Abstract
Fibroblasts provide a structural framework for multiple organs and are essential for wound repair and fibrotic processes. Here, we demonstrate functional roles of FOXL1 (forkhead box L1), a transcription factor that characterizes the pulmonary origin of lung fibroblasts. We detected high FOXL1 transcripts associated with DNA hypomethylation and super-enhancer formation in lung fibroblasts, which is in contrast with fibroblasts derived from other organs. RNA in situ hybridization and immunohistochemistry in normal lung tissue indicated that FOXL1 mRNA and protein are expressed in submucosal interstitial cells together with airway epithelial cells. Transcriptome analysis revealed that FOXL1 could control a broad array of genes that potentiate fibroblast function, including TAZ (transcriptional coactivator with PDZ-binding motif)/YAP (Yes-associated protein) signature genes and PDGFRα (platelet-derived growth factor receptor-α). FOXL1 silencing in lung fibroblasts attenuated cell growth and collagen gel contraction capacity, underscoring the functional importance of FOXL1 in fibroproliferative reactions. Of clinical importance, increased FOXL1 mRNA expression was found in fibroblasts of idiopathic pulmonary fibrosis lung tissue. Our observations suggest that FOXL1 regulates multiple functional aspects of lung fibroblasts as a key transcription factor and is involved in idiopathic pulmonary fibrosis pathogenesis.
Keywords: fibroblast, FOXL1, cap analysis of gene expression, super-enhancer, idiopathic pulmonary fibrosis
Fibroblasts are ubiquitous mesenchymal cells that regulate extracellular matrix (ECM) turnover, support the structural framework of various tissues, and modulate cell–cell or cell–ECM interactions, thereby playing a crucial role in wound healing and tissue regeneration (1). Fibroblast populations are heterogeneous (2) and display different gene expression patterns in different organs (3, 4).
We previously analyzed transcriptome data from 45 primary cultured human fibroblast lines derived from different organs and identified 14 transcription factors that showed significantly higher expression in lung-derived fibroblast lines (5). We also determined that eight of these transcription factors, including TBX4 (T-box transcription factor 4), are associated with super-enhancers crucial for the establishment of cell identity (6–8). Of these genes, we focused on FOXL1 (forkhead box L1) in the present study and analyzed its functional roles in lung fibroblasts.
FOXL1 is an evolutionarily conserved transcription factor of the FOX gene family containing a winged helix DNA-binding domain (9). FOXL1 is located in the FOX gene cluster on chromosome 16q24.1, which also contains FOXC2 and FOXF1. Early investigations of mouse fetal development demonstrated that Foxl1 (the mouse homolog of FOXL1) is expressed in the mesenchyme surrounding the anterior gut and lung (10). Targeted disruption of Foxl1 in mice led to aberrant cell positioning, dysregulated epithelial proliferation, and distorted tissue architecture of the stomach and small intestine, indicating its essential role in the development of the gastrointestinal tract (11). One possible mechanism for the gastrointestinal defects was the reduced expression of Bmp2 (bone morphogenetic protein 2) and Bmp4 observed in Foxl1-deficient mice (11). More recently, it was demonstrated that Foxl1-positive subepithelial fibroblasts constitute the intestinal stem cell niche that produces Wnt signals and maintains intestinal stem cells (12, 13).
According to gene expression profiling databases, FOXL1 is expressed in multiple human organs at the transcript level. Although FOXL1 is expressed in the human lung (14), its significance in lung physiology has not been explored, and it remains uncertain whether FOXL1 is functionally relevant as a transcription factor in human lung fibroblasts.
Recent advances in lineage tracing in mice have revealed that fibroblasts regulate alveolar epithelial cell growth and differentiation (15), and PDGF-A (platelet-derived growth factor A)/PDGFRα (PDGF receptor-α) signaling is required for alveologenesis (16). Most recently, single-cell RNA-seq (RNA-sequencing) studies identified an Axin2+Pdgfra+ mesenchymal cell subpopulation that supports lung regeneration (17). In analogy with the intestinal tract, FOXL1-positive fibroblasts in the lungs may be involved in epithelial cell growth and differentiation (12, 13).
In addition to physiological roles in tissue homeostasis, lung fibroblasts play central roles in the pathogenesis of pulmonary diseases (18). In particular, lung fibroblasts are key effector cells of fibrotic responses in idiopathic pulmonary fibrosis (IPF) (19, 20), a chronic fibrosing interstitial pneumonia of unknown cause with a median survival of 2–4 years (21). Pathological features of IPF include alveolar epithelial regenerative failure, accumulation of activated fibroblasts, and remodeling of the ECM (22). It is believed that clusters of fibroblasts, termed “fibroblastic foci,” are responsible for the fibrotic processes in IPF lung tissue (23).
TAZ (transcriptional coactivator with PDZ binding motif) and its homolog YAP (Yes-associated protein) are downstream effectors of the Hippo pathway. They interact with TEA domain (TEAD) family transcription factors in the nucleus to activate genes associated with cell proliferation and differentiation (24). We previously reported that TAZ is highly expressed in the fibroblastic foci of IPF lung tissue (25). We further showed that TAZ regulates a subset of profibrotic genes, including CTGF (connective tissue growth factor), in lung fibroblasts (25).
In the present study, we determined that FOXL1 mRNA is highly expressed in lung fibroblasts, possibly through epigenetic mechanisms, and demonstrated that FOXL1 controls cell growth and collagen gel contraction capacity. Transcriptome analysis revealed that FOXL1 could regulate an array of genes important for fibroblast function, including BMP ligands, PDGFRα, and TAZ/YAP signature genes such as CTGF. Of pathological importance, we identified increased FOXL1 expression in fibroblasts of IPF lung tissue, suggesting its involvement in IPF pathogenesis.
Materials and Methods
Public Data
Public datasets used in this study are summarized in Table E1 in the data supplement. Mapped sequence data were visualized using Integrative Genomics Viewer (26). DAVID 6.8 functional annotation tool was used for gene ontology analysis. Gene set enrichment analysis was performed as described previously (27). Single-cell RNA-seq data were reanalyzed using Seurat version 3 (17, 28). Identification of super-enhancers (8) and visualization of ChIP-seq (chromatin immunoprecipitation-sequencing) profiles were performed as previously described (5–7).
