Significance Statement
Reliable, digital, automated detection of interstitial fibrosis and tubular atrophy (IFTA) has not yet been developed. Machine learning (ML) can reproduce the renal pathologist’s visual assessment of IFTA and glomerulosclerosis. Well-trained ML methods not only showed similar agreement to that seen among renal pathologists for the assessment of IFTA and glomerulosclerosis, but also equivalent statistical association with patient outcome. These methods can help expedite research on very large digital archives of renal biopsy specimens, and may also benefit clinical practice by acting as a stand-in reading for pathology scenarios where renal expertise is limited or unavailable.
Keywords: interstitial fibrosis, tubular atrophy, glomerulosclerosis, prognostication, convolutional neural network, whole slide segmentation, diabetes, transplant, eGFR
Visual Abstract
Abstract
Background
Interstitial fibrosis, tubular atrophy (IFTA), and glomerulosclerosis are indicators of irrecoverable kidney injury. Modern machine learning (ML) tools have enabled robust, automated identification of image structures that can be comparable with analysis by human experts. ML algorithms were developed and tested for the ability to replicate the detection and quantification of IFTA and glomerulosclerosis that renal pathologists perform.
Methods
A renal pathologist annotated renal biopsy specimens from 116 whole-slide images (WSIs) for IFTA and glomerulosclerosis. A total of 79 WSIs were used for training different configurations of a convolutional neural network (CNN), and 17 and 20 WSIs were used as internal and external testing cases, respectively. The best model was compared against the input of four renal pathologists on 20 new testing slides. Further, for 87 testing biopsy specimens, IFTA and glomerulosclerosis measurements made by pathologists and the CNN were correlated to patient outcome using classic statistical tools.
Results
The best average performance across all image classes came from a DeepLab version 2 network trained at 40× magnification. IFTA and glomerulosclerosis percentages derived from this CNN achieved high levels of agreement with four renal pathologists. The pathologist- and CNN-based analyses of IFTA and glomerulosclerosis showed statistically significant and equivalent correlation with all patient-outcome variables.
Conclusions
ML algorithms can be trained to replicate the IFTA and glomerulosclerosis assessment performed by renal pathologists. This suggests computational methods may be able to provide a standardized approach to evaluate the extent of chronic kidney injury in situations in which renal-pathologist time is restricted or unavailable.
Histopathologic changes observed in kidney biopsy specimens are not only vital for the diagnosis of kidney disease, but also serve as valuable prognostic and predictive markers. The primary histologic indicators of irreparable renal injury include interstitial fibrosis, tubular atrophy (IFTA), and glomerulosclerosis. IFTA is the best morphologic prognostic marker of CKD progression and outcome, irrespective of the etiology of the disease.1–10 However, IFTA assessment can be variable between pathologists11–15 because it is determined on the basis of visual estimation of overall percent renal cortical area involvement, rather than counting discrete, individual structures, as is the case for glomeruli. Given that IFTA typically occurs in a patchy fashion, it can be difficult for the pathologist to mentally aggregate the regions. Furthermore, IFTA occurs as a continuous morphologic spectrum, which is often variable between pathologists in terms of each pathologist’s threshold for binary classification of IFTA (i.e., scarred versus unscarred renal cortex). For these reasons, IFTA assessment can suffer from suboptimal interpathologist reliability.
Contemporary digital-image analysis has shown promise to accurately recognize and measure specific features on biopsy specimens that may be of clinical value. Major advantages of such technology include the ability to at least partially automate biopsy specimen analysis and to provide more precision and accuracy in morphometric quantification of specific histologic findings. This, in turn, enables the severity of features, such as IFTA, to be reported as continuous variables rather than traditional categoric variables, which may show improved clinical utility.
In this study, we sought to automate the identification and quantification of IFTA and glomerulosclerosis on whole-slide images (WSIs) with machine learning (ML). Several previous studies have applied various morphometric methods to improve the reproducibility and accuracy of IFTA assessment.16–19 Additionally, ML algorithms have already been successfully applied to glomerular segmentation by us and others.18,20–23 However, to date, no studies have focused specifically on a whole-slide classifier to directly replicate a pathologist’s assessment of IFTA and glomerulosclerosis on renal biopsy specimens. In this study, we developed such a classifier and assessed its performance, reliability, and prognostic capability.
Methods
Select technical terms that have a more extensive explanation are defined in the glossary of terms in Supplemental Table 1. Our study consists of three main components that address the following: (1) performance of convolutional neural networks (CNNs) for segmentation of IFTA and glomerulosclerosis, (2) interobserver reliability between the best performing CNN and multiple renal pathologists, and (3) statistical association of CNN-based IFTA and glomerulosclerosis quantification with patient outcome. These components will be referred to as substudy 1, substudy 2, and substudy 3, respectively, throughout the rest of the manuscript. The study was approved by the institutional review boards at the University of California at Davis, University of Buffalo, Seoul National University Hospital, Coimbra Hospital and University Center, and Johns Hopkins University. The study design for human samples from Seoul National University Hospital complied with the 2013 Declaration of Helsinki.
For reproducibility, we released our best performing segmentation model for other researchers to use, along with several WSIs and their network segmented outputs. The network models can be used with our H-AI-L (human–artificial intelligence–loop) algorithm22 to segment new WSIs. Information on codes for WSI segmentation can be found at our Github repository (see https://github.com/SarderLab/IFTA_segmentation). The shared whole-slide data, segmented output, and trained network model can be found at https://bit.ly/3eywm0J.
Image Data
WSIs of renal biopsy specimens from 205 patients were used in this study, collected from six different institutions. In no particular order to preserve anonymity, these institutions were the Kidney Translational Research Core at the Washington University School of Medicine in Saint Louis, University of California at Davis, Vanderbilt University Medical Center, Seoul National University Hospital, the Nephrology Unit at Coimbra Hospital and University Center, and Johns Hopkins University. Tissue sections were prepared at 2–3 µm thickness and stained with Periodic acid–Schiff (PAS). Where possible, depending on the source of the WSI, an adjacent trichrome section was also acquired for pathologists to reference. Slides were scanned using a brightfield microscopy whole-slide scanner (Aperio; Leica) at 40× magnification, resulting in an apparent resolution of 0.25 µm/pixel.
Case Selection
IFTA is a highly complex, histopathologic feature that can manifest as a spectrum of morphologies. In this study, we were interested in specifically studying the influence of chronic injury on kidney function. Therefore, while selecting the cases for all three substudies, a renal pathologist reviewed the cases to ensure the following selection criteria were met: (1) the amount of early or evolving IFTA with variable intermixed edema was minimized, and (2) cases were selected to represent the full range of IFTA severity. All types of IFTA, including classic, endocrinization, and thyroidization types, were included in the analysis, without distinguishing between the types.
Table 1 shows a summary of the study population. The specific descriptions of the cases used for each of our three substudies follow (Supplemental Table 2). For substudy 1, on 116 PAS-stained WSIs of renal tissue sections (113 biopsy specimens and three specimens from nephrectomies), IFTA, nonsclerotic glomeruli, and sclerotic glomeruli were annotated by a single renal pathologist who had the most experience among the annotators in our study (RP1). These slides were split into a training set (n=79), an internal testing set (n=17), and an external testing set (n=20). WSIs from five of the institutes were used to train the CNNs (training set) and to measure their performance on data produced in the same institutions (internal testing set). WSIs from a sixth institute were completely withheld from training and only used for performance evaluation (external testing set). Of the total slides, 62 were native kidney biopsy specimens with diabetic nephropathy (DN), 38 were transplant kidney biopsy specimens, 13 were deceased-donor kidney biopsy specimens, and three were nephrectomies from parenchyma uninvolved by renal cell carcinoma. The training and testing WSIs were selected so that a range of IFTA severity was sampled from each institute. For substudy 2, 20 new biopsy specimens were selected from four of the six institutes (five cases from each), resulting in a total of ten DN and ten transplant WSIs. These biopsy specimens were assessed for IFTA and glomerulosclerosis by four renal pathologists (RP1, RP2, RP3, RP4), as discussed below in Renal Pathologist and CNN Agreement. For substudy 3, 69 new biopsy specimens from patients with serial eGFR measurements available (25 transplant, 44 DN) were selected from the same four institutions in substudy 2. Three biopsy specimens from the internal testing set of substudy 1 and 15 biopsy specimens from substudy 2 also had sufficient serial eGFR measurements to be included, bringing the total number of WSIs to 30 transplant and 57 DN cases, for a total of 87 WSIs. These slides were assessed by four renal pathologists, as described below in Substudy 3: Association of Histologic Scores with Patient Outcome.
Table 1.
Summary of study population
| Substudy 1 | Substudy 2 | Substudy 3 | ||||
|---|---|---|---|---|---|---|
| DN | Tx | DN | Tx | DN | Tx | |
| n | 81 | 35 | 10 | 10 | 57 | 30 |
| Age (yr), mean (SD) | 51.2 (15) | 51.1 (15) | 54.3 (11) | 54.3 (17) | 52.9 (14) | 48.6 (16) |
| Female sex, % | 46.9 | 37.0 | 30.0 | 10.0 | 42.1 | 36.7 |
| Black, % | 23.5 | 17.1 | 50.0 | 10.0 | 17.5 | 10.0 |
| White, % | 49.4 | — | 0.0 | 50.0 | 28.1 | — |
| Asian, % | 17.3 | — | 50.0 | — | 54.4 | — |
| Race other/unknown, % | 9.9 | 82.9 | 0.0 | 40.0 | 0.0 | 90.0 |
DN, diabetic nephropathy; Tx, transplant.
For transplant cases used in substudy 1, biopsy specimens were sampled from various time points post-transplantation to represent a sufficient range of chronic injury in the training and test sets. For transplant cases in substudy 3, 1-year surveillance biopsy specimens without any history of acute changes (i.e., acute tubular injury, acute rejection, etc.) in prior biopsy specimens were selected. For DN cases used in all studies, the slides were selected from all possible CKD stages.
