Abstract
Rationale: A link among sphingolipids, 17q21 genetic variants, and childhood asthma has been suggested, but the underlying mechanisms and characteristics of such an asthma endotype remain to be elucidated.
Objectives: To study the sphingolipid-associated childhood asthma endotype using multiomic data.
Methods: We used untargeted liquid chromatography–mass spectrometry plasma metabolomic profiles at the ages of 6 months and 6 years from more than 500 children in the COPSAC2010 (Copenhagen Prospective Studies on Asthma in Childhood) birth cohort focusing on sphingolipids, and we integrated the 17q21 genotype and nasal gene expression of SPT (serine palmitoyl-CoA transferase) (i.e., the rate-limiting enzyme in de novo sphingolipid synthesis) in relation to asthma development and lung function traits from infancy until the age 6 years. Replication was sought in the independent VDAART (Vitamin D Antenatal Asthma Reduction Trial) cohort.
Measurements and Main Results: Lower concentrations of ceramides and sphingomyelins at the age of 6 months were associated with an increased risk of developing asthma before age 3, which was also observed in VDAART. At the age of 6 years, lower concentrations of key phosphosphingolipids (e.g., sphinganine-1-phosphate) were associated with increased airway resistance. This relationship was dependent on the 17q21 genotype and nasal SPT gene expression, with significant interactions occurring between the genotype and the phosphosphingolipid concentrations and between the genotype and SPT expression, in which lower phosphosphingolipid concentrations and reduced SPT expression were associated with increasing numbers of at-risk alleles. However, the findings did not pass the false discovery rate threshold of <0.05.
Conclusions: This exploratory study suggests the existence of a childhood asthma endotype with early onset and increased airway resistance that is characterized by reduced sphingolipid concentrations, which are associated with 17q21 genetic variants and expression of the SPT enzyme.
Keywords: sphingolipids, childhood asthma, lung function
At a Glance Commentary
Scientific Knowledge on the Subject
Experimental studies have suggested that sphingolipid synthesis is implicated in asthma pathogenesis through genetically regulated activity of SPT (serine palmitoyl-CoA transferase) (i.e., the rate-limiting enzyme in de novo sphingolipid synthesis). Recently, a study of school-aged children with asthma showed a lower amount of sphingolipid formation, in which 17q21 asthma risk variants were shown to decrease sphingolipid synthesis. These findings suggest that sphingolipid synthesis is affected genetically in childhood asthma, but the underlying mechanisms and clinical characteristics remain to be elucidated.
What This Study Adds to the Field
This exploratory study consisting of a discovery cohort and an independent replication cohort used metabolomic data to investigate sphingolipid concentrations in early life and integrated 17q21 genotype data and nasal SPT gene expression data to study the development of childhood asthma and lung function traits. The findings suggest the existence of a sphingolipid-associated childhood asthma endotype with an early onset of symptoms and increased airway resistance by the age of 6 years, which is characterized by a reduction in sphingolipid concentrations that is already present in infancy and is associated with 17q21 genetic variants and expression of the SPT enzyme.
Asthma is a chronic inflammatory airway disease that has increased in prevalence among children in Westernized societies (1). The disease is believed to originate in early life from gene–environment interactions (2) that lead to the onset of chronic inflammatory processes, which manifest as intermittent or more persistent troublesome respiratory symptoms later in childhood (3, 4). It may therefore be useful to understand the perturbed pathways and biochemical mechanisms involved in early life before the debut of symptoms to aid in both primary and secondary prevention.
Metabolomic data are suitable for studying early-onset disease mechanisms because they represent a snapshot of the ongoing biological pathways and metabolic status of an individual, which enables uncovering subtle phenotype differences and perturbed metabolic pathways that are essential for biomarker discovery and development of novel treatment modalities. Metabolomics has previously been applied in epidemiological studies of asthma in children and adults (5, 6), showing alterations in lipids, steroids, amino acids, bile acids, and metabolites related to immune responses and oxidative stress (5, 7–13). Longitudinal birth cohort studies with metabolomic data are particularly well suited to investigating the early-life origins of asthma, especially in the context of using discovery and replication cohorts.
A particular class of lipids containing a backbone of sphingoid bases (i.e., the sphingolipids), which includes phosphosphingolipids, ceramides, and sphingomyelins, has gained attention as an important player in asthma pathogenesis (14). A recent study showed decreased sphingolipid concentrations in children 5–17 years of age with nonallergic asthma, in whom 17q21 asthma risk variants were shown to decrease de novo sphingolipids synthesis (15). Furthermore, we previously found that high-dose vitamin D supplementation during pregnancy in VDAART (Vitamin D Antenatal Asthma Reduction Trial) was associated with increased plasma concentrations of sphingosine-1-phosphate in children with the low-risk 17q21 genotype at the age of 3 years. Furthermore, higher sphingolipid concentrations were associated with a decreased risk of asthma, with evidence of interactions among the sphingosine-1-phosphate concentration, the vitamin D intervention, and the 17q21 genotype (16). These findings suggest the existence of a sphingolipid-associated childhood asthma endotype, but the underlying mechanisms and characteristics of such an endotype remain to be elucidated.
Here, we performed global metabolomic profiling of plasma samples from children in the population-based COPSAC2010 (Copenhagen Prospective Studies on Asthma in Childhood) mother–child cohort at the ages of 6 months and 6 years and investigated the association with the development of asthma and lung function traits to obtain mechanistic insights into asthma pathogenesis, focusing on the sphingolipid pathway. We integrated 17q21 genotype data and gene expression data for the SPT (serine palmitoyl transferase) enzyme to explore the regulation of the biochemical findings. Replication was sought in the independent VDAART birth cohort (17).
Methods
Study Populations
COPSAC2010
The COPSAC2010 population-based, mother–child cohort of 738 pregnant women and their 700 children has previously been described in detail (18). The pregnant women participated in two nested, double-blind, randomized control trials (RCTs) with n-3 long-chain polyunsaturated fatty acids and high-dose vitamin D. The study was approved by The National Committee on Health Research Ethics (H-B-2008-093) and the Danish Data Protection Agency (2015-41-3696), with oral and written consent being provided by parents before enrollment. The details and results of the RCTs have been previously published (19, 20).
VDAART
The VDAART mother–child cohort of 881 women with a history of asthma, eczema, or allergic rhinitis and their 810 children also has an embedded RCT of high-dose vitamin D supplementation during pregnancy (17). The children were prospectively followed for asthma/recurrent wheezing until age 3. Plasma samples from a subset of children at the age of 1 year were used for metabolomic profiling analysis by Metabolon, Inc., and were used for replication (16).
Data Collection
Clinical outcomes
The COPSAC2010 children were followed prospectively for asthma/recurrent wheezing, lower respiratory tract infections, wheezing episodes, and acute severe exacerbations until the age of 6 years, at which time spirometry (FEV1, FEV1/FVC ratio), whole-body plethysmography (specific airway resistance [sRaw]), and bronchial responsiveness to methacholine (PD20) were measured. In addition, the parents filled a daily diary from birth, recording troublesome lung symptoms and use of antiasthmatic treatments.
Metabolomics
Blood samples for plasma metabolomic analysis were collected in ethylenediaminetetraacetic acid tubes at the scheduled clinic visits at 6 months of age and 6 years of age. The sample extracts were analyzed using four liquid chromatography–mass spectrometry methods: two separated reverse-phase ultraperformance liquid chromatography electrospray ionization tandem mass spectrometry methods optimized for hydrophilic and hydrophobic compounds, one reverse-phase ultraperformance liquid chromatography tandem mass spectrometry method using basic optimized conditions, and one hydrophilic interaction liquid chromatography/ultraperformance liquid chromatography tandem mass spectrometry method. The metabolomic analysis of the plasma samples was performed by Metabolon, Inc., using their HD4 platform, which was also applied to the VDAART samples. The identification level follows the criteria described by Sumner and colleagues (21).
