Abstract
Introduction
The pandemic of a novel coronavirus disease 2019 (COVID-19) caused by a severe acute respiratory coronavirus 2 (SARS-CoV-2) infection has been problematic worldwide. A new SARS-CoV-2 antigen test (LUMIPULSEⓇ) was licensed and widely used in Japan since May 2020. We conducted this study intending to whether the automated quantitative CLEIA antigen test using a saliva sample is effective and valid for the diagnosis of COVID-19.
Patients and methods
We analyzed and compared the diagnostic accuracy of both the automated quantitative CLEIA antigen test and real-time RT-PCR (rRT-PCR) using a saliva sample from individuals suspected as having COVID-19.
Results
A total of 305 samples were collected and tested in Aichi Medical University Hospital and affiliated facilities from December 2020 until January 2021 at our institute. Using reverse-transcription PCR as a reference, the AUROC of the automated quantitative CLEIA antigen test was 0.903 (95% confidential interval 0.845–0.962, p < 0.001). The appropriate cut-off antigen level was 4.0 pg/mL and had a sensitivity of 77.8%, a specificity of 99.6%, a positive predictive value of 98%, and a negative predictive value of 94.5%. On the other hand, the diagnostic accuracy of the antigen test decreased among patients among patients with COVID-19 with threshold cycle (Ct-value)≥27, which shows the AUROC was 0.795 (95%CI 0.687–0.907, p < 0.001).
Conclusion
While the automated quantitative CLEIA antigen test from saliva specimen could be one of the most useful diagnostic tests for the diagnosis of COVID-19 in general practice, clinicians should know the limitations of the antigen test.
Keywords: SARS-CoV-2, Saliva, Quantitative chemiluminescent enzyme immunoassay, RT-PCR, COVID-19
1. Introduction
Emergence of a novel coronavirus in Wuhan, China, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the pandemic of COVID-19 (coronavirus disease 2019) caused a severe crisis in healthcare as well as the economy worldwide, and still are threatening at the time of January 2021 [[1], [2], [3]]. While the diagnosis as having COVID-19 has been made by reverse-transcription-polymerase chain reaction (RT-PCR), it is costly and requires a trained laboratory technician and medical equipment to perform, taking 3–4 h per assay [4]. A newly developed SARS-CoV-2 antigen test system, LUMIPULSE has been licensed since May 2020 in Japan [5], and has been used in general practice. We performed this retrospective study to report the efficacy and validity of automated quantitative chemiluminescence enzyme immunoassay (CLEIA) antigen test (LUMIPULSE antigen test) for the diagnosis of COVID-19. As far as we had searched, this is the first report documenting the diagnostic accuracy of the automated quantitative chemiluminescence enzyme immunoassay (CLEIA) antigen test by using saliva samples for the diagnosis of COVID-19.
2. Patients and methods
2.1. Patients and samples
We collected 305 saliva clinical specimens from individuals from Dec 2020 until Jan 2021 at Aichi Medical University Hospital and affiliated facilities. All patients were suspected to have COVID-19 based on their clinical symptoms (within 9 days from the onset) or met the definition of close contact with COVID-19 patients. Saliva was collected based on the standard protocol in the same manner as the previous study [6]. Using the samples, we performed rRT-PCR as well as the automated quantitative CLEIA antigen test in diagnosing COVID-19 patients. Then, we analyzed the diagnostic characteristics such as diagnostic accuracy and ROC curves, threshold cycle (Ct) value of the rRT-PCR, and compared the results of the two methods. This study was approved by the Institutional Review Board of Aichi Medical University Hospital.
2.2. SARS-CoV-2 antigen test
As for the SARS-CoV-2 antigen test, we used a newly developed SARS-CoV-2 antigen test system, LUMIPULSE SARS-CoV-2 antigen kit (Fujirebio, Japan), based on CLEIA, following the manufacture’s protocol. Based on the package insert, a cut-off value of 0.67 pg/mL was used. The performance of the automated quantitative CLEIA antigen test for the detection of SARS-CoV-2 antigen was evaluated by comparing the results with those from rRT-PCR experiment. Qualitative results (number of positive, negative, or invalid) and quantitative results (antigen quantities or threshold cycle value; used a smaller value from N1 and N2) were compared between the automated quantitative CLEIA antigen test and rRT-PCR.