Cell Cultures
The protocol of lung fibroblast isolation was approved by the University of Tokyo Ethics Committee or by the National Hospital Organization Tokyo National Hospital Institutional Review Board (5). All patients gave written informed consent. Lung tissue was cut into 1-mm3 fragments aseptically, and fibroblasts proliferating from these specimens were grown to 80% confluence and then passaged. Lung fibroblast lines derived from patients with IPF were purchased from Lonza. Details are shown in Table E2.
Knockdown of FOXL1
siRNA against human FOXL1 (Silencer Select siRNA) was purchased from Life Technologies (Table E3). Cells were transfected with 20 nM siRNA using Lipofectamine RNAiMAX (Life Technologies).
RNA-Seq Analysis
RNA-seq reads were analyzed using CLC Genomics workbench software (Qiagen) (5). The data are deposited in the Gene Expression Omnibus database (GSE137823). Motif analysis was performed using the HOMER findMotifs.pl program (5).
Quantitative RT-PCR
Detailed procedures were described previously (29). PCR primers are shown in Table E3.
Immunoblot Analysis
Detailed procedures were described previously (29). Mouse monoclonal antibodies for α-tubulin, CTGF, and PDGFRα were from Sigma-Aldrich, Santa Cruz Biotechnology, and Cell Signaling, respectively.
Collagen Gel Contraction Assay
Three-dimensional collagen gel cultures were carried out as described previously (30).
Migration Assay
A thin-membrane 96-well plate ChemoTx chemotaxis system (#116-8; Neuro Probe Inc.) was used. The migrated cells were treated with WST-8 solution (Nacalai Tesque).
Human Tissue Samples and Immunohistochemistry
Formalin-fixed paraffin-embedded human lung tissue samples (Table E4) were used for immunohistochemistry. The protocol was approved by the Ethics Committee at the National Hospital Organization Tokyo National Hospital Institutional Review Board (5). Antigen retrieval was accomplished in citrate buffer (pH of 6.0 at 98°C for 40 min). Anti-FOXL1 rabbit polyclonal antibody (ab190226; Abcam) or anti-vimentin (V9) mouse monoclonal antibody (IR630; Dako) was used. Tissue sections were processed with the Vectastain Elite ABC Kit (Vector Laboratories).
RNA in Situ Hybridization
RNA in situ hybridization (ISH) was performed on human lung tissue sections obtained from three transplant donors (Table E4) who were previously healthy, as described previously (31). The protocol to procure human lung tissue was approved by the University of North Carolina Institutional Review Board (03-1396). The probes and reagents were used according to the RNAscope protocol from Advanced Cell Diagnostics.
Statistical Analysis
Differences were examined by Dunnett test with JMP version 13 (SAS Institute Inc.). Pearson’s correlation coefficient (r) and decision coefficient (R2) were calculated for correlation analysis.
Results
Higher Expression of FOXL1 Transcripts in Lung Fibroblasts
Human fibroblasts isolated from different anatomical sites are heterogeneous in terms of their gene expression patterns (3). To obtain a gene list that defines the pulmonary origin of lung fibroblasts, we used the Cap Analysis of Gene Expression (CAGE) dataset from the FANTOM5 (Functional ANnoTation Of the Mammalian genome 5) consortium (32) and analyzed transcriptome data from 45 different fibroblast lines (5) in search of CAGE-defined transcription start sites (also termed “promoters” in the FANTOM5 project) that are selectively activated in lung fibroblasts. By comparing the CAGE profiles among three human lung–derived fibroblast lines and 42 other fibroblast lines, we identified 189 promoters (including those of 88 protein-coding genes) with lung-specific expression (5). Among these, 37 promoters were annotated to transcription factors; TBX4, TBX5, FOXL1, HOXA5, and HOXB5 were among the top 10 promoters ranked by false discovery rate (Table E5).
We recently explored the functional roles of TBX4 and other T-box family genes (TBX2 and TBX5) in lung fibroblasts (5). The crucial importance of Hox5 genes (including Hoxa5 and Hoxb5) in lung morphogenesis and in lung fibroblast function has also been described (33). However, no reports have described the role of FOXL1 in the lungs or lung fibroblasts; thus, we selected the FOXL1 gene for further analysis.
There are two transcripts of human FOXL1 (designated FOXL1-201 and FOXL1-202); the major transcript (FOXL1-201), encoding full-length FOXL1, is composed of a single exon with multiple transcription start sites. In the analyzed CAGE dataset, alternative promoters for the same gene are ranked by their expression levels and numbered (e.g., p1, p2, and p3) (32). Four different promoters annotated to FOXL1-201 (p2, p3, p4, and p5) were included in the above-mentioned 189 promoters. To ascertain the specific activity of FOXL1 promoters in lung fibroblasts, we compared CAGE count data among three groups, including 16 lung epithelial cell lines, three lung fibroblast lines, and 42 other fibroblast lines. We confirmed that FOXL1 expression from p2, p3, p4, and p5 promoters was prominent in lung fibroblast lines, and p2 promoter–derived transcripts were the most abundant. On the other hand, p1 promoter showed activation in all three groups (Figure 1A). Thus, the major transcription start site for FOXL1 in lung fibroblasts (p2 promoter) was distinct from that in other cell types (p1 promoter).
