The prevalence of allergy and asthma has increased significantly over the past several decades, particularly so in industrialized nations where environmental exposures and lifestyles have rapidly diverged from those with which humans evolved. Multifactorial and heterogenous, there is an urgent need to understand the origins of allergy and asthma and develop effective interventions for precision treatment and prevention. Epidemiological studies have identified prenatal and early-life exposures that prevent or promote disease in childhood, a number of which exert strong selective pressures on the types, functions, epigenetic modifications, and development of cellular populations (microbial and immune) critical to immune training, function, and asthma pathogenesis. Technological advances in the fields of human genetics, epigenetics, immunology, and microbiome research have more recently expanded our view of human biology. Studies employing these tools individually have uncovered key relationships with allergy and asthma development across temporal and geospatial gradients. These observations suggest that although these factors are important in determining asthma risk, their influence is likely integrative, contextual, and cumulative across temporal gradients. Clearly, a range of extrinsic and intrinsic factors collude to promote disease development. Central to the integration and phenotypic translation of these exposures are cellular populations, in particular, maternal prenatal, and early-life postnatal microbial and immune cells, whose activities and interactions set in motion successional programming events that govern developmental trajectories and downstream health outcomes. This Perspective discusses the most salient of these events and proposes an actionable research scenario that integrates and contextualizes them.
Environmental Microbiota and Asthma
Although a wealth of epidemiologic data point to strong effects of environmental microbial exposures on asthma risk (1–3), probably few examples are as eloquent as the “farm effect.” Children raised on traditional farms are strongly protected from asthma (4), and this protection is largely explained by the child’s contact with farm animals and their microbiota in early life (5–7). The strength and pervasiveness of the farm effect are well illustrated by studies in Amish and Hutterite children who share genetic ancestry and lifestyles but follow distinct farming practices (traditional among the Amish, industrialized among the Hutterites) and have sharply different amounts of asthma prevalence (four-times lower in the Amish) (8, 9). In parallel with these distinct lifestyles, concentrations of house dust endotoxin (bacterial cell-associated polysaccharides) were almost seven-fold higher in Amish households, and profound differences in the proportions, phenotypes, and functions of innate and adaptive immune cells were detected in these populations (9). Moreover, inhalation of Amish, but not Hutterite, house dust was sufficient to protect mice against cardinal allergic asthma phenotypes, and these effects required innate immunity (8). These data strongly support a role of environmental exposure, particularly increased microbial exposures, in asthma protection. A similar association between increased exposure to environmental microbiota and asthma protection has also been described in American inner-city populations (2, 3, 10) and in Finnish versus Russian Karelia (11). The latter represent two socioeconomically distinct but geoclimatically similar regions. Allergies were more common in Finnish than in Russian children, and increased microbial load in house dust and drinking water was associated with allergy protection in Russia (11). Comparable observations have also been made in two populations of Mexican ancestry resident only 70 miles apart on either side of the United States/Mexico border but significantly different in both their rates of childhood asthma (four-fold lower in Mexico) and their house dust and bacterial load (strikingly higher in Mexico) (12). Thus, across a variety of environments, diminished environmental microbial exposure associated with Westernized lifestyles is consistently associated with increased risk of asthma. How these altered environmental microbial exposures may mediate their effects requires an understanding of the human microbiome, which develops in early life, significantly expands the functional capacity of the human host (13), and encodes activities and products that shape (14, 15) and are shaped by human immunity.
Early-Life Microbiomes and Asthma Development
Mammalian development is intimately linked to early-life microbiological development, particularly in the gut microbiome, which houses the largest diversity and burden of microbes in the human host (16). Reflective of coevolution, gut microbial products influence immune function and physiological development both locally in the gut and at remote organ and mucosal sites (17). Moreover, the gut microbiome integrates environmental exposures, including microbial (18), dietary (16), and antibiotic (19) encounters that exert both acute and pervasive effects on microbiome membership, productivity, and development. Thus, an emerging hypothesis for the origins of childhood asthma focuses on the prenatal and early postnatal period, which represents an intergenerational inflection point in microbiome development and immune training. This hypothesis posits that environmental exposures shape prenatal maternal microbiomes and immunity, which in turn influence vertical transmission, development, and activity of microbiomes and training of host immune responses during the critical postnatal period of immune training and physiological maturation (Figure 1). These events are likely influenced by a combination of human genetics (20) and early-life exposures, including maternal and infant nutrition and antimicrobial administration (21), which sculpt the nascent microbial colonization landscape, and by epigenetic modifications that are both intergenerationally and transgenerationally inherited (22) and postnatally forged by microbial-derived metabolic products (23). We note that the interactions between human genetics and the environment remain proverbially difficult to decipher, especially in a genome-wide context. Thus, candidate gene studies have shown that variants in the asthma-associated chromosome 17q21 locus, which encodes genes involved in pyroptosis (cellular defense against intracellular microbes), influence risk of asthma and allergy development by interacting with specific microbial exposures (24, 25). On the other hand, the results obtained using unbiased microbial genome-wide association studies to investigate microbiome–host genome associations are currently less clear; associations are typically weak and distributed across multiple variants, similar to the signals typically observed in classical genome-wide association studies of complex traits. Progress will likely occur once uniform data and reporting formats are adopted to facilitate replication and meta-analysis efforts, stringent statistical criteria are enforced to reduce the number of false-positive findings, and sample sizes increase by orders of magnitude (26). Overall, the emerging theory is that exogenous and endogenous factors that deplete coevolved microbial species critical to training asthma-protective immunity result in pathogenic microbiome development in early life, skewed immune function, and asthma outcomes in childhood.
