reliability evaluation by using the pooled variant of Cohen κ, along with quantity and allocation disagreements and their confidence intervals, are summarized in Table 3 for overlap greater than 0 and Tables E8 (supplement) and E9 (supplement) for overlap greater than 0.25 and greater than 0.5, respectively.
Table 3:
Detection Reliability at Minimum Overlap Greater than 0
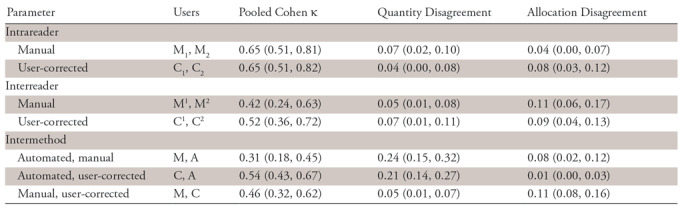
Note.—Data in parentheses are 95% confidence intervals. Segmentations are automated (A), user-corrected (C), and manual (M). Ci and Mi are both replicates of user-corrected or manual segmentation by reader i ϵ {1, 2}. Cj or Mj is the jth replicate of user-corrected or manual segmentations by both readers.
At overlap greater than 0, intrareader agreement on detection appeared higher than interreader agreement (κ = 0.65 vs κ= 0.42) for manual segmentation. Agreement between manual and automated segmentations (κ = 0.31) and between manual and user-corrected segmentations (κ = 0.46) appeared similar to interreader agreement. The lower κ for the former was due to a significantly higher quantity disagreement (0.24 vs 0.05). User correction resolved the quantity disagreement by correcting missed lesion detections.
Repeatability and Reproducibility
Intrareader repeatability and interreader and intermethod reproducibility for lesion volume estimation calculated by using Bland-Altman analyses are summarized in Table 4 and Table E10 (supplement). The volume mean was 7.4 mL ± 17.3 (range, 0.002–113.8 mL), and the diameter mean was 19.6 mm ± 15.3 (range, 0.7–79.5 mm). Intermethod variation was higher than that of manual segmentation, as shown by significantly (P < .001) higher repeatability coefficients. Both intrareader and interreader coefficients of repeatability were significantly (P < .001) higher for corrections of automated segmentations than for manual segmentations. The Bland-Altman plots in Figure E1a and E1c (supplement) reveal a wider volume dispersion for small lesions, especially for manual segmentation.
Table 4:
Lesion Volume: Intrareader, Interreader, and Intermethod Agreement at Minimum Overlap Greater than 0 Using Bland-Altman Analyses
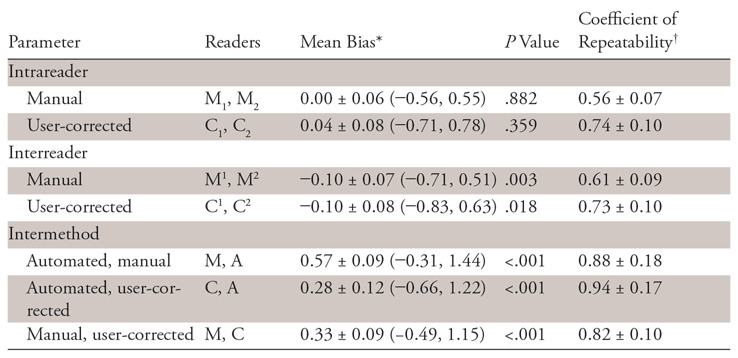
Note.—Segmentations are automated (A), user-corrected (C), and manual (M). Ci and Mi are both replicates of user-corrected or manual segmentation by reader i ϵ {1, 2}. Cj or Mj is the jth replicate of user-corrected or manual segmentations by both readers.
*Data are means ± 95% confidence intervals, with 95% limits of agreement in parentheses.
†Data are coefficients of repeatability ± 95% confidence intervals.