CNN Development
To facilitate WSI annotation and subsequent CNN segmentation on WSIs, we used our previously published, publicly available, H-AI-L pipeline,22 which consists of a library of functions that allows investigators to train CNNs directly from annotations made within the WSI viewer Aperio ImageScope (Figure 1). For training the CNNs, all annotations were performed directly on WSIs using ImageScope (Figure 1A). H-AI-L converts ML predictions on image patches of a WSI (Figure 1D) into viewable boundaries in ImageScope (Figure 1, E and F), which can be used to qualitatively and quantitatively assess the CNN performance at the WSI level. The segmentation CNN model used in our study was DeepLab V2,24 built in Tensorflow.25(preprint) This network can be configured to be larger or smaller, depending on the encoder that is used. The two DeepLab version 2 variants that were studied included the standard DeepLab version 2 with DeepLab encoder, and a smaller variant using a ResNet-50 encoder. Larger network architectures are typically capable of yielding higher performance than smaller ones, but they are more prone to overfitting. Therefore, we tested both CNN variants using a low-magnification, high-magnification, and combined low- and high-magnification approach (i.e., identify regions of interest in low magnification, segment precise boundaries in high magnification).22
Figure 1.
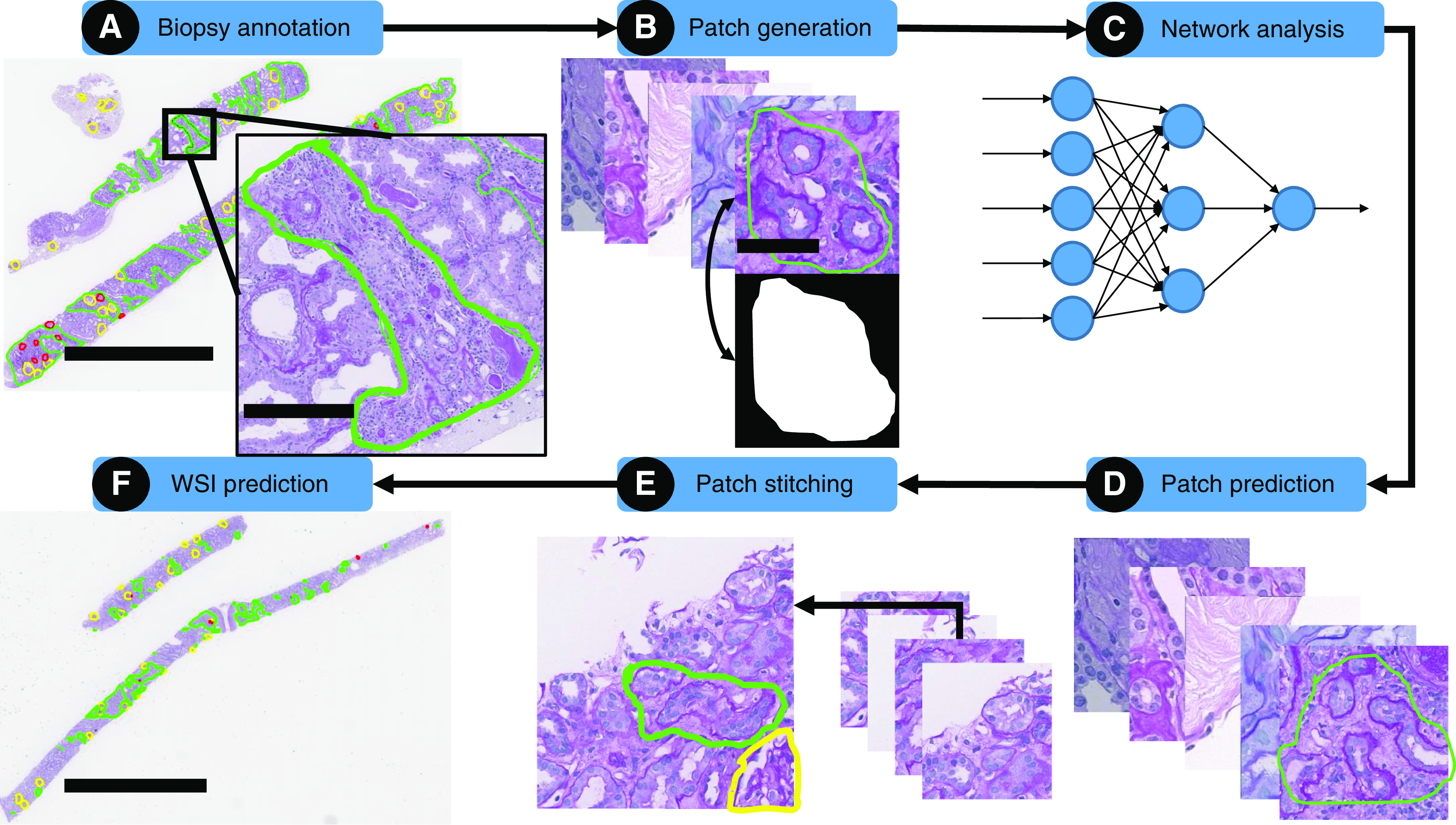
Schematic overview of CNN analysis using the H-AI-L pipeline. (A) Biopsy specimens were annotated by a pathologist who circled regions of interest; IFTA is shown in green, nonsclerotic glomeruli in yellow, and sclerotic glomeruli in red. Scale bars, whole biopsy specimen, 3 mm; zoomed region, 300 µm. (B) WSIs were chopped into image patches and masks were created for each image patch on the basis of the annotations. Scale bar, 50 µm. (C) Image patches and their corresponding masks were used to train a CNN model. (D) The CNN model was used to segment patches from testing WSIs. (E) Test images and their corresponding segmentation were stitched back together into a whole-slide segmentation mask. (F) Whole-slide, CNN-predicted regions of interest were overlaid on the test WSIs. Scale bar, 4 mm.
Network Training
Low-magnification CNN variants were trained using 40× magnification image patches downsampled four times in each dimension (resolution, 1 µm/pixel; equivalent to 10× magnification). High-magnification variants were trained directly using image patches at 40× (resolution, 0.25 µm/pixel). To convert WSIs into image patches suitable for input to a CNN, the H-AI-L algorithm starts by detecting usable tissue regions of the WSI from a low-resolution thumbnail. First, the low resolution RGB (red, green, blue) image was transformed to the HSV (hue, saturation, value) space, blurred with a Gaussian filter with standard deviation 20, and thresholded at 0.05, resulting in a binary mask detailing the tissue boundaries. A sliding-window technique was used to chop input image patches from these regions to a size of 560×560 pixels. Each patch overlapped with neighboring patches by 50% of the image width in both x and y directions. Image augmentation, the strategy of creating duplicated, slightly modified versions of the input images to increase dataset size, was used for low-magnification training, but not for high-magnification training, because down-sampling the images four times in each dimension reduced the overall data magnitude by 16 times, significantly reducing the number of training patches (approximately 35,000 low-magnification versus approximately 560,000 high-magnification patches). Details of the augmentation strategy for each image patch is available in Supplemental Appendix 1. When training a CNN, a critical parameter to select carefully is the number of steps for which the network will train. In our work, we used the concept of epochs to determine how many steps to train the CNNs. One epoch is equivalent to the CNN having trained enough steps to see all of the images in the training set one time. This value also depends on the number of images the network uses for each step of training, also referred to as the training batch size (we used batch size 2). In this study, to best compare the different network configurations, each CNN variant was trained for a similar number of steps. The high-magnification dataset had approximately 560,000 images and saw two images in each training step; thus, specifying four epochs yielded 1.1 million training steps. The low-magnification dataset after augmentation contained approximately 220,000 images and also saw two images per step; thus, a total of ten epochs also yielded 1.1 million training steps. Both networks were trained with initial learning rate 2.5×10−4, power 0.9, momentum 0.9, and weight decay 0.0005. The DeepLab encoded model was initialized on a checkpoint pretrained on the Microsoft Common Objects in COntext (MSCOCO) dataset.26 The ResNet-50–encoded model was initialized on a checkpoint pretrained on the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) dataset.27
Network Testing
A unique property of fully convolutional networks, such as DeepLab, is that, although fixed-size input images are required for training, the size of image used for segmentation does not need to match that used for training. In fact, the images can be arbitrarily sized and are only constrained by available graphical processing unit (GPU) memory. Entire WSI images cannot fit within GPU memory; therefore, similar to training, the WSI must be tiled into patches and segmented separately. Subsequently, the segmentations can be stitched back into a whole-slide prediction. For all testing conducted in this work, unless otherwise specified, we used a sliding window of size 3000×3000 pixels and allowed 50% overlap between the chopped patches (the equivalent image size for low-magnification testing was 750×750 pixels). A 3000×3000 pixel image patch centered on the tissue core was sufficiently large such that the entire biopsy specimen width was captured in each patch. For predictions using a combined low- and high-magnification approach, regions were first detected by one network trained at low magnification (10×), and then extracted and passed to a second network trained at high magnification (40×) to refine the image boundaries.