Figure 1.
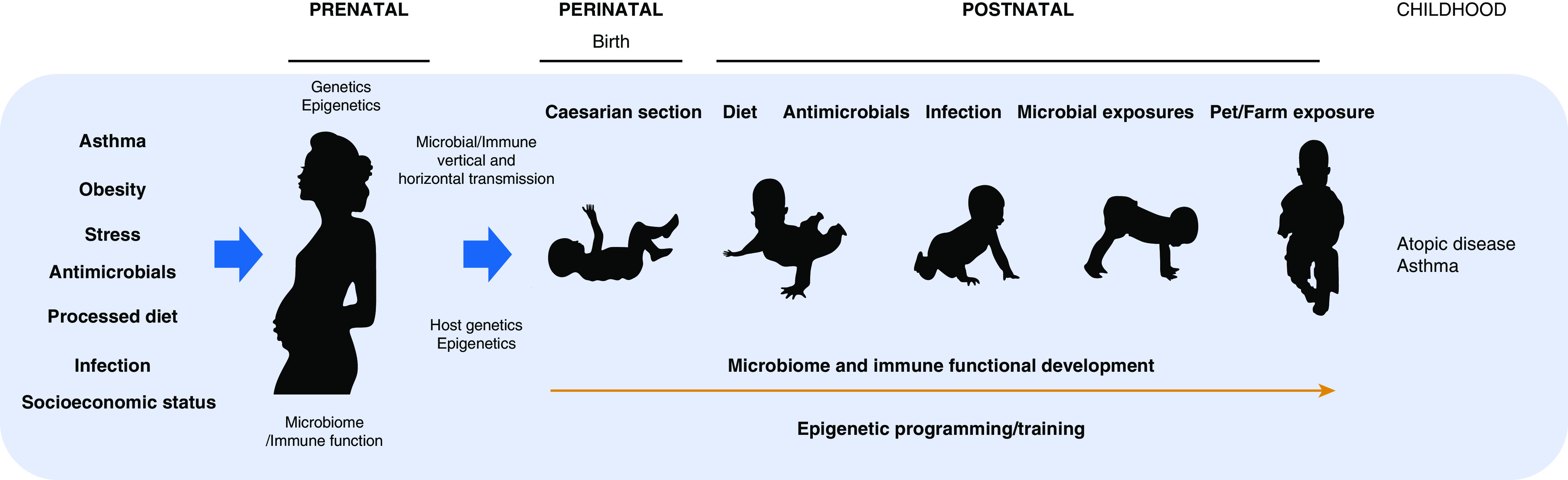
Integrative model for the origin of childhood atopy and asthma focuses on exposures that shape prenatal microbial and immune development and influence epigenetic imprinting that affect microbial and immune vertical transmission, postnatal microbial and immune development, and clinical outcomes in later childhood.
Consistent with this hypothesis, independent birth cohorts based in the United States and Canada have demonstrated that early-life (at birth, 1 mo, or 3 mo) gut microbiota perturbation is associated with increased risk of atopy/asthma in later childhood (27–29). Of note, in two of these cohorts (27, 28), the high-risk gut microbiota was associated with a lack of exposure to dogs in early life, indicating that environmental exposures associated with reduced risk of childhood asthma may mediate their effect by altering the composition and functional properties of the infant gut microbiome. Early-life gut bacterial depletions related to increased asthma risk are relatively consistent across these studies and associated with fecal metabolic dysfunction. Cell-free extracts of these perturbed high-risk neonatal gut microbiomes increase CD4+ IL-4+ cell frequency and activity and reduce CD4+ FOXP3+ cell frequency in vitro (28), suggesting that the products of the gut microbiome of infants who subsequently develop atopy or asthma may promote intestinal inflammation and thus alter microbial and immune development in these subjects. Consistent with this hypothesis, specific metabolites, such as 12,13 DiHOME, identified as enriched in the gut microbiomes of 1-month-old infants at significantly higher risk of atopy and asthma in childhood, have been shown to decrease regulatory T-cell frequency and IL-10 secretion (28). In a mouse model of allergic airway sensitization, introduction into the gut microbiome of bacterial epoxide hydrolase genes responsible for the production of 12,13 DiHOME is sufficient to increase circulating concentrations of this monohydroxy fatty acid, reduce airway regulatory T-cell populations, and exacerbate airway allergic inflammation (30), confirming a role for this gut microbial–derived lipid in asthma-associated immune dysfunction. In other studies, mice fed a high-fiber diet, which is exclusively fermented by gut microbes to produce antiinflammatory short chain fatty acids (SCFAs), exhibited significantly reduced airway allergic inflammation after antigen challenge (31). Thus, these data indicate that gut microbial–derived metabolic products modulate airway mucosal cellular populations critical to prevention of allergic inflammation and that diet influences microbial activities involved in such processes.