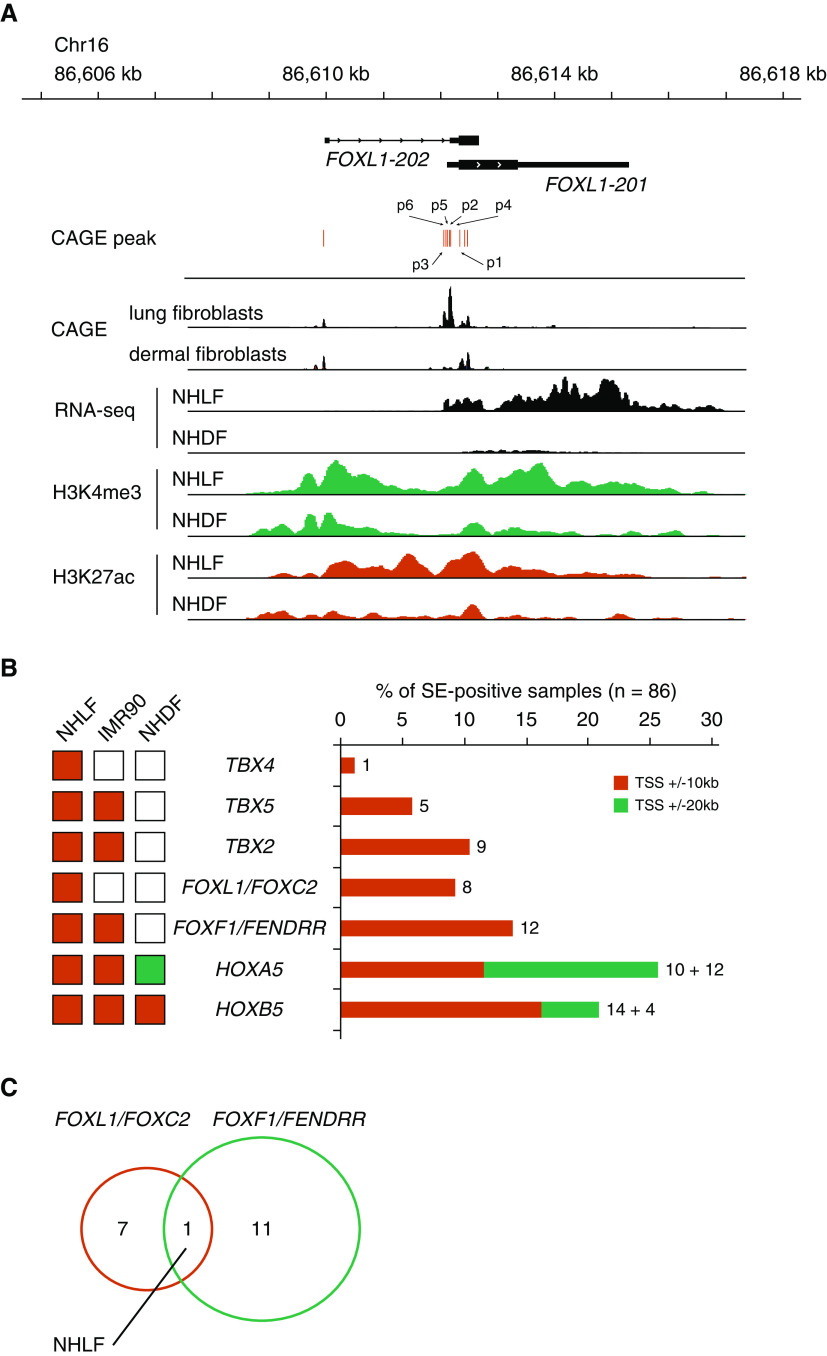
Figure 1.
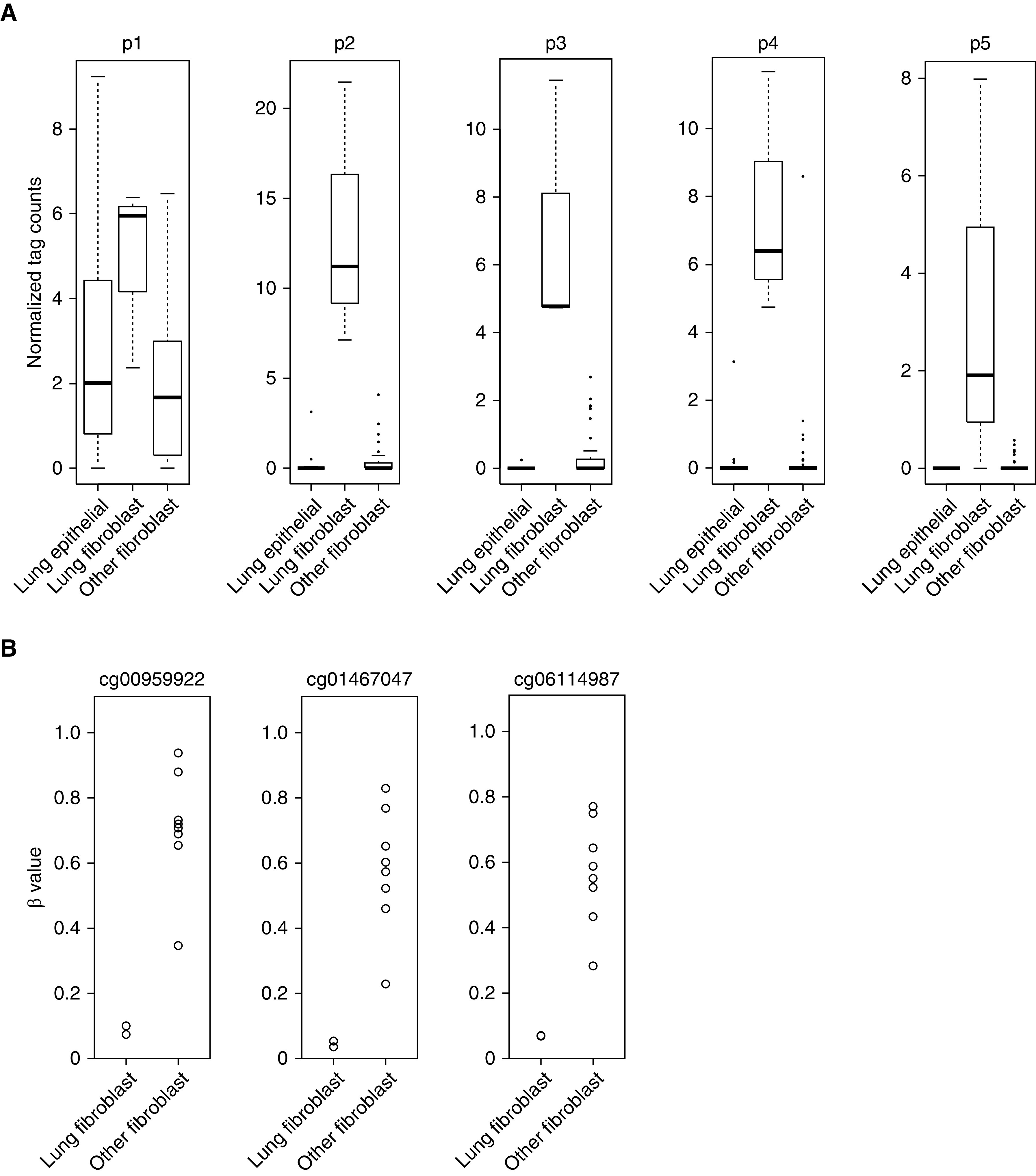
Higher expression of FOXL1 (forkhead box L1) in lung fibroblasts. (A) Box plot showing normalized cap analysis of gene expression (CAGE) tag counts of p1, p2, p3, p4, and p5 promoters annotated to FOXL1. CAGE data were compared among the following three groups: 16 lung epithelial cell lines (alveolar, small airway, bronchial, and tracheal epithelial cells), three lung fibroblast lines, and 42 other fibroblast lines that included gingival fibroblasts (n = 6), periodontal ligament fibroblasts (n = 6), cardiac fibroblasts (n = 6), choroid plexus fibroblasts (n = 3), conjunctival fibroblasts (n = 2), dermal fibroblasts (n = 6), aortic adventitial fibroblasts (n = 3), lymphatic fibroblasts (n = 3), mammary fibroblasts (n = 3), pulmonary artery fibroblast (n = 1), and villous mesenchymal fibroblasts (n = 3). The central line in the box indicates the median value. (B) DNA methylation levels shown as β values (the GSE40699 dataset) were compared between lung fibroblast lines (n = 2) and other fibroblast lines (n = 8). CpG sites located upstream from the transcription start site (TSS) are indicated (cg00959922, cg01467047, and cg06114987).
We next compared DNA methylation levels (β values) of the FOXL1 gene between two human lung fibroblast lines and eight other fibroblast lines using the GSE40699 dataset (Infinium 450K methylation array) from the ENCODE (Encyclopedia of DNA Elements) project (34, 35). In agreement with the finding of higher FOXL1 expression in lung fibroblasts, most of the 11 methylation probes annotated to FOXL1 showed lower β values in lung fibroblast lines than in other fibroblast lines. Of particular note, CpG sites located in the promoter region upstream from the transcription start site (cg00959922, cg01467047, and cg06114987) were evidently hypomethylated in lung fibroblast lines in clear contrast to other fibroblast lines (Figure 1B).
FOXL1 Is Associated with Super-Enhancers in Lung Fibroblasts
To investigate the epigenetic regulation of FOXL1, we compared expression levels (RNA-seq) and histone H3 modifications between normal human lung fibroblasts NHLF (normal human lung fibroblasts) and NHDF (normal human dermal fibroblasts) using the ENCODE datasets (34, 35). Consistent with our observations from the CAGE dataset, FOXL1 transcripts were higher in NHLF, and the FOXL1 locus was enriched in active histone marks, histone H3 acetylation at lysine 27 (H3K27ac), and H3 methylation at lysine 4 (H3K4me3) (Figure 2A).
Figure 2.