Genotyping
Variation in the 17q21 region was assessed by genotyping the SNPs rs12936231, rs2305480, rs4065275, and rs7216389 using the Illumina Infinium HumanOmniExpressExome BeadChip Kit at the AROS Applied Biotechnology A/S center.
Gene expression
Nasal-brushing transcriptomic data at 6 years of age (22) were generated from RNA extracted using the SMART-Seq v4 Ultra Low Input RNA Kit (Takara Bio Inc.), and cDNA libraries were created with the Illumina Nextera XT kit using the default instructions. The Agilent 2100 Bioanalyzer was used to determine concentrations. FastQC and MultiQC (Babraham Bioinformatics) were used to assess the read quality. The RNA-sequencing reads were mapped to the genome by using STAR (version 2.5.1) aligner software (23).
Data Analysis
The COPSAC2010 6-month metabolomic data set included 577 children and 1,068 metabolites, whereas the 6-years data set comprised 513 children and 1,076 metabolites. Missing values were imputed metabolite-wise with half of the minimum value, and data were autoscaled before analysis. Data preprocessing is detailed in the online supplement.
Univariate regression analyses, consisting of linear regression, Cox proportional hazards regression, and logistic and quasi-Poisson modeling, were employed to relate the metabolite levels at 6 months of age and 6 years of age with the clinical outcomes by applying a multiple-testing false discovery rate (FDR) < 0.05 significance threshold. Metabolites in the sphingolipid pathway in COPSAC2010 were sought and replicated in VDAART in terms of the direction of association and significance at an FDR < 0.05 level.
Supervised multivariate analysis, specifically partial least squares discriminant analysis (PLS-DA), was employed to explore the association between the metabolite concentrations and sRaw. The sRaw measurements were split into quartiles and used for the analysis as a two-class problem comparing the lower and the upper quartile.
All models were adjusted for breastfeeding duration, z-scored child body mass index, and the pregnancy n-3 long-chain polyunsaturated fatty acid and high-dose vitamin D interventions.
Further details are outlined in the online supplement.
Results
Baseline characteristics and clinical outcome measures of the children with available plasma metabolomic profiling data by the ages of 6 months (n = 577) and 6 years (n = 513) are shown in Table E1 in the online supplement.
Metabolomic Profile at the Age of 6 Months and Clinical Outcomes
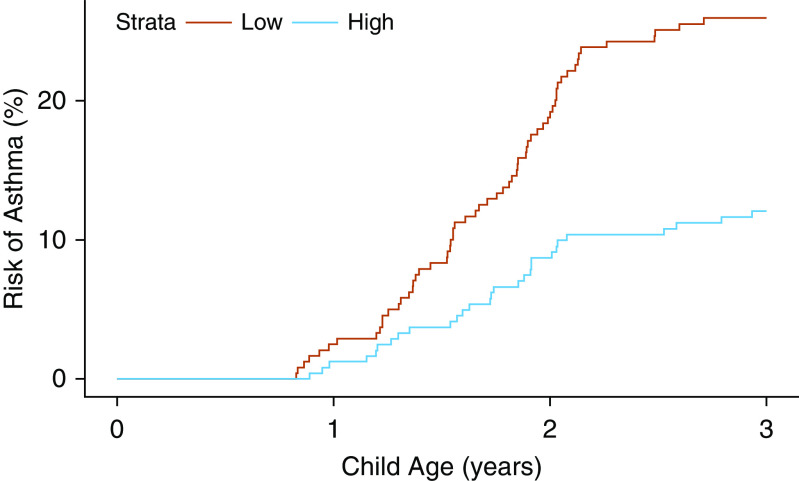
The plasma metabolomic profile at the age of 6 months was analyzed in relation to development of an early-onset asthma phenotype (i.e., symptom debut in the first 3 years of life) and in relation to asthma status by the age of 6 years (24). Using Cox regression analysis of asthma development until 3 years of age, we observed 150 out of 1,076 metabolites passing nominal significance (P ≤ 0.05), with 27 of these 150 metabolites being sphingolipids. The nominally significant sphingolipids were ceramides and sphingomyelins containing long-chain saturated and monounsaturated fatty acids, and they were negatively associated with asthma development (i.e., lower concentrations increased the risk of developing early-onset asthma). However, neither of the sphingolipids passed a multiple-testing FDR < 0.05 threshold (Table 1). The sphingolipid that was most strongly associated with asthma was the ceramide glycosyl-N-stearoyl-sphingosine (d18:1/18:0): β-estimate, −0.52 (95% confidence interval [CI], −0.78 to −0.25), P < 0.001, FDR = 0.09 (see Kaplan-Meier curve in Figure 1).
Table 1.
Sphingolipids at the Age of 6 Months Associated with Early-Onset Asthma Development before 3 Years of Age: Discovery in COPSAC2010 and Replication in VDAART
| Metabolite | Subpathway | β-Estimate for COPSAC2010 | CI | P Value | FDR P Value | β-Estimate for VDAART | P Value | FDR P Value |
|---|---|---|---|---|---|---|---|---|
| Glycosyl-N-stearoyl-sphingosine (d18:1/18:0) | Ceramides | −0.516 | −0.777 to −0.254 | 0.0001 | 0.09 | −0.598 | 0.03 | 0.93 |
| Glycosyl ceramide (d18:1/20:0, d16:1/22:0)* | −0.448 | −0.687 to −0.208 | 0.0003 | 0.24 | −0.409 | 0.15 | 0.93 | |
| Glycosyl-N-behenoyl-sphingadienine (d18:2/22:0)* | −0.319 | −0.558 to −0.081 | 0.009 | 1 | — | — | — | |
| Glycosyl-N-(2-hydroxynervonoyl)-sphingosine (d18:1/24:1)* | −0.454 | −0.726 to −0.182 | 0.001 | 1 | — | — | — | |
| N-stearoyl-sphingosine (d18:1/18:0)* | −0.282 | −0.515 to −0.048 | 0.02 | 1 | −0.115 | 0.50 | 0.95 | |
| Ceramide (d16:1/24:1, d18:1/22:1)* | −0.225 | −0.437 to −0.012 | 0.04 | 1 | — | — | — | |
| Sphingomyelin (d18:1/19:0, d19:1/18:0)* | Sphingomyelins | −0.428 | −0.647 to −0.21 | 0.0001 | 0.12 | −0.286 | 0.13 | 0.93 |
| Sphingomyelin (d18:2/21:0, d16:2/23:0)* | −0.421 | −0.643 to −0.199 | 0.0002 | 0.19 | −0.290 | 0.17 | 0.93 | |
| Stearoyl sphingomyelin (d18:1/18:0) | −0.391 | −0.61 to −0.172 | 0.0005 | 0.41 | −0.951 | 0.02 | 0.93 | |
| Sphingomyelin (d18:1/17:0, d17:1/18:0, d19:1/16:0) | −0.368 | −0.603 to −0.133 | 0.002 | 1 | −0.526 | 0.06 | 0.93 | |
| Sphingomyelin (d18:1/20:0, d16:1/22:0)* | −0.330 | −0.568 to −0.092 | 0.007 | 1 | −0.333 | 0.38 | 0.93 | |
| Sphingomyelin (d18:1/20:1, d18:2/20:0)* | −0.293 | −0.505 to −0.081 | 0.007 | 1 | −1.487 | 0.005 | 0.71 | |
| Sphingomyelin (d18:1/18:1, d18:2/18:0) | −0.311 | −0.541 to −0.08 | 0.008 | 1 | −0.994 | 0.02 | 0.93 | |
| Sphingomyelin (d18:2/18:1)* | −0.303 | −0.53 to −0.075 | 0.009 | 1 | −0.622 | 0.08 | 0.93 | |
| Sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1)* | −0.273 | −0.482 to −0.064 | 0.01 | 1 | −0.449 | 0.17 | 0.93 | |