Statistically significant biases were observed with intermethod analyses (P <.001) with the largest bias (0.57 ± 0.09) observed when comparing automated segmentations to manual and a smaller bias observed when comparing automated segmentations to corrected automated segmentations (0.28 ± 0.12) or corrected to manual (0.33 ± 0.09). As can be seen in Figure E1f (supplement), corrections were mostly either addition of lesions missed by the automated method or corrections of undersegmented lesions. These corrections were not sufficient to remove systematic bias compared with manual segmentations. Interreader bias was small in magnitude, at −0.10 ± 0.07 (P =.003) for manual segmentation and −0.10 ± 0.08 (P = .018) for corrections of automated segmentations.
Efficiency
Mean interaction time was 7.7 minutes ± 2.4 per case for manual segmentation and 4.8 minutes ± 2.1 per case for user-corrected segmentation, with automated run time at around 1 second. Interaction time was significantly shorter for user-corrected and fully automated segmentation than for manual segmentation (P <.001).
Discussion
We found that deep learning with a pair of convolutional neural networks could be used for lesion detection and segmentation in patients with CLMs at contrast-enhanced CT; however, automated results are not yet at the level of trained annotators with a training set of 261 lesions annotated from CT studies. We further explored manual correction of automated segmentation results, thus achieving in less time similar per-patient detection performance as entirely manual segmentation and higher detection performance than fully automated segmentation. Overall, sensitivity and PPV increased for larger lesion size. A fully automated lesion detection method performed well, approaching manual and user-corrected performance for lesions larger than 20 mm. A prior study and meta-analysis reporting the per-lesion sensitivity of manual CLM detection at CT and using histopathologic findings as the reference standard showed overall sensitivities ranging from 74% to 81% but also found lower sensitivities of 8%–55% for detection of small CLMs (≤10 mm) (36,37). In addition, a meta-analysis on CLMs depicted by using imaging commented that the reference standard in several studies tended to be suboptimal because small metastases were excluded from analysis, which led to inflation of detection rates (38). A convolutional neural network–based fully automated method had a reported sensitivity of 86% for liver metastasis detection, but lesion size was not reported (39).
In our study, we found that a user-corrected segmentation method improved segmentation performance compared with automated segmentation, especially for small lesions (<10 mm), achieving an overall performance similar to that of manual segmentation. Our results for automated segmentation of lesions larger than 20 mm were within the Dice coefficient range of 65%–94% reported in previous studies using fully automated methods (40–42). Lower segmentation accuracy for small lesions observed in our study was consistent with prior studies, including one using an FCN-based algorithm for lesion segmentation (41,42). Except for studies using the 3D Image Reconstruction for Comparison of Algorithm Database (3Dircadb) (24), lesion size is often not reported in datasets, an approach consistent with the Response Evaluation Criteria in Solid Tumors policy of excluding lesions smaller than 10 mm (42,43). Alternative approaches such as Mask R-CNN (44) may not be optimal for this data, as it targets instance segmentation where different object classes overlap and compete for pixel classification.
Intrareader reliability of lesion detection was substantial and identical for manual and user-corrected segmentation. Interreader reliability for user-corrected segmentation was substantial and higher than for manual segmentation. Interreader reliability for manual segmentation was moderate and similar to reported agreement of CLM detection at CT among five observers of different experience described previously (45) and to the reported disagreement for segmentation among five radiologists on the MIDAS dataset of 9.8% (43).
Intra- and interreader agreement of lesion volume estimation was better for manual segmentation than user-corrected automated segmentation. Similarly, segmentation scores were improved from automated segmentation after user correction but remained below that of manual segmentation. The automated method is repeatable by design; however, automated segmentation introduced a substantial proportional bias in segmentation volume, tending to undersegment lesions. Although user correction successfully resolved deficiencies in lesion detection by the automated method, it reduced but failed to remove this segmentation bias. This may be because user correction is biased by the segmentations being corrected. It is thus more important to reduce segmentation bias in the automated segmentation method than to improve detection, when user correction is available.