CNN Segmentation Performance
Segmentation performances for substudy 1 were calculated by pixel-level comparison of entire WSI predictions against manually annotated ground-truth WSI annotations performed by RP1. Reported performance metrics include sensitivity, specificity, and the Matthew correlation coefficient (MCC).28 MCC is a measure of the overall correlation between the predicted and known pixel classification labels, and ranges between −1 (perfect pixel-wise disagreement with the annotator), 0 (no better than random chance), and +1 (perfect pixel-wise agreement). The MCC is regarded as the best single-binary performance metric29,30 because it accounts for all possible outcomes of the binary classification (i.e., true positives, false positives, true negatives, and false negatives). This metric makes it less likely to be misleading in cases where the class distributions are highly imbalanced, which is a very common and significant problem in whole-slide analysis. Because the MCC is a contingency matrix method of calculating the Pearson correlation coefficient,29 the strength of the values can be interpreted the same. With regards to any MCC strengths referenced in our results, we followed the conventions outlined for Pearson coefficients by Chan.31,32
Renal Pathologist and CNN Agreement
To study the reliability of our CNN algorithm, its segmented IFTA and glomerulosclerosis were compared against the agreement of four renal pathologists using the cases detailed in Image Data for substudy 2. Each renal pathologist assessed all biopsy specimens in two passes. For the first pass, the pathologists visually counted the number of (1) nonsclerotic glomeruli, (2) globally sclerotic glomeruli, and (3) segmentally sclerotic glomeruli, and then visually estimated the percentage of IFTA involving the renal cortex. In the second pass, the pathologists annotated the boundaries of IFTA on the PAS-stained WSI of each renal biopsy specimen. Trichrome slides were available for reference for both passes. Each pathologist recorded the amount of time taken for each pass. Viewing and annotation of PAS- and trichrome-stained WSIs by multiple pathologists was accomplished using the Digital Slide Archive.33
Association of Histologic Scores with Patient Outcome
The cases detailed in Image Data for substudy 3 were divided among the four pathologists. Each pathologist assessed their share of biopsy specimens using the method described in Renal Pathologist and CNN Agreement, again using trichrome for reference where needed. Linear and logistic regression analyses were used to model the relationship between patient outcome and either pathologist- or CNN-based estimations of IFTA fraction and glomerulosclerosis percentage. A single predictor variable was used to compare the visual estimation of IFTA by pathologists, annotation-based IFTA by pathologists, IFTA assessed by the CNN, glomerulosclerosis assessed by pathologists, and glomerulosclerosis assessed by the CNN. For patients who had received a transplant, outcome variables were eGFR at time of biopsy as well as 1 and 2 years after biopsy. For patients with DN, outcome variables were the same, but also included whether the patient progressed to ESKD at any time after biopsy. DN WSIs were pooled from three multinational institutions and, due to this, not all patients had all outcome markers available. A total of 57 patients had eGFR at biopsy, 30 patients had eGFR at 1 year after biopsy, 23 patients had eGFR at 2 years after biopsy, and 57 patients had ESKD status after biopsy.
Calculation of IFTA and Glomerulosclerosis Percentages
Calculation of IFTA percentages from renal-pathologist annotations and CNN segmentations was performed by dividing the total area of encircled IFTA by the total area of the cortex. To calculate the cortical region of each biopsy specimen, the regions were marked manually. Glomerulosclerosis percentage was calculated by dividing the number of globally sclerotic glomeruli by the total number of glomeruli. The CNN output for glomerulosclerosis does not provide direct counts of glomeruli, but rather a pixel-level map of predicted sclerotic and nonsclerotic parts of each glomerulus. To enumerate glomerular counts from this pixel map, first, all of the pixels in the WSI predicted as nonsclerotic and sclerotic glomeruli were added together into a single binary mask depicting classless glomerular regions. Next, any doublet-detected glomeruli in this mask were split using the watershed algorithm,34,35 and binary regions with size <1500 µm2 were removed from the whole-slide mask. Finally, to count whether each detected glomerular region was either nonsclerotic or globally sclerotic, we used a voting procedure. First, the total number of pixels in each glomerulus of the WSI was tallied. Second, for each glomerulus, the number of pixels classified as sclerotic and nonsclerotic by the network were tallied. Then, using these values, the percentages of predicted sclerotic area and predicted nonsclerotic area were calculated for each glomerulus. Using these percentages, if a glomerular region consisted of >60% nonsclerotic pixels, it was counted as nonsclerotic, and, if a glomerular region had >60% sclerotic pixels, it was counted as globally sclerotic.
Pathologist versus CNN Time Comparison
For the biopsy specimen assessment performed in substudy 2 and 3, pathologists recorded the amount of time taken to visually estimate IFTA and glomerulosclerosis, and the amount of time taken to manually annotate IFTA. The amount of time taken for the CNN to segment IFTA and glomeruli on each WSI was also recorded. For this assessment, 0% overlap between chopped image patches was used, resulting in the fastest predictions.
Hardware
Computational processing was performed on a Linux distribution (Ubuntu 16.04) computer with an Intel Xeon E5-2630 CPU with 40 cores at 2.20 GHz, 64 GB of RAM, and 64 GB of swap memory. Network training and predictions were performed either on an NVIDIA (Santa Clara, CA) Titan X GPU (12 GB of memory), a Geforce RTX 2080 Ti (11 GB memory), or GeForce GTX 1080 (8 GB memory). For comparing assessment time between pathologists and the CNN, computational segmentation was performed on a Linux distribution (CentOS 7) computer with Intel Xeon Gold 6130 CPU (32 cores at 2.1 GHz), 192 GB of RAM, and an NVIDIA Tesla V100 GPU (16 GB memory).
Statistical Analyses
Intraclass correlation coefficient (ICC) values were calculated using the Real Statistics Resource Pack software.36 Binary whole-slide Cohen κ was calculated in Python using the standard formula.37 Regressions on eGFR value were performed using simple linear regression, and the F-statistics of the respective ANOVA was used to assess the significance of each predictor variable’s association with outcome variables. The R2 correlation of the respective regression models for each pair of predictor variables were compared for significance using the Z-score method,38 as discussed previously,39 and implemented in R.40 For the binary ESKD outcome, logistic regression was used, with a chi-squared test comparing the simple linear regression against a null model.41 Significance level for all analysis was 0.05, and all statistical-regression computations were performed using the R programming language.
Results
Quantitative Segmentation Performance and Model Selection (Substudy 1)
To generate a network with optimal performance, we tested the effect of image magnification (low, high, or a combination of both) on the computational detection of IFTA and glomerulosclerosis. Two different versions of the DeepLab version 2 CNN, one smaller model encoded by ResNet-50 and one larger model, the standard DeepLab version 2 model, were also compared. These models were trained on 79 slides from five institutions, and quantitative performance was evaluated on 17 test slides from these institutions (i.e., internal testing set) and 20 slides from an institution the CNN did not see in training (i.e., external testing set). Ground truth for this substudy was determined on the basis of annotations from one renal pathologist. The performance was evaluated on the basis of MCC, given that sensitivity and specificity in the context of whole-slide analysis can be misleading because the (frequently imbalanced) distribution of image classes in each WSI is not retained when averaging across a batch of WSIs.
In general, the CNNs performed best in detecting nonsclerotic glomeruli, followed by sclerotic glomeruli, and finally followed by IFTA (Figure 2). Only small performance differences were seen between the various model types. The DeepLab version 2 network operating at high magnification (D-HR) achieved the highest MCC for IFTA (0.74), nonsclerotic glomeruli (0.94), and glomerulosclerosis (0.8) detection in the internal testing set. In the external testing set, it achieved the second highest MCC for IFTA (D-HR, 0.67 versus 0.68 by ResNet-50 which combined low and high resolution), the highest MCC for nonsclerotic glomeruli detection (0.89), and the third highest MCC for glomerulosclerosis detection (D-HR, 0.66 versus 0.67 [ResNet-50 combined low and high resolution] and 0.68 [ResNet-50 low resolution]). Overall, because D-HR scored the highest average MCC on four of the six possible categories, it was selected as our optimal model for the remainder of this study.
Figure 2.
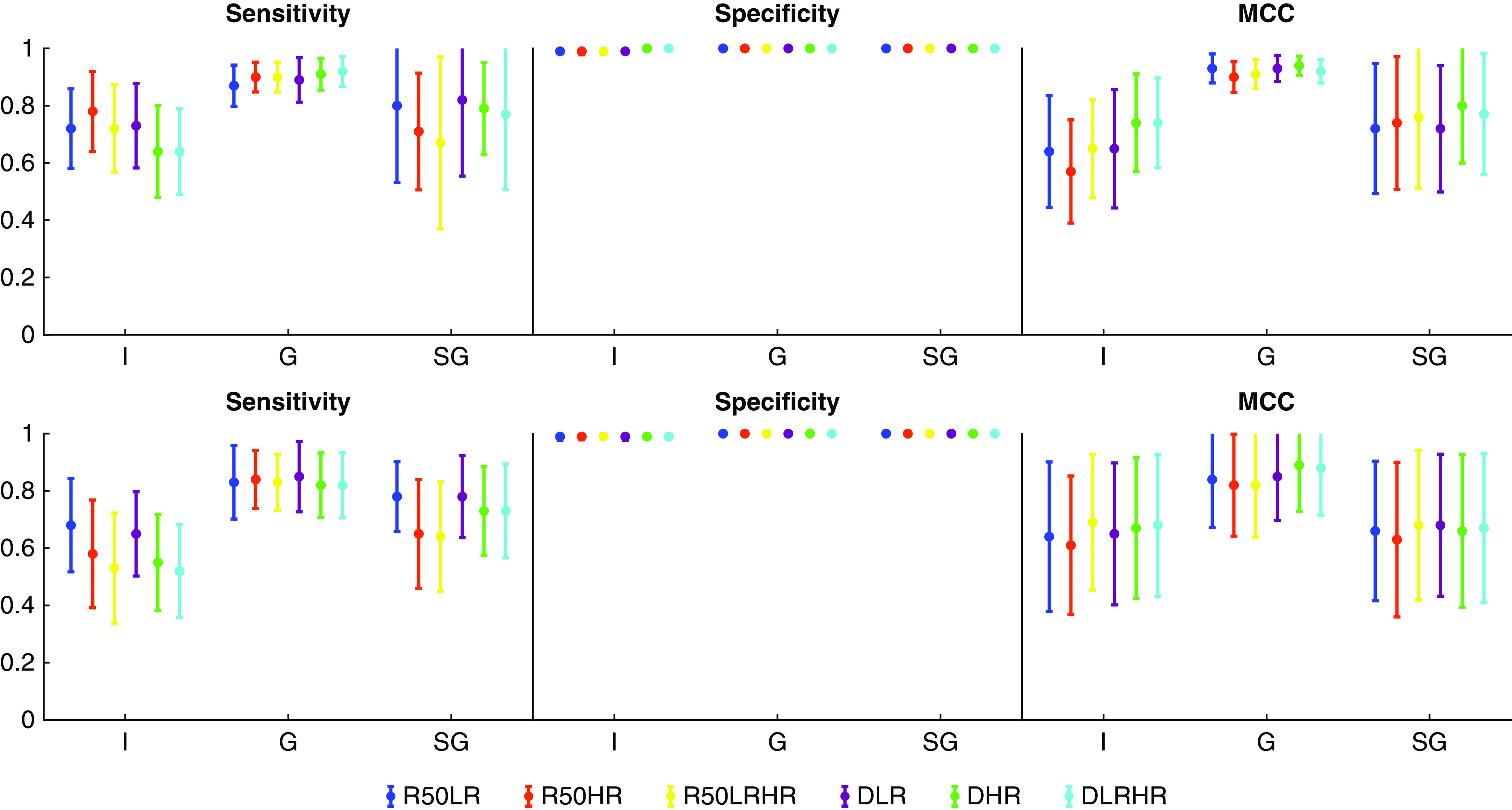
Binary segmentation performance of six different CNN configurations suggests DeepLab version 2 network operating at high magnification offers best performance. Sensitivity, specificity, and MCC for segmentation of IFTA (I), glomeruli (G), and globally sclerotic glomeruli (SG) in the internal (top row) and external (bottom row) testing sets are shown. CNN configurations were based on either a ResNet-50–encoded (R50) or DeepLab-encoded (D) network, using either low-resolution (LR), high-resolution (HR), or combined low- and high-resolution (LRHR) image patches.