| Sphingomyelin (d17:1/16:0, d18:1/15:0, d16:1/17:0)* | −0.269 | −0.477 to −0.062 | 0.01 | 1 | −0.248 | 0.35 | 0.93 | |
| Sphingomyelin (d18:1/21:0, d17:1/22:0, d16:1/23:0)* | −0.307 | −0.556 to −0.057 | 0.02 | 1 | −0.168 | 0.35 | 0.93 | |
| Sphingomyelin (d17:2/16:0, d18:2/15:0)* | −0.276 | −0.505 to −0.047 | 0.02 | 1 | −0.053 | 0.76 | 0.98 | |
| Sphingomyelin (d18:2/23:1)* | −0.266 | −0.491 to −0.042 | 0.02 | 1 | −0.247 | 0.33 | 0.93 | |
| N-palmitoyl-sphinganine (d18:0/16:0) | 0.219 | 0.032 to 0.407 | 0.02 | 1 | −0.146 | 0.32 | 0.93 | |
| Sphingomyelin (d18:1/14:0, d16:1/16:0)* | −0.253 | −0.471 to −0.036 | 0.02 | 1 | −0.111 | 0.70 | 0.97 | |
| Sphingomyelin (d18:1/22:1, d18:2/22:0, d16:1/24:1)* | −0.245 | −0.456 to −0.034 | 0.02 | 1 | −0.443 | 0.40 | 0.93 | |
| Sphingomyelin (d18:0/20:0, d16:0/22:0)* | −0.266 | −0.501 to −0.031 | 0.03 | 1 | −0.269 | 0.19 | 0.93 | |
| Sphingomyelin (d18:1/20:2, d18:2/20:1, d16:1/22:2)* | −0.232 | −0.449 to −0.016 | 0.04 | 1 | −0.417 | 0.14 | 0.93 | |
| Sphingomyelin (d18:0/18:0, d19:0/17:0)* | −0.266 | −0.519 to −0.013 | 0.04 | 1 | −0.275 | 0.07 | 0.93 | |
| Palmitoyl sphingomyelin (d18:1/16:0) | −0.210 | −0.413 to −0.007 | 0.04 | 1 | −1.301 | 0.06 | 0.93 | |
| Sphingomyelin (d18:1/25:0, d19:0/24:1, d20:1/23:0, d19:1/24:0)* | −0.245 | −0.488 to −0.001 | 0.05 | 1 | −0.208 | 0.14 | 0.93 |
Definition of abbreviations: CI = confidence interval; COPSAC2010 = Copenhagen Prospective Studies on Asthma in Childhood; FDR = false discovery rate; VDAART = Vitamin D Antenatal Asthma Reduction Trial.
Figure 1.
Kaplan-Meier curve of the association between the ceramide glycosyl-N-stearoyl sphingosine (d18:1/18:0) dichotomized by the median (low and high) and the development of asthma in the first 3 years of life.
In the VDAART plasma metabolomic data set for the age 1 year (n = 469), 23 of the 27 sphingolipids that were nominally significant in COPSAC2010 were detected. As shown in Table 1, 4 out of the 23 sphingolipids were significantly associated with development of asthma/wheezing before the age of 3 years (P ≤ 0.05), and a further 4 showed a trend of association (P ≤ 0.10). All of these sphingolipids showed the same direction of association as in COPSAC2010 (i.e., increasing risk with lower concentrations). The sphingolipids included the ceramide glycosyl-N-stearoyl-sphingosine (d18:1/18:0) and seven sphingomyelins, including stearoyl sphingomyelin (d18:1/18:0), sphingomyelin (d18:1/20:1, d18:2/20:0)*, sphingomyelin (d18:1/18:1, d18:2/18:0), sphingomyelin (d18:1/17:0, d17:1/18:0, d19:1/16:0), sphingomyelin (d18:0/18:0, d19:0/17:0)*, sphingomyelin (d18:2/18:1)*, and palmitoyl sphingomyelin (d18:1/16:0). However, none of these sphingolipids were replicated in VDAART at a multiple-testing FDR < 0.05 threshold.
Apart from sphingolipids, homocitrulline showed a strong positive association with development of asthma before the age of 3 years in COPSAC2010 (β-estimate, 0.28 [95% CI, 0.16–0.4], P < 0.001, FDR = 0.01), but homocitrulline was not associated with early-onset asthma/wheezing in VDAART (P = 0.99, FDR = 1).
There was no association between the metabolome at the age of 6 months and asthma by the age 6 of years and the number of wheezing episodes, lower respiratory tract infections, acute exacerbations of wheezing, or any of the lung function outcomes (see Figure E1).
Metabolomic Profile at 6 Years and Clinical Outcomes
The plasma metabolomic profiles at the age of 6 years were investigated in relation to asthma status and lung function measurements at the same time point.
The univariate cross-sectional analyses of the association between the metabolome and asthma at 6 years showed no significant associations (Figure E2). Furthermore, in a Spearman correlation analysis, the plasma concentrations of the 8 sphingolipids at the age of 6 months, which showed similar results in relation to early-onset asthma in COPSAC2010 and VDAART, were not correlated with the corresponding plasma concentrations at the age of 6 years (Table E2).
Among the lung function measurements, the univariate analyses showed significant associations for sRaw but not for FEV1, the FEV1/FVC ratio, or the PD20 of methacholine (Figure E3). For sRaw (n = 495), a total of 73 metabolites were nominally significant (P ≤ 0.05), with 2 of these being sphingolipids. The significant sphingolipids were among the top six metabolites most strongly associated with sRaw in the univariate linear models. They were both phosphosphingolipids and showed an inverse association, with lower concentrations resulting in increasing airway resistance (sphinganine-1-phosphate: β-estimate, −0.04 [95% CI, −0.06 to −0.02], P < 0.001, FDR = 0.16; sphingosine-1-phosphate: β-estimate, −0.03 [95% CI, −0.05 to −0.01], P = 0.002, FDR = 1).
To validate the univariate findings, a multivariate PLS-DA model was applied to investigate the association between the overall patterns of metabolites and sRaw, which was analyzed by comparing children with an sRaw measurement in the first and fourth quartiles. The final PLS-DA model consisted of two components and 37 metabolites, had a cross-validated area under the curve = 0.85, a cross-validated classification error = 0.23, and a P value = 0.005 (based on 100 permutations) for separating children with low versus high sRaw (Figures 2 and E4). The PLS-DA model showed results comparable to those of the univariate analyses, with lower amounts of the phosphosphingolipids sphinganine-1-phosphate and sphingosine-1-phosphate in children with higher sRaw.
Figure 2.