The overall user interaction time was significantly reduced (approximately by half) by correction of automated predictions compared with manual segmentation, although user correction of the predicted lesions remained the most time-consuming step in the proposed method. When the method was used as fully automated and left uncorrected, segmentation time was further reduced to less than 1 second. Our per-case runtime for the user-corrected segmentation method (4.8 minutes per case) was shorter than a fully automated method reported by Moghbel et al (16 minutes per case) (43). Lesion detection and segmentation being a necessary but often time-consuming step in a clinical setting, a 38% improvement in efficiency would facilitate the clinical workflow. The trade-off for faster segmentation is usually losses in repeatability and accuracy (46).
We expect that a semiautomated strategy that involves human interaction as part of the automation model’s inputs to yield greater efficiency than what we have observed with user-correction of automated segmentation. A well-established traditional approach of semiautomated segmentation is interactive graph cuts, where a user specifies some pixels of the foreground and background to guide the automated graph-cuts optimization (47). Semiautomated segmentation has also been effectively combined with efficient user correction for liver segmentation (48). Segmentation efficiency could be improved with the development of a semiautomated method, taking advantage of recent advances with deep neural networks, in combination with improved user correction tools.
Our study had limitations. First, all patients in the training and testing datasets had liver lesions. The lack of patients without hepatic lesions may have biased the performance of the model toward pathologic liver conditions. Future studies are needed to evaluate the model on patients with a spectrum of liver disease from healthy to steatotic liver parenchyma. Second, our detection performance and segmentation accuracy were calculated based on manual segmentation as the reference standard, which may have led to variability. However, because these data combine volumes and segmentations from seven sites, these data likewise combine segmentations from multiple radiologists, and could generalize across the different segmentation examples, on different volumes, from different radiologists. This is a common limitation of data available for training segmentation models—especially when seeking to collect a sufficiently large quantity of this data. Because reference segmentations are not perfect, because training data do not typically contain repeated segmentations for each case, and because we hope that the model would generalize across cases, we seek to evaluate both the reliability of manual segmentation labels (which are used so faithfully as a reference for models) and the reliability of a model trained as described previously in this article. Therefore, in addition, we have assessed interreader agreement and repeatability to demonstrate the strength of our method in comparison to manual segmentation. Third, the training and validation stages were performed on the LiTS challenge dataset that included patients with non-CLM, whereas the testing dataset included only CLMs. Fourth, interreader agreement was only assessed by two readers. Finally, the training set is composed of only 261 lesions from 115 patient CT scans, which is an order of magnitude less than that of other deep learning applications in radiology, which includes thousands of images.
In conclusion, a deep learning method shows promise for facilitating detection and segmentation of CLMs, thus augmenting the work of radiologists while improving efficiency and providing similar variability. User correction of automated segmentations can generally resolve deficiencies of fully automated segmentation for small metastases and is faster than manual segmentation.
APPENDIX
SUPPLEMENTAL FIGURES
Acknowledgments
We acknowledge the contribution of Walid El Abyad and Assia Belblidia, Department of Radiology, University of Montreal.
S.K. supported by The Canada Research Chair in Medical Imaging and Assisted Interventions (950-228359). A.T. supported by Fondation de l’Association des Radiologistes du Québec and Fonds de Recherche du Québec-Santé (34939). S.T. supported by Roger Des Groseillers Hepato-Pancreatico-Biliary Surgical Oncology Research Chair. E.V. supported by Natural Sciences and Engineering Research Council of Canada. P.R. supported by PREMIER (previously COPSE) Internship Award sponsored by the Institut du Cancer de Montréal. Study supported by Imagia (co-funding to MEDTEQ operating grant), MEDTEQ, Polytechnique Montréal, and MITACS-Cluster Accelerate (IT05356).
Author contributions: Guarantors of integrity of entire study, E.V., M.C., L.D.J., R.L., A.T.; study concepts/study design or data acquisition or data analysis/interpretation, all authors; manuscript drafting or manuscript revision for important intellectual content, all authors; approval of final version of submitted manuscript, all authors; agrees to ensure any questions related to the work are appropriately resolved, all authors; literature research, E.V., M.C., P.R., A.T.; clinical studies, M.C., L.D.J., S.T., S.K.; statistical analysis, E.V., P.R., L.D.J., C.J.P., S.K.; and manuscript editing, all authors
E.V. and M.C. contributed equally to this work.