FOXL1 is associated with super-enhancers in lung fibroblasts. (A) Genomic coordinates, FOXL1 transcripts, and CAGE peaks are indicated. CAGE-sequencing data were merged from three lung fibroblast lines or six dermal fibroblast lines. RNA-sequencing (RNA-seq) signals and ChIP (chromatin immunoprecipitation) peaks of H3K4me3 and H3K27ac in normal human lung fibroblasts (NHLF) or normal human dermal fibroblasts (NHDF) are shown. Note that CAGE peaks, RNA-seq signals, and active histone marks (H3K4me3 and H3K27ac) for FOXL1 are more apparent in lung fibroblasts than in dermal fibroblasts. The same y-axis scales are used for the comparisons between NHLF and NHDF. (B) Left: associations of transcription factors with super-enhancers among NHLF, IMR90, and NHDF. Super-enhancers were identified as reported previously (8). Right: associations of transcription factors with super-enhancers among 86 human cell line and tissue samples. Super-enhancers overlapping with ±10 or 20 kb from the TSSs of the indicated genes were identified. Numbers of samples positive for super-enhancers are indicated. (C) Venn diagram showing that NHLF is the only sample in which both FOXL1/FOXC2 and FOXF1/FENDRR are associated with super-enhancers. Chr = chromosome; FENDRR = FOXF1 adjacent non-coding developmental regulatory RNA; HOXA5 = homeobox A5; SE-positive = super-enhancer–positive; TBX4 = T-box transcription factor 4.
Super-enhancers are large genomic regions defined by exceptional enrichment of Mediator complexes and master transcription factors or by high concentrations of H3K27ac that induce the expression of cell identity genes (6–8). We explored the presence of super-enhancers at TBX2, TBX4, TBX5, HOXA5, HOXB5, FOXL1/FOXC2, and FOXF1/FENDRR gene loci across 86 human cell line and tissue samples that included IMR90 (human fetal lung fibroblasts), NHLF, and NHDF (Table E6). FOXL1/FOXC2 genes were found to be associated with super-enhancers in eight samples, including NHLF, human umbilical vein endothelial cells, astrocytes, and osteoblasts (Figure 2B). Notably, among 86 samples, both FOXL1/FOXC2 and FOXF1/FENDRR were associated with super-enhancers only in NHLF, suggesting that a highly activated FOX gene cluster is a hallmark of lung fibroblasts (Figure 2C). Visualization of H3K27ac ChIP-seq signals in different cell line and tissue samples showed that broad peaks in both FOXL1/FOXC2 and FOXF1/FENDRR gene loci were characteristic of lung fibroblasts (NHLF and IMR90) and appeared associated with cellular identity (see Figure E1 in the data supplement).
