The central dogma of molecular biology has retained its explanatory power since it was first articulated in 1957 by Francis Crick. Since then, advances in technology have made it easier to obtain molecular information, but more data do not always lead to more knowledge. Understanding kidney function, and therapeutic approaches to kidney disease, rest on knowledge derived from decades of work, using a variety of approaches, from clearance to gene knockout. Global transcriptional analyses are now adding significantly to this knowledge base, but only if placed within the context of established truth.1 Two papers in this issue of JASN successfully combine established knowledge about the kidney with transcriptomic approaches to derive new insights into kidney function.2,3 They join a growing list of papers4–6 that not only describe cell characteristics, but provide user-friendly interfaces so investigators can see the results with their own eyes (see https://cello.shinyapps.io/kidneycellexplorer/ and websites referenced in these papers). In doing so, they generate a cornucopia of information that not only broadens and deepens our understanding, but also provides investigators and clinicians new tools to analyze and treat kidney disease in the future. When applied to humans, this is part of the promise of the Kidney Precision Medicine Project, sponsored by the National Institute of Diabetes and Digestive and Kidney Disease.
In the first paper, Knepper and colleagues dissected single nephron segments from mice, and used RNA sequencing to map the transcript expression across 14 nephron segments. This approach, on the basis of the tubule dissection method developed by Burg and colleagues, relied on prior work documenting “gold standard” markers for various nephron segments, which could be used to validate the accuracy of segment identification.1 This allowed the investigators to confirm many fundamental observations regarding kidney gene expression and make several striking new observations, and generated new questions that will need to be addressed experimentally in the future.
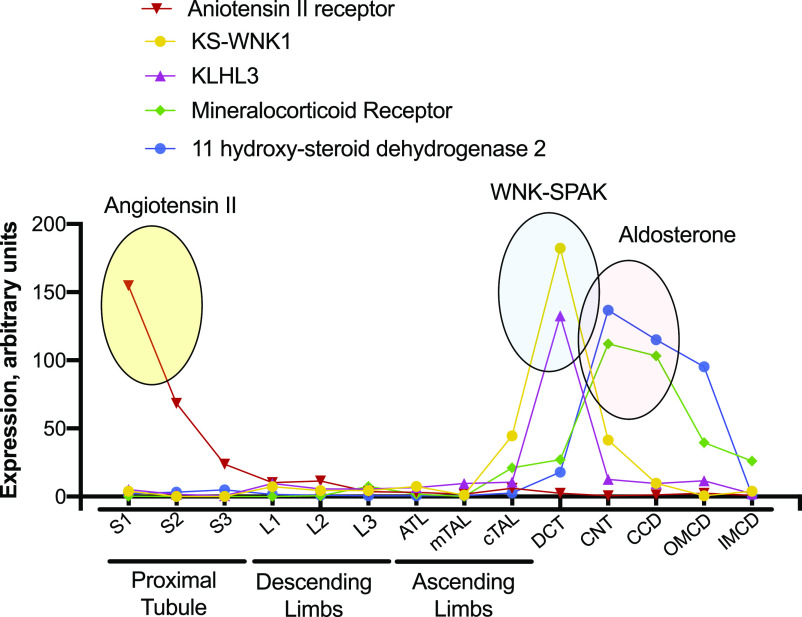
There is so much information here that one hesitates to highlight only a portion, but several observations deserve special comment; these are shown graphically in Figure 1. The first concerns the localization of genes of the renin-angiotensin-aldosterone signaling pathway, perhaps the dominant renal regulatory system. A vast literature has highlighted effects of angiotensin II (AngII) on glomerular hemodynamics and on tubule salt transport, with actions described in nearly every nephron segment, and in mesangial and smooth muscle cells. This ubiquity of effect along the nephron challenges older physiologic models that suggested AngII tends to shift sodium reabsorption from more distal to more proximal segments. It is not surprising that the predominant stimulatory receptor for AngII, Agtr1a, is most highly expressed by glomeruli; additionally, there is substantial expression along proximal tubules, where activation is known to stimulate sodium reabsorption.7 What is surprising, however, is the near absence of these receptors in segments beyond the proximal tubule, a result in agreement with other recent RNA sequencing results.4 Current dogma suggests AngII activates thiazide-sensitive sodium chloride cotransport along the distal convoluted tubule and activates the sodium channel, ENaC, along the collecting duct. Three possibilities may explain the discrepancy between expression and function: (1) receptors are present at the protein level, despite a nearly absent message; (2) AngII effects are secondary to actions in other nephron segments or other tissues (such as the central nervous system); or (3) effects are mediated by other receptors. These questions are now begging to be addressed experimentally.
