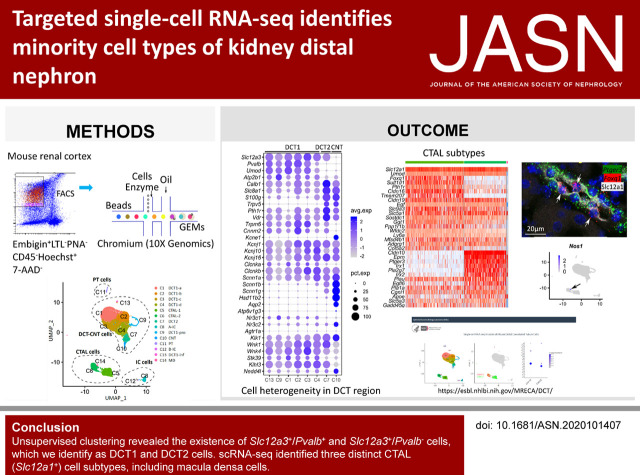
Significance Statement
A major objective in modern biology is generation of comprehensive atlases of various organs that identify all cell types and their expressed genes. In the kidney, extensive data describe proximal tubule and collecting duct cells but not the rarer intermediate epithelial cell types. Coupling of a cell enrichment protocol with single-cell RNA-seq analysis resolved the cellular composition and transcriptional profiles of the minority epithelial cell types of mouse kidney distal nephron. These data are provided in user-friendly websites that enable the mapping and comparison of genes of interest among cell types and renal tubule epithelia.
Keywords: scRNA-seq, distal convoluted tubule, thick ascending limb, macula densa
Visual Abstract
Abstract
Background
Proximal tubule cells dominate the kidney parenchyma numerically, although less abundant cell types of the distal nephron have disproportionate roles in water and electrolyte balance.
Methods
Coupling of a FACS-based enrichment protocol with single-cell RNA-seq profiled the transcriptomes of 9099 cells from the thick ascending limb (CTAL)/distal convoluted tubule (DCT) region of the mouse nephron.
Results
Unsupervised clustering revealed Slc12a3+/Pvalb+ and Slc12a3+/Pvalb− cells, identified as DCT1 and DCT2 cells, respectively. DCT1 cells appear to be heterogeneous, with orthogonally variable expression of Slc8a1, Calb1, and Ckb. An additional DCT1 subcluster showed marked enrichment of cell cycle–/cell proliferation–associated mRNAs (e.g., Mki67, Stmn1, and Top2a), which fit with the known plasticity of DCT cells. No DCT2-specific transcripts were found. DCT2 cells contrast with DCT1 cells by expression of epithelial sodium channel β- and γ-subunits and much stronger expression of transcripts associated with calcium transport (Trpv5, Calb1, S100g, and Slc8a1). Additionally, scRNA-seq identified three distinct CTAL (Slc12a1+) cell subtypes. One of these expressed Nos1 and Avpr1a, consistent with macula densa cells. The other two CTAL clusters were distinguished by Cldn10 and Ptger3 in one and Cldn16 and Foxq1 in the other. These two CTAL cell types were also distinguished by expression of alternative Iroquois homeobox transcription factors, with Irx1 and Irx2 in the Cldn10+ CTAL cells and Irx3 in the Cldn16+ CTAL cells.
Conclusions
Single-cell transcriptomics revealed unexpected diversity among the cells of the distal nephron in mouse. Web-based data resources are provided for the single-cell data.
The mammalian kidneys play a crucial role in regulation of body fluid composition and BP. These functions are largely achieved by control of sodium reabsorption across the epithelia of renal tubules.1,2 There are at least 14 different renal tubule segments, each with characteristic cell types with distinct gene expression profiles and function.3,4 Of particular importance is the distal convoluted tubule (DCT), a short segment that mediates fine regulation of sodium ion transport.5 The apical component of sodium transport across the DCT occurs via the Na+-Cl− cotransporter (NCC; Slc12a3), which is exclusively expressed in DCT cells and is the chief target of thiazide diuretics,6 a staple in the treatment of many forms of arterial hypertension. The DCT is believed to be heterogeneous and is separated into at least two subsegments, DCT1 and DCT2.7–9 However, this DCT classification is largely defined on the basis of immunocytochemical results. It remains unclear how many cell types make up the DCT and whether DCT1 and DCT2 are distinct segments or represent two ends of a continuum.
NaCl transport across the DCT was originally thought to be affected in part by binding of circulating aldosterone to the mineralocorticoid receptor (MR) in DCT cells.10,11 Aldosterone-MR signaling requires enzyme 11-β-hydroxysteroid dehydrogenase 2 (Hsd11b2) to deactivate the more abundant circulating corticosteroid cortisol (or corticosterone in rodents), which would otherwise bind to the MR, resulting in constitutive activation. However, immunocytochemical and in situ hybridization methods to localize Hsd11b2 in DCT cells have been indicative of variable expression along the DCT with detectable levels only in the distal-most DCT.12,13 These findings raise doubts about a possible direct role of aldosterone in the early DCT and whether the Hsd11b2-positive DCT corresponds to DCT2. Recent studies further suggest that Slc12a3 (NCC) abundance is regulated by aldosterone in the DCT only indirectly via its propensity to cause hypokalemia.5,14 Activation of NCC by low extracellular potassium activity is thought to be dependent on basolateral potassium channels (Kcnj10 and Kcnj16), chloride channels (Clcnkb and possibly Clcnka), ubiquitin ligase components (Nedd4l, Klhl3, and Cul3), and protein kinases (Wnk1, Wnk4, and Stk39 [SPAK]).5,14 However, a systematic mapping of these elements in DCT is currently not available.
The progressive development of recent single-cell RNA-seq (scRNA-seq) techniques in kidney15–22 has provided a deep analysis of gene expression in kidney epithelial cell types. The tubule microdissection method,23 however, is unable to effectively isolate the DCT1 and DCT2 due to the overall shortness of the DCT.24 Despite great progress of comprehensive “shotgun” scRNA-seq in the kidney, it devotes most of the sequencing reads to more abundant proximal tubule cells and nonepithelial cells, leading to analysis of only limited numbers of minority epithelial cell types. Thus, the heterogeneity of cell types like DCT cells as well as various cell types in the loop of Henle (for example, macula densa [MD] and cortical thick ascending limb [CTAL] cells) is not completely resolved. A solution to this dilemma is to enrich the target cells before analysis as previously introduced for scRNA-seq analysis of collecting duct cell types.16
Here, we exploit a FACS-based enrichment protocol coupled to scRNA-seq analysis to identify transcriptomes of epithelial cell types from the mouse nephron region spanning from the CTAL of Henle to the DCT. We also provide web-based resources that allow users to explore and download data for future studies.