2.3. rRT-PCR
rRT-PCR was performed by using BD MAX system (a fully-integrated, automated platform that performs nucleic acid extraction and real-time PCR) (Japan Becton Dickinson and Company, Japan). The re-suspended saliva was centrifuged at 500×g for 1 min, and the volumes of 750 μL supernatant fluid were assayed on the BD MAX system using the BD SARS-CoV-2 reagents for BD MAX System. In these reagents, the primer and double-quencher probe sets were based on the United States Centers for Disease Control and Prevention (US CDC) assay for specific detection of SARS-CoV-2 by amplifying two unique regions of the N gene (i.e., N1 and N2), and the human RNase P gene as an internal control.
2.4. Statistical analysis
Statistical analyses were performed using SPSS Version 26 for Windows (SPSS Inc., Chicago, IL, USA). P-values<0.05 were considered statistically significant.
3. Results
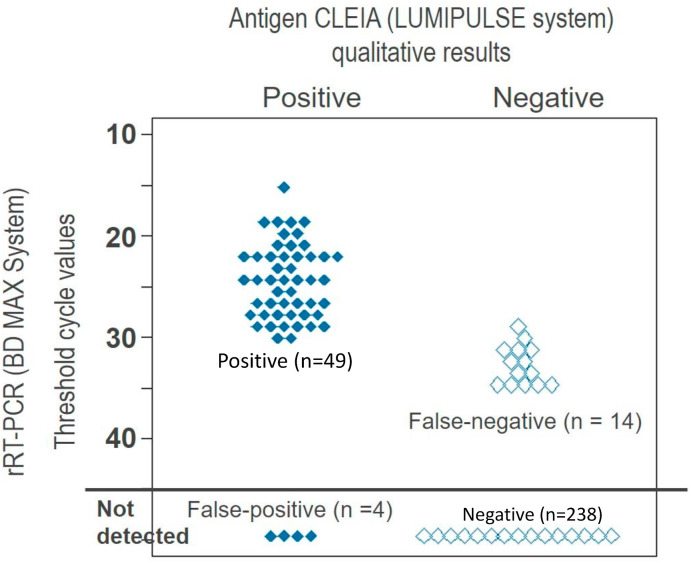
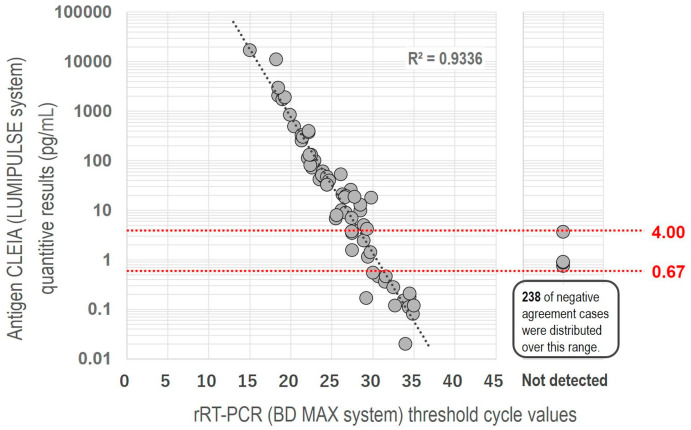
A total of 305 saliva samples were tested and the rRT-PCR results were 63 (20.7%) positive and 242 (79.3%) negative. The median antigen level of the positive rRT-PCR samples was 18.6 pg/ml (range 1.14–17046.6) and the median Ct-value was 26.6 cycles (range 15.5–36.2). We had 4 false-positive (positive-automated quantitative CLEIA antigen test, but negative-rRT-PCR) and 14 false-negative (negative-automated quantitative CLEIA antigen test, but positive-rRT-PCR). All samples of false-negative showed a Ct-value of 29.2–35.0 by rRT-PCR. False-positive samples revealed the antigen test ranging 0.75–3.66 pg/ml. Compared to the positive and negative results of rRT-PCR, the mean Ct-value was much lower in the positive samples than in the negative ones (24.9 v.s. 44.5 cycles, p < 0.001 by Mann-Whitney U test) (Fig. 1 ). The mean antigen level of positive rRT-PCR samples was much higher than that of the negative rRT-PCR samples (652.9 v.s. 0.2 pg/ml, p < 0.001 by Mann-Whitney U test). Fig. 2 shows the correlation between the antigen levels and the Ct-values of the rRT-PCR (R2 = 0.9336).