Figure 1.
Sites of relative gene expression along the nephron. Ovals emphasize predominant expression sites for components of signaling pathways. Note the preponderance of AngII receptors along the proximal tubule (yellow oval), the expression of KLHL3 and KS-WNK1 along the DCT (blue oval), and the expression of 11 β-hydroxysteroid dehydrogenase 2 along the connecting tubule and cortical collecting duct (red oval). S1–S3, segments of the proximal tubule, L1–L3, segments of the descending limbs, ATL, ascending thin limb; TAL, thick ascending limb; CNT, connecting tubule; CCD, cortical collecting duct; OMCD, outer medullary collecting duct; IMCD, inner medullary collecting duct. Redrawn from https://esbl.nhlbi.nih.gov/MRECA/Nephron/.
A similar surprise derives from the expression levels of transcripts related to steroid hormone signaling. Cells that are responsive to aldosterone must express mineralocorticoid receptors (MR), but they also need to express the metabolic enzyme 11 β-hydroxysteroid dehydrogenase 2. This enzyme “protects” the receptors from glucocorticoid hormones, which bind to these receptors with high affinity and circulate at high levels. Thus, the signature of a classic aldosterone-responsive cell is the mutual expression of the receptor and the enzyme. Two striking findings emerge from the nephron segment data, the first being that 11 β-hydroxysteroid dehydrogenase 2 expression is very low along distal convoluted tubules (DCT), although MRs are expressed there (Figure 1); this finding is similar to data reported recently by others.4 The low expression is surprising because it is widely accepted that DCT cells respond to aldosterone.8 One intriguing possibility is that MRs in the DCT are typically bound by cortisol (or, in rodents, corticosterone), rendering them active even when aldosterone levels are low. Nevertheless, the current data suggest the ability of aldosterone to stimulate thiazide-sensitive NaCl cotransport is primarily indirect, likely mediated by changes in potassium concentration.9,10
The companion manuscript makes complementary observations, but they are just as intriguing. Here, Chen, Chou, and Knepper developed a method to digest cells in the cold, then enrich specifically for cells from the distal nephron. It has been suggested there are two molecularly distinct portions of DCTs, which are of special interest because of their unique roles in human diseases, in modulating salt and potassium balance, and in determining calcium and magnesium excretion. Yet, these subsegments were not apparent using the segment dissection approach.
By using this molecular “magnifying glass,” one driven by prior knowledge of kidney anatomy and physiology, the authors opened a window on cell populations that might otherwise be opaque. This approach yielded another rich dataset, and suggested a host of new hypotheses, but several observations deserve special mention. First, the investigators found three distinct populations of thick ascending limb cells. One comprises cells of the macula densa; these cells coexpress neuronal nitric oxide synthase, a classic macula densa marker. Interestingly, however, these cells also express other genes that are typically found in neurons, including genes involved in axon guidance and synaptic adhesion, leading the investigators to label them as “neuroepithelial cells.” They speculate that these genes may play a role in homing cells to the vascular pole.
The two other classes of thick ascending limb cell provide additional support for recent suggestions that cations traverse unique parallel paracellular pathways, one for sodium (claudin 10), and the other for calcium and magnesium (claudin 16).11 Strikingly, the two thick ascending limb populations also express different homeobox transcription factors, and only the claudin 10 population expresses receptors for prostaglandins (PGs). This suggests the effects of PGs to alter sodium chloride transport along the thick ascending limb may be cell-type specific.