We also reanalyzed a recently reported single-cell RNA-seq dataset of mouse lung mesenchymal cells (17) (Figure E2A) and found that Foxl1 was preferentially expressed in the Axin2+Pdgfra+ mesenchymal cell subpopulation that was reportedly involved in lung regeneration (Figure E2B). These observations suggested that FOXL1 might have important roles both in human lung fibroblasts and in mouse lung mesenchymal cells.
Expression of FOXL1 in Human Lung Tissue
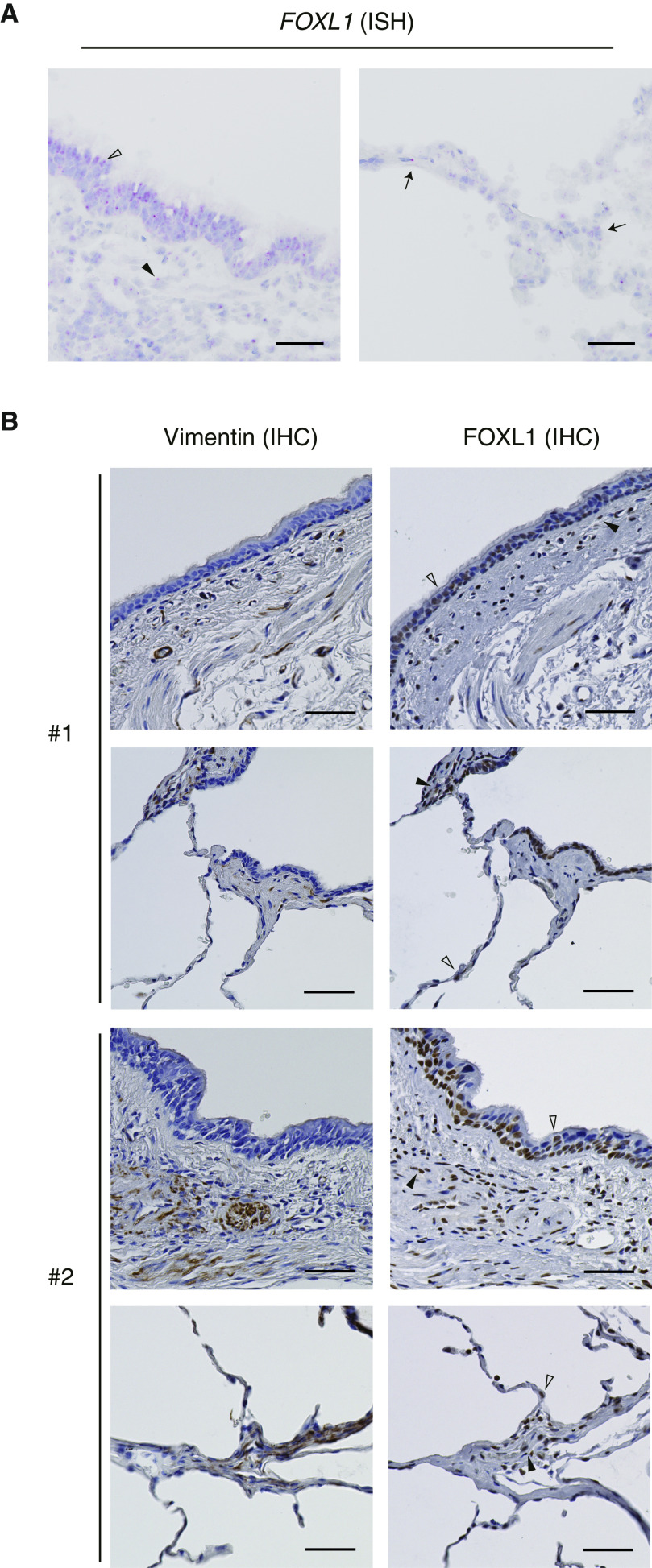
To characterize topographic distribution of FOXL1 expression in the lungs, we performed RNA ISH and immunohistochemistry. FOXL1 mRNA (Figures 3A and E3) and protein (Figure 3B) showed concordant expression patterns throughout the airway, and nuclear positivity for FOXL1 was detected in the airway epithelium. FOXL1-positive cells were also found in the submucosal interstitium, where fibroblasts reside. Positive staining of FOXL1 in interstitial cells was commonly observed among the analyzed lung specimens. FOXL1 expression was also observed in the nuclei of alveolar cells by both RNA ISH (Figure 3A) and immunohistochemistry (Figure 3B), although it was hard to identify each cell type. Importantly, immunohistochemical studies indicated that FOXL1-positive interstitial cells were also stained for vimentin (Figure 3B), suggesting the presence of resident fibroblasts expressing FOXL1.
Figure 3.
Expression of FOXL1 in human lung tissue. (A) FOXL1 expression detected by RNA ISH in normal lung tissue. Left: FOXL1 was positively stained in the nuclei of both epithelial cells (open arrowhead) and interstitial cells (solid arrowhead) in the airway. Right: Nuclear FOXL1 positivity was found in the alveolar cells (arrows). Scale bars, 50 μm. (B) Immunohistochemistry (IHC) for vimentin (left) and FOXL1 (right) in normal lung tissue. Two different subjects (#1 and #2) are presented. FOXL1 was positively stained in the nuclei of both epithelial cells (open arrowheads) and interstitial cells (solid arrowheads) in the airway (top) and the alveolus (bottom). Note that nuclear FOXL1 staining is observed in the interstitial cells positive for vimentin. Scale bars, 50 µm. ISH = in situ hybridization.
RNA-Seq Analysis of Lung Fibroblasts after FOXL1 Knockdown
To identify genes possibly regulated by FOXL1, we performed an RNA-seq analysis of NHLF treated with FOXL1 siRNA (siFOXL1). Using a threshold of reads per kilobase of transcript per million mapped reads of greater than one in negative control siRNA (siNC)–treated cells and a log2 fold change of less than −0.5 both in the siFOXL1 groups 1 and 2, we found 273 downregulated transcripts after FOXL1 knockdown (Table E7). These transcripts defined a siFOXL1-downregulated gene signature, which was used for further analysis.