Whole-Slide Segmentations (Substudy 1)
IFTA
Examples of segmentations with the D-HR network on testing WSIs are shown in Figure 3, with each panel representing a different institution. At low magnification, obvious differences in PAS stain color, saturation, and darkness were apparent between institutions. IFTA detection was moderate in the transplant (mean MCC, 0.69) and DN (mean MCC, 0.74) biopsy specimens for internal testing cases. IFTA detection was very strong in deceased-donor tissue (mean MCC, 0.82). The network also generalized, with moderate strength, to the external testing set of DN biopsy specimens (mean MCC, 0.67), showing similar performance as compared with the internal testing set.
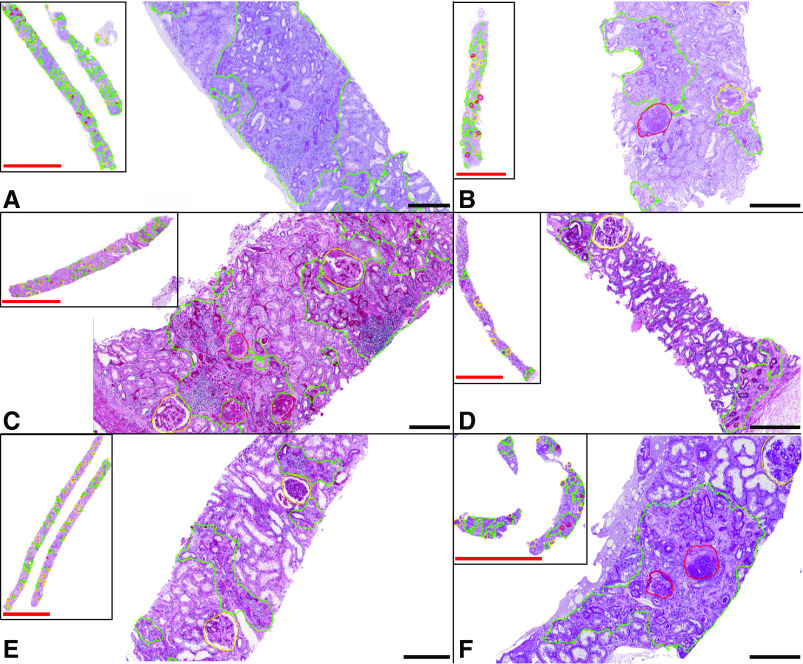
Figure 3.
CNN segmentation performance of IFTA in transplant and DN on WSIs from six different institutions was found to be moderate to very high. Whole-slide snapshots and magnified fields show IFTA (green outline), nonsclerotic glomeruli (yellow outline), and sclerotic glomeruli (red outline) segmented by the DeepLab version 2 CNN using high-resolution image patches. Each biopsy specimen shown was processed and stained at a different institution. Internal testing cases include (A) transplant biopsy specimen, (B–D) native biopsy specimens with DN, and (E) deceased-donor kidney biopsy specimen. (F) The external testing case represents native biopsy specimens with DN from an institution on which the CNN was not trained. Scale bars, 3 mm (red) and 300 µm (black).
Glomerulosclerosis
Examples of glomerular detection by the D-HR network are shown in Figure 4, where yellow boundaries indicate nonsclerotic glomeruli and red boundaries indicate sclerotic glomeruli. Again, each panel is from a different institution. The performance for detecting nonsclerotic glomeruli was very strong for all types of biopsy specimens examined, including those from transplants (mean MCC, 0.91), native kidneys with DN (mean MCC, 0.92), and deceased-donor kidneys (mean MCC, 0.95) from the internal testing set. Detection of nonsclerotic glomeruli showed similar performance in the external testing set of DN biopsy specimens, with a mean MCC of 0.89.
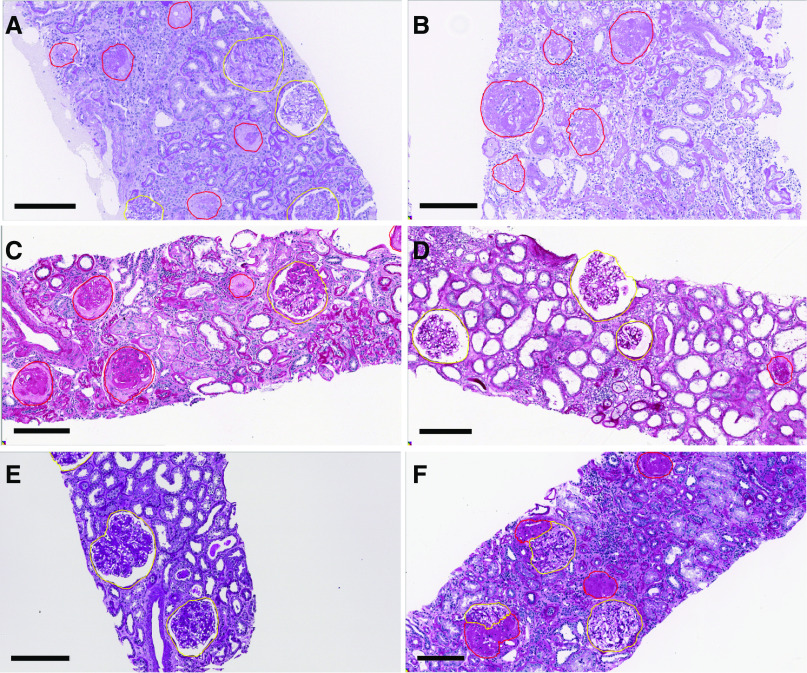
Figure 4.
CNN segmentation performance for nonsclerotic glomeruli was found to be very high, and for globally sclerotic glomeruli, moderate to very high. Nonsclerotic glomeruli (yellow) and globally sclerotic glomeruli (red) as predicted by the DeepLab version 2 CNN are shown. Each biopsy specimen was processed and stained at a different institution. Internal testing cases include (A) transplant biopsy specimen, (B and C) native biopsy specimen with DN, and (D) deceased-donor biopsy specimen. External testing cases that are shown include a native biopsy specimen with (E) DN and (F) FSGS from institutions on which the CNN was not trained, the latter of which demonstrates an example of CNN self-learned prediction of segmental sclerosis. Scale bar, 200 µm.
Detection of globally sclerotic glomeruli was moderate in transplant biopsy specimens (mean MCC, 0.66) and very strong in native kidney biopsy specimens with DN (mean MCC, 0.81) and deceased-donor tissues (mean MCC, 0.92) in the internal testing set. In the external testing set of DN biopsy specimens, the average sclerotic glomerulus MCC was moderate (MCC, 0.66).
An interesting, unexpected result was that, despite not being trained on segmentally sclerotic glomeruli, the CNN was able to identify some glomeruli as segmentally sclerotic. To test the network’s segmental glomerulosclerosis predictions in a more typical case showing segmentally sclerotic glomeruli, a single biopsy specimen with confirmed FSGS was processed by the CNN. The output of the network’s prediction is shown in Figure 4F, where the network identified regions of open glomerular capillary loops as nonsclerotic glomerulus, and adjacent regions with sclerosis as sclerotic glomerulus. Although the quantitative performance assessment of segmental sclerosis was not the focus of our study, the qualitative segmentation result suggests convolutional approaches may be able to help objectify determination of segmental sclerosis in future studies.
Interpathologist/CNN Reliability (Substudy 2)
In substudy 1, ground truth used for training and performance evaluation was generated by a single renal pathologist (RP1). However, manual annotation of IFTA is imprecise due to its complex morphologic definition. Past studies have shown clear disagreement between pathologists for the grading of IFTA.11,15,42,43 Therefore, in substudy 2, the interpathologist/CNN reliability was assessed. Four renal pathologists examined 20 WSIs and (1) visually estimated IFTA percentage, (2) visually counted nonsclerotic and sclerotic glomeruli, and (3) directly annotated IFTA on the WSIs. These data were compared with the D-HR CNN assessment.