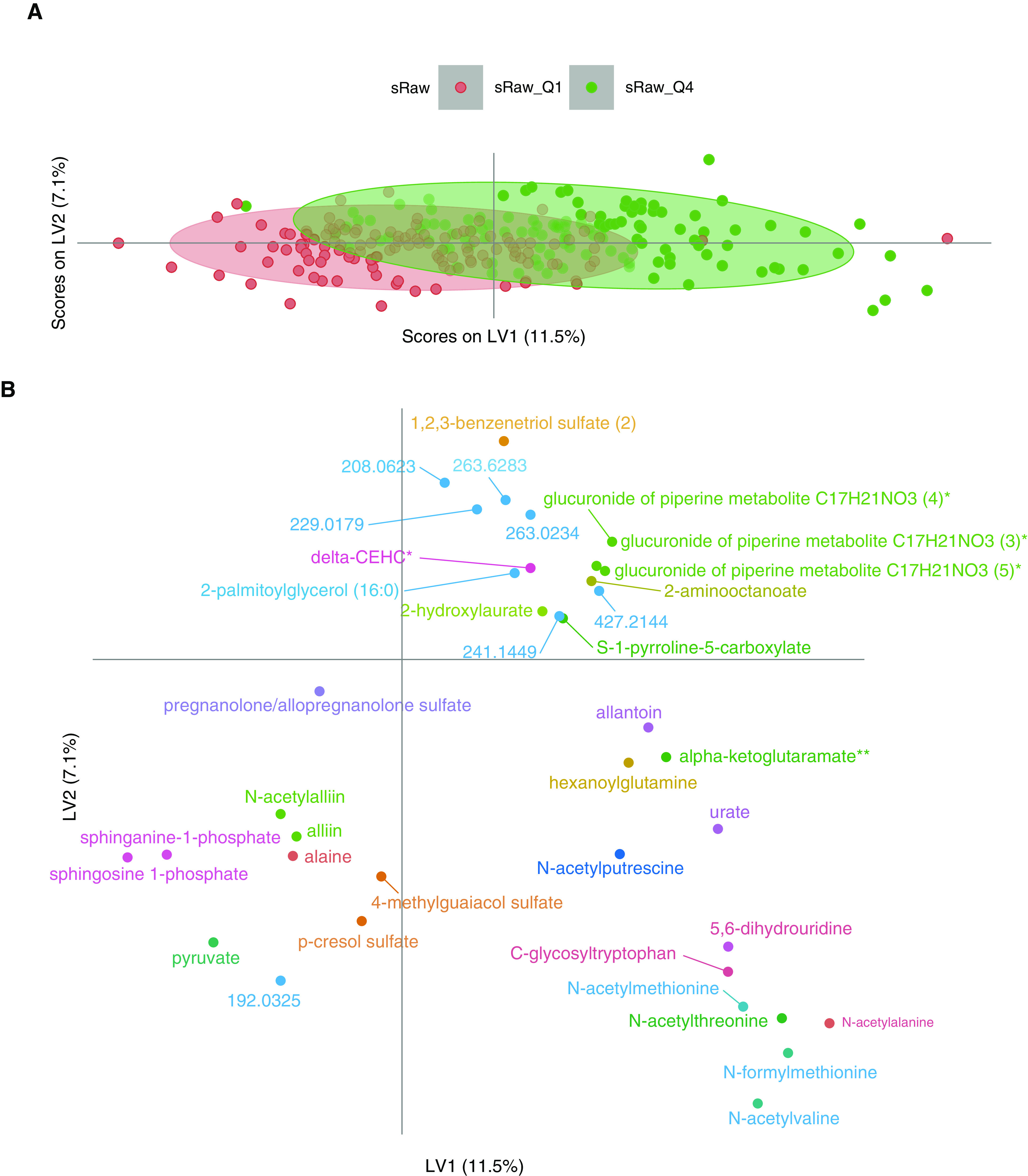
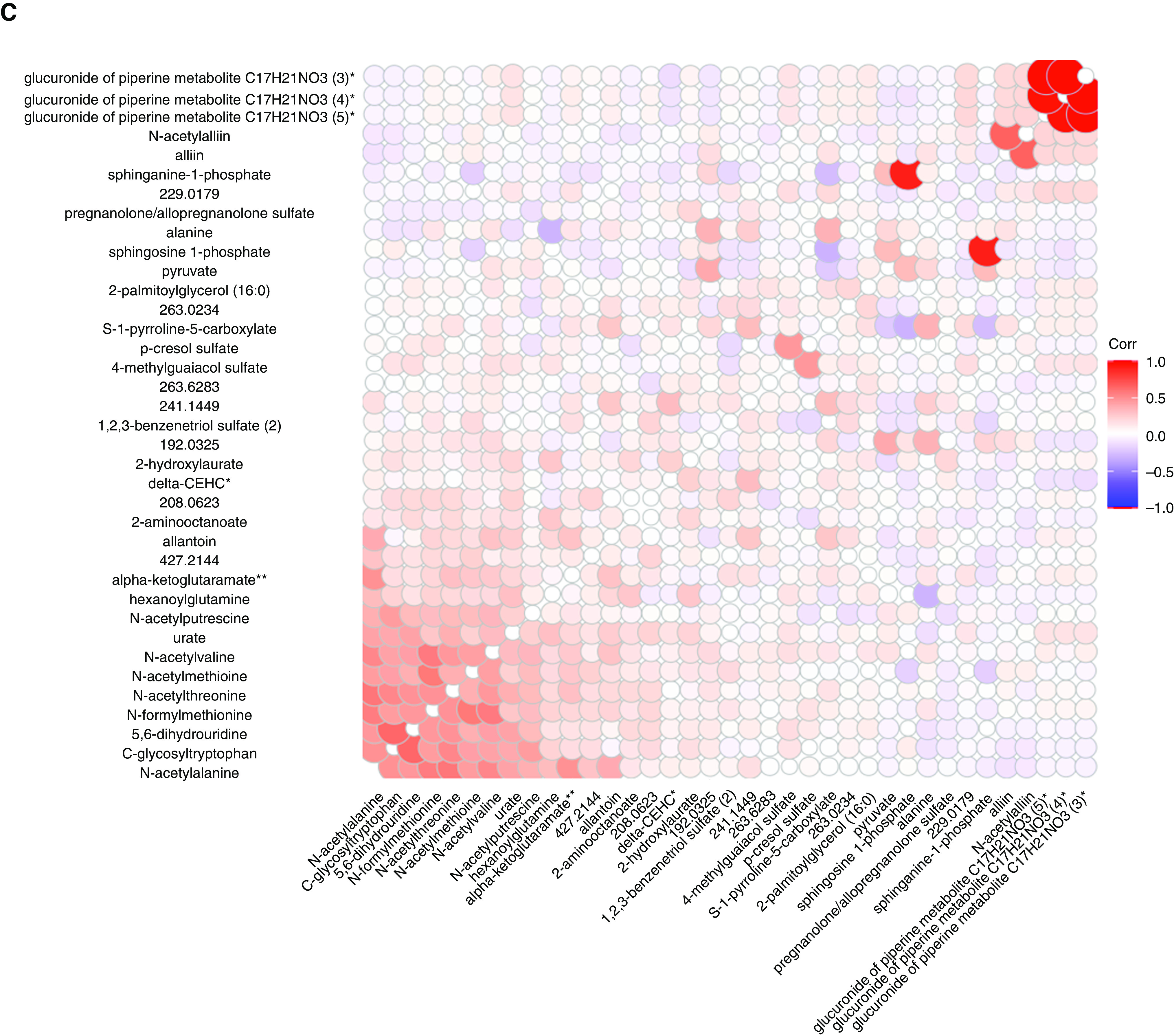
Partial least squares discriminant analysis of the metabolome at the age 6 years versus the sRaw at the age of 6 years. (A) Scores plot. (B) Loadings plot. (C) Spearman Corr map of the metabolites at the age of 6 years selected in the partial least squares discriminant analysis model. Corr = correlation; delta-CEHC = C14H18O4; LV = latent variable; Q = quartile; sRaw = specific airway resistance.
sRaw, Phosphosphingolipids, 17q21 Genotype, and Nasal Gene Expression of SPT
It has been shown that 17q21 SNPs associated with childhood asthma outcomes are linked to an increased transcription of ORMDL3 (25, 26), which is believed to affect the enzyme activity of SPT in de novo sphingolipid synthesis, particularly involving phosphosphingolipids. Therefore, we investigated this relationship using four SNPs in the 17q21 region previously shown to increase expression of ORMDL3 (27): rs12936231 (28), rs7216389 (29), rs2305480 (29), and rs4065275 (28). Subsequently, we used transcriptomic data from nasal brushings to evaluate possible interactions among the expression of the SPTLC1, SPTLC2, SPTLC3, SPTSSA, and SPTSSB genes coding for the SPT enzyme, the expression of the ORMDL3 gene encoding the ORMDL3 protein, phosphosphingolipid levels, and 17q21 genotypes.