Gene ontology analysis showed that the siFOXL1-downregulated gene signature was enriched with genes involved in inflammatory response, cell–cell signaling, and regulation of cell growth, suggesting that FOXL1 is involved in various fibroblast functions (Figure 4A and Table E8).
Figure 4.
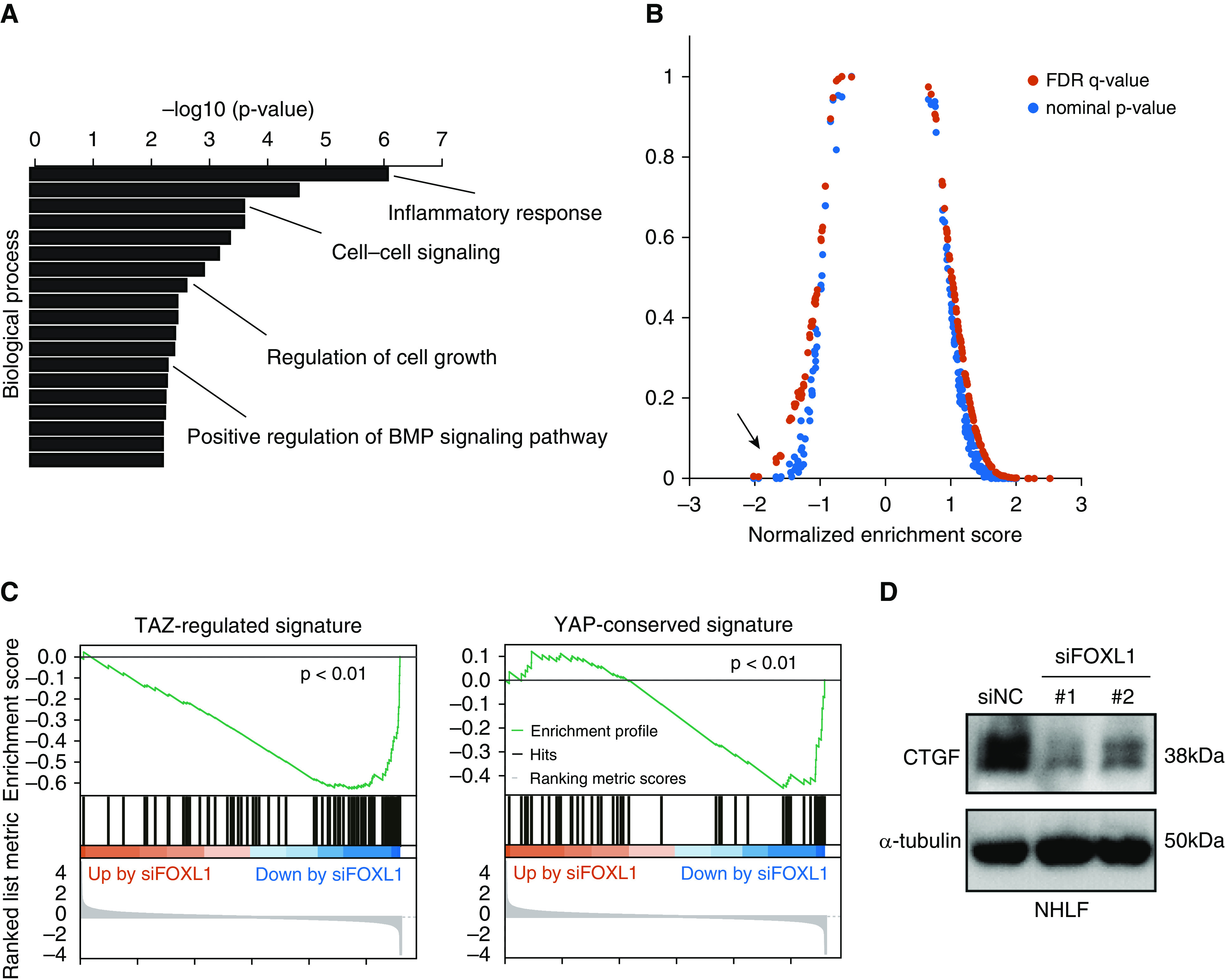
RNA-seq analysis of lung fibroblasts after FOXL1 knockdown. (A) Gene ontology analysis was performed using the FOXL1 siRNA (siFOXL1)-downregulated gene signature (273 genes). Enriched gene ontology terms for biological process are sorted by −log10 (P value). (B) Gene set enrichment analysis was performed using the RNA-seq result and different gene signatures derived from the C6 oncogenic signature collection of the Molecular Signatures Database. YAP (Yes-associated protein)-conserved signature indicated by the arrow showed a high association with the genes downregulated by FOXL1 knockdown. (C) Gene set enrichment analysis was performed using the RNA-seq data in NHLF with FOXL1 knockdown using TAZ (transcriptional coactivator with PDZ-binding motif)-regulated signature defined by our previous study (left) or YAP-conserved signature from the Molecular Signatures Database (right). (D) IB for CTGF (connective tissue growth factor) in NHLF treated with negative control siRNA (siNC) or siFOXL1 (#1 or #2). α-tubulin was used as the loading control. BMP = bone morphogenetic protein; FDR = false discovery rate.
FOXL1 Is Related to TAZ/YAP Signaling in Lung Fibroblasts
Next, we performed gene set enrichment analysis using different gene signatures derived from the C6 oncogenic signature collection of the Molecular Signatures Database (version 7.0). Among them, “YAP-conserved signature” (70 genes) showed a high association with the genes downregulated by FOXL1 knockdown (Figure 4B). In line with this finding, motif analysis of the promoter regions of the siFOXL1-downregulated gene signature revealed that TEAD-binding consensus sequences were enriched, together with those for FOX genes (Figure E4). In our previous study, we defined “TAZ-regulated signature” (94 genes) in lung fibroblasts (25). Gene set enrichment analysis indicated that TAZ-regulated signature and YAP-conserved signature were enriched among the genes downregulated by FOXL1 knockdown in lung fibroblasts (25, 27, 36) (Figure 4C).