IFTA Agreement
Each renal pathologist’s estimated IFTA percentage was highly correlated with their IFTA percentage measured by annotation (RP1, R=0.96; 95% CI, 0.87 to 0.99; RP2, R=0.97; 95% CI, 0.91 to 0.99; RP3, R=0.98; 95% CI, 0.93 to 0.99; RP4, R=0.95; 95% CI, 0.85 to 0.98). To measure agreement of IFTA annotations, Cohen κ was calculated pairwise between renal pathologists, and pairwise between the CNN and each pathologist, at the whole-slide pixel level; all achieved moderate agreement (Table 2). The Cohen κ between any two renal pathologists ranged from 0.41 to 0.58, and the Cohen κ for any CNN-pathologist pair was similar in range (from 0.45 to 0.55). These results indicate the average interobserver reliability of IFTA segmentation made by the CNN was quantitatively indistinguishable from annotations made by the renal pathologists.
Table 2.
Interpathologist/CNN reliability
| Annotator | RP1 | RP2 | RP3 | RP4 | C |
|---|---|---|---|---|---|
| RP1 | 0.58 | 0.56 | 0.49 | 0.55 | |
| RP2 | 0.49 | 0.44 | 0.51 | ||
| RP3 | 0.41 | 0.48 | |||
| RP4 | 0.45 |
Average whole-slide Cohen κ calculated at the pixel level for IFTA annotation between each pair of renal pathologists and the computer are shown. RP, renal pathologist; C, computer.
To further evaluate CNN reliability, we compared the IFTA percentages determined by pathologists and the CNN using the ICC. We first measured the ICC among renal pathologists alone, and then measured the ICC again but included the CNN as a fifth observer (Figure 5). IFTA percentage determined by visual estimation showed high agreement among the pathologists (ICC, 0.95; 95% CI, 0.90 to 0.98), and the agreement was nearly identical when including the CNN IFTA assessment (ICC, 0.94; 95% CI, 0.89 to 0.97). Agreement among pathologists using IFTA percentages based on annotations was even higher (ICC, 0.97; 95% CI, 0.93 to 0.99), and, again, was essentially identical when the CNN assessment was included (ICC, 0.97; 95% CI, 0.94 to 0.99). These data indicate that our CNN-based algorithm was able to show similar reliability in performance as renal pathologists.
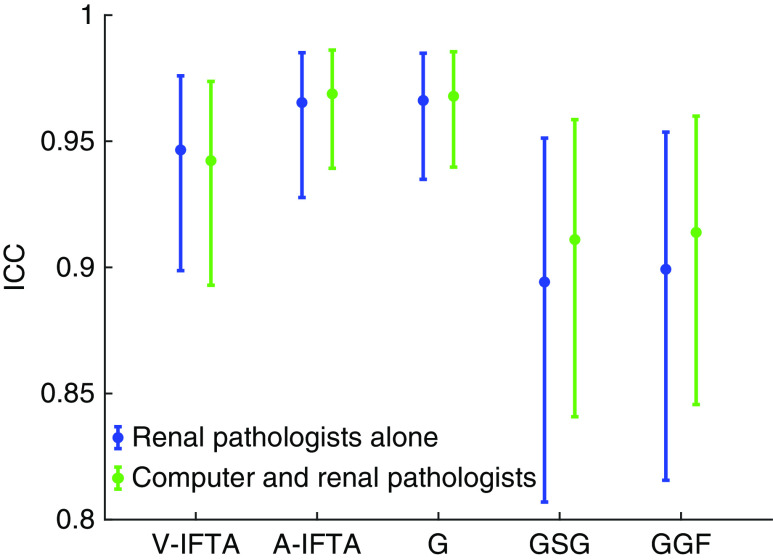
Figure 5.
Comparison of ICCs for detection of IFTA and glomerulosclerosis indicates that our CNN-based algorithm and renal pathologists have similar reliability. ICCs with 95% confidence intervals are shown for visual estimation of IFTA (V-IFTA), annotation-based IFTA (A-IFTA), nonsclerotic glomeruli (G), globally sclerotic glomeruli (GSG), and glomerulosclerosis percentage (GGF). ICCs are compared on the basis of renal pathologists alone as a group (blue) versus renal pathologists and the CNN-based assessment (green).
Glomerulosclerosis Agreement
A similar ICC analysis was used to evaluate the CNN’s reliability in detecting glomeruli. Detection of nonsclerotic glomeruli had the highest, and essentially identical, ICC for renal pathologists alone (0.97; 95% CI, 0.93 to 0.98) and when the CNN assessment was included (0.97; 95% CI, 0.94 to 0.99). The agreement for globally sclerotic glomeruli was slightly lower with a renal pathologist ICC of 0.89 (95% CI, 0.81 to 0.95) and a renal pathologist with CNN ICC of 0.91 (95% CI, 0.84 to 0.96). As expected, the ICC for percentage of global glomerulosclerosis fell between the ICC values observed for its associated constituents (i.e., globally sclerotic and nonsclerotic glomeruli) with an ICC of 0.90 (95% CI, 0.82 to 0.95) for renal pathologists alone and an ICC of 0.91 (95% CI, 0.85 to 0.96) for renal pathologists with CNN assessment.
Correlation of CNN-Based IFTA and Glomerulosclerosis Detection with Patient Outcome (Substudy 3)
The percentage of IFTA and globally sclerotic glomeruli based on pathologists’ assessment of biopsy tissue are important prognostic markers of CKDs.2,44,45 Thus, we evaluated whether CNN-based IFTA and glomerulosclerosis quantification preserved this well-established relationship between these histologic manifestations of chronic kidney injury and patient outcome. Regression analysis was performed for CNN-based biopsy assessment and compared with pathologists’ biopsy specimen evaluation using patient eGFR at biopsy, after biopsy, and ESKD status as independent variables. Regression analysis was carried out in two separate cohorts, one consisting of 30 1-year post-transplant surveillance biopsy specimens and the other consisting of 57 native kidney biopsy specimens with DN. For all 87 biopsy specimens, IFTA and glomerulosclerosis were measured by the CNN. The biopsy specimens were also divided up among the renal pathologists so that each case was examined by one of the four renal pathologists.
Transplant Biopsy Specimens
Regression analysis for transplant biopsy specimens is shown in Table 3. All predictor variables (pathologist’s visual estimation of IFTA, pathologist’s annotation-based IFTA, pathologist’s glomerulosclerosis percentage, and CNN-based IFTA and glomerulosclerosis percentage) were significantly associated with eGFR at time of biopsy and with eGFR at 2 years after biopsy. All predictors, except for pathologist-based glomerulosclerosis percentage, were significantly associated with eGFR at 1 year after biopsy. Pairwise comparison of R2 values for the models of IFTA and glomerulosclerosis detection revealed no significant differences for all eGFR time points. This indicated both pathologist- and CNN-based IFTA fraction and glomerulosclerosis percentage were similar in their ability to predict patient outcome (Table 4).
Table 3.
Correlation of IFTA and glomerulosclerosis detection by pathologists and CNN to transplant patient eGFRs
| Variable | Biopsy | 1 Year | 2 Years | |||
|---|---|---|---|---|---|---|
| R2 | P | R2 | P | R2 | P | |
| V-IFTA (pathologist) | 0.25 | 0.005 | 0.24 | 0.007 | 0.21 | 0.01 |
| A-IFTA (pathologist) | 0.29 | 0.002 | 0.25 | 0.005 | 0.28 | 0.003 |
| C-IFTA (CNN) | 0.29 | 0.002 | 0.31 | 0.002 | 0.34 | <0.001 |
| GS (pathologist) | 0.17 | 0.02 | 0.11 | 0.07 | 0.15 | 0.04 |
| GS (CNN) | 0.27 | 0.003 | 0.18 | 0.02 | 0.24 | 0.006 |
R2 and P value of the F-statistics of simple linear regression are shown for time of biopsy (biopsy) and 1 year and 2 years after time of biopsy. V-IFTA, visual estimation of IFTA; A-IFTA, annotation-based IFTA; GS, glomerulosclerosis.
Table 4.
eGFR-dependent comparison of R2 for IFTA and glomerulosclerosis as performed by pathologists and the CNN in transplant patients
| Comparative Variables | Biopsy | 1 Year | 2 Years | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Z | P | 95% CI | Z | P | 95% CI | Z | P | 95% CI | |
| V-IFTA versus A-IFTA | 0.40 | 0.69 | −0.20 to 0.30 | 0.16 | 0.88 | −0.22 to 0.26 | 0.75 | 0.45 | −0.15 to 0.34 |
| C-IFTA versus V-IFTA | 0.45 | 0.65 | −0.18 to 0.29 | 0.78 | 0.44 | −0.14 to 0.33 | 1.41 | 0.16 | −0.07 to 0.41 |
| C-IFTA versus A-IFTA | 0.06 | 0.95 | −0.11 to 0.12 | 1.28 | 0.20 | −0.04 to 0.19 | 1.30 | 0.19 | −0.04 to 0.19 |
| C-GS versus P-GS | 1.67 | 0.09 | −0.03 to 0.32 | 1.26 | 0.21 | −0.06 to 0.27 | 1.52 | 0.13 | −0.04 to 0.30 |
Comparisons are shown for time of biopsy (biopsy) and 1 year and 2 years after time of biopsy.
V-IFTA, visual estimation of IFTA by pathologist; A-IFTA, annotation-based IFTA by pathologist; C-IFTA, IFTA assessed by CNN; C-GS, glomerulosclerosis assessed by CNN; P-GS, glomerulosclerosis assessed by pathologist.