Fig. 1.
shows the comparison between qualitative results of antigen CLEIA and threshold cycle values of rRT-PCR.
Fig. 2.
shows the correlation the antigen levels and the Ct-values of rRT-PCR relationship.
3.1. Diagnostic accuracy of the automated quantitative CLEIA antigen test by saliva sample
Using the result of rRT-PCR as a reference, the area under receiver-operating characteristic (AUROC) curve is 0.903 [p < 0.001, 95% confidential interval (CI) 0.845–0.962]. The cut-off is 0.67, which was recommended by Fujirebio, and had a sensitivity of 77.8%, a specificity of 98.3%, a positive predictive value of 77.8%, and a negative predictive value of 94.4%. The appropriate cut-off antigen level was 4.0 pg/mL and had a sensitivity of 77.8%, a specificity of 99.6%, a positive predictive value of 98%, and a negative predictive value of 94.5% as shown in Table 1 . The cut-off was set based on the Youden Index [7]. Regarding cases with Ct-value≥27, the AUROC of the antigen test was 0.797 (p < 0.001, 95%CI 0.687–0.907). The diagnostic accuracy decreased as shown in Table 1 and Fig. 1. The appropriate cut-off antigen level was 0.67 pg/mL and had a sensitivity of 53.3%, a specificity of 94.4%, a positive predictive value of 80%, and a negative predictive value of 98.3% as shown in Table 1.
Table 1.
Diagnostic accuracy of LUMIPULSE for the diagnosis if COVID-19.
| Cut-off (pg/mL) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | YI |
|---|---|---|---|---|---|
| All cases (n = 305) | |||||
| 0.67 | 77.8 | 98.3 | 92.5 | 94.4 | 0.76 |
| 1.0 | 77.8 | 99.6 | 98 | 94.5 | 0.77 |
| 4.0 | 68.3 | 100 | 100 | 92.4 | 0.68 |
| Cases showing Ct-value≥27 (n = 272) | |||||
| 0.67 | 53.3 | 98.3 | 80 | 94.4 | 0.52 |
| 1.0 | 50 | 99.6 | 93.8 | 94.1 | 0.5 |
| 4.0 | 33.3 | 100 | 100 | 92.4 | 0.33 |
COVID-19, coronavirus disease 2019; Ct, threshold cycle; PPV, positive predictive value; NPV, negative predictive value; YI, Youden index.
4. Discussion
We found that the automated quantitative CLEIA antigen test using a saliva sample was effective and valid for the diagnosis of COVID-19. Some already reported the effectiveness of the automated quantitative CLEIA antigen test for the diagnosis of COVID-19 using a nasopharyngeal sample [7,8]. Compared to the studies, the automated quantitative CLEIA antigen test using a saliva sample could be one of the most effective diagnostic tests, which is acceptable. This test does not require any specific instruments or trained technicians. The automated quantitative CLEIA antigen test is less expensive and faster to obtain the result than RT-PCR, so it could replace RT-PCR for the near future in a medical institute that has no equipment for PCR. The automated quantitative CLEIA antigen test is already used widely for the diagnosis of seasonal influenza virus infection and viral hepatitis [9]. In contrast to rRT-PCR which needs 3–4 h, the automated quantitative CLEIA antigen test needs just 30–45 min. The general cost of the antigen test is much cheaper, which is one-third (6000 JPY = 56USD) in comparison to rRT-PCR (18,000JPY = 168USD) [10]. Moreover, obtaining saliva samples is safer and easier than nasopharyngeal samples for the prevention of secondary transmission from patients to medical staff. On the other hand, the diagnostic accuracy of the automated quantitative CLEIA antigen test was lower in patients with Ct ≧ 27 cycles than those with Ct < 27 cycles. We had 4 false-positive cases, exhibiting values of 0.75–3.66 pg/ml by the automated quantitative CLEIA antigen test. As written in the package insert, the diagnosis should be made with clinical information among the patients with a value of 0.75–3.66 pg/ml from the saliva sample. While a recent study demonstrated that patients with Ct-value≧34 would not be extremely contagious [11], patients with a value of 27–34 could be a cause of secondary transmission to others. Clinicians should be aware of the limitations of the antigen test.
We previously reported that 47% of COVID-19 pneumonia patients were asymptomatic, even when radiological findings on chest CT were confirmed [3]. The severity of COVID-19 could be correlated with the viral load as well as the Ct-value of RT-PCR [12,13]. Patients who show a high antigen level of the automated quantitative CLEIA antigen test should receive a chest CT scan, particularly those with underlying disease. Otherwise, pneumonia could be missed, resulting in an unexpected death.