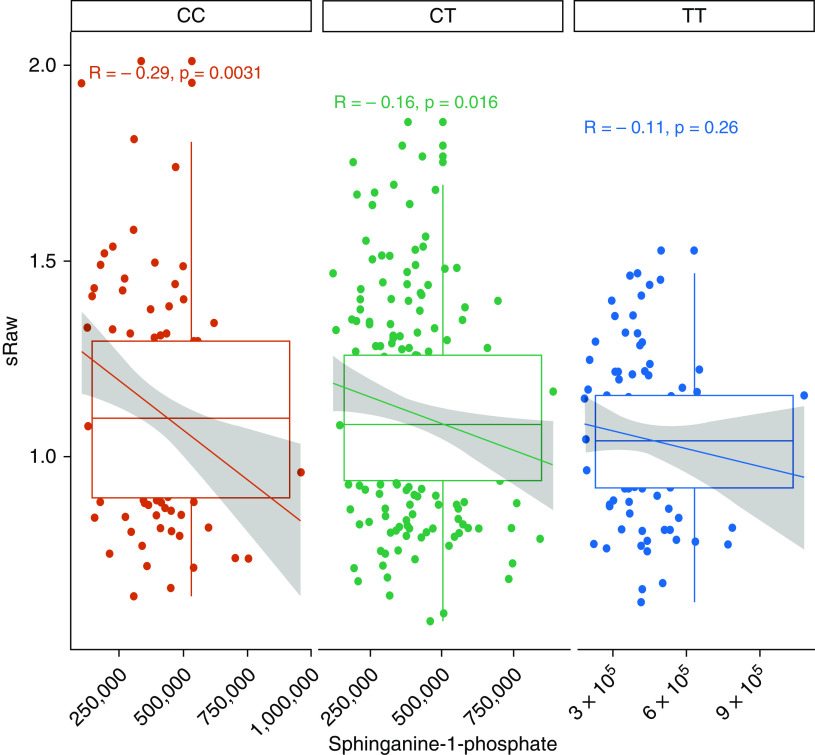
First, when analyzing for an interaction between 17q21 genotypes and the amounts of sphinganine-1-phosphate and sphingosine-1-phosphate in relation to sRaw at the age of 6 years, we observed that rs12936231 showed a significant interaction with sphinganine-1-phosphate (P = 0.043) (Table E3). Our rs12936231 genotype–stratified analysis showed that this interaction was due to lower sphinganine-1-phosphate concentrations in children carrying increasing numbers of the high-risk allele (Figure 3).
Figure 3.
Association between sphinganine-1-phosphate and sRaw stratified by rs1293623 genotype (i.e., CC, CT, and TT). Each box represents the median with a 95% confidence interval. The line and the text above each group represents the Spearman correlation value. sRaw = specific airway resistance.
Second, we investigated whether this relationship could be observed in the gene expression levels for genes related to SPT and ORMDL3, phosphosphingolipid levels, and sRaw. The interaction models between the 17q21 genotypes and SPT gene expression in relation to sRaw showed significant interactions between SPTLC1 expression and all four of the 17q21 SNPs. In addition, SPTSSA expression was lower in children carrying rs12936231 risk alleles (Figure 4A). Models stratified by 17q21 genotype showed that this interaction was explained by increasing sRaw with decreasing SPT expression. These associations were significant between SPTLC1 expression and 17q21 genotypes with two risk alleles, with similar patterns of association occurring between SPTSSA and SPTLC3 expression and 17q21 genotypes (Figure 4B).
Figure 4.
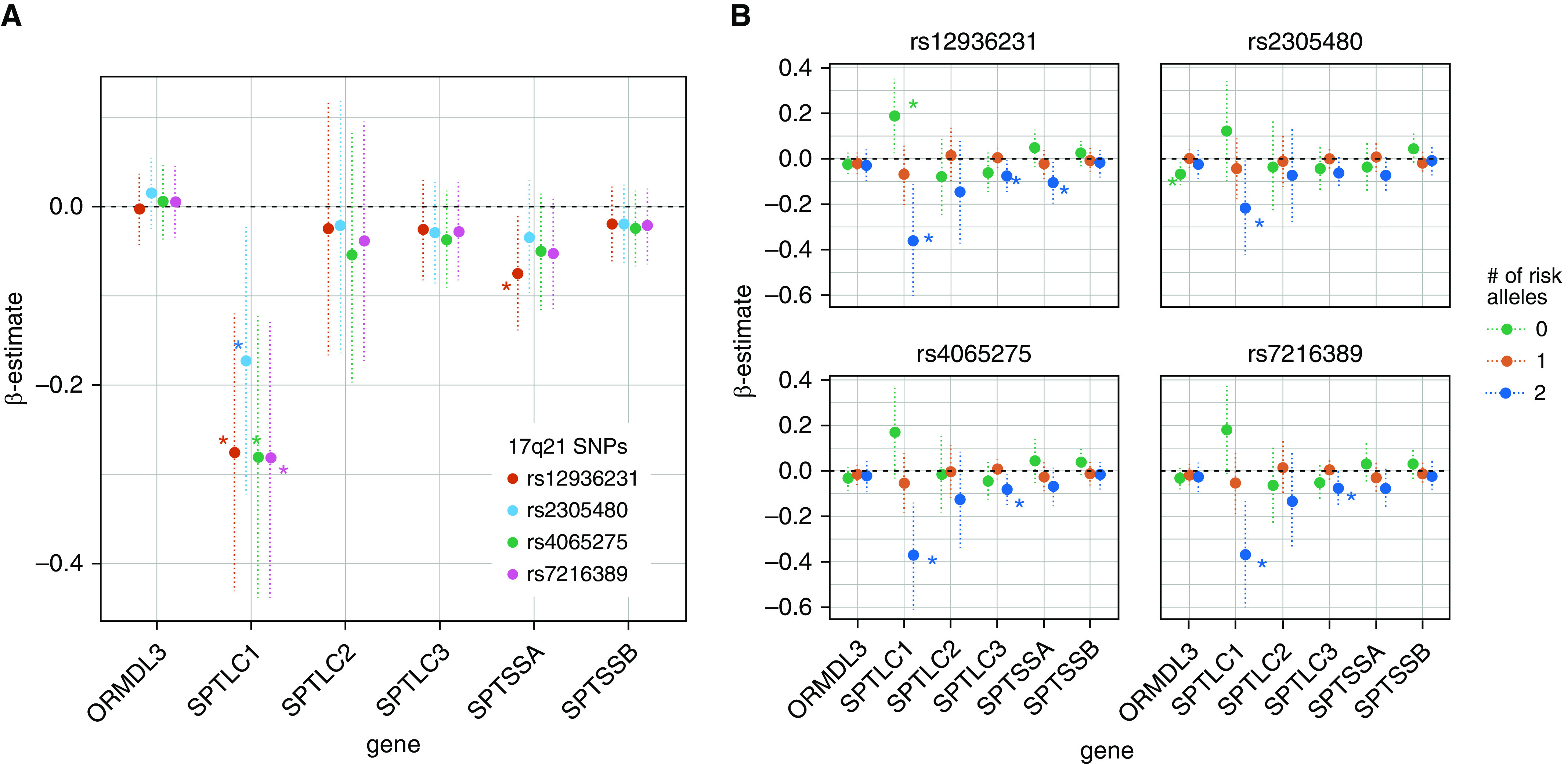
(A) Interaction model between nasal SPT (serine palmitoyl-CoA transferase) and ORMDL3 gene expression (x-axes) and 17q21 genotypes in relation to specific airway resistance (sRaw). (B) Association between nasal SPT and ORMDL3 gene expression (x-axes) and sRaw stratified by 17q21 genotypes. Each point represents the β-estimate of the association, and the dotted vertical lines represent the confidence intervals. *P ≤ 0.05.
Finally, we investigated whether concentrations of sphinganine-1-phosphate and sphingosine-1-phosphate were affected by SPT expression and 17q21 genotype, which could explain the negative association between sRaw and phosphosphingolipid levels. The analyses overall showed significant interactions between SPTSSB expression and the 17q21 SNPs (Figure 5A). In 17q21 genotype–stratified models, we observed that the interactions were explained by an association between phosphosphingolipid concentrations and SPTSSB expression only among children without risk alleles. A similar trend was observed for the SPTLC2 expression (Figure 5B).