A comparison between the siFOXL1-downregulated gene signature (273 genes) and TAZ-regulated signature (25) revealed 12 common genes, including CTGF, GREM1 (gremlin 1), and FSTL1 (follistatin-like 1). For further validation, we performed IB for CTGF in lung fibroblasts treated with siNC or siFOXL1 and confirmed its decreased expression at the protein level after FOXL1 silencing (Figure 4D).
FOXL1 Is Related to BMP Signaling in Lung Fibroblasts
Recent studies indicated that BMP signaling has a substantial impact on epithelial–mesenchymal interactions and is important for murine lung homeostasis (37, 38). As noted above, BMP signaling antagonists (GREM1 and FSTL1) were downregulated by FOXL1 knockdown (38). In addition, we found that the expression of BMP ligands (BMP2 and BMP4) was also decreased after FOXL1 knockdown (Table E7).
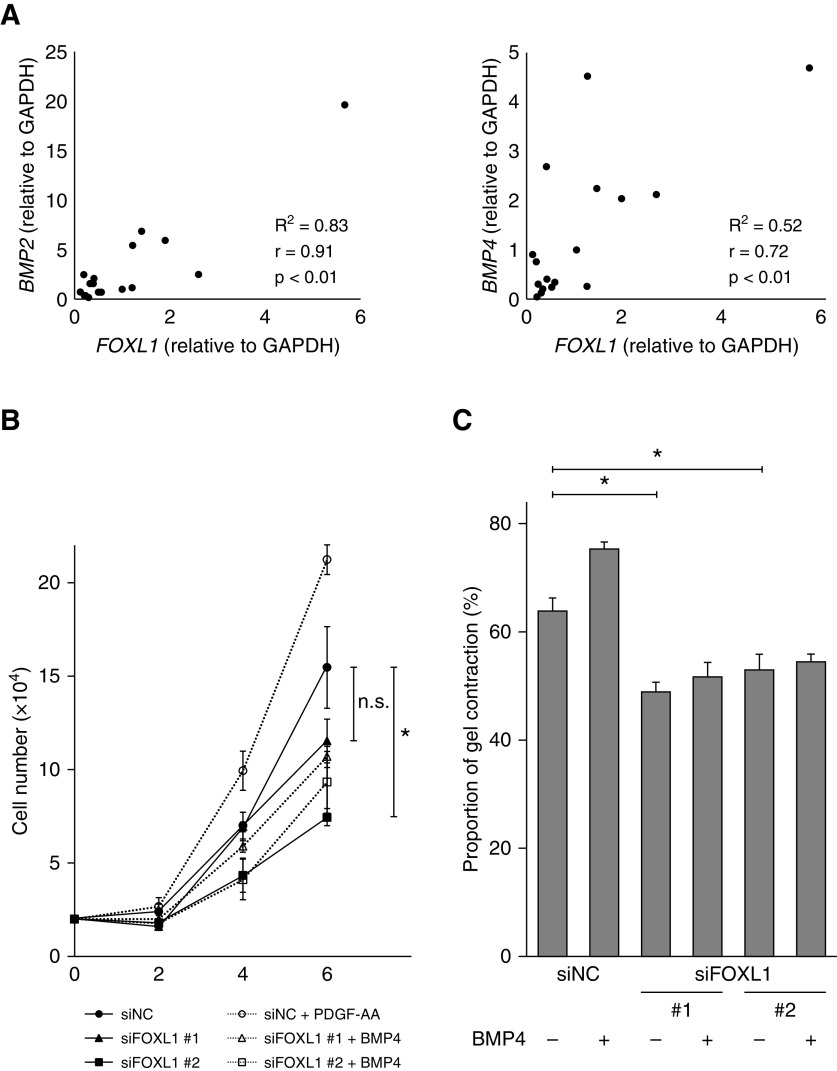
To further explore the relationships between FOXL1 and BMP ligands/antagonists (BMP2, BMP4, GREM1, and FSTL1), we compared their expression levels by quantitative RT-PCR in 17 different human lung fibroblast lines (Table E2). In line with our RNA-seq result, FOXL1 expression levels were positively correlated with BMP2 (r = 0.91), BMP4 (r = 0.72), and GREM1 (r = 0.54), suggesting that FOXL1 positively regulates BMP ligands and antagonists, and thereby tunes BMP signaling (Figures 5A and E5A).
Figure 5.
FOXL1 is related to BMP signaling in lung fibroblasts. (A) Associations of expression levels between FOXL1 and BMP ligands (BMP2 and BMP4) in lung fibroblast lines (n = 17) were evaluated by quantitative RT-PCR. The expression levels were normalized to that of GAPDH. (B) Cell numbers of NHLF transfected with siNC or siFOXL1 (#1 or #2) in the presence or absence of BMP4 (20 ng/ml) were counted 2, 4, and 6 days after seeding. The experiments were performed in triplicate. Error bars represent SEs. The cells transfected with siFOXL1 (#1 or #2) were compared with the siNC-transfected cells in the absence of BMP4. *P < 0.05. (C) NHLF transfected with siNC or siFOXL1 (#1 or #2) in the presence or absence of BMP4 (20 ng/ml) were embedded in collagen gels (n = 6). After 72 hours, the area of each gel was quantified. The percentage of contracted gel area was measured and compared with the initial size. Error bars represent SEs. The cells transfected with siFOXL1 (#1 or #2) were compared with the siNC-transfected cells in the absence of BMP4. *P < 0.05. n.s. = not significant.
Next, we explored the functional relevance of FOXL1 in association with BMP signaling in lung fibroblasts. BMP4 treatment enhanced cell growth in NHLF (Figure 5B). On the other hand, FOXL1 knockdown tended to suppress cell growth (Figure 5B). Three-dimensional collagen gels embedded with fibroblasts are used as a model of fibroblast-mediated tissue contraction (30). Exogenous BMP4 promoted contraction of collagen gels embedded with NHLF, whereas FOXL1 knockdown inhibited collagen gel contraction (Figure 5C).