There are several limitations in our study. First, we tested only for SARS-CoV-2 from saliva samples and not for coinfection with any other viruses. Second, there was no clinical information of the patients. Therefore, we could not evaluate any correlation between clinical manifestations and Ct-value.
In conclusion, the automated quantitative CLEIA antigen test of SARS-CoV-2 by saliva sample is effective for the diagnosis of COVID-19 in general practice and could replace r-RT-PCR. However, clinicians should know that the diagnostic accuracy among patients with Ct-value≧27 cycles could be lower than those with Ct-value<27 cycles.
Author contributions
DS, NA, AK, and HM designed this study. NA and DS drafted this manuscript. DS, AN, YK, NM, TO, AY, IK, HS performed the LUMIPULSE antigen test and rRT-PCR. NA, DS, WO, AN, HK, AS, YY and MH contributed to the data collection, data analysis. HM supervised, reviewed, and edited this manuscript.
Declaration of competing interest
H. Mikamo received grant support from Asahi Kasei Pharma Corporation, Shionogi & Co. Ltd., Daiichi Sankyo Co., Ltd., Sumitomo Dainippon Pharma Co., Ltd., Pfizer Japan Inc. and FUJIFILM Toyama Chemical Co., Ltd., payment for lectures from Astellas Pharma Inc., MSD K.K., Daiichi Sankyo Co., Ltd., Sumitomo Dainippon Pharma Co., Ltd., MIYARISAN Pharmaceutical Co., Ltd. Becton, Dickinson and Company Japan, and FUJIFILM Toyama Chemical Co. Ltd. The other authors declare that they have no conflicts of interest.
Acknowledgments
We are grateful for the diligent and thorough critical reading of our manuscript by Dr. Yoshihiro Ohkuni, Chief Physician, Taiyo and Mr. John Wocher, Advisor, Kameda Medical Center (Japan). We thank technicians, Ms. Akiko Takao, Mr. Tatsuo Terada, Ms. Maki Okayasu for performing the testing.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai N., Sakanashi D., Nakamura A., Kishino T., Kato H., Hagihara M., et al. Clinical manifestations and radiological features by chest computed tomographic findings of a novel coronavirus disease-19 pneumonia among 92 patients in Japan. J Microbiol Immunol Infect. 2020;S1684–1182(20):30168–30177. doi: 10.1016/j.jmii.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai C.C., Wang C.Y., Ko W.C., Hsueh P.R. In vitro diagnostics of coronavirus disease 2019: technologies and application. J Microbiol Immunol Infect. 2020 doi: 10.1016/j.jmii.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoki K., Nagasawa T., Ishii Y., Yagi S., Okuma S., Kashiwagi K., et al. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. J Infect Chemother. 2020;S1341–321X(20):30428–30431. doi: 10.1016/j.jiac.2020.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakanashi D., Asai N., Nakamura A., Miyazaki N., Kawamoto Y., Ohno T., et al. Comparative evaluation of nasopharyngeal swab and saliva specimens for the molecular detection of SARS-CoV-2 RNA in Japanese patients with COVID-19. J Infect Chemother. 2021;27:126–129. doi: 10.1016/j.jiac.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youden W.J. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 8.Aoki K., Nagasawa T., Ishii Y., Yagi S., Okuma S., Kashiwagi K., et al. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. J Infect Chemother. 2020;S1341–321X(20):30428–30431. doi: 10.1016/j.jiac.2020.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue T., Ohike T., Ohne K., Sato S., Goto T., Tanaka Y. Clinical evaluation of a newly developed chemiluminescent enzyme immunoassay for hepatitis C virus core antigen in Japan. Jpn J Infect Dis. 2019;72:285–291. doi: 10.7883/yoken.JJID.2018.472. [DOI] [PubMed] [Google Scholar]
- 10.SRL, Inc SRL news. http://www.srl-group.co.jp/ 8 Mar 2021.
- 11.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int J Infect Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asai N., Sakanashi D., Ohashi W., Nakamura A., Yamada A., Kawamoto Y., et al. Could threshold cycle value correctly reflect the severity of novel coronavirus disease 2019 (COVID-19)? J Infect Chemother. 2021;27:117–119. doi: 10.1016/j.jiac.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]