DN Biopsy Specimens
Regression analysis for the patients with DN is shown in Table 5. Again, all predictor variables were significantly associated with eGFR at time of biopsy and with eGFR at 1 year after biopsy. All IFTA predictors were significantly associated with eGFR at 2 years after biopsy, but neither CNN nor pathologist’s glomerulosclerosis percentages were significant for eGFR at this time point. Again, pairwise comparison of R2 values for the modes of IFTA and glomerulosclerosis detection revealed no significant differences for all eGFR time points (Table 6). Furthermore, all predictor variables were found to be significantly associated with ESKD status (Table 7).
Table 5.
Correlation of IFTA and glomerulosclerosis detection by pathologists and CNN to the eGFRs of patients with DN
| Variable | Biopsy | 1 Year | 2 Years | |||
|---|---|---|---|---|---|---|
| R2 | P | R2 | P | R2 | P | |
| V-IFTA (pathologist) | 0.45 | <0.001 | 0.47 | <0.001 | 0.39 | 0.001 |
| A-IFTA (pathologist) | 0.41 | <0.001 | 0.42 | <0.001 | 0.43 | 0.001 |
| C-IFTA (CNN) | 0.43 | <0.001 | 0.47 | <0.001 | 0.44 | 0.001 |
| GS (pathologist) | 0.21 | <0.001 | 0.21 | 0.01 | 0.07 | 0.24 |
| GS (CNN) | 0.23 | <0.001 | 0.23 | 0.01 | 0.06 | 0.28 |
R2 and P value of the F-statistics of simple linear regression are shown for time of biopsy (biopsy) and 1 year and 2 years after time of biopsy. V-IFTA, visual estimation of IFTA; A-IFTA, annotation-based IFTA; C-IFTA, IFTA assessed by CNN; GS, glomerulosclerosis.
Table 6.
eGFR-dependent comparison of R2 for IFTA and glomerulosclerosis as performed by pathologists and the CNN in patients with DN
| Comparative Variables | Biopsy | 1 Year | 2 Years | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Z | P | 95% CI | Z | P | 95% CI | Z | P | 95% CI | |
| V-IFTA versus A-IFTA | 0.57 | 0.57 | −0.12 to 0.23 | −0.57 | 0.57 | −0.23 to 0.12 | 0.32 | 0.76 | −0.25 to 0.34 |
| C-IFTA versus V-IFTA | −0.20 | 0.84 | −0.22 to 0.18 | 0.01 | 0.99 | −0.31 to 0.32 | 0.37 | 0.71 | −0.28 to 0.42 |
| C-IFTA versus A-IFTA | 0.08 | 0.94 | −0.08 to 0.08 | 0.68 | 0.50 | −0.15 to 0.30 | 0.16 | 0.87 | −0.24 to 0.28 |
| C-GS versus P-GS | 0.51 | 0.61 | −0.08 to 0.13 | 0.50 | 0.62 | −0.10 to 0.17 | −0.30 | 0.76 | −0.16 to 0.12 |
Comparisons are shown for time of biopsy (biopsy) and 1 year and 2 years after time of biopsy. V-IFTA, visual estimation of IFTA by pathologist; A-IFTA, annotation-based IFTA by pathologist; C-IFTA, IFTA assessed by CNN; C-GS, glomerulosclerosis assessed by CNN; P-GS, glomerulosclerosis assessed by pathologist.
Table 7.
Logistic regression results for IFTA and glomerulosclerosis versus ESKD status any time after biopsy in patients with DN
| Variable | Coefficient | 95% CI | P Value of Wald Test | Chi-Square Score of Model Fit (df=1) | P Value of Chi-Square Score |
|---|---|---|---|---|---|
| C-IFTA | 6.11 | 17 to 11959.91 | <0.001 | 19.21 | 0 |
| V-IFTA | 4.65 | 9.13 to 1192.34 | <0.001 | 18.84 | 0 |
| A-IFTA | 5.77 | 12.21 to 8343.14 | <0.001 | 16.36 | <0.001 |
| C-GS | 2.88 | 1.79 to 176.89 | 0.01 | 6.86 | 0.01 |
| P-GS | 3.3 | 2.52 to 292.38 | 0.01 | 8.78 | 0.003 |
C-IFTA, IFTA assessed by CNN; V-IFTA, visual estimation of IFTA by pathologist; A-IFTA, annotation-based IFTA by pathologist; C-GS, glomerulosclerosis assessed by CNN; P-GS, glomerulosclerosis assessed by pathologist; df, degrees of freedom.
On the basis of comparison of R2 values, all IFTA estimates, independent of whether they were determined by pathologists or CNN, were comparable in terms of correlating to patient outcomes, for both transplant and DN biopsy specimens. Similarly, both pathologist- and CNN-based glomerulosclerosis percentages were comparable in correlating to patient outcomes. IFTA estimates were overall better predictors of patient outcome than glomerulosclerosis percentage for both transplant and DN biopsy specimens.
Comparison of CNN versus Pathologist Biopsy Specimen Assessment Speed
For substudy 2 and substudy 3, the pathologists recorded the amount of time taken for visual estimation of IFTA and enumeration of nonsclerotic and sclerotic glomeruli, and separately for annotation of IFTA borders. We compared this against the time taken by the CNN to segment IFTA and glomerulosclerosis for the same set of biopsy specimens. For the fastest WSI segmentation times, we used 0% overlap between the image patches chopped for processing by the CNN. In the previous three substudies, we had used 50% overlap between patches for denser prediction. To ensure that reducing the patch overlap would not significantly reduce the performance of the CNN, we first reran substudy 1 using our best CNN model (D-HR), using 0% overlap between image prediction patches. With 50% overlap, the average MCCs were 0.74 (IFTA), 0.94 (nonsclerotic glomeruli), and 0.80 (sclerotic glomeruli). Using 0% overlap, the average MCCs were 0.75 (IFTA), 0.93 (nonsclerotic glomeruli), and 0.79 (sclerotic glomeruli), indicating essentially identical performance.
The amount of time taken by the CNN and by the pathologists is shown in Figure 6. The median time taken per biopsy specimen by pathologists for the combined visual estimation of IFTA percentages and enumeration of glomeruli in substudy 2 ranged from 64 to 256 seconds. Annotation-based IFTA assessment by pathologists took significantly longer, with median times ranging between 98 and 602 seconds. In comparison, CNN-based segmentation of both IFTA and glomerulosclerosis took a median time of 155 seconds per biopsy specimen, comparable with pathologists’ visual estimation times. For the biopsy specimens of substudy 3, the median time taken per biopsy specimen for the group of pathologists to perform visual IFTA estimation and glomerulus enumeration was 120 seconds, and for annotation of IFTA, 270 seconds. In comparison, the CNN median time was 151 seconds for segmentation of both IFTA and glomerulosclerosis. For annotation, the CNN showed a much lower 75th percentile time than pathologists, although the pathologist 25th percentile values were lower (Figure 6). This finding was observed because, in the pathologists’ annotation process, slides that have minimal IFTA require short annotation times, whereas slides with patchy IFTA take relatively longer to detail the complex boundaries. The CNN, on the other hand, processes all image patches in an equal amount of time, regardless of their content, leading to slightly higher times taken than the pathologist, on average, but showing a much smaller range of times taken.
Figure 6.
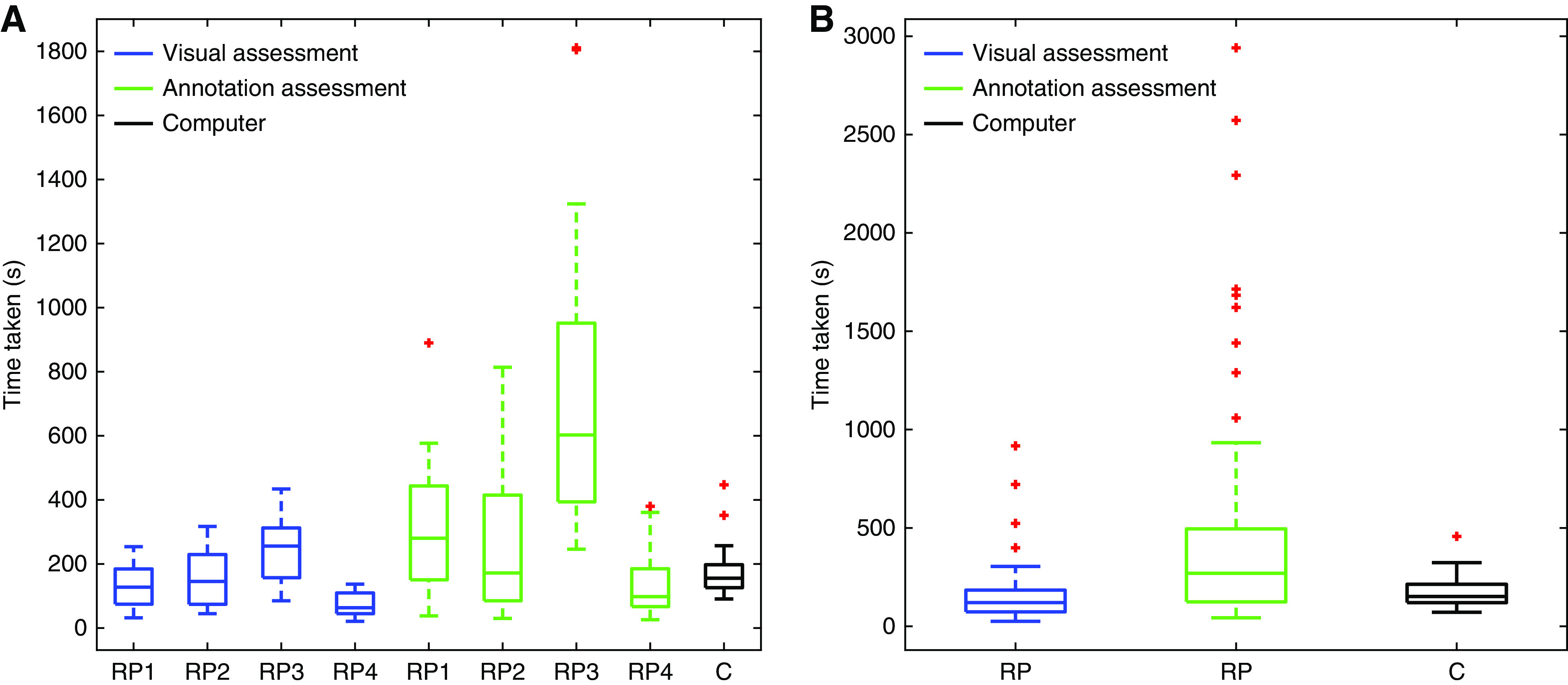
Comparison of time taken for detection of IFTA by renal pathologists and CNN suggests that the CNN can annotate IFTA and glomeruli with comparable average speed to human visual assessment. (A) Time taken by individual renal pathologists (RP) in substudy 2 to visually estimate IFTA and glomerulosclerosis (blue) and manually annotate IFTA (green) as compared with the time taken by the CNN (C; black) to segment IFTA and glomerulosclerosis. (B) Time taken by the group of renal pathologists in substudy 3 to visually estimate IFTA and glomerulosclerosis (blue) and manually annotate IFTA (green) as compared with the time taken by the CNN (C; black) to segment and quantify IFTA and glomerulosclerosis.