That such early-life microbial activities are important for disease development is reflected by findings in two independent birth cohorts showing that for every doubling of bacterial epoxide hydrolase gene copy number or nanogram increase in 12,13 DiHOME concentration in 1-month-old infant feces, the odds ratio for atopy or asthma in childhood significantly increased (30). Though only a single example of a more complex and dynamic microbiome–host interaction, these data offer a mechanistic link between early-life gut microbiome metabolic productivity and airway immune dysfunction associated with allergy and asthma development. Moreover, they indicate that microbial activities in the gut can influence airway mucosal immune responses with the potential to shape respiratory microbial colonization events at this site in early life.
The possibility that early-life events and influences set in motion distinct trajectories of microbial development is supported by observations made in a longitudinal study of infants defined as having high or low risk for asthma on the basis of parental asthma status. Meconium microbiota of high-risk infants was distinct from those at lower risk of disease (27), supporting the hypothesis that the maternal prenatal microbiome, which is shaped by intrinsic and extrinsic factors, influences the pattern of microbial vertical transmission. These high-risk infants followed a significantly distinct trajectory of gut microbiota development, exhibiting a decreased rate of gut bacterial diversification over the first year of life (27). Essentially, high-risk infants failed to accumulate microbial species from their environment at the same rate as their healthy counterparts, thus developing a genetically distinct and functionally depleted gut microbiome. Meconium microbiota by definition form in utero and are sourced from maternal microbiomes. Given the observation that meconium microbiota are distinct in infants at high and low risk for asthma, the focus on early-life microbiomes has now broadened to encompass the prenatal period, maternal microbiomes, and immune status. Indeed, as further discussed below, our recent studies have identified viable immunomodulatory bacteria in the human fetal intestine (32) and shown that the maternal prenatal immune profile (expressed by the IFN-γ/IL-13 ratio in the supernatant of mitogen-stimulated peripheral blood mononuclear cells isolated during the third trimester of pregnancy) is a robust predictor of asthma in the child (33).
Trained Immunity and Epigenetics as Mediators of Early Exposure Effects on Immune Development
Functional human adaptive immunity is evident at birth and includes antigen-experienced memory T- and B-cell populations capable of responding to microbial stimuli (34, 35), indicating that priming and programming of human immunity begins in utero. That vertically inherited microbial species are important for early-life intestinal immune development is reinforced by a recent study of a cohort of 50 human fetal (16–22 wk gestation) meconium samples. Subsets of human fetal meconium were relatively enriched for either Lactobacillus or Micrococcus (32). The presence of these species related to both the proportion of lamina propria innate-like PLZF+ CD161+ CD4+ T-memory cells and to divergent epithelial cell layer transcriptomes. Fetal intestinal Lactobacillus and Micrococcus were viable as evidenced by scanning electron microscopy but could only be isolated from fetal meconium when cultured under conditions that mimicked the in utero milieu (i.e., supplementation with pregnancy hormones or monocyte feeder cells) (32), indicating that specialized signals provided in the human fetal intestine support the presence of very specific immunomodulatory microbial strains. In vitro assays of human fetal antigen-presenting/T-cell cocultures revealed that fetal intestinal Lactobacillus jensenii isolates reduced antigen-presenting cell activation and promoted IL-17F production, whereas a fetal Micrococcus luteus isolate reduced TNF-α production by antigen-presenting cells and inhibited IFN-γ production by memory PLZF+ T cells (32). These data indicate that prenatal adaptive immunity, which shapes the mucosal landscape for postnatal microbial colonization, can be influenced by the presence of viable fetal intestinal bacteria that are maternally sourced and likely dependent on the presence of these strains in the maternal prenatal microbiome.