Methods
Adult Mouse Kidney Cell Isolation and scRNA-seq
This protocol was performed as described previously.21 The kidneys were perfused directly via the left ventricle with perfusion buffer (5 mM HEPES, 120 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2.5 mM Na2HPO4, 1.2 mM MgSO4, 5.5 mM glucose, and 5 mM Na acetate in H2O at pH 7.4) to remove blood cells, followed by perfusion with the same perfusion buffer supplemented with 1 mg/ml collagenase B (11088831001; Sigma). Kidneys were immediately removed and placed in ice-cold PBS. The cortical region of kidney tissue was then dissected and minced on ice, followed by dissociation in 20 ml cold perfusion buffer consisting of collagenase B (2.5 mg ml−1), Dispase II (1.2 U ml−1; Roche), 7.5 mg/ml Bacillus licheniformis protease (P5380; Sigma), and DNase I (LS002058, 125 Kunitz ml−1; Worthington) for 25–30 minutes at 10°C with frequent agitation every 5 minutes. In the last 10 minutes before the end of digestion, the suspension was passed through a 20-gauge needle. Digestion enzymes were neutralized by adding 20 ml 20% FBS, and the suspensions were then centrifuged at 250×g at 4°C for 3 minutes. The pelleted cells were resuspended briefly in 5 ml lysis buffer (118–156–101; Quality Biologic) for 30 seconds to remove red blood cells followed by dilution in 45 ml PBS. After centrifugation at 250×g for 3 minutes, the pellet was resuspended and washed again with 50 ml PBS. The final cell pellet was resuspended in 30 ml FACS buffer (perfusion buffer with 0.05% BSA) and then passed sequentially through 100-, 70-, and 40-μm cell strainers (VWR) to obtain a suspension of isolated cells (single-cell suspension). The single-cell suspension was pelleted at 250×g for 3 minutes and resuspended in 1 ml FACS buffer; 120 µl FcR blocking reagent (130–092–575; Miltenyi Biotec) was added to 1 ml single-cell suspension together with 1:500 Hoechst 33342 (H3570; Invitrogen) for cell staining, 1:200 lotus tetragonolobus lectin (LTL)–FITC (FL-1321–2; Vector Laboratories) for labeling of proximal tubule cells,25 1:500 PNA-Cy5 (CL-1075–1; Vector Laboratories) for labeling type B intercalated cells,26,27 1:200 CD45 (103149; BioLegend) for labeling of immune cells,28 and 1:10 Emb-PE (130–107–948; Miltenyi Biotec) for labeling of non-PC cells16 from DCT region. The staining was done in the dark at 4°C with agitation. After 30 minutes of staining, the cells were washed twice with FACS buffer and resuspended in 2 ml FACS buffer before sorting; 7-AAD (559925; BD) at 1:10 was added to the cells before FACS. The cells were sorted in 700 µl FACS buffer with a low flow rate and pressure to ensure high viability. Thirty to forty thousand cells were collected and centrifuged at 250×g for 3 minutes in a swinging bucket at 4°C. The top supernatant was carefully removed, and 30 µl volume was left to resuspend the cell pellet. Ten microliters of cells were mixed with 10 µl Trypan Blue (17–942E; Lonza) and then loaded on the Countess II Automated Cell Counter (Invitrogen) for counting and viability test. Cells at 700–1200 cells per microliter were directly loaded to 10× chips for Gel Bead-In Emulsions (GEM) generation.
scRNA-seq Data Analysis
Lysed or dead cells in droplet-based scRNA-seq assays release ambient RNA or cell-free RNA, resulting in low fraction reads in cells (i.e., reads are not 100% associated with a valid cell barcode in GEM). Such background levels of contamination vary between batches and cell types. SoupX (https://doi.org/10.1101/303727) estimates such ambient RNA by pooling of droplets with low numbers of UMI (ten UMI or less). As the majority of generated GEMs contain no cells, this provides means to estimate the background noise. The bimodal distribution of transcripts guides the contamination fraction calculation for each cell. The counts are then adjusted for downstream analysis.
The 10× Chromium raw sequencing data were first processed with cellranger (version 3.0.2). SoupX (version 0.3.1) was then applied for background correction. Kap and Slc12a1 were identified and used for expression profile correction. Three batches were independently estimated, corrected, and merged in R with Seurat (version 3.0) package. Initially, cells with <200 features (genes) and features expressed in fewer than ten cells were discarded. Cells were further filtered on the basis of nFeature_RNA>1000, nFeature_RNA<5800, percent.mt<35, percent.mt >5, and nCount_RNA<30,000. A small cluster of cells showing extremely low mitochondria reads and UMI was removed. The resulting Seurat object contains 17,655 features across 9099 samples. To correct batch effects for three independent preparations, the integration workflow was applied to the combined object. The combined object was split and normalized, and the top 2000 highly variable features were identified. Similar biologic states for individual cells (anchors) between datasets were identified using the FindIntegrationAnchors function, which used the first 30 Canonical Correlation Analysis dimensions (https://satijalab.org/seurat/v3.0/integration.html). Datasets were then integrated on the basis of the anchors found before.
Standard Seurat analysis was then applied to the integrated data. Data were scaled, and mitochondria variation was regressed out. The first 15 principal components were chosen for downstream clustering after linear dimensional reduction (RunPCA). Cell clusters were determined by FindNeighbors and FindClusters. Uniform Manifold Approximation and Projection (UMAP) was used throughout the paper to visualize the single-cell data. Differentially expressed genes in each cluster were calculated by FindAllMarkers function. For better visualization, sometimes subset function was used to extract certain clusters. Other R packages, including ggplot2 and dplyr, were used for final figure preparation.
The scRNA-seq data published by Ransick et al.21 were downloaded from the Gene Expression Omnibus (accession no. GSE129798) and processed with Seurat using parameters mentioned in the original paper. Briefly, we removed the nonepithelial cell data from the Seurat object on the basis of lack of Cdh16 expression after clustering. Slc12a1+ cells were further selected and were projected on UMAP. We identified Slc12a1+/Nos1+ cells as MD cells.3
Immunohistochemistry and RNAscope In Situ Hybridization
The mouse kidney tissue was prepared as described previously.16 Mice were cervically dislocated and perfused with ice-cold DPBS followed by 4% PFA in DPBS. Kidneys were then fixed for 2 hours in 4% PFA before transferring to 20% sucrose at 4°C overnight. The blocks were further processed in the National Heart, Lung, and Blood Institute (NHLBI) Pathology Facility. Six-micrometer-thick sections were cut. Frozen sections were thawed at room temperature for 10–20 minutes and rehydrated in PBS for 10 minutes. After blocking for 30 minutes with 1% BSA and 0.2% gelatin, primary antibodies were applied overnight at 4°C. Sections were washed for 3×5 minutes in PBS. The secondary antibody incubation was carried out for 1 hour at room temperature.
RNAscope was performed on fixed frozen tissue sections according to the RNAscope Multiplex Fluorescent Reagent Kit v2 user manual (document 323100-USM). TSA Plus fluorophores at a concentration of 1:1500 were used to develop signals. After the signals were developed for each channel, standard immunohistochemistry protocol was followed for antibody staining. The confocal images were taken in the NHLBI Light Microscopy Core.
Immunohistochemistry Antibodies and RNAscope Probes
Immunolabeling reagent includes several antibodies generated in the laboratory of M.A.K., including rabbit anti-Slc12a3 (4375), chicken anti-Slc12a1 (C20), chicken anti-Scnn1g (LC38), and chicken anti-Scnn1b (LC40). Commercial antibodies were Pvalb (GP72; Swant) and Cldn10 (catalog no. 38–8400; Thermo Fisher Scientific). Probes used in this study are Foxq1 (catalog no. 504801; Advanced Cell Diagnostics) and Ptger3 (catalog no. 501831-C3; Advanced Cell Diagnostics). Fluorescently labeled lectins were LTL (FL-1321; Vector Laboratories) and PNA (CL-1073 or CL-1075; Vector Laboratories).
Websites
Websites for sharing of curated data were built around the Shiny package in R. The website https://esbl.nhlbi.nih.gov/MRECA/DCT/ was developed in Shiny to directly visualize scRNA-seq data. Interactive UMAP and dot plots were created using Seurat and modified by ggplot.
Results
scRNA-seq of DCT Cells
To characterize the cellular composition and transcriptional profiles of the cells in the CTAL-DCT-CNT region, we developed the FACS protocol described below and carried out scRNA-seq analysis of these cells (Figure 1A). The resulting data can be browsed at https://esbl.nhlbi.nih.gov/MRECA/DCT/.
Figure 1.