Discussion
With recent progressive technologic advances, it is now feasible to train ML algorithms built on CNNs to classify pixel-based image data with unprecedented robustness. In this study, we developed CNN-based algorithms to detect IFTA and glomerulosclerosis in renal biopsy specimen WSIs. Our CNN was trained on slides from multiple institutions and encompassed various types of biopsy specimens including transplant, native kidneys with DN, and deceased-donor kidneys. As a result, the CNN performance was robust and generalizable to WSIs originating from an institution that was not observed in training. Although our CNN was trained only on one pathologist’s annotations, the network performance on secondary testing showed similar interobserver reliability as other individual renal pathologists. Thus, the CNN appeared to replicate the performance of individual renal pathologists for detection of IFTA and glomerulosclerosis. Finally, we found that both renal pathologist and CNN-based assessment of IFTA and glomerulosclerosis were significantly and similarly associated with patient outcome in terms of eGFR and ESKD status. Overall, these results indicate that, when sufficiently trained, CNNs are able to detect IFTA and glomerulosclerosis accurately and reliably as compared with renal pathologists, and can generalize to biopsy specimen data from unseen institutions.
We also found the time taken by our CNN to detect and quantitate IFTA and glomerulosclerosis was comparable with the equivalent visual assessment by pathologists, and both of these methods were much more time efficient than a pathologist’s manual annotation. Thus, reliable CNN annotation offers a significant advantage over manual annotation, not only by saving time per case, but also CNNs can be easily replicated and run on multiple computers, and do not require rest like humans do. Thus, massive annotation loads can be created in a relatively short period of time, which is ideal for research purposes.
Limitations to our study include inherent imprecision of digital annotation and the lack of a gold-standard definition for IFTA boundaries in the biopsy specimen. The former issue is a result of the imperfect ability of the computer mouse to precisely replicate the intended mental action of its operator. In fact, it would take an infeasible amount of time to generate a set of pixel-level, “perfectly” annotated data. Even with such a dataset, it would be unlikely that the increased annotation precision would result in any significant improvements in network performance. Instead, the main source of imprecision in IFTA assessment stems from the fact that there is no consensus in the community over a quantifiable definition or biologic marker that can be used to clearly delineate IFTA. For instance, to what degree must the interstitium be expanded by collagen accumulation for that area of the renal parenchyma to be designated interstitial fibrosis? Also, regions of IFTA typically do not have clear-cut borders, and annotation by pathologists can be quite subjective when marking borders of these regions. Ground truth generated through molecular markers, such as type-3 collagen immunohistochemistry (IHC), would likely provide the most exact localization and quantification of collagen, but what degree of collagen accumulation would be considered pathologic remains unclear.11,46 Additionally, using an IHC marker for ground-truth annotation requires the same IHC tissue section to be histochemically stained (e.g., with PAS), while preserving underlying tissue architecture, which can present technical challenges. Reproducible image registration to align the histochemical-stained images with the IHC-stained images can present additional problems. Because of these limitations, we resorted to annotations by renal pathologists as the gold standard in this study. However, given the variable morphologic presentation of IFTA, especially in early or evolving IFTA with wispy collagen accumulation and intermixed edema, we chose to limit the scope of our study to cases with well-developed morphologic manifestations of IFTA (i.e., classic, endocrinization, and thyroidization types), which is the most common form seen in chronic kidney injury. Future studies examining early or evolving IFTA may be helpful to determine whether specific morphologic features can predict progression to well-developed IFTA or repair with recovery.
The finding of equivalence between our CNN and renal pathologist performance is not particularly surprising, because it is intuitive that the performance of the algorithm would be limited to the performance of its ground truth. However, given equivalent performance, the computational method may be used as a “stand-in” for renal pathologists in scenarios where the availability of a renal pathologist’s assessment or time is infeasible. One example may be for areas of the world that do not have access to renal-pathologist expertise. Another example would be in research, where large datasets of hundreds or thousands of slides require annotation. Finally, although this application of CNNs did not exceed the renal pathologists’ performance in detecting IFTA and glomerulosclerosis, future work to examine further details in quantification analysis of specific morphometric features and/or subvisual characteristics in an automated fashion could possibly lead to discovery of image features that could be superior, diagnostically or prognostically, to mere IFTA quantitation.
In conclusion, the application of ML algorithms for the analysis of renal biopsy specimens has significant merit for reproducing certain aspects of assessment by renal pathologists. In particular, detection and quantification of IFTA and glomerulosclerosis were shown in this study. Such automated processes have the potential to provide invaluable access to large-scale image data that, in the future, may be further mined for possible novel or improved prognostic indicators of renal outcome.
Disclosures
C. Parikh serves as a data and safety monitoring board member for Genfit; has consultancy agreements with Genfit Biopharmaceutical Company; reports receiving research funding from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and National Heart, Lung, and Blood Institute; and is a member of the advisory board of, and owns equity in, RenalytixAI. A. Rosenberg reports being a scientific advisor for or member of Escala; receiving honoraria from Georgetown University, Ichilov Hospital (Tel Aviv, Israel), and Stony Brook University; and receiving research funding from the National Kidney Foundation and NIH. All remaining authors have nothing to disclose.
Funding
The project was supported by NIDDK Diabetic Complications Consortium grants U24 DK076169, R01 DK114485, CKD Biomarker Consortium grant U01 DK103225, Kidney Precision Medicine Project grant U2C DK114886, and NIH Office of Director (OD) grant R01 DK114485 (02S1 and 03S1). The deceased-donor study cohort is supported by NIDDK grant R01 DK093770.
Supplementary Material
Acknowledgments
The authors thank Ms. Ellen Donnert for her assistance in selecting DN biopsy specimens from the Vanderbilt University Medical Center collection. The authors are grateful to Ms. Diane Salamon, for assistance in identifying cases for the study, and for the support from the Kidney Translational Research Center and the Division of Nephrology at the Washington University School of Medicine in St. Louis.
The authors acknowledge the assistance of the Histology Core Laboratory and Multispectral Imaging Suite in the Department of Pathology and Anatomical Sciences, Jacobs School of Medicine and Biomedical Sciences, University at Buffalo. The authors thank NVIDIA Corporation for the donation of the Titan X Pascal GPU used for this research. The authors acknowledge the assistance of the Seoul National University Hospital Human Biobank (a member of the National Biobank of Korea, which is supported by the Ministry of Health and Welfare, Republic of Korea) for provision of human biospecimens used. The authors acknowledge support provided by the Center for Computational Research at the University at Buffalo.47
Mr. Brandon Ginley conceptualized and performed the quantitative analyses, designed and conducted the computational methods, interpreted the results, and wrote the manuscript; Dr. Kuang-Yu Jen conceived the IFTA research scheme, provided the transplant biopsy specimen data, provided ground-truth annotation for substudy 1, provided IFTA annotation and biopsy specimen visual assessment for studies 2 and 3, provided clinical feedback to optimize the design of the computational strategy, and cowrote the manuscript; Dr. Seung Seok Han, Dr. Donghwan Yun, and Dr. Kyung Chul Moon provided biopsy specimen and clinical data for patients with diabetes; Dr. Luís Rodrigues provided biopsy specimen data for patients with diabetes and those who have received transplants, and IFTA annotation and biopsy specimen visual assessment; Dr. Sanjay Jain and Dr. Agnes B. Fogo provided DN cases, and gave overall advice and input on the manuscript; Dr. Jonathan Zuckerman and Dr. Vighnesh Walavalkar provided IFTA annotation and biopsy specimen visual assessment; Dr. Jeffrey C. Miecznikowski performed statistical analysis and provided interpretation of the results; Dr. Yumeng Wen and Dr. Chirag Parikh gathered biopsy specimen data from deceased-donor kidneys for network training; Ms. Felicia Yen selected proper cases and clinical data from the University of California at Davis archival records; Dr. Avi Rosenberg provided clinical feedback on the work; Dr. Pinaki Sarder conceived the overall research scheme, orchestrated the study and study team, conducted statistical analysis and critically analyzed the results, and assisted in manuscript preparation.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Automated Quantification of Chronic Changes in the Kidney Biopsy: Another Step in the Right Direction,” on pages 767–768.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2020050652/-/DCSupplemental.
Supplemental Table 1. Supplementary term definitions.
Supplemental Table 2. Collection of slides from each institution.
Supplemental Appendix 1. Augmentation strategy.