What other processes and mechanisms may allow prenatal maternal immune responses to exert a long-term influence on the child’s immune development and her/his trajectory to asthma? Evidence emerging from our most recent work points to an important role of trained immunity, a (relatively) novel way to think about innate immune responses. First, some context: in a seminal paper published in 1997, Ruslan Medzhitov and Charlie Janeway, who had just transformed the innate immunity field by cloning a human homolog of the Drosophila Toll receptor responsible for innate immune responses in flies (36), extolled the virtues of innate immunity as a nonclonal, germline-encoded system capable of ensuring virtually flawless discrimination between self and nonself. In this scenario, the ability to activate naive T cells when and only when a pathogen is present relies on Toll-like receptors (TLRs), a family of molecules expressed primarily on myeloid lineages. Being germline-encoded and thus free from the vagaries of on-demand genomic rearrangements, TLRs are shaped by evolution to ensure that innate immune recognition be directed toward invariant targets that are shared by multiple microorganisms and are typically necessary for those microorganisms to thrive. On the downside, compared with the clonal, rearrangement-based machinery of adaptive immunity, germline-encoded innate immune receptors cannot provide long-term antigen-specific immunological memory; the process of self-/nonself-discrimination restarts at every encounter rather than being stored in rearranged T- and/or B-cell receptor–bearing lymphocytes that rapidly expand to face renewed insults. This is why, the argument went, innate and adaptive responses need one another to provide adequate, measured responses against microbes.
Although still firmly in place, this scenario has undergone some important adjustments. Given the evolutionary success of organisms lacking adaptive immunity (up to 97% of the total biodiversity on our planet) (37), immunological memory is unlikely to have evolved only in vertebrates. Thus, it is hardly surprising that the dichotomy between innate and adaptive immune memory has become increasingly blurred over the past two decades. Indeed, mounting evidence shows that the immune system of plants and invertebrates may be “primed” by an initial infection in a manner that protects against subsequent infections (reviewed in Reference 37). Likewise, functions akin to memory have been recently described in the innate immune system of vertebrates and referred to as “trained immunity,” a de facto innate immune memory characterized by a heightened reaction to secondary infections or sterile triggers of inflammation (37, 38).
Unlike adaptive immune memory, which depends on de novo rearrangements of antigen receptor genes in T and B lymphocytes, trained immunity stems from the long-term functional reprogramming of innate myeloid cells, such as monocytes and dendritic cells, activated in response to exogenous or endogenous stimuli (38). The demonstration that trained immunity resides in mature myeloid cells initially resulted in a conundrum because these cells are typically short lived (39, 40), but trained immunity can be maintained in the myeloid compartment for several months and years (41). As further discussed below, though, trained immunity involves not only blood monocytes and tissue macrophages (peripheral trained immunity) but also bone marrow progenitor cells (central trained immunity), a property that makes it possible for trained immunity to persist.
The induction of trained immunity appears to be supported by the convergence of multiple regulatory epigenetic mechanisms, including changes in chromatin architecture at topologically associated domains, transcription of long noncoding RNAs, and DNA methylation (38). Stimulation of innate immune cells can leave an “epigenetic scar” in target genes, changing long-term cellular responsiveness by promoting a functional trained immunity program. The following two key epigenetic marks accompany trained immunity: the acquisition of histone 3 lysine 27 acetylation marks at distal enhancers decorated with histone 3 lysine 4 methylation, and the consolidation of histone 3 lysine 4 trimethylation marks at the promoters of stimulated genes. Newer studies suggest that changes in DNA methylation patterns discriminate between individuals who do or do not develop trained immunity in response to stimuli such as Bacillus Calmette-Guerin (BCG). For instance, individuals who responded to BCG vaccination by more effectively containing M. tuberculosis replication exhibited a wide loss of DNA methylation at immune gene promoters compared with nonresponders (42). Methylation at 43 genes was found to significantly differ in BCG-naive responders and nonresponders and was proposed to predict responsiveness to trained immunity–inducing stimuli (43).
In the asthma field, our most recent work has identified distinct neonatal DNA methylation modules that are associated with the capacity to mount innate immune responses linked with childhood asthma risk. Interestingly, in our longitudinal study, which included both pregnant mothers and their children followed to the age of 9 years, the exposure(s) seemingly responsible for training the neonate’s innate immune response and influencing her/his asthma risk occurred through the mother, in utero. Our findings reinforce recent evidence suggesting that the fetal innate immune system can be trained by microbial exposures during pregnancy (44, 45). For instance, infants exposed to, but not infected by, HIV, malaria, or hepatitis B virus during pregnancy showed increased cytokine production by TLR agonist–stimulated monocytes and enhanced monocyte activation and maturity (reviewed in Reference 45). Innate immune responses in offspring can also be heightened by maternal vaccination during pregnancy, as evidenced by an association between maternal BCG scar size and infant proinflammatory cytokine production elicited by TLR stimulation (46).