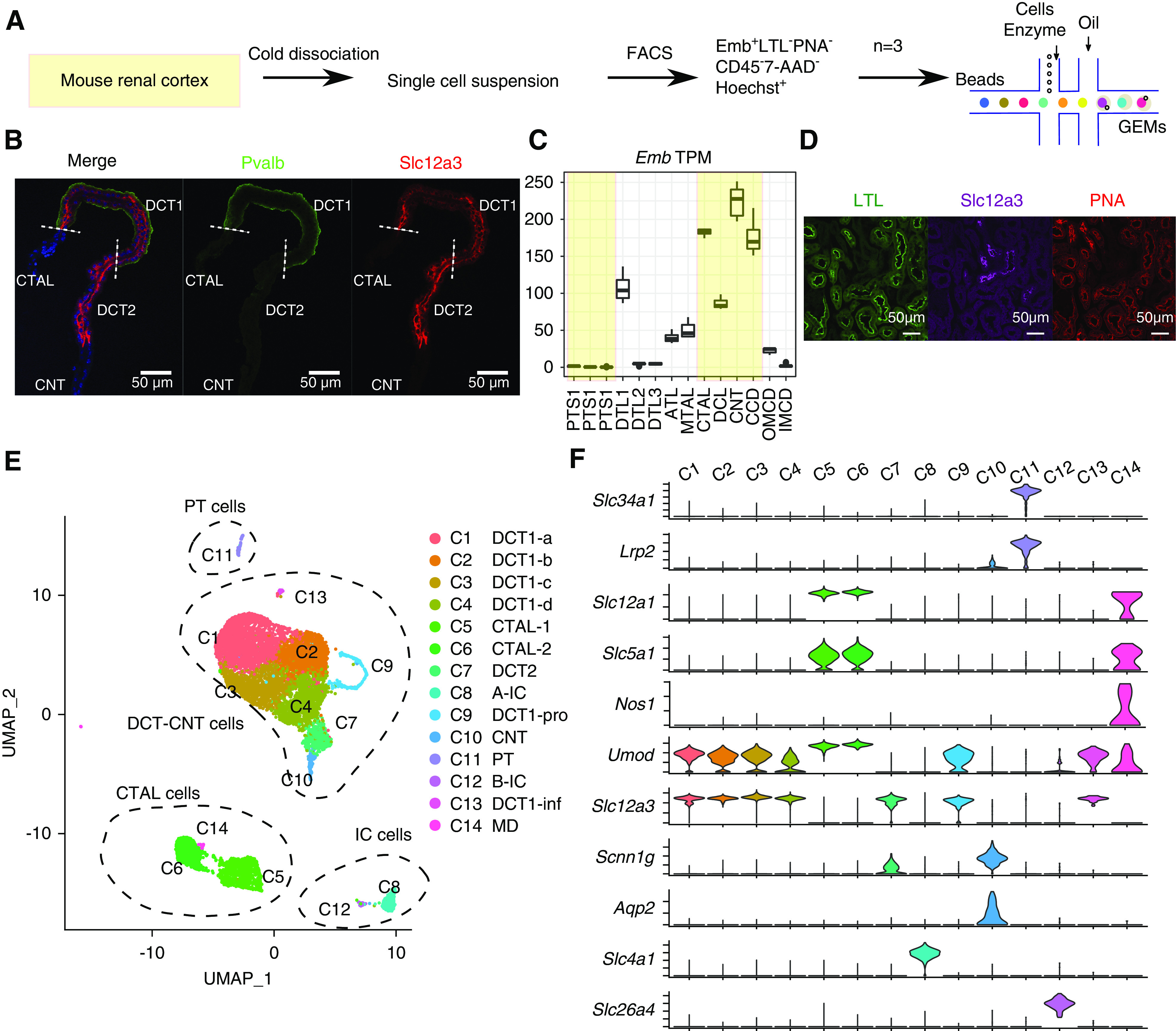
scRNA-seq of DCT cells. (A) Overview of protocol for the enrichment of DCT cells for single-cell analysis. (B) Immunostaining showing the distribution of Slc12a3 (red) and Pvalb (green) in microdissected CTAL to CNT region. Nuclei are stained using DAPI (blue). Scale bars, 50 μm. DAPI, 4′,6-diamidino-2-phenylindole. (C) Distribution pattern of Embigin transcript along the mouse renal nephron segments.24 The tubules in the renal cortex are shown in shaded yellow. TPM, Transcripts Per Million. (D) Embigin was used as a positive surface marker for DCT and thick ascending limb cells and a negative marker for proximal tubule cells and principal cells. (E) UMAP projection of cells from all three 10× Chromium datasets. Different clusters are colored and labeled. Four major clusters are highlighted by dashed circles and annotated on the basis of (F). (F) Violin plots of renal cell marker genes across all clusters. Cluster information is shown at the top.
The DCT is generally subdivided into DCT1 and DCT2.7–9 Both express Slc12a3. DCT1 but not DCT2 expresses Pvalb (Figure 1B), whereas DCT2 selectively expresses the epithelial sodium channel (see below). The fairly short nature of the DCT2 and ill-defined DCT1/DCT2 and DCT2/CNT boundaries (Figure 1B) prevented us from identifying a DCT2 transcriptome in microdissected tubules.24 Thus, we sought to use a FACS approach for the enrichment of DCT cells for scRNA-seq (Figure 1A) on the basis of a strategy similar to one successfully applied to study heterogeneity of CD cells.16 We began by dissociating mouse renal cortical tissue into single-cell suspensions using a cold-active protease.15 The FACS sorting used positive selection with an antibody to Embigin (Emb), which encodes a single-pass type 1 membrane protein present in all cortical renal tubule cells except for proximal tubule (Figure 1C), and principal cells,16,18 thus largely eliminating abundant PT cells29 and PC cells while enriching DCT cells. We also used lectins for negative selection, namely LTL25 and PNA (peanut lectin),26,27 to remove remaining proximal cells and β-intercalated cells (Figure 1D). scRNA-seq utilized the 10× Chromium platform (Figure 1A). We carried out three independent runs with 3000–6000 cells each as input. After data quality control and batch effects removal (Methods, Supplemental Figure 1A), we detected 17,655 genes across 9099 cells (Supplemental Figure 1B).
Unbiased clustering and UMAP visualization revealed 14 cell clusters corresponding to five major cell types, namely PT (C11); CTAL (C5, C6, and C14); DCT (C1, C2, C3, C4, C7, C9, and C13); IC (C8 and C12); and CNT (C10) (Figure 1E). The transcriptomes of each cell cluster are provided in Supplemental Table 1. The clusters were annotated on the basis of classic renal tubule markers (Figure 1F) and differentially expressed genes for each cell cluster (Supplemental Table 2). On average, >3000 genes per cell were detected in this analysis, and most cells were from DCT + CNT (77.3%), indicating the success of the enrichment process (Supplemental Figure 1B). Cells sometimes suffer from stress responses during single-cell dissociation characterized by overexpression of immediate early genes, confounding the biologic interpretation of scRNA-seq data.30 As seen in Supplemental Figure 1C, no evidence of expression of the immediate early genes Fos, Nr4a1, and Junb was found, demonstrating a relative lack of stress artifact in this dataset. Additional analyses of proximal tubule and intercalated cell clusters (A-IC and B-IC) are provided in Supplemental Figure 1D. Here, we focus first on DCT cell types followed by CTAL subtypes.
Heterogeneity of DCT1 Cells
The scRNA-seq analysis has identified six tightly arranged clusters of cells expressing both Slc12a3 and Pvalb, which we identify as DCT1 cells (C1, C2, C3, C4, C9, and C13) (Figure 2, A and B). As shown in Figure 2C and Supplemental Figure 2, four of these (C1–C4) expressed the same basic set of genes and are discriminated on the basis of gradients of gene expression (e.g., creatine kinase B [Ckb] decreasing from C1 toward C4; Na+/Ca2+ exchanger [Slc8a1] and calbindin-1 [Calb1] decreasing from C2 to C3) (Figure 2C). The general similarity of the cells in these four clusters leads us to conclude that all of these cells can be classified as a single-cell type (“DCT1 cell”) with subtypes DCT1a–d (Figure 2C). Two other clusters, C9 and C13, also strongly expressed Slc12a3 and Pvalb but exhibited superimposed gene signatures that distinguish them from the C1–C4 DCT1 cells. The C9 cells showed marked enrichment of cell cycle– and cell proliferation–associated mRNAs (for example, Stmn1, Mki67, Cenpf, and Top2a) (Figure 3A, Supplemental Figure 2). This proliferative profile is consistent with the idea that DCT1 cells are “plastic” in the sense that they can rapidly undergo increases and decreases in cell number and/or volume in response to physiologic stimuli.31–34 The C13 cells appeared to be DCT1 cells with a superimposed “innate immune signature” with an excess expression of IFN regulatory factor transcription factors and many of the genes that they regulate (e.g., MHC class 1 antigen transcripts and β2-microglobulin) (Supplemental Figure 2). These genes have been associated with damage-associated molecular patterns triggered by various signals, including foreign nucleic acids.35 This signature was not seen with other cell types. Thus, the expression patterns seen may be unique to DCT1. In addition to parvalbumin (Pvalb), Lrrc52 (Figure 3A, right panel), a subunit of Maxi-K channel36,37 that was selectively found in microdissected DCT in small sample RNA-seq,24 appeared to be expressed in DCT1 cells. Collectively, these data suggest an underlying variation in gene expression among DCT1 cells representing different physiologic states of a single cell type. The specific pattern of DCT1 cell subtypes in space and time remains to be elucidated.