References
- 1.Okada T, Nagao T, Matsumoto H, Nagaoka Y, Wada T, Nakao T: Histological predictors for renal prognosis in diabetic nephropathy in diabetes mellitus type 2 patients with overt proteinuria. Nephrology (Carlton) 17: 68–75, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Tervaert TW, Mooyaart AL, Amann K, Cohen AH, Cook HT, Drachenberg CB, et al.; Renal Pathology Society: Pathologic classification of diabetic nephropathy. J Am Soc Nephrol 21: 556–563, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Weening JJ, D’Agati VD, Schwartz MM, Seshan SV, Alpers CE, Appel GB, et al.; International Society of Nephrology Working Group on the Classification of Lupus Nephritis; Renal Pathology Society Working Group on the Classification of Lupus Nephritis: The classification of glomerulonephritis in systemic lupus erythematosus revisited [published correction appears in Kidney Int 65: 1132, 2004]. Kidney Int 65: 521–530, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Roberts IS, Cook HT, Troyanov S, Alpers CE, Amore A, Barratt J, et al.; Working Group of the International IgA Nephropathy Network and the Renal Pathology Society: The Oxford classification of IgA nephropathy: Pathology definitions, correlations, and reproducibility. Kidney Int 76: 546–556, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Sethi S, D’Agati VD, Nast CC, Fogo AB, De Vriese AS, Markowitz GS, et al.: A proposal for standardized grading of chronic changes in native kidney biopsy specimens. Kidney Int 91: 787–789, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Srivastava A, Palsson R, Kaze AD, Chen ME, Palacios P, Sabbisetti V, et al.: The prognostic value of histopathologic lesions in native kidney biopsy specimens: Results from the Boston kidney biopsy cohort study. J Am Soc Nephrol 29: 2213–2224, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosio FG, El Ters M, Cornell LD, Schinstock CA, Stegall MD: Changing kidney allograft histology early posttransplant: Prognostic implications of 1-year protocol biopsies. Am J Transplant 16: 194–203, 2016 [DOI] [PubMed] [Google Scholar]
- 8.Serón D, Moreso F: Protocol biopsies in renal transplantation: Prognostic value of structural monitoring. Kidney Int 72: 690–697, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Myllymäki J, Saha H, Mustonen J, Helin H, Pasternack A: IgM nephropathy: Clinical picture and long-term prognosis. Am J Kidney Dis 41: 343–350, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Bohle A, Wehrmann M, Bogenschütz O, Batz C, Müller CA, Müller GA: The pathogenesis of chronic renal failure in diabetic nephropathy. Investigation of 488 cases of diabetic glomerulosclerosis. Pathol Res Pract 187: 251–259, 1991 [DOI] [PubMed] [Google Scholar]
- 11.Farris AB, Chan S, Climenhaga J, Adam B, Bellamy CO, Serón D, et al.: Banff fibrosis study: Multicenter visual assessment and computerized analysis of interstitial fibrosis in kidney biopsies. Am J Transplant 14: 897–907, 2014 [DOI] [PubMed] [Google Scholar]
- 12.Snoeijs MG, Boonstra LA, Buurman WA, Goldschmeding R, van Suylen RJ, van Heurn LW, et al.: Histological assessment of pre-transplant kidney biopsies is reproducible and representative. Histopathology 56: 198–202, 2010 [DOI] [PubMed] [Google Scholar]
- 13.Grootscholten C, Bajema IM, Florquin S, Steenbergen EJ, Peutz-Kootstra CJ, Goldschmeding R, et al.: Interobserver agreement of scoring of histopathological characteristics and classification of lupus nephritis. Nephrol Dial Transplant 23: 223–230, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Gough J, Rush D, Jeffery J, Nickerson P, McKenna R, Solez K, et al.: Reproducibility of the Banff schema in reporting protocol biopsies of stable renal allografts. Nephrol Dial Transplant 17: 1081–1084, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Furness PN, Taub N, Assmann KJ, Banfi G, Cosyns JP, Dorman AM, et al.: International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol 27: 805–810, 2003 [DOI] [PubMed] [Google Scholar]
- 16.Farris AB, Adams CD, Brousaides N, Della Pelle PA, Collins AB, Moradi E, et al.: Morphometric and visual evaluation of fibrosis in renal biopsies. J Am Soc Nephrol 22: 176–186, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Servais A, Meas-Yedid V, Noël LH, Martinez F, Panterne C, Kreis H, et al.: Interstitial fibrosis evolution on early sequential screening renal allograft biopsies using quantitative image analysis. Am J Transplant 11: 1456–1463, 2011 [DOI] [PubMed] [Google Scholar]
- 18.Hermsen M, de Bel T, den Boer M, Steenbergen EJ, Kers J, Florquin S, et al.: Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol 30: 1968–1979, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolachalama VB, Singh P, Lin CQ, Mun D, Belghasem ME, Henderson JM, et al.: Association of pathological fibrosis with renal survival using deep neural networks. Kidney Int Rep 3: 464–475, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bukowy JD, Dayton A, Cloutier D, Manis AD, Staruschenko A, Lombard JH, et al.: Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol 29: 2081–2088, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedraza A, Gallego J, Lopez S, Gonzalez L, Laurinavicius A, Bueno G: Glomerulus classification with convolutional neural networks. In: Medical Image Understanding and Analysis: 21st Annual Conference, MIUA 2017, Edinburgh, United Kingdom, July 11–13, 2017, Proceedings, edited by Valdés Hernández M, González-Castro V. Cham, Switzerland, Springer International Publishing, 2017, pp 839–849 [Google Scholar]
- 22.Lutnick B, Ginley B, Govind D, McGarry SD, LaViolette PS, Yacoub R, et al.: An integrated iterative annotation technique for easing neural network training in medical image analysis. Nat Mach Intell 1: 112–119, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginley B, Lutnick B, Jen KY, Fogo AB, Jain S, Rosenberg A, et al.: Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol 30: 1953–1967, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Ji S: Smoothed dilated convolutions for improved dense prediction. Presented at the 2018 24th Association for Computing Machinery Special Interest Group on Knowledge Discovery in Data International Conference on Knowledge Discovery and Data Mining, London, August 19–23, 2018
- 25.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al.: Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv. 1603.04467 (Preprint posted March 14, 2016)
- 26.Lin TY, Maire M, Belongie S, Hays J, Perona P, Ramanan D, et al.: Microsoft COCO: Common objects in context. Lect Notes Comput Sci 8693: 740–755, 2014 [Google Scholar]
- 27.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al.: ImageNet large scale visual recognition challenge. Int J Comput Vis 115: 211–252, 2015 [Google Scholar]
- 28.Matthews BW: Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta 405: 442–451, 1975 [DOI] [PubMed] [Google Scholar]
- 29.Powers DMW: Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. J Mach Learn Technol 2: 37–63, 2011 [Google Scholar]
- 30.Chicco D, Jurman G: The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 21: 6, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan YH: Biostatistics 104: Correlational analysis. Singapore Med J 44: 614–619, 2003 [PubMed] [Google Scholar]
- 32.Akoglu H: User’s guide to correlation coefficients. Turk J Emerg Med 18: 91–93, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutman DA, Khalilia M, Lee S, Nalisnik M, Mullen Z, Beezley J, et al.: The digital slide archive: A software platform for management, integration, and analysis of histology for cancer research. Cancer Res 77: e75–e78, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer F, Beucher S: Morphological segmentation. J Vis Commun Image Represent 1: 21–46, 1990 [Google Scholar]
- 35.Serge Beucher FM: The morphological approach to segmentation: The watershed transformation. In: Mathematical Morphology in Image Processing, edited by Dougherty ED, Boca Raton, FL, CRC Press, 1993, pp 433–481 [Google Scholar]
- 36.Zaiontz C: Real statistics resource pack software (release 7.2), 2020. Available at: https://www.real-statistics.com/. Accessed January 7, 2020
- 37.Landis JR, Koch GG: The measurement of observer agreement for categorical data. Biometrics 33: 159–174, 1977 [PubMed] [Google Scholar]
- 38.Meng XL, Rosenthal R, Rubin DB: Comparing correlated correlation-coefficients. Psychol Bull 111: 172–175, 1992 [Google Scholar]
- 39.Diedenhofen B, Musch J: cocor: A comprehensive solution for the statistical comparison of correlations [published correction appears in PLoS One 10: e0131499, 2015 10.1371/journal.pone.0131499]. PLoS One 10: e0121945, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team: A language and environment for statistical computing, 2020. Available at: https://www.R-project.org/. Accessed January 7, 2020
- 41.Hosmer DW, Lemeshow S, Sturdivant RX: Applied Logistic Regression, 3rd Ed., Hoboken, NJ, Wiley, 2013 [Google Scholar]
- 42.Jen KY, Olson JL, Brodsky S, Zhou XJ, Nadasdy T, Laszik ZG: Reliability of whole slide images as a diagnostic modality for renal allograft biopsies. Hum Pathol 44: 888–894, 2013 [DOI] [PubMed] [Google Scholar]
- 43.Ozluk Y, Blanco PL, Mengel M, Solez K, Halloran PF, Sis B: Superiority of virtual microscopy versus light microscopy in transplantation pathology. Clin Transplant 26: 336–344, 2012 [DOI] [PubMed] [Google Scholar]
- 44.Austin HA 3rd, Boumpas DT, Vaughan EM, Balow JE: Predicting renal outcomes in severe lupus nephritis: Contributions of clinical and histologic data. Kidney Int 45: 544–550, 1994 [DOI] [PubMed] [Google Scholar]
- 45.Roufosse C, Simmonds N, Clahsen-van Groningen M, Haas M, Henriksen KJ, Horsfield C, et al.: A 2018 reference guide to the Banff classification of renal allograft pathology. Transplantation 102: 1795–1814, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farris AB, Alpers CE. What is the best way to measure renal fibrosis?: A pathologist’s perspective. Kidney Int Suppl (2011) 4: 9–15, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.University at Buffalo : Center for Computational Research. Available at: http://www.buffalo.edu/ccr.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.