That training of the innate immune system in infants can occur prenatally underscores the ability of the fetal immune system to respond to signals generated by maternal inflammation, infection, or, more generally, maternal microbiota and/or their products (45). Although direct movement of maternal microbes to the fetal intestine can occur and relates to immune cell function (32), fetal exposure to maternal microbial metabolites (direct or through maternal antibody-bound microbial molecules) can also influence fetal immune development. Elegant mouse studies have shown that SCFAs, gut microbial–derived metabolites produced in pregnant mice fed a high-fiber diet, can directly enter the fetal circulation and influence fetal immune cell production, function, and, ultimately, offspring immunity (47, 48). Specifically, the development of allergic airway disease was dampened by suppressing the expression of mouse fetal lung genes linked to both human and mouse allergic asthma (47). Interestingly, the impact of SCFAs on allergic airway disease susceptibility may be mediated by direct effects on hemopoiesis (31). Indeed, treatment of adult mice with the SCFA propionate led to alterations in bone marrow hemopoiesis characterized by enhanced generation of macrophage and dendritic cell precursors and subsequent seeding of the lungs by dendritic cells with a high phagocytic capacity but an impaired ability to promote type-2 cell effector function. The effects of propionate on allergic inflammation were specifically dependent on GPR41 (G protein-coupled receptor 41)/FFAR3 (free fatty acid receptor 3) (31). Our studies of gut microbiome manipulation in a model of respiratory viral infection demonstrate that these effects extend beyond SCFAs to include a broader range of immunomodulatory lipids. Oral supplementation with a Lactobacillus species before respiratory syncytial virus infection resulted in airway protection, which was associated with a distinct circulating metabolic environment, including enrichment of antiinflammatory polyunsaturated fatty acids (49). In vitro, plasma from these animals, or exposure to specific enriched polyunsaturated fatty acids, reprogrammed bone marrow–derived dendritic cells, reducing their inflammatory response to respiratory syncytial virus infection (49). Though the longevity of this response is unknown, human studies in pregnant mothers indicate that prenatal DHA supplementation results in global epigenetic modifications in infant blood at birth, a number of which are maintained in circulating leukocytes at 5 years of age (50). Overall, these data are consistent with the notion that maternal prenatal microbiomes whose metabolic activities are presumably largely shaped by diet are sensed by fetal bone marrow progenitors and affect the immune system in the neonatal period through central trained immunity, with long-lasting effects on the child’s immune function.
One critical way in which maternal influences may train the child’s innate immune system is by enabling rapid responsiveness to maturation-inducing microbial signals received at birth. To the extent that delayed immune maturation contributes to the allergy/asthma trajectory (1, 51), immune training in utero likely contributes to averting this risk. The lessons learned from studying asthma-protected farm children (52) are very relevant in this respect. Cord blood mononuclear cells from farm children demonstrated prenatal priming evidenced by precocious upregulation of host defense functions such as microbial-sensing TLRs (53, 54) and cord blood innate cytokines (55). These findings suggest that maternal prenatal microbial exposures calibrate the development and maturation of immune responses in the offspring both quantitatively and temporally, ultimately enhancing asthma resistance (51, 52).
Where We Are Now…
Our current working model integrates several of the research themes discussed above. Findings by us and others provide strong support for the notion that in utero exposure to maternal microbial and immune signals relies on epigenetic mechanisms to influence fetal immune responses to postnatal exposures. The picture is not complete, but several components already stand out. First, we demonstrated for the first time that neonates who will not become asthmatic by the age of 9 years carry DNA methylation profiles in their immune cord blood cells that are distinct from those of neonates who will develop asthma, suggesting that epigenetic mechanisms may be involved in asthma inception and that an epigenetic trajectory to asthma may be in place already at birth (56). Next, we moved one step back and focused on the mother-to-child asthma trajectory, finding that the maternal prenatal immune profile associated with childhood asthma also affects the neonatal DNA methylation landscape. Indeed, neonates born to mothers with the lowest IFN-γ/IL-13 ratio carry both the highest asthma risk and distinct DNA methylation profiles in their immune cells. Most importantly perhaps, neonatal differential methylation at a large module (cluster) of genomic sites was associated not only with the maternal IFN-γ/IL-13 ratio but also with an enhanced capacity of neonatal cord blood mononuclear cells to produce innate cytokines in response to microbial stimuli (LPS) and with childhood asthma by the age of 9 years. In combination, these findings suggest that maternal prenatal immune profiles shape the neonatal epigenome in ways that influence innate immune responses to postnatal exposures and ultimately asthma risk (Devries and colleagues, unpublished data).
Critically, we interpret an enhanced capacity of neonatal cord blood mononuclear cells to produce innate cytokines in response to microbial stimuli as functional evidence for trained innate immune memory. Because asthma rates between 5 and 9 years of age were drastically reduced among neonates with high innate cytokine-producing capacity, we infer that epigenetically trained innate immunity, manifested as an increased neonatal immune response to microbial stimuli, protects from asthma. A heightened ability to respond to microbial stimuli already at birth likely reflects accelerated, more efficient immune maturation and thus may counteract the immune maturation delay that promotes type 2 responses and has been linked to increased risk for asthma and allergy (1, 51).