Figure 5.
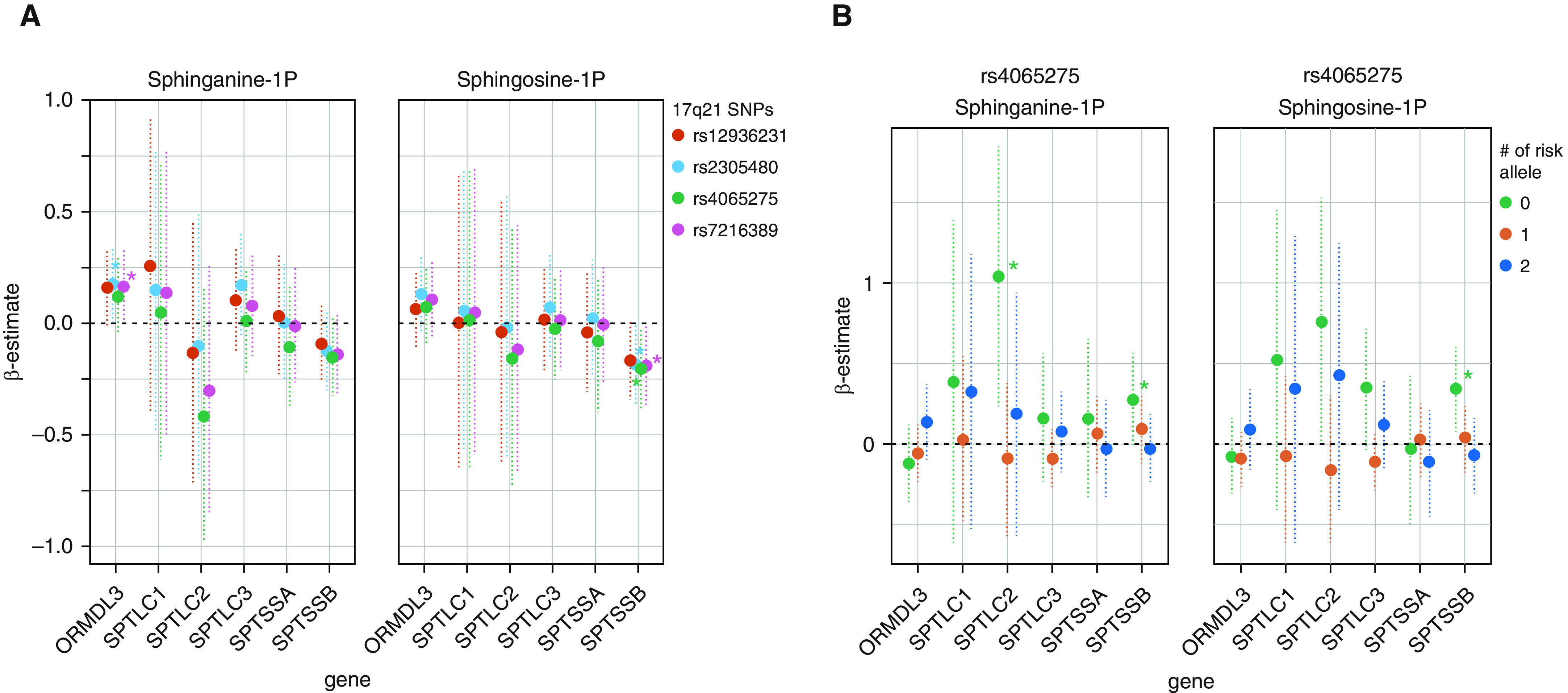
(A) Interaction model between nasal SPT (serine palmitoyl-CoA transferase) and ORMDL3 gene expression (x-axes) and 17q21 genotypes in relation to plasma phosphosphingolipid levels. (B) Association between nasal SPT and ORMDL3 gene expression (x-axes) and phosphosphingolipid levels stratified by 17q21 genotypes. Each point represents the β-estimate of the association, the dotted vertical lines represent the confidence intervals, and each color indicates the genotype stratum expressed as the number of risk alleles. *P ≤ 0.05.
Discussion
This exploratory study suggests that changes in the sphingolipid pathway across two time points in early life (i.e., 6 months and 6 years) are associated with the risk of developing early-onset asthma and increased airway resistance, which involves both the de novo and the salvage sphingolipid pathways. The findings were specific for airway resistance measured by plethysmography and were not present for FEV1 or bronchial hyperreactivity, which may suggest that a perturbed sphingolipid metabolism is associated with inflammation, particularly in the smaller airways in young children.
For the age of 6 months, we found that decreased amounts of ceramides and sphingomyelins were associated with the development of asthma/wheezing before the age of 3 years. This is possibly explained by the formation/degradation of complex sphingolipids from the action of specific enzymes, such as kinases, phosphatases, and lyases, present in the sphingolipid pathway. However, we cannot discriminate whether the observed phenomenon is due to a lower formation of ceramides from the de novo pathway or whether it is due to a general effect on the salvage pathway because none of the significant sphingolipids that we observed to be associated with early-onset asthma/wheezing are produced solely in the de novo pathway. On the other hand, for children at the age of 6 years, we observed that children with higher airway resistance had lower plasma concentrations of the phosphosphingolipids (i.e., sphinganine-1-phosphate and sphingosine-1-phosphate), which are present in the de novo and salvage pathways, respectively. This latter observation may indicate suboptimal sphingolipid production in susceptible individuals, which is similar to what we observed during infancy at the age of 6 months before disease onset. However, the phosphosphingolipids associated with airway resistance by the age of 6 years are different from the sphingolipids at the age of 6 months that were associated with an early onset of symptoms. Furthermore, despite our observation of similar associations in the discovery and replication cohorts, the sphingolipids did not pass an FDR < 0.05 multiple-testing threshold, and these results should therefore be interpreted with caution.
Sphingolipid metabolites are involved in several cellular functions, including immune responses, growth, and differentiation (30, 31). Studies of sphingolipids in different cellular and animal airway models, particularly those investigating the role of the phosphosphingolipid sphingosine-1-phosphate, have shown diverging results. A study in mice showed that increasing amounts of sphingosine-1-phosphate by systemic administration was linked to increasing airway hyperresponsiveness (32), and another study using a mast cell–dependent murine model of allergic asthma showed that lowering the concentration of sphingosine-1-phosphate by using a sphingosine kinase 1 inhibitor attenuated airway hyperresponsiveness and inflammation (33) (i.e., better asthma outcomes in mice with lower phosphosphingolipid concentrations). A study by Ammit and colleagues showed that increasing the amount of sphingosine-1-phosphate in the BAL fluid of allergen-challenged individuals with asthma modulated airway smooth muscle cell functions that promote inflammation and airway remodeling (34). Furthermore, decreased sphingolipid synthesis, including in the phosphosphingolipids sphinganine-1-phosphate and sphingosine-1-phosphate, has been observed in mice overexpressing ORMDL3 (35) as well as in children with 17q21 asthma risk genotypes (15), which was associated with asthma and airway remodeling (32) (i.e., worse asthma outcomes with lower phosphosphingolipid concentrations in genetically asthma-susceptible mice and children).
A lower amount of sphingolipid formation has previously been linked to the homeostatic regulation of ORMDL3 on the SPT enzyme, as first demonstrated in yeast (36) and then in mice (37) as well as human cell lines (38, 39). The SNPs in the 17q21 region, which are associated with development of childhood asthma, have been shown to be strongly associated with increased expression of the ORMDL3 gene (25, 40). Furthermore, in mouse models and human cell line studies, the 17q21 risk allele increases the transcription of the ORMDL3 protein, which blocks the SPT enzyme, leading to lower amounts of sphingolipid formation (14). Our exploratory study shows associations that support the existence of such mechanisms in the inception of a particular childhood asthma endotype with early onset.