Figure 2.
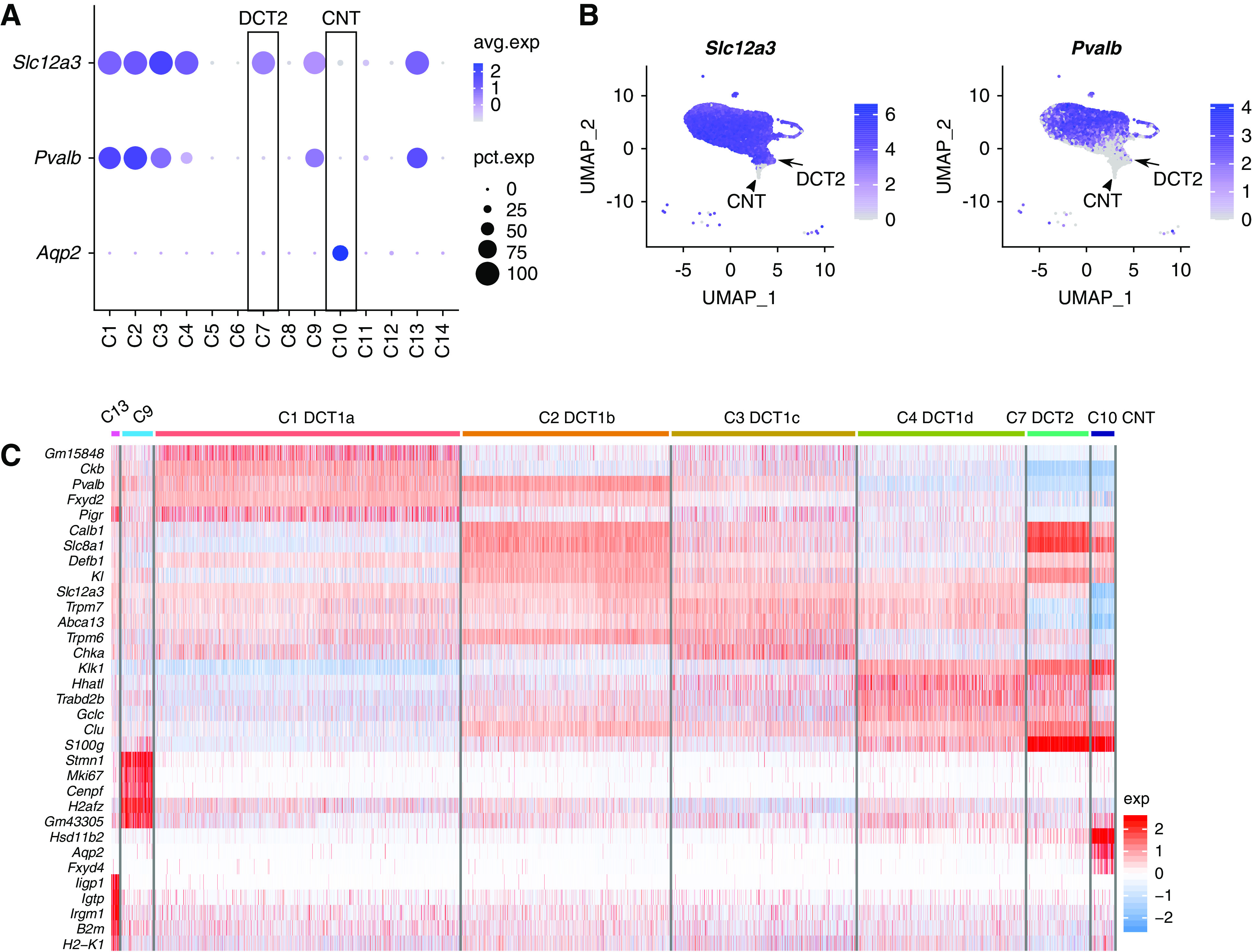
Heterogeneity of DCT cells revealed by scRNA-seq. (A) Dot plot showing the distribution of Slc12a3, Pvalb, and Aqp2 expression across the clusters. Data are normalized and scaled (z score) to examine relative expression across the cell clusters; “avg.exp” is the z score of the average gene expression of all cells within a cluster (scaled values), and “pct.exp” is the percentage of cells with nonzero gene expression. (B) UMAP projection for Slc12a3 and Pvalb expression. Arrows point to DCT2 cells; arrowheads point to CNT cells. (C) Heat map showing the top five genes in DCT and CNT cell clusters. Data are scaled (z score). A similar analysis is also provided in Supplemental Figure 2.
Figure 3.
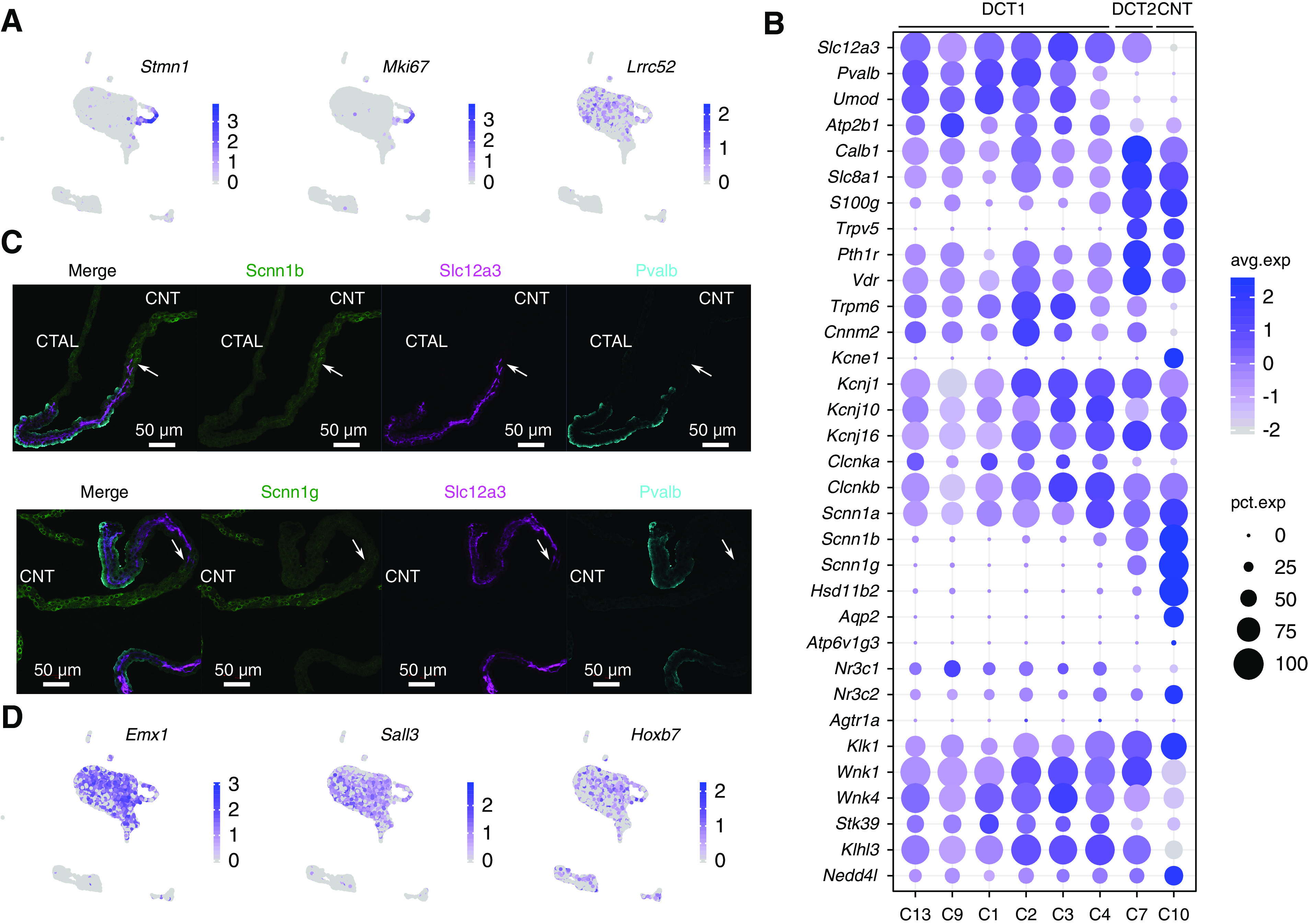
Distributions of transcripts associated with major transport pathways in DCT cells. (A) UMAP projection for Stmn1, Mki67, and Lrrc52. (B) Distribution of major transport pathways in DCT cells; “avg.exp” is the z score of the average gene expression of all cells within a cluster (scaled values), and “pct.exp” is the percentage of cells with nonzero gene expression. (C) Immunostaining showing Scnn1b and Scnn1g expression in microdissected tubule from the CTAL-DCT-CNT region. Scnn1b and Scnn1g are in green, Slc12a3 is in purple, Pvalb is in cyan, and DNA is in blue. Arrows point to regions at the point of initial expression of Scnn1b and Scnn1g. Scale bars, 50 µm. (D) UMAP projection for Emx1, Sall3, and Hoxb7.