Disclosures of Conflicts of Interest: E.V. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employed by Microsoft Research Montreal (unrelated research internship); received grant from NSER PGS-D; conference travel expenses paid by the lab; received stipend from Imagia (contribution under MEDTEQ academic-industrial collaboration). Other relationships: disclosed no relevant relationships. M.C. disclosed no relevant relationships. P.R. disclosed no relevant relationships. L.D.J. Activities related to the present article: employed at Imagia. Activities not related to the present article: employed at Imagia. Other relationships: disclosed no relevant relationships. C.J.P. Activities related to the present article: institution received grants from MEDTECH and IVADO. Activities not related to the present article: employed part-time at Element AI (company does not work on biomedical applications of AI); receives royalties from Morgan Kauffman/Elsevier; gave presentation to MBA class at University of Montreal but did not apply paperwork to receive payment; has stock options with Element AI. Other relationships: disclosed no relevant relationships. R.L. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: advisory board member for IPSEN Canada, Novartis Canada, and Amgen; consultant for IPSEN Canada and Amgen; received payment for lectures from IPSEN Canada and Amgen Canada; received payment for travel expenses from Amgen and Novartis Canada. Other relationships: disclosed no relevant relationships. F.V. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: received payment for travel from Novartis (IHPBA 2016 meeting) and Merck (Optimising outcomes in Gastrointestinal Surgery Colorectal, MATTU, Guildford, Surrey, United Kingdom). Other relationships: disclosed no relevant relationships. S.T. Activities related to the present article: received grant from IVADO (coinvestigator on grant); received grant from MEDTEQ (coinvestigator on grant). Activities not related to the present article: consultant for TVM Life Sciences Management; grant from Bristol-Myers Squibb; received payment for lecture from Celgene; payment for development of educational presentations from AstraZeneca Canada. Other relationships: disclosed no relevant relationships. S.K. Activities related to the present article: institution received grant from MEDTEQ (with industry support from Imagia). Activities not related to the present article: consultant for NuVasive; expert testimony for CIHR, NSERC, CFI, Canada Research Chairs, MITACS. Other relationships: disclosed no relevant relationships. A.T. Activities related to the present article: institution received grant from MEDTEQ and Imagia; received research scholarship from FRQ-S and FARQ clinical research scholarship junior 2 salary award (34939). Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships.
Abbreviations:
- CLM
- colorectal liver metastasis
- FCN
- fully convolutional network
- LiTS
- Liver Tumor Segmentation
- PPV
- positive predictive value
References
- 1.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67(3):177–193. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136(5):E359–E386. [DOI] [PubMed] [Google Scholar]
- 3.Donadon M, Ribero D, Morris-Stiff G, Abdalla EK, Vauthey JN. New paradigm in the management of liver-only metastases from colorectal cancer. Gastrointest Cancer Res 2007;1(1):20–27. [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 2012;62(4):220–241. [DOI] [PubMed] [Google Scholar]
- 5.Adam R, de Gramont A, Figueras J, et al. Managing synchronous liver metastases from colorectal cancer: a multidisciplinary international consensus. Cancer Treat Rev 2015;41(9):729–741. [DOI] [PubMed] [Google Scholar]
- 6.Hwang M, Jayakrishnan TT, Green DE, et al. Systematic review of outcomes of patients undergoing resection for colorectal liver metastases in the setting of extra hepatic disease. Eur J Cancer 2014;50(10):1747–1757. [DOI] [PubMed] [Google Scholar]
- 7.Tomlinson JS, Jarnagin WR, DeMatteo RP, et al. Actual 10-year survival after resection of colorectal liver metastases defines cure. J Clin Oncol 2007;25(29):4575–4580. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher DJ, Kemeny N. Metastatic colorectal cancer: from improved survival to potential cure. Oncology 2010;78(3-4):237–248. [DOI] [PubMed] [Google Scholar]