The enrichment approach allowed the authors to confirm there are indeed two types of DCT cells, one expressing the thiazide-sensitive NaCl cotransporter alone (DCT1 cells), and the other expressing the cotransporter with the epithelial sodium channel (DCT2 cells). Yet the authors also report subtypes of DCT1 cells; strikingly, one population expressed markers typical for proliferative cells (e.g., Mki67, which encodes Ki67), supporting a growing body of work suggesting the DCT undergoes remodeling under stress; a second population of DCT1 cells expresses an “innate immune signature.”
Finally, the investigators provide new insights into the localization and function of the WNK-SPAK-NCC signaling pathway, the pathway that, when mutated in humans, causes high BP and hyperkalemia. Several components of this pathway, including WNK4, SPAK, and cullin 3, are widely expressed, but two components are essentially restricted to the DCT (Figure 1). These are KLHL3, the adaptor protein involved in WNK kinase degradation, and the still mysterious kidney-specific kinase-deficient form of WNK1, called KS-WNK1. Given that the disease phenotype is recapitulated by activating thiazide-sensitive NaCl cotransport in the DCT1 alone,12 this suggests key roles for these unique signaling molecules.
In summary, by combining old and new techniques, and by leveraging established knowledge concerning anatomy and physiology, the authors have advanced our understanding of the kidney in health and disease, and opened new avenues to explore.
Disclosures
D.H. Ellison reports Scientific Advisor/Membership as an Author, UpToDate; Consulting Editor, Hypertension; and Editorial Board Am. J Physiology Renal. He is also on the Council of the American Society of Nephrology.
Funding
None.
Acknowledgments
The content of this article reflects the personal experience and views of the author(s) and should not be considered medical advice or recommendations. The content does not reflect the views or opinions of the American Society of Nephrology (ASN) or JASN. Responsibility for the information and views expressed herein lies entirely with the author(s).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Chen L, Clark JZ, Nelson JW, Kaissling B, Ellison DH, Knepper MA: Renal-tubule epithelial cell nomenclature for single-cell RNA-sequencing studies [published correction appears in J Am Soc Nephrol 30: 2475, 2019]. J Am Soc Nephrol 30: 1358–1364, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, Chou C-L, Knepper MA: A comprehensive map of mRNAs and their isoforms across all 14 renal tubule segments of mouse. J Am Soc Nephrol 32: 897–912, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Chou C-L, Knepper MA: Targeted single-cell RNA-seq identifies minority cell types of kidney distal nephron. J Am Soc Nephrol 32: 886–896, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ransick A, Lindström NO, Liu J, Zhu Q, Guo JJ, Alvarado GF, et al.: Single-cell profiling reveals sex, lineage, and regional diversity in the mouse kidney. Dev Cell 51: 399–413.e7, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, et al.: Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360: 758–763, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, Kirita Y, Donnelly EL, Humphreys BD: Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 30: 23–32, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurley SB, Riquier-Brison ADM, Schnermann J, Sparks MA, Allen AM, Haase VH, et al.: AT1A angiotensin receptors in the renal proximal tubule regulate blood pressure. Cell Metab 13: 469–475, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rojas-Vega L, Gamba G: Mini-review: Regulation of the renal NaCl cotransporter by hormones. Am J Physiol Renal Physiol 310: F10–F14, 2016 [DOI] [PubMed] [Google Scholar]
- 9.Terker AS, Yarbrough B, Ferdaus MZ, Lazelle RA, Erspamer KJ, Meermeier NP, et al.: Direct and indirect mineralocorticoid effects determine distal salt transport. J Am Soc Nephrol 27: 2436–2445, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canonica J, Sergi C, Maillard M, Klusonova P, Odermatt A, Koesters R, et al.: Adult nephron-specific MR-deficient mice develop a severe renal PHA-1 phenotype. Pflugers Arch 468: 895–908, 2016 [DOI] [PubMed] [Google Scholar]
- 11.Milatz S, Himmerkus N, Wulfmeyer VC, Drewell H, Mutig K, Hou J, et al.: Mosaic expression of claudins in thick ascending limbs of Henle results in spatial separation of paracellular Na+ and Mg2+ transport. Proc Natl Acad Sci U S A 114: E219–E227, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm PR, Coleman R, Delpire E, Welling PA: Constitutively active SPAK causes hyperkalemia by activating NCC and remodeling distal tubules. J Am Soc Nephrol 28: 2597–2606, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]