FOXL1 Is Related to PDGF Signaling in Lung Fibroblasts
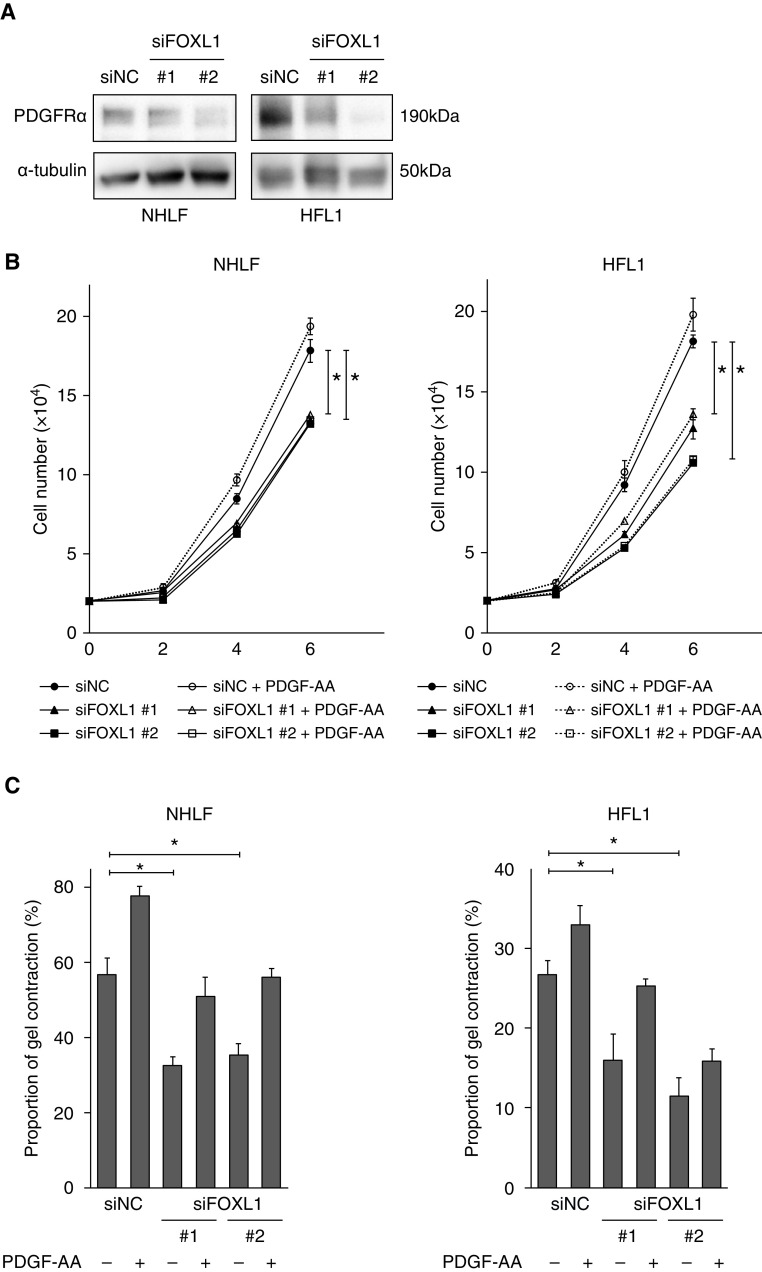
PDGF signaling in the mesenchyme is essential for lung morphogenesis (16) and Pdgfra+ lung fibroblasts contribute to lung regeneration in mice (15, 39). We noted that PDGFRA was downregulated by FOXL1 knockdown in our RNA-seq dataset (Table E7), and this effect was further confirmed at the protein level in two different lung fibroblast lines, NHLF and HFL1 (human fetal lung fibroblasts) (Figure 6A). This observation suggested that FOXL1 could regulate lung fibroblast functions by modulating PDGF signaling (40).
Figure 6.
FOXL1 is related to PDGF (platelet-derived growth factor) signaling in lung fibroblasts. (A) IB for PDGFRα (PDGF receptor-α) in NHLF and HFL1 treated with siNC or siFOXL1 (#1 or #2). α-tubulin was used as the loading control. (B) Cell numbers of NHLF or HFL1 (human fetal lung fibroblasts) transfected with siNC or siFOXL1 (#1 or #2) in the presence or absence of PDGF-AA (10 ng/ml) were counted 2, 4, and 6 days after seeding. The experiments were performed in quadruplicate. Error bars represent SEs. The cells transfected with siFOXL1 (#1 or #2) were compared with the siNC-transfected cells in the absence of PDGF-AA. *P < 0.05. (C) NHLF or HFL1 transfected with siNC or siFOXL1 (#1 or #2) in the presence or absence of PDGF-AA (10 ng/ml) were embedded in collagen gels. After 72 hours, the area of each gel was quantified. Percentage of contracted gel area was measured and compared with the initial size. The experiments were performed in triplicate. Error bars represent SEs. The cells transfected with siFOXL1 (#1 or #2) were compared with the siNC-transfected cells in the absence of PDGF-AA. *P < 0.05.
We tested whether FOXL1 silencing could affect cell growth in the presence or absence of PDGF-AA stimulation. PDGF-AA tended to promote cell growth of both NHLF and HFL1, whereas FOXL1 knockdown suppressed it (Figure 6B). Similarly, PDGF-AA tended to enhance collagen gel contraction in both fibroblast lines, whereas FOXL1 knockdown inhibited it (Figure 6C). In addition, FOXL1 knockdown inhibited the migration of NHLF (Figure E6).
PDGF-AA can only activate PDGFRα homodimers, not PDGFRα/PDGFRβ heterodimers or PDGFRβ homodimers (41). Given that FOXL1 knockdown leads to PDGFRα downregulation (Figure 6A), the effects of FOXL1 knockdown could be partly attributed to attenuated PDGF signaling.
Collectively, our results showed that FOXL1 governs a subset of genes related to key signaling (CTGF, PDGFRA, BMP2, and BMP4) in lung fibroblasts. We further explored FOXL1 and TEAD4 binding sequences (Figure E7A) in the promoter regions of these genes and identified multiple putative recognition sites (Figure E7B). These observations suggested that these genes could be partly regulated in a direct manner by FOXL1 or TAZ/YAP signaling.
FOXL1 Is Expressed in Fibroblasts of IPF Lung Tissue