- 9.Tirumani SH, Kim KW, Nishino M, et al. Update on the role of imaging in management of metastatic colorectal cancer. RadioGraphics 2014;34(7):1908–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai K, Allard MA, Benitez CC, et al. Early Recurrence after hepatectomy for colorectal liver metastases: what optimal definition and what predictive factors? Oncologist 2016;21(7):887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz RE, Abdalla EK, Aloia TA, Vauthey JN. AHPBA/SSO/SSAT sponsored consensus conference on the multidisciplinary treatment of colorectal cancer metastases. HPB 2013;15(2):89–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 13.Mantatzis M, Kakolyris S, Amarantidis K, Karayiannakis A, Prassopoulos P. Treatment response classification of liver metastatic disease evaluated on imaging: are RECIST unidimensional measurements accurate? Eur Radiol 2009;19(7):1809–1816. [DOI] [PubMed] [Google Scholar]
- 14.Rothe JH, Grieser C, Lehmkuhl L, et al. Size determination and response assessment of liver metastases with computed tomography—comparison of RECIST and volumetric algorithms. Eur J Radiol 2013;82(11):1831–1839. [DOI] [PubMed] [Google Scholar]
- 15.Odland A, Server A, Saxhaug C, et al. Volumetric glioma quantification: comparison of manual and semi-automatic tumor segmentation for the quantification of tumor growth. Acta Radiol 2015;56(11):1396–1403. [DOI] [PubMed] [Google Scholar]
- 16.van Heeswijk MM, Lambregts DM, van Griethuysen JJ, et al. Automated and semiautomated segmentation of rectal tumor volumes on diffusion-weighted MRI: can it replace manual volumetry? Int J Radiat Oncol Biol Phys 2016;94(4):824–831. [DOI] [PubMed] [Google Scholar]
- 17.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. arXiv170205747. [preprint] https://arxiv.org/abs/1702.05747. Posted February 19, 2017. Revised June 4, 2017. Assessed April 20, 2018.
- 18.Moghbel M, Mashohor S, Mahmud R, Saripan MIB. Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artif Intell Rev 2017;50(4):497–537. [Google Scholar]
- 19.Gotra A, Sivakumaran L, Chartrand G, et al. Liver segmentation: indications, techniques and future directions. Insights Imaging 2017;8(4):377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norman B, Pedoia V, Majumdar S. Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 2018;288(1):177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasaka K, Akai H, Abe O, Kiryu S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 2018;286(3):887–896. [DOI] [PubMed] [Google Scholar]
- 22.Chartrand G, Cheng PM, Vorontsov E, et al. Deep learning: a primer for radiologists. RadioGraphics 2017;37(7):2113–2131. [DOI] [PubMed] [Google Scholar]
- 23.MIDAS liver tumor dataset from National Library of Medicines Imaging Methods Assessment and Reporting (IMAR) project (MIDAS dataset 2016). https://www.insight-journal.org/midas/collection/view/38. Accessed March 8, 2019.
- 24.IRCAD France . 3Dircadb dataset from Research Institute against Digestive Cancer (IRCAD dataset 2016). https://www.ircad.fr/research/3d-ircadb-01/. Accessed January 27, 2019.
- 25.LiTS: Liver Tumor Segmentation challenge. https://competitions.codalab.org/competitions/17094. Accessed April 28, 2018.
- 26.TCIA . Cancer imaging archive of the Frederick National Laboratory for Cancer Research CIR dataset. 2016. http://www.cancerimagingarchive.net. Accessed May 9, 2018.
- 27.Sliver’07. Dataset from The Medical Image Computing and Computer Assisted Intervention Society MICCAI liver segmentation challenge (Sliver07 dataset 2016). http://www.sliver07.org/. Accessed May 9, 2018.
- 28.Canadian Tissue Repository Network . http://www.ctrnet.ca/. Accessed January 24, 2019.
- 29.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical image computing and computer-assisted intervention – MICCAI 2015. MICCAI 2015. Lecture notes in computer science, vol 9351. Cham, Switzerland: Springer, 2015. [Google Scholar]
- 30.Vorontsov E, Tang A, Pal C, Kadoury S. Liver lesion segmentation informed by joint liver segmentation. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), April 20, 2018.