Using nasal-brushing transcriptomic data, we assessed the role of the SPT enzyme and 17q21 risk variants in plasma phosphosphingolipid concentrations. The SPT enzyme is encoded by the SPTLC1, SPTLC2 (41), and SPTLC3 (42) genes and by the two small subunits SPTSSA and SPTSSB. SPT1 and SPT2 are the main active forms of the enzyme, but either the SPTSSA or the SPTSSB protein confers full enzyme activity (43). We hypothesize that a significant association with one or more of the subunits expressing the SPT enzyme might be sufficient to affect de novo sphingolipid production, as they are all involved in full enzyme activity. Therefore, we performed our analysis using all five subunits and thereby identified an inverse association between lower amounts of phosphosphingolipids and increasing sRaw by the age of 6 years. This relationship was significantly affected by the 17q21 genotype, which interacted with the expression of the SPT enzyme and decreased the phosphosphingolipid concentrations (e.g., significant for sphingosine-1-phosphate when interacting with SPTSSB).
We did not observe a significant interaction between 17q21 genotypes and the ORMDL3 expression in relation to sRaw, but in children without any risk alleles, there was a significant negative association for rs2305480 and a positive association between sRaw and SPTLC1. Both phenomena are plausible in children with a wild-type genotype, due to other biological conditions, which could affect the expression of ORMDL3, translating to a suboptimal protein amount and then leading to worse lung function. Similar hypotheses could also explain the increased SPT expression also affecting the phosphosphingolipid production in a nonoptimal way. The significant interaction between ORMDL3 and the SNPs rs2305480 and rs7216389 was not present in the stratified model, which could indicate a spurious finding.
Another important point to consider is ORMDL3/SPT stoichiometry playing a role in the regulation of the SPT enzyme activity. In fact, it has been demonstrated that overexpression of ORMDL3 in the lung epithelium increased ceramides concentrations, potentially with a feedback mechanism from the salvage pathway (44). On the other hand, it was also shown that a small increase in ORMDL3 expression decreased ceramides concentrations (44), which might explain our findings.
Our study is an exploratory study, which deals with the intrinsic data collinearity that is present in all untargeted metabolomic data sets (45). This collinearity is due to biological factors (i.e., the fact that similar metabolites behave similarly, particularly in the same pathway, as in the sphingolipids) but is also due to analytical factors (e.g., chromatographic coelution, charge competition, etc.). We chose a setup with a discovery and replication cohort, which showed similar findings for the early metabolomic time point in relation to early-onset asthma/wheezing, but the results did not survive multiple-testing correction. As we had no replication for the metabolomic data at the age of 6 years, we validated the univariate findings in a multivariate PLS-DA model, which showed similar results.
Conclusions
This exploratory study suggests the existence of a childhood asthma endotype with early onset of symptoms and increased airway resistance, which is characterized by reduced sphingolipid concentrations that are associated with 17q21 genetic variants and expression of the SPT enzyme.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the children and families of the COPSAC2010 cohort study for all their support and commitment. They also thank the COPSAC research team for their efforts and thank Prof. Carole Ober for intellectual input on the manuscript.
Footnotes
COPSAC (Copenhagen Prospective Studies on Asthma in Childhood) is funded by private and public research funds, which are all listed on www.copsac.com. The Lundbeck Foundation, Danish State Budget, Danish Council for Strategic Research, Danish Council for Independent Research, and The Capital Region Research Foundation have provided core support for COPSAC. The study is further supported by the following NIH grants: R01 HL129735 and R01 HL141826. C.E.W. was supported by the Swedish Heart–Lung Foundation (Hjärt–Lungfonden 20170734; 20180290). This project has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 946228).
Author Contributions: The guarantor of the study is H.B., who is responsible for the integrity of the work as a whole, from conception and design of the study to conduct of the study and the acquisition of data, the analysis and interpretation of data, and the writing of the manuscript. H.B. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. D.R. and B.L.C. were responsible for data analysis and wrote the first draft of the manuscript. C.-E.T.P., M.H., R.S.K., G.G., N.B., H.K., K.A.L.-S., A.M., M.A.R., J.S., K.B., A.A.L., C.E.W., S.T.W., and J.L.-S. contributed to the design of the study, interpretation of the data, and writing of the manuscript. C.-E.T.P. and M.H. contributed to the data analysis. All co-authors have contributed substantially to the analyses and/or interpretation of the data and have provided important intellectual input and approval of the final version to be published. The lead author (H.B.) affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned have been explained.
Data sharing statement: The COPSAC (Copenhagen Prospective Studies on Asthma in Childhood) biobank is publicly available at the Danish National Biobank (www.biobankdenmark.dk), and data will become available in the Danish Data Archive (www.sa.dk) upon request to the corresponding author.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202008-3206OC on February 3, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355:2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 2.Wheelock CE, Rappaport SM. The role of gene-environment interactions in lung disease: the urgent need for the exposome. Eur Respir J. 2020;55:1902064. doi: 10.1183/13993003.02064-2019. [DOI] [PubMed] [Google Scholar]
- 3.Prescott SL. Early-life environmental determinants of allergic diseases and the wider pandemic of inflammatory noncommunicable diseases. J Allergy Clin Immunol. 2013;131:23–30. doi: 10.1016/j.jaci.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Prescott S, Allen KJ. Food allergy: riding the second wave of the allergy epidemic. Pediatr Allergy Immunol. 2011;22:155–160. doi: 10.1111/j.1399-3038.2011.01145.x. [DOI] [PubMed] [Google Scholar]
- 5.Chiu C-Y, Lin G, Cheng ML, Chiang MH, Tsai MH, Su KW, et al. Longitudinal urinary metabolomic profiling reveals metabolites for asthma development in early childhood. Pediatr Allergy Immunol. 2018;29:496–503. doi: 10.1111/pai.12909. [DOI] [PubMed] [Google Scholar]
- 6.Chawes BL, Giordano G, Pirillo P, Rago D, Rasmussen MA, Stokholm J, et al. Neonatal urine metabolic profiling and development of childhood asthma. Metabolites. 2019;9:185. doi: 10.3390/metabo9090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly RS, Virkud Y, Giorgio R, Celedón JC, Weiss ST, Lasky-Su J. Metabolomic profiling of lung function in Costa-Rican children with asthma. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1590–1595. doi: 10.1016/j.bbadis.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blighe K, Chawes BL, Kelly RS, Mirzakhani H, McGeachie M, Litonjua AA, et al. Vitamin D prenatal programming of childhood metabolomics profiles at age 3 y. Am J Clin Nutr. 2017;106:1092–1099. doi: 10.3945/ajcn.117.158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly RS, Chawes BL, Blighe K, Virkud YV, Croteau-Chonka DC, McGeachie MJ, et al. An integrative transcriptomic and metabolomic study of lung function in children with asthma. Chest. 2018;154:335–348. doi: 10.1016/j.chest.2018.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee-Sarwar KA, Kelly RS, Lasky-Su J, Zeiger RS, O’Connor GT, Sandel MT, et al. Integrative analysis of the intestinal metabolome of childhood asthma. J Allergy Clin Immunol. 2019;144:442–454. doi: 10.1016/j.jaci.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly RS, Dahlin A, McGeachie MJ, Qiu W, Sordillo J, Wan ES, et al. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest. 2017;151:262–277. doi: 10.1016/j.chest.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saude EJ, Skappak CD, Regush S, Cook K, Ben-Zvi A, Becker A, et al. Metabolomic profiling of asthma: diagnostic utility of urine nuclear magnetic resonance spectroscopy. J Allergy Clin Immunol. 2011;127:757–764, e1–e6. doi: 10.1016/j.jaci.2010.12.1077. [DOI] [PubMed] [Google Scholar]