… And Where We Are Going: an Integrated Framework for Future Research
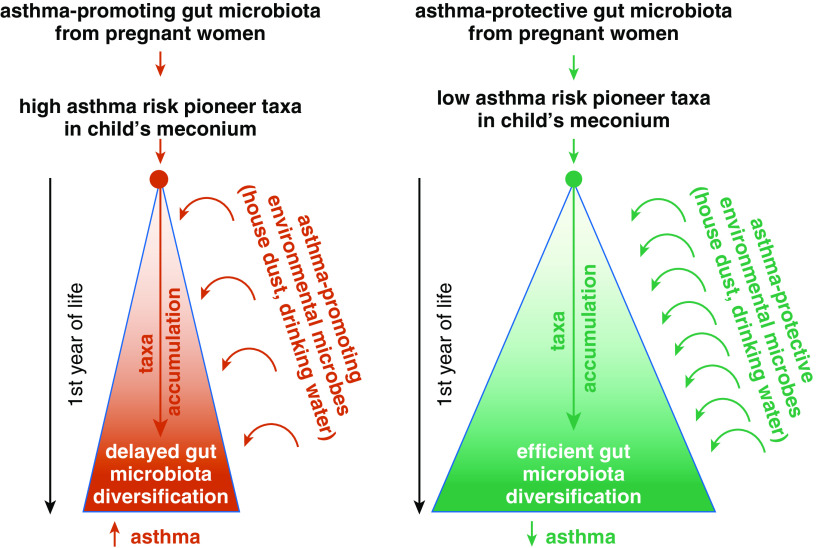
Building on multiple results from our laboratories and others, we propose a transgenerational framework for future studies that seeks to decisively advance our understanding of the origins of asthma and allergy pathogenesis by integrating microbiome, immunology, genetics, epigenetics, and nutrition research in human populations and model systems. Specifically, we suggest that environmental exposures during pregnancy, including maternal diet, shape maternal microbiomes and immune function, which in turn influence the immune and microbial status of the fetus by interacting with each individual’s genetic makeup. Relying in part on epigenetic mechanisms, these interacting influences train the neonate’s innate immune system and regulate its ability to respond to the stimuli provided by microbes that are vertically transmitted from the mother and initially colonize neonatal body habitats. Depending on their composition and functional properties, as suggested by community ecology models (57), pioneer microbes and their activities (governed in large part by species–species interactions and nutrient availability) shape the host’s immune function and the rate and types of exogenous microbes accumulated into these body habitats during the first year of life, thereby determining trajectories of microbiota development (58, 59), innate and adaptive immune development, and ultimately, asthma risk (Figure 2). We note that in this scenario both environmental and personal gut microbiota are critical for the asthma trajectory. Yet, they are typically studied separately. Thus, one implication of the integrated framework we propose is that these hitherto parallel research tracks should converge to determine how very early-life microbes in the context of extrinsic (e.g., early-life nutrition) and intrinsic (e.g., genetic) factors direct the accumulation of environmental microbes in early life and how the composition and metabolic capacity of the child’s microbiome shaped by these interacting influences controls immune development and asthma risk.
Figure 2.
The interaction between vertically transmitted maternal pioneer microbiota and the accumulation of environmental microbiota shapes the diversification and timing of the child’s gut microbiome and her/his immune development.
Supplementary Material
Footnotes
S.V.L. and D.V. are supported by NIH/National Institute of Allergy and Infectious Diseases awards AI148104 and AI133765. S.V.L. is also supported by AI114271, OD023282, AI104317, and AI089473. D.V. is also supported by AI144722.