- 31.The Medical Imaging Interaction Toolkit (MITK). http://mitk.org/wiki/The_Medical_Imaging_Interaction_Toolkit(MITK). Accessed April 20, 2018.
- 32.Silverman PM. Liver metastases: imaging considerations for protocol development with multislice CT (MSCT). Cancer Imaging 2006;6(1):175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotra A, Chartrand G, Massicotte-Tisluck K, et al. Validation of a semiautomated liver segmentation method using CT for accurate volumetry. Acad Radiol 2015;22(9):1088–1098. [DOI] [PubMed] [Google Scholar]
- 34.De Vries H, Elliott MN, Kanouse DE, Teleki SS. Using pooled kappa to summarize interrater agreement across many items. Field Methods 2008;20(3):272–282. [Google Scholar]
- 35.Zou GY. Confidence interval estimation for the Bland-Altman limits of agreement with multiple observations per individual. Stat Methods Med Res 2013;22(6):630–642. [DOI] [PubMed] [Google Scholar]
- 36.Ko Y, Kim J, Park JK, et al. Limited detection of small (≤ 10 mm) colorectal liver metastasis at preoperative CT in patients undergoing liver resection. PLoS One 2017;12(12):e0189797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niekel MC, Bipat S, Stoker J. Diagnostic imaging of colorectal liver metastases with CT, MR imaging, FDG PET, and/or FDG PET/CT: a meta-analysis of prospective studies including patients who have not previously undergone treatment. Radiology 2010;257(3):674–684. [DOI] [PubMed] [Google Scholar]
- 38.van Erkel AR, Pijl ME, van den Berg-Huysmans AA, Wasser MN, van de Velde CJ, Bloem JL. Hepatic metastases in patients with colorectal cancer: relationship between size of metastases, standard of reference, and detection rates. Radiology 2002;224(2):404–409. [DOI] [PubMed] [Google Scholar]
- 39.Ben-Cohen A, Diamant I, Klang E, Amitai M, Greenspan H. Fully convolutional network for liver segmentation and lesions detection. International Workshop on Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. Cham, Switzerland: Springer, 2016; 77–85. [Google Scholar]
- 40.Linguraru MG, Richbourg WJ, Liu J, et al. Tumor burden analysis on computed tomography by automated liver and tumor segmentation. IEEE Trans Med Imaging 2012;31(10):1965–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Jia F, Hu Q. Automatic segmentation of liver tumor in CT images with deep convolutional neural networks. J Comput Commun 2015;3(11):146–151. [Google Scholar]
- 42.Christ PF, Ettlinger F, Grün F, et al. Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. arXiv 170205970. [preprint] https://arxiv.org/abs/1702.05970. Posted February 20, 2017. Revised February 23, 2017. Accessed April 20, 2018.
- 43.Moghbel M, Mashohor S, Mahmud R, Saripan MI. Automatic liver tumor segmentation on computed tomography for patient treatment planning and monitoring. EXCLI J 2016;15:406–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. arXiv 170306870. [preprint] https://arxiv.org/abs/1703.06870. Posted March 20, 2017. Revised January 24, 2018.
- 45.Albrecht MH, Wichmann JL, Müller C, et al. Assessment of colorectal liver metastases using MRI and CT: impact of observer experience on diagnostic performance and inter-observer reproducibility with histopathological correlation. Eur J Radiol 2014;83(10):1752–1758. [DOI] [PubMed] [Google Scholar]
- 46.Udupa JK, Leblanc VR, Zhuge Y, et al. A framework for evaluating image segmentation algorithms. Comput Med Imaging Graph 2006;30(2):75–87. [DOI] [PubMed] [Google Scholar]
- 47.Boykov YY, Jolly MP. Interactive graph cuts for optimal boundary & region segmentation of objects in N-D images. International Conference on Computer Vision, 2001; 105–112. [Google Scholar]
- 48.Chartrand G, Cresson T, Chav R, Gotra A, Tang A, De Guise JA. Liver segmentation on CT and MR using Laplacian mesh optimization. IEEE Trans Biomed Eng 2017;64(9):2110–2121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.