DCT2 Cells and Transport Pathways
We identify cluster 7 as DCT2 cells. These cells show Slc12a3 expression without Pvalb (Figure 2, A and B) but express epithelial sodium channel subunits (see below). These DCT2 cells can also be distinguished from DCT1 cells in part on the basis of a relative lack of Umod38 (Figure 3B). DCT2 cells appear to account for a minority of the DCT as a whole. No transcripts were identified that are expressed in DCT2 (i.e., cluster 7) but are not expressed in either DCT1 or CNT (Figure 3B, Supplemental Figure 2). That is, no DCT2-specific markers were identified in this study. Instead, the cells appear to represent hybrid, transitional cells. DCT2 cells strongly express transcripts involved in transcellular calcium reabsorption, including two major receptors (Pth1r and Vdr), an apical calcium channel Trpv5, the intracellular calcium-binding proteins Calb1 and S100g, and the basolateral sodium/calcium exchanger Slc8a1 as well as plasma membrane calcium-transporting ATPase 1 (Atp2b1) (Figure 3B). In this regard, DCT2 cells resemble CNT cells and differ from DCT1 in mediating calcium transport.9,39 On the other hand, DCT2 resembles DCT1, and not CNT, with regard to the expression of two magnesium transport–related transcripts (Trpm6 and Cnnm2) (Figure 3B), suggesting that both DCT1 and DCT2 are involved in regulation of transcellular magnesium reabsorption.40
Sodium reabsorption in DCT is achieved largely through Slc12a3 (NCC), which is regulated by a signaling pathway involving two Wnk kinases (Wnk1 and Wnk4) and Stk39 (SPAK).41 This signaling pathway is believed to be regulated in part by angiotensin II, potassium, intracellular chloride concentration, and possibly aldosterone.5,10,11,14 As shown in Figure 3B, the transcripts coding for major steroid hormone receptors (Nr3c1 and Nr3c2), potassium channels (Kcnj1, Kcnj10, and Kcnj16), chloride channels (Clcnka and Clcnkb), and regulatory factors (Cul3, Klhl3, and Nedd4l) are expressed in both DCT1 and DCT2 cells. Angiotensin II receptor mRNAs (Agtr1a and Agtr1b), which were undetectable in microdissected DCTs,24 were also undetectable in DCT1 and DCT2 single cells. DCT2 but not DCT1 cells express Scnn1b and Scnn1g (Figure 3, B and C). As discussed above with regard to divalent cation transport, the DCT2 transcriptome is not simply the union of the DCT1 and CNT transcriptomes. It lacks expression of the water channel Aqp2 and Kcne1, both of which are strongly expressed in CNT (Figure 3B). Although CNT cells show the highest expression of glucocorticoid-metabolizing enzyme Hsd11b2, considerably smaller amounts were found in DCT2 cells. Specifically, 96.3% of CNT cells had detectible Hsd11b2, whereas only 23.3% of DCT2 cells had detectable levels, and DCT1 cells (C1–C4) had barely measurable levels (Figure 3B).
An alternative to a purely physiologic role for DCT2 cells is the possibility that the role of these cells is to join the nephron and collecting duct, which are derived from different developmental progenitors.42 Some clues to a possible such role could be derived from patterns of expression of homeobox genes that are generally involved in tissue segmentation during development. One homeobox gene (Emx1) was found to be highly selectively expressed in DCT in our small sample RNA-seq profiling,24 whereas the single-cell analysis did not show any selectivity in DCT2 versus DCT1 cells (Figure 3D, left panel). Emx1 has previously been identified in zebrafish as an important regulator of distal nephron development.43 Sall3, a zinc-finger protein that was identified as DCT selective in the small sample RNA-seq studies,24 is also not selective for DCT2 versus DCT1 (Figure 3D, center panel). The homeobox gene most associated with ureteric bud differentiation is Hoxb7,44 which is approximately equally expressed in all DCT cell subtypes, CNT, and intercalated cells (Figure 3D, right panel).
Mosaic Cell–Type Pattern in CTAL
scRNA-seq analysis identified three distinct CTAL (Slc12a1+) cell subtypes (C5, C6, and C14) (Figure 4A). One of these (C14) expressed genes characteristic of MD cells, namely Nos145,46 and Avpr1a47 (Figure 4B, Supplemental Figure 3A). We found that Pappa2 was also strongly expressed in MD cells (Figure 4B). This gene has been previously linked to salt-sensitive hypertension in Dahl S rats.48 Strikingly, these cells expressed multiple neuronal genes aside from Nos1, including several involved with axon guidance and synaptic adhesion (Robo2, Ncam1, Sema6a, and Auts). This suggests the characterization of MD cells as “neuroepithelial cells.” We speculate that the axon-guidance proteins could be part of the homing mechanism that localizes MD cells to the vascular pole of the glomerulus. Interestingly, MD cells selectively express Bmp3 (Supplemental Figure 3C), a secreted protein that could be involved with communication with other cell types, including DCT cells, if secreted apically or renin-secreting juxtaglomerular cells (JG cells) if secreted basolaterally. In addition, there are multiple transcripts that are strongly expressed in the CTAL but are very low in MD, including uromodulin (Umod)49 and Egf (Egf)50 (Figure 4C, Supplemental Figure 3A). To confirm these observations, we downloaded and analyzed data from another scRNA-seq paper21 (Methods, Supplemental Table 3), which also showed coexpression of Nos1, Avpr1a, and Pappa2 in Slc12a1-positive cells (Supplemental Figure 3B).
Figure 4.