- 13.Mattarucchi E, Baraldi E, Guillou C. Metabolomics applied to urine samples in childhood asthma; differentiation between asthma phenotypes and identification of relevant metabolites. Biomed Chromatogr. 2012;26:89–94. doi: 10.1002/bmc.1631. [DOI] [PubMed] [Google Scholar]
- 14.Worgall TS, Veerappan A, Sung B, Kim BI, Weiner E, Bholah R, et al. Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci Transl Med. 2013;5:186ra67. doi: 10.1126/scitranslmed.3005765. [DOI] [PubMed] [Google Scholar]
- 15.Ono JG, Kim BI, Zhao Y, Christos PJ, Tesfaigzi Y, Worgall TS, et al. Decreased sphingolipid synthesis in children with 17q21 asthma-risk genotypes. J Clin Invest. 2020;130:921–926. doi: 10.1172/JCI130860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly RS, Chawes BL, Guo F, Zhang L, Blighe K, Litonjua AA, et al. The role of the 17q21 genotype in the prevention of early childhood asthma and recurrent wheeze by vitamin D. Eur Respir J. 2019;54:1900761. doi: 10.1183/13993003.00761-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litonjua AA, Lange NE, Carey VJ, Brown S, Laranjo N, Harshfield BJ, et al. The Vitamin D Antenatal Asthma Reduction Trial (VDAART): rationale, design, and methods of a randomized, controlled trial of vitamin D supplementation in pregnancy for the primary prevention of asthma and allergies in children. Contemp Clin Trials. 2014;38:37–50. doi: 10.1016/j.cct.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bisgaard H, Vissing NH, Carson CG, Bischoff AL, Følsgaard NV, Kreiner-Møller E, et al. Deep phenotyping of the unselected COPSAC2010 birth cohort study. Clin Exp Allergy. 2013;43:1384–1394. doi: 10.1111/cea.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bisgaard H, Stokholm J, Chawes BL, Vissing NH, Bjarnadóttir E, Schoos AM, et al. Fish oil-derived fatty acids in pregnancy and wheeze and asthma in offspring. N Engl J Med. 2016;375:2530–2539. doi: 10.1056/NEJMoa1503734. [DOI] [PubMed] [Google Scholar]
- 20.Chawes BL, Bønnelykke K, Stokholm J, Vissing NH, Bjarnadóttir E, Schoos AM, et al. Effect of vitamin D3 supplementation during pregnancy on risk of persistent wheeze in the offspring: a randomized clinical trial. JAMA. 2016;315:353–361. doi: 10.1001/jama.2015.18318. [DOI] [PubMed] [Google Scholar]
- 21.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin A, McKennan CG, Pedersen CT, Stokholm J, Chawes BL, Malby Schoos AM, et al. Epigenetic landscape links upper airway microbiota in infancy with allergic rhinitis at 6 years of age. J Allergy Clin Immunol. 2020;146:1358–1366. doi: 10.1016/j.jaci.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thysen AH, Waage J, Larsen JM, Rasmussen MA, Stokholm J, Chawes B, et al. Distinct immune phenotypes in infants developing asthma during childhood. Sci Transl Med. 2020;12:eaaw0258. doi: 10.1126/scitranslmed.aaw0258. [DOI] [PubMed] [Google Scholar]
- 25.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 26.Stein MM, Thompson EE, Schoettler N, Helling BA, Magnaye KM, Stanhope C, et al. A decade of research on the 17q12-21 asthma locus: piecing together the puzzle. J Allergy Clin Immunol. 2018;142:749–764, e3. doi: 10.1016/j.jaci.2017.12.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acevedo N, Reinius LE, Greco D, Gref A, Orsmark-Pietras C, Persson H, et al. Risk of childhood asthma is associated with CpG-site polymorphisms, regional DNA methylation and mRNA levels at the GSDMB/ORMDL3 locus. Hum Mol Genet. 2015;24:875–890. doi: 10.1093/hmg/ddu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmiedel BJ, Seumois G, Samaniego-Castruita D, Cayford J, Schulten V, Chavez L, et al. 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nat Commun. 2016;7:13426. doi: 10.1038/ncomms13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GTEx Consortium. Human genomics: the Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breslow DK, Weissman JS. Membranes in balance: mechanisms of sphingolipid homeostasis. Mol Cell. 2010;40:267–279. doi: 10.1016/j.molcel.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roviezzo F, D’Agostino B, Brancaleone V, De Gruttola L, Bucci M, De Dominicis G, et al. Systemic administration of sphingosine-1-phosphate increases bronchial hyperresponsiveness in the mouse. Am J Respir Cell Mol Biol. 2010;42:572–577. doi: 10.1165/rcmb.2009-0108OC. [DOI] [PubMed] [Google Scholar]
- 33.Price MM, Oskeritzian CA, Falanga YT, Harikumar KB, Allegood JC, Alvarez SE, et al. A specific sphingosine kinase 1 inhibitor attenuates airway hyperresponsiveness and inflammation in a mast cell-dependent murine model of allergic asthma. J Allergy Clin Immunol. 2013;131:501–511, e1. doi: 10.1016/j.jaci.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ammit AJ, Hastie AT, Edsall LC, Hoffman RK, Amrani Y, Krymskaya VP, et al. Sphingosine 1-phosphate modulates human airway smooth muscle cell functions that promote inflammation and airway remodeling in asthma. FASEB J. 2001;15:1212–1214. doi: 10.1096/fj.00-0742fje. [DOI] [PubMed] [Google Scholar]
- 35.Miller M, Rosenthal P, Beppu A, Gordillo R, Broide DH. Oroscomucoid like protein 3 (ORMDL3) transgenic mice have reduced levels of sphingolipids including sphingosine-1-phosphate and ceramide. J Allergy Clin Immunol. 2017;139:1373–1376, e4. doi: 10.1016/j.jaci.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serlachius E, Svennilson J, Schalling M, Aperia A. Protein kinase C in the developing kidney: isoform expression and effects of ceramide and PKC inhibitors. Kidney Int. 1997;52:901–910. doi: 10.1038/ki.1997.411. [DOI] [PubMed] [Google Scholar]
- 38.Siow D, Sunkara M, Dunn TM, Morris AJ, Wattenberg B. ORMDL/serine palmitoyltransferase stoichiometry determines effects of ORMDL3 expression on sphingolipid biosynthesis. J Lipid Res. 2015;56:898–908. doi: 10.1194/jlr.M057539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta SD, Gable K, Alexaki A, Chandris P, Proia RL, Dunn TM, et al. Expression of the ORMDLS, modulators of serine palmitoyltransferase, is regulated by sphingolipids in mammalian cells. J Biol Chem. 2015;290:90–98. doi: 10.1074/jbc.M114.588236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wills-Karp M. At last: linking ORMDL3 polymorphisms, decreased sphingolipid synthesis, and asthma susceptibility. J Clin Invest. 2020;130:604–607. doi: 10.1172/JCI134333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hojjati MR, Li Z, Jiang X-C. Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim Biophys Acta. 2005;1737:44–51. doi: 10.1016/j.bbalip.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Hornemann T, Richard S, Rütti MF, Wei Y, von Eckardstein A. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J Biol Chem. 2006;281:37275–37281. doi: 10.1074/jbc.M608066200. [DOI] [PubMed] [Google Scholar]
- 43.Han G, Gupta SD, Gable K, Niranjanakumari S, Moitra P, Eichler F, et al. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc Natl Acad Sci U S A. 2009;106:8186–8191. doi: 10.1073/pnas.0811269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oyeniran C, Sturgill JL, Hait NC, Huang WC, Avni D, Maceyka M, et al. Aberrant ORM (yeast)-like protein isoform 3 (ORMDL3) expression dysregulates ceramide homeostasis in cells and ceramide exacerbates allergic asthma in mice. J Allergy Clin Immunol. 2015;136:1035–46.e6. doi: 10.1016/j.jaci.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Playdon MC, Joshi AD, Tabung FK, Cheng S, Henglin M, Kim A, et al. Metabolomics analytics workflow for epidemiological research: perspectives from the Consortium of Metabolomics Studies (COMETS) Metabolites. 2019;9:145. doi: 10.3390/metabo9070145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.