Originally Published in Press as DOI: 10.1164/rccm.202010-3779PP on January 25, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.von Mutius E, Smits HH. Primary prevention of asthma: from risk and protective factors to targeted strategies for prevention. Lancet. 2020;396:854–866. doi: 10.1016/S0140-6736(20)31861-4. [DOI] [PubMed] [Google Scholar]
- 2.O’Connor GT, Lynch SV, Bloomberg GR, Kattan M, Wood RA, Gergen PJ, et al. Early-life home environment and risk of asthma among inner-city children. J Allergy Clin Immunol. 2018;141:1468–1475. doi: 10.1016/j.jaci.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch SV, Wood RA, Boushey H, Bacharier LB, Bloomberg GR, Kattan M, et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol. 2014;134:593–601, e12. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 5.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrländer C, et al. GABRIELA Transregio 22 Study Group. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 6.Illi S, Depner M, Genuneit J, Horak E, Loss G, Strunz-Lehner C, et al. GABRIELA Study Group. Protection from childhood asthma and allergy in Alpine farm environments-the GABRIEL advanced studies. J Allergy Clin Immunol. 2012;129:1470–1477, e6. doi: 10.1016/j.jaci.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Wlasiuk G, Vercelli D. The farm effect, or: when, what and how a farming environment protects from asthma and allergic disease. Curr Opin Allergy Clin Immunol. 2012;12:461–466. doi: 10.1097/ACI.0b013e328357a3bc. [DOI] [PubMed] [Google Scholar]
- 8.Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, et al. Innate immunity and asthma risk in amish and hutterite farm children. N Engl J Med. 2016;375:411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrusch CL, Stein MM, Gozdz J, Holbreich M, von Mutius E, Vercelli D, et al. T-cell phenotypes are associated with serum IgE levels in Amish and Hutterite children. J Allergy Clin Immunol. 2019;144:1391–1401, e10. doi: 10.1016/j.jaci.2019.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacharier LB, Beigelman A, Calatroni A, Jackson DJ, Gergen PJ, O’Connor GT, et al. NIAID sponsored Inner-City Asthma Consortium. Longitudinal phenotypes of respiratory health in a high-risk urban birth cohort. Am J Respir Crit Care Med. 2019;199:71–82. doi: 10.1164/rccm.201801-0190OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci USA. 2012;109:8334–8339. doi: 10.1073/pnas.1205624109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carr TF, Beamer PI, Rothers J, Stern DA, Gerald LB, Rosales CB, et al. Prevalence of asthma in school children on the Arizona-sonora border. J Allergy Clin Immunol Pract. 2017;5:114–120, e2. doi: 10.1016/j.jaip.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. MetaHIT Consortium; MetaHIT Consortium. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 14.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanoue T, Atarashi K, Honda K. Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol. 2016;16:295–309. doi: 10.1038/nri.2016.36. [DOI] [PubMed] [Google Scholar]
- 16.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharon G, Cruz NJ, Kang D-W, Gandal MJ, Wang B, Kim Y-M, et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell. 2019;177:1600–1618, e17. doi: 10.1016/j.cell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein MM, Thompson EE, Schoettler N, Helling BA, Magnaye KM, Stanhope C, et al. A decade of research on the 17q12-21 asthma locus: piecing together the puzzle. J Allergy Clin Immunol. 2018;142:749–764, e3. doi: 10.1016/j.jaci.2017.12.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Ryan CA, Boyaval P, Dempsey EM, Ross RP, Stanton C. Maternal vertical transmission affecting early-life microbiota development. Trends Microbiol. 2020;28:28–45. doi: 10.1016/j.tim.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Perez MF, Lehner B. Intergenerational and transgenerational epigenetic inheritance in animals. Nat Cell Biol. 2019;21:143–151. doi: 10.1038/s41556-018-0242-9. [DOI] [PubMed] [Google Scholar]
- 23.Felizardo RJF, de Almeida DC, Pereira RL, Watanabe IKM, Doimo NTS, Ribeiro WR, et al. Gut microbial metabolite butyrate protects against proteinuric kidney disease through epigenetic- and GPR109a-mediated mechanisms. FASEB J. 2019;33:11894–11908. doi: 10.1096/fj.201901080R. [DOI] [PubMed] [Google Scholar]
- 24.Calışkan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loss GJ, Depner M, Hose AJ, Genuneit J, Karvonen AM, Hyvärinen A, et al. PASTURE (Protection against Allergy Study in Rural Environments) Study Group. The early development of wheeze. environmental determinants and genetic susceptibility at 17q21. Am J Respir Crit Care Med. 2016;193:889–897. doi: 10.1164/rccm.201507-1493OC. [DOI] [PubMed] [Google Scholar]
- 26.Weissbrod O, Rothschild D, Barkan E, Segal E. Host genetics and microbiome associations through the lens of genome wide association studies. Curr Opin Microbiol. 2018;44:9–19. doi: 10.1016/j.mib.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Durack J, Kimes NE, Lin DL, Rauch M, McKean M, McCauley K, et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat Commun. 2018;9:707. doi: 10.1038/s41467-018-03157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. CHILD Study Investigators. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 30.Levan SR, Stamnes KA, Lin DL, Panzer AR, Fukui E, McCauley K, et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol. 2019;4:1851–1861. doi: 10.1038/s41564-019-0498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 32.Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med. 2020;26:599–607. doi: 10.1038/s41591-020-0761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothers J, Stern DA, Lohman IC, Spangenberg A, Wright AL, DeVries A, et al. Maternal cytokine profiles during pregnancy predict asthma in children of mothers without asthma. Am J Respir Cell Mol Biol. 2018;59:592–600. doi: 10.1165/rcmb.2017-0410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halkias J, Rackaityte E, Hillman SL, Aran D, Mendoza VF, Marshall LR, et al. CD161 contributes to prenatal immune suppression of IFNγ-producing PLZF+ T cells. J Clin Invest. 2019;129:3562–3577. doi: 10.1172/JCI125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen JW, Rice TA, Bannock JM, Bielecka AA, Strauss JD, Catanzaro JR, et al. Autoreactivity in naïve human fetal B cells is associated with commensal bacteria recognition. Science. 2020;369:320–325. doi: 10.1126/science.aay9733. [DOI] [PubMed] [Google Scholar]
- 36.Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 37.Purvis A, Hector A. Getting the measure of biodiversity. Nature. 2000;405:212–219. doi: 10.1038/35012221. [DOI] [PubMed] [Google Scholar]
- 38.Netea MG, Domínguez-Andrés J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol. 2020;20:375–388. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rieckmann A, Villumsen M, Sørup S, Haugaard LK, Ravn H, Roth A, et al. Vaccinations against smallpox and tuberculosis are associated with better long-term survival: a Danish case-cohort study 1971-2010. Int J Epidemiol. 2017;46:695–705. doi: 10.1093/ije/dyw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verma D, Parasa VR, Raffetseder J, Martis M, Mehta RB, Netea M, et al. Anti-mycobacterial activity correlates with altered DNA methylation pattern in immune cells from BCG-vaccinated subjects. Sci Rep. 2017;7:12305. doi: 10.1038/s41598-017-12110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das J, Verma D, Gustafsson M, Lerm M. Identification of DNA methylation patterns predisposing for an efficient response to BCG vaccination in healthy BCG-naïve subjects. Epigenetics. 2019;14:589–601. doi: 10.1080/15592294.2019.1603963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy O, Wynn JL. A prime time for trained immunity: innate immune memory in newborns and infants. Neonatology. 2014;105:136–141. doi: 10.1159/000356035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Apostol AC, Jensen KDC, Beaudin AE. Training the fetal immune system through maternal inflammation-a layered hygiene hypothesis. Front Immunol. 2020;11:123. doi: 10.3389/fimmu.2020.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mawa PA, Webb EL, Filali-Mouhim A, Nkurunungi G, Sekaly RP, Lule SA, et al. Maternal BCG scar is associated with increased infant proinflammatory immune responses. Vaccine. 2017;35:273–282. doi: 10.1016/j.vaccine.2016.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 48.Kimura I, Miyamoto J, Ohue-Kitano R, Watanabe K, Yamada T, Onuki M, et al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science. 2020;367:eaaw8429. doi: 10.1126/science.aaw8429. [DOI] [PubMed] [Google Scholar]
- 49.Fonseca W, Lucey K, Jang S, Fujimura KE, Rasky A, Ting HA, et al. Lactobacillus johnsonii supplementation attenuates respiratory viral infection via metabolic reprogramming and immune cell modulation. Mucosal Immunol. 2017;10:1569–1580. doi: 10.1038/mi.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Dijk SJ, Zhou J, Peters TJ, Buckley M, Sutcliffe B, Oytam Y, et al. Effect of prenatal DHA supplementation on the infant epigenome: results from a randomized controlled trial. Clin Epigenetics. 2016;8:114. doi: 10.1186/s13148-016-0281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holt PG, Strickland DH, Custovic A. Targeting maternal immune function during pregnancy for asthma prevention in offspring: harnessing the “farm effect”? J Allergy Clin Immunol. 2020;146:270–272. doi: 10.1016/j.jaci.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Ober C, Sperling AI, von Mutius E, Vercelli D. Immune development and environment: lessons from Amish and Hutterite children. Curr Opin Immunol. 2017;48:51–60. doi: 10.1016/j.coi.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ege MJ, Bieli C, Frei R, van Strien RT, Riedler J, Ublagger E, et al. Parsifal Study team. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol. 2006;117:817–823. doi: 10.1016/j.jaci.2005.12.1307. [DOI] [PubMed] [Google Scholar]
- 54.Loss G, Bitter S, Wohlgensinger J, Frei R, Roduit C, Genuneit J, et al. PASTURE study group. Prenatal and early-life exposures alter expression of innate immunity genes: the PASTURE cohort study. J Allergy Clin Immunol. 2012;130:523–530, e9. doi: 10.1016/j.jaci.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 55.Pfefferle PI, Buchele G, Blumer N, Roponen M, Ege MJ, Krauss-Etschmann S, et al. Cord blood cytokines are modulated by maternal farming activities and consumption of farm dairy products during pregnancy: the PASTURE Study. J Allergy Clin Immunol. 2010;125:108–115, e1–e3. doi: 10.1016/j.jaci.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 56.DeVries A, Vercelli D. The neonatal methylome as a gatekeeper in the trajectory to childhood asthma. Epigenomics. 2017;9:585–593. doi: 10.2217/epi-2016-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilbert JA, Lynch SV. Community ecology as a framework for human microbiome research. Nat Med. 2019;25:884–889. doi: 10.1038/s41591-019-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fierer N, Nemergut D, Knight R, Craine JM. Changes through time: integrating microorganisms into the study of succession. Res Microbiol. 2010;161:635–642. doi: 10.1016/j.resmic.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.