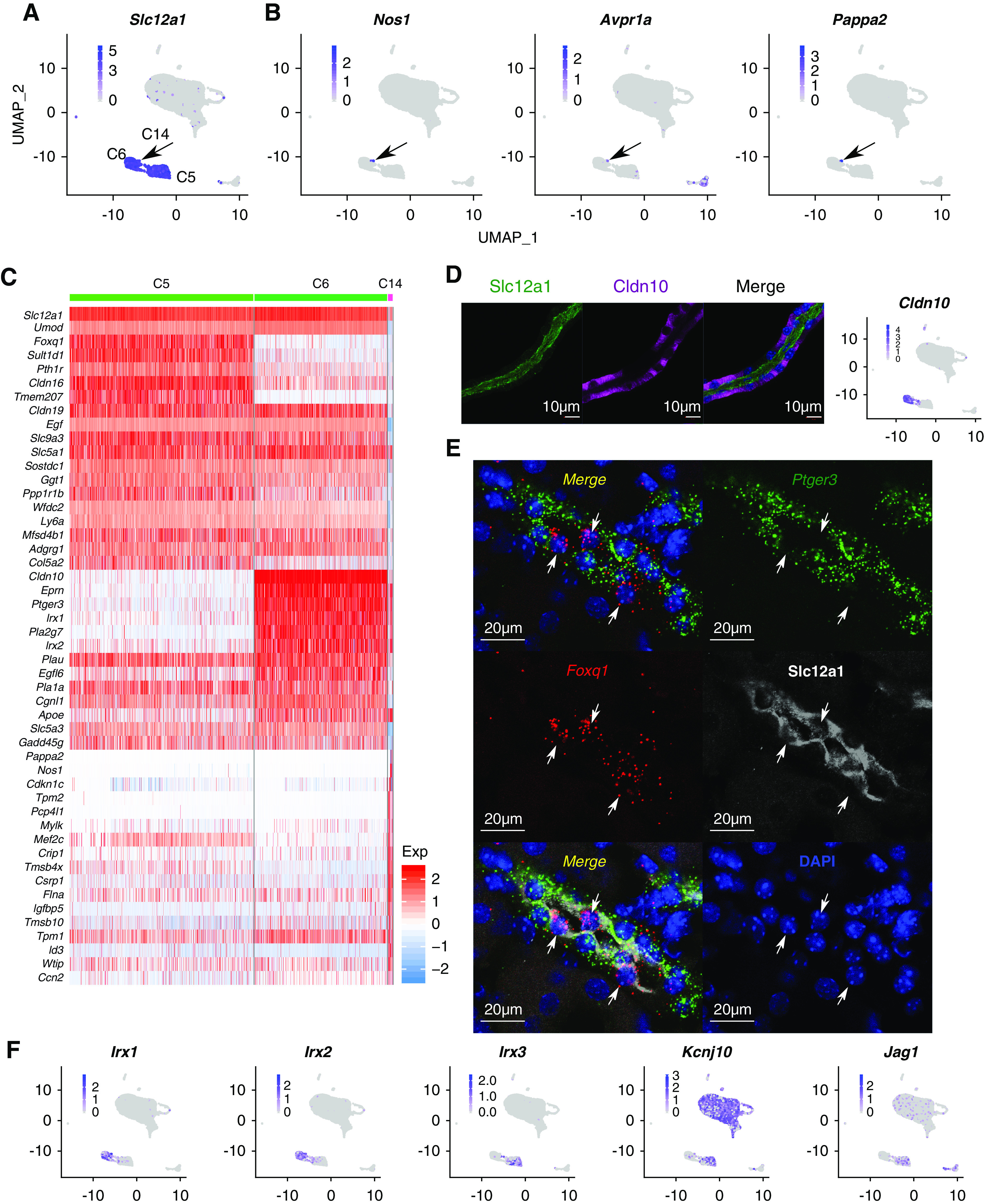
Single-cell analysis of CTAL cells. (A) UMAP visualization of Slc12a1. (B) UMAP plot for Nos1, Avpr1a, and Pappa2. (C) Heat map showing the top 20 marker genes in C5, C6, and C14. Each column indicates a cell. Colors indicate the expression level (z score). (D) Immunostaining showing Slc12a1 (green) and Cldn10 (purple) on a microdissected mouse TAL. Nuclei are stained with DAPI (blue). DAPI, 4′,6-diamidino-2-phenylindole. The feature plot for Cldn10 is shown in the right panel. Scale bars, 10 µm. (E) Immunostaining coupled with in situ hybridization (RNAscope) showing Slc12a1 (white), Ptger3 (green), and Foxq1 (red) on mouse kidney section. Nuclei are stained with DAPI (blue). Arrows point to Foxq1+-positive cells (red). Lower-magnification views are provided in Supplemental Figure 4 and can be viewed at https://esbl.nhlbi.nih.gov/MRECA/Supplement/. Scale bars, 20 µm. (F) UMAP visualization of Irx1, Irx2, Irx3, Kcnj10, and Jag1.
The other two CTAL clusters were distinguished by Cldn10 and Ptger3 in one (C6) and Cldn16 and Foxq1 in the other (C5). As shown in Figure 4D, some CTAL cells lack Cldn10 as reported before.51 Multiplex localization of Slc12a1 (immunofluorescence) with Foxq1 and Ptger3 mRNA (in situ hybridization with RNAScope) in mouse kidney sections highlighted the mutually exclusive pattern of Foxq1 and Ptger3 in TAL (Figure 4E, Supplemental Figure 4). In addition, these two CTAL cell types were distinguished by alternative expression of Iroquois homeobox transcription factors, with Irx1 and Irx2 in the Cldn10+ CTAL cells (C6) and Irx3 in the Foxq1+ CTAL cells (C5) (Figure 4F). The subsequent analysis showed that Jag1 (ligand for Notch) was predominantly expressed in C5 (Figure 4E), whereas Hes1 expression was greatest in C6 (Supplemental Figure 3D), suggesting a potential binary cell fate between these two CTAL cell types regulated by Notch signaling.52 Interestingly, Kcnj1 and Kcnj16 were strongly expressed in both C5 and C6 clusters (Supplemental Figure 3E); however, there is little or no Kcnj10 in C6 (Figure 4F). Kcnj10 and Kcnj16 are expressed in other distal cell types, where they coassemble into heterodimeric channels.53
Discussion
Here, we combined cell enrichment and scRNA-seq to investigate the transcriptome landscape of Slc12a3+-DCT cells (https://esbl.nhlbi.nih.gov/MRECA/DCT/). Our data demonstrate that DCT cells are subdivided into Slc12a3+/Pvalb+ DCT1 cells and Slc12a3+/Pvalb− DCT2 cells. We observed that DCT1 cells are heterogeneous. This heterogeneity appears to be associated with variable expression of Slc8a1, Calb1, and Ckb among other mRNAs. The relevance of such topology of gene expression to the spatial and temporal arrangement of DCT1 cells, as well as DCT function, remains for further investigation. How can the heterogeneity of DCT1 cells or the hybrid property of DCT2 cells (mentioned below) be explained? Because the DCT and downstream collecting duct system are derived from two separate progenitors, one possible explanation for the heterogeneity could be reciprocal mutual inductions between nephron progenitors and ureteric bud during kidney development.42 Among DCT1 cells, we also identified a small population of cells that showed marked enrichment of cell cycle– and cell proliferation–associated mRNAs (e.g., Pcna, Mki67, Cdk1, and Top2a). These cells are rare but unlikely to be artificial doublets as these proliferation markers are not seen in other epithelial cells (PT, CTAL, and ICs) studied here. This finding suggests that certain DCT1 cells might possess a proliferative potential, even in normal kidneys under normal physiologic conditions. Consistent with this conclusion is that the DCT is known to undergo extensive remodeling under different stimuli.31–34 A future direction would be to assess whether this proliferative-type DCT1 cell undergoes changes in abundance in response to these stimuli and to identify the mechanism.
Identification of unique DCT2 markers could potentially be the basis of future methods for isolation, identification, and targeting of DCT2 cells. However, no unique transcripts were found in DCT2 cells. DCT2 cells may instead be hybrid cells between DCT1 and CNT cells. Our data suggest that similar to DCT1 cells, DCT2 cells are involved in magnesium reabsorption, whereas similar to CNT cells, DCT2 cells are involved in calcium absorption.9,39
Our study revealed that DCT2 cells had detectable but much lower levels of Hsd11b2 mRNA than CNT. However, the experiments do not reveal protein levels or enzyme activity in DCT2 cells. In a recent study using protein mass spectrometry in microdissected tubules of rat, we found readily measurable Hsd11b2 protein levels in DCT (DCT1 + DCT2).54 Thus, we cannot rule out the possibility that there is enough Hsd11b2 in DCT1/DCT2 to metabolize glucocorticoids. Furthermore, Figure 3B shows significant MR (Nr3c2) expression in all DCT clusters.
We found three types of CTAL cells, including a small cell cluster with a gene expression pattern consistent with MD cells. These cells express the classic MD cell marker, neuronal nitric oxide synthase (Nos1),45,46 as well as a number of neuronal genes, indicating that these cells are “neuroepithelial” in character. Identification of transcripts coding for secreted proteins, like Pappa2, Gdf15, and Bmp3, may shed light on the ways that MD cells communicate with neighboring cell types. The other two CTAL cell types were more abundant. Previous studies found two morphologically distinguishable cell types in rat TAL: rough-surfaced cells with well-developed microvilli and smooth-surfaced cells.55 A striking observation from this study is the identification of two CTAL subtypes on the basis of gene expression that might correspond to the morphologically identified cell types, namely cells in the C5 cluster (Foxq1+Cldn10−Ptger3−) and the C6 cluster (Foxq1−Cldn10+Ptger3+). C6 was enriched in Kcnj16 but not Kcnj10. The role of Kcnj16, independent of Kcnj10, in this CTAL subtype requires further investigation.56,57 Heterogeneous protein expression of Kcnj1 was reported in CTAL58; however, we found no difference in Kcnj1 transcript expression between C5 and C6 cell clusters. Our data further showed Jag1 was expressed in C5 while absent in C6. Such a mosaic cell pattern mimics collecting duct epithelium (i.e., PC and IC), where cell specification is regulated by the Notch signaling pathway.59 Whether the Notch signaling pathway regulates the fate of these two cell types in TAL remains unknown. Also, the differential expression of Irx1/Irx2 versus Irx3 in these two cell types could play roles in their differentiation and physiologic function.
In summary, systematic analyses of cellular composition and gene expression provide insights into kidney functions and diseases. We provide our data in a comprehensive website and expect that this study will be an essential asset for future kidney studies. Because technologic advances enable us to profile different properties of tissues or individual cells, such as the genome, transcriptome, epigenome, and proteome, or even multiomes,60 we expect these approaches will open avenues for further kidney research.
Disclosures
Because M.A. Knepper is an editor of JASN, he was not involved in the peer review process for this manuscript. A guest editor oversaw the peer review and decision-making process for this manuscript. All remaining authors have nothing to disclose.
Funding
The work was primarily funded by Division of Intramural Research, National Heart, Lung, and Blood Institute projects ZIA-HL001285 (to M.A. Knepper) and ZIA-HL006129 (to M.A. Knepper).
Data Sharing Statement
Sequencing data have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150338).
Supplementary Material
Acknowledgments
The authors thank Dr. Jurgen Schnermann and Dr. Robert Hoover for helpful discussions regarding macula densa cells and DCT2 cells, respectively.
Next-generation sequencing was done in the NHLBI DNA Sequencing Core Facility (Dr. Yuesheng Li, Director). Confocal images were taken in the NHLBI Confocal Microscopy Core Facility (Dr. Christian Combs, Director). Cell sorting was performed in the NHLBI Flow Cytometry Core (Dr. J. Philip McCoy, Director). Cell viability assay and counting were done in the NHLBI Induced Pluripotent Stem Cells Core (Dr. Jizhong Zhou, Director). Tissue sections were prepared in the NHBLI Pathology Facility (Dr. Zu-Xi Yu, Director).
L. Chen and M.A. Knepper designed research, analyzed data, and made figures; L. Chen performed experiments and designed websites; and L. Chen, C.-L. Chou, and M.A. Knepper contributed reagents, discussed the results, and drafted the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2020101407/-/DCSupplemental.
Supplemental Figure 1. Single-cell RNA-seq data quality control.
Supplemental Figure 2. Heterogenous gene expression among DCT cells.
Supplemental Figure 3. Additional single-cell RNA-seq analysis of CTAL cell types.
Supplemental Figure 4. Heterogeneity of cortical thick ascending limb cells.
Supplemental Table 1. Average expression.
Supplemental Table 2. Cluster markers.
Supplemental Table 3. MD average expression.
References
- 1.Palmer LG, Schnermann J: Integrated control of Na transport along the nephron. Clin J Am Soc Nephrol 10: 676–687, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reilly RF, Ellison DH: Mammalian distal tubule: Physiology, pathophysiology, and molecular anatomy. Physiol Rev 80: 277–313, 2000 [DOI] [PubMed] [Google Scholar]
- 3.Kriz W, Bankir L: A standard nomenclature for structures of the kidney.The Renal Commission of the International Union of Physiological Sciences (IUPS). Kidney Int 33: 1–7, 1988 [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Clark JZ, Nelson JW, Kaissling B, Ellison DH, Knepper MA: Renal-tubule epithelial cell nomenclature for single-cell RNA-sequencing studies. J Am Soc Nephrol 30: 1358–1364, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCormick JA, Ellison DH: Distal convoluted tubule. Compr Physiol 5: 45–98, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamba G, Saltzberg SN, Lombardi M, Miyanoshita A, Lytton J, Hediger MA, et al.: Primary structure and functional expression of a cDNA encoding the thiazide-sensitive, electroneutral sodium-chloride cotransporter. Proc Natl Acad Sci U S A 90: 2749–2753, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obermüller N, Bernstein P, Velázquez H, Reilly R, Moser D, Ellison DH, et al.: Expression of the thiazide-sensitive Na-Cl cotransporter in rat and human kidney. Am J Physiol 269: F900–F910, 1995 [DOI] [PubMed] [Google Scholar]
- 8.Schmitt R, Ellison DH, Farman N, Rossier BC, Reilly RF, Reeves WB, et al.: Developmental expression of sodium entry pathways in rat nephron. Am J Physiol 276: F367–F381, 1999 [DOI] [PubMed] [Google Scholar]
- 9.Loffing J, Loffing-Cueni D, Valderrabano V, Kläusli L, Hebert SC, Rossier BC, et al.: Distribution of transcellular calcium and sodium transport pathways along mouse distal nephron. Am J Physiol Renal Physiol 281: F1021–F1027, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Velázquez H, Bartiss A, Bernstein P, Ellison DH: Adrenal steroids stimulate thiazide-sensitive NaCl transport by rat renal distal tubules. Am J Physiol 270: F211–F219, 1996 [DOI] [PubMed] [Google Scholar]
- 11.Kim GH, Masilamani S, Turner R, Mitchell C, Wade JB, Knepper MA: The thiazide-sensitive Na-Cl cotransporter is an aldosterone-induced protein. Proc Natl Acad Sci U S A 95: 14552–14557, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bostanjoglo M, Reeves WB, Reilly RF, Velázquez H, Robertson N, Litwack G, et al.: 11Beta-hydroxysteroid dehydrogenase, mineralocorticoid receptor, and thiazide-sensitive Na-Cl cotransporter expression by distal tubules [published correction appears in J Am Soc Nephrol 9: 2179, 1998]. J Am Soc Nephrol 9: 1347–1358, 1998 [DOI] [PubMed] [Google Scholar]
- 13.Câmpean V, Kricke J, Ellison D, Luft FC, Bachmann S: Localization of thiazide-sensitive Na(+)-Cl(-) cotransport and associated gene products in mouse DCT. Am J Physiol Renal Physiol 281: F1028–F1035, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Welling PA: Roles and regulation of renal K channels. Annu Rev Physiol 78: 415–435, 2016 [DOI] [PubMed] [Google Scholar]
- 15.Adam M, Potter AS, Potter SS: Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: A molecular atlas of kidney development. Development 144: 3625–3632, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Lee JW, Chou CL, Nair AV, Battistone MA, Păunescu TG, et al.: Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc Natl Acad Sci U S A 114: E9989–E9998, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karaiskos N, Rahmatollahi M, Boltengagen A, Liu H, Hoehne M, Rinschen M, et al.: A single-cell transcriptome atlas of the mouse glomerulus. J Am Soc Nephrol 29: 2060–2068, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Kirita Y, Donnelly EL, Humphreys BD: Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 30: 23–32, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, et al.: Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360: 758–763, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young MD, Mitchell TJ, Vieira Braga FA, Tran MGB, Stewart BJ, Ferdinand JR, et al.: Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361: 594–599, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ransick A, Lindström NO, Liu J, Zhu Q, Guo JJ, Alvarado GF, et al.: Single-cell profiling reveals sex, lineage, and regional diversity in the mouse kidney. Dev Cell 51: 399–413.e7, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lake BB, Chen S, Hoshi M, Plongthongkum N, Salamon D, Knoten A, et al.: A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun 10: 2832, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burg MB: Isolated perfused tubule. Introduction: Background and development of microperfusion technique. Kidney Int 22: 417–424, 1982 [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Chou C-L, Knepper MA: A comprehensive map of mRNAs and their isoforms across all 14 renal tubule segments of mouse. J Am Soc Nephrol 32: 897–912, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusaba T, Lalli M, Kramann R, Kobayashi A, Humphreys BD: Differentiated kidney epithelial cells repair injured proximal tubule. Proc Natl Acad Sci U S A 111: 1527–1532, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambrey R, Kurth I, Peti-Peterdi J, Houillier P, Purkerson JM, Leviel F, et al.: Renal intercalated cells are rather energized by a proton than a sodium pump. Proc Natl Acad Sci U S A 110: 7928–7933, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S, Park J, Qiu C, Chung KW, Li SY, Sirin Y, et al.: Jagged1/Notch2 controls kidney fibrosis via Tfam-mediated metabolic reprogramming. PLoS Biol 16: e2005233, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry MR, Mathews RJ, Ferdinand JR, Jing C, Loudon KW, Wlodek E, et al.: Renal sodium gradient orchestrates a dynamic antibacterial defense zone. Cell 170: 860–874.e19, 2017 [DOI] [PubMed] [Google Scholar]
- 29.Clark JZ, Chen L, Chou CL, Jung HJ, Lee JW, Knepper MA: Representation and relative abundance of cell-type selective markers in whole-kidney RNA-Seq data. Kidney Int 95: 787–796, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Flanagan CH, Campbell KR, Zhang AW, Kabeer F, Lim JLP, Biele J, et al.; CRUK IMAXT Grand Challenge Team: Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol 20: 210, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaissling B, Bachmann S, Kriz W: Structural adaptation of the distal convoluted tubule to prolonged furosemide treatment. Am J Physiol 248: F374–F381, 1985 [DOI] [PubMed] [Google Scholar]
- 32.Ellison DH, Velázquez H, Wright FS: Adaptation of the distal convoluted tubule of the rat. Structural and functional effects of dietary salt intake and chronic diuretic infusion. J Clin Invest 83: 113–126, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimm PR, Coleman R, Delpire E, Welling PA: Constitutively active SPAK causes hyperkalemia by activating NCC and remodeling distal tubules. J Am Soc Nephrol 28: 2597–2606, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saritas T, Puelles VG, Su XT, McCormick JA, Welling PA, Ellison DH: Optical clearing in the kidney reveals potassium-mediated tubule remodeling. Cell Rep 25: 2668–2675.e3, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong T, Liu L, Jiang W, Zhou R: DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat Rev Immunol 20: 95–112, 2020 [DOI] [PubMed] [Google Scholar]
- 36.Yan J, Aldrich RW: BK potassium channel modulation by leucine-rich repeat-containing proteins. Proc Natl Acad Sci U S A 109: 7917–7922, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng XH, Yang C, Xia XM, Liu M, Lingle CJ: SLO3 auxiliary subunit LRRC52 controls gating of sperm KSPER currents and is critical for normal fertility. Proc Natl Acad Sci U S A 112: 2599–2604, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokonami N, Takata T, Beyeler J, Ehrbar I, Yoshifuji A, Christensen EI, et al.: Uromodulin is expressed in the distal convoluted tubule, where it is critical for regulation of the sodium chloride cotransporter NCC. Kidney Int 94: 701–715, 2018 [DOI] [PubMed] [Google Scholar]
- 39.Dimke H, Hoenderop JG, Bindels RJ: Molecular basis of epithelial Ca2+ and Mg2+ transport: Insights from the TRP channel family. J Physiol 589: 1535–1542, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai LJ, Ritchie G, Kerstan D, Kang HS, Cole DE, Quamme GA: Magnesium transport in the renal distal convoluted tubule. Physiol Rev 81: 51–84, 2001 [DOI] [PubMed] [Google Scholar]
- 41.Hoorn EJ, Nelson JH, McCormick JA, Ellison DH: The WNK kinase network regulating sodium, potassium, and blood pressure. J Am Soc Nephrol 22: 605–614, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costantini F, Kopan R: Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev Cell 18: 698–712, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morales EE, Handa N, Drummond BE, Chambers JM, Marra AN, Addiego A, et al.: Homeogene emx1 is required for nephron distal segment development in zebrafish. Sci Rep 8: 18038, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivas S, Goldberg MR, Watanabe T, D’Agati V, al-Awqati Q, Costantini F: Expression of green fluorescent protein in the ureteric bud of transgenic mice: A new tool for the analysis of ureteric bud morphogenesis. Dev Genet 24: 241–251, 1999 [DOI] [PubMed] [Google Scholar]
- 45.Wilcox CS, Welch WJ, Murad F, Gross SS, Taylor G, Levi R, et al.: Nitric oxide synthase in macula densa regulates glomerular capillary pressure. Proc Natl Acad Sci U S A 89: 11993–11997, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mundel P, Bachmann S, Bader M, Fischer A, Kummer W, Mayer B, et al.: Expression of nitric oxide synthase in kidney macula densa cells. Kidney Int 42: 1017–1019, 1992 [DOI] [PubMed] [Google Scholar]
- 47.Aoyagi T, Izumi Y, Hiroyama M, Matsuzaki T, Yasuoka Y, Sanbe A, et al.: Vasopressin regulates the renin-angiotensin-aldosterone system via V1a receptors in macula densa cells. Am J Physiol Renal Physiol 295: F100–F107, 2008 [DOI] [PubMed] [Google Scholar]
- 48.Cowley AW Jr., Yang C, Kumar V, Lazar J, Jacob H, Geurts AM, et al.: Pappa2 is linked to salt-sensitive hypertension in Dahl S rats. Physiol Genomics 48: 62–72, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoyer JR, Sisson SP, Vernier RL: Tamm-Horsfall glycoprotein: Ultrastructural immunoperoxidase localization in rat kidney. Lab Invest 41: 168–173, 1979 [PubMed] [Google Scholar]
- 50.Salido EC, Barajas L, Lechago J, Laborde NP, Fisher DA: Immunocytochemical localization of epidermal growth factor in mouse kidney. J Histochem Cytochem 34: 1155–1160, 1986 [DOI] [PubMed] [Google Scholar]
- 51.Milatz S, Himmerkus N, Wulfmeyer VC, Drewell H, Mutig K, Hou J, et al.: Mosaic expression of claudins in thick ascending limbs of Henle results in spatial separation of paracellular Na+ and Mg2+ transport. Proc Natl Acad Sci U S A 114: E219–E227, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bray SJ: Notch signalling in context. Nat Rev Mol Cell Biol 17: 722–735, 2016 [DOI] [PubMed] [Google Scholar]
- 53.Palygin O, Pochynyuk O, Staruschenko A: Role and mechanisms of regulation of the basolateral Kir 4.1/Kir 5.1K+ channels in the distal tubules. Acta Physiol (Oxf) 219: 260–273, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Limbutara K, Chou CL, Knepper MA: Quantitative proteomics of all 14 renal tubule segments in rat. J Am Soc Nephrol 31: 1255–1266, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen F, Tisher CC: Morphology of the ascending thick limb of Henle. Kidney Int 9: 8–22, 1976 [DOI] [PubMed] [Google Scholar]
- 56.Zhang C, Wang L, Su XT, Lin DH, Wang WH: KCNJ10 (Kir4.1) is expressed in the basolateral membrane of the cortical thick ascending limb. Am J Physiol Renal Physiol 308: F1288–F1296, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C, Wang L, Zhang J, Su XT, Lin DH, Scholl UI, et al.: KCNJ10 determines the expression of the apical Na-Cl cotransporter (NCC) in the early distal convoluted tubule (DCT1). Proc Natl Acad Sci U S A 111: 11864–11869, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mennitt PA, Wade JB, Ecelbarger CA, Palmer LG, Frindt G: Localization of ROMK channels in the rat kidney. J Am Soc Nephrol 8: 1823–1830, 1997 [DOI] [PubMed] [Google Scholar]
- 59.Jeong HW, Jeon US, Koo BK, Kim WY, Im SK, Shin J, et al.: Inactivation of Notch signaling in the renal collecting duct causes nephrogenic diabetes insipidus in mice. J Clin Invest 119: 3290–3300, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macaulay IC, Ponting CP, Voet T: Single-cell multiomics: Multiple measurements from single cells. Trends Genet 33: 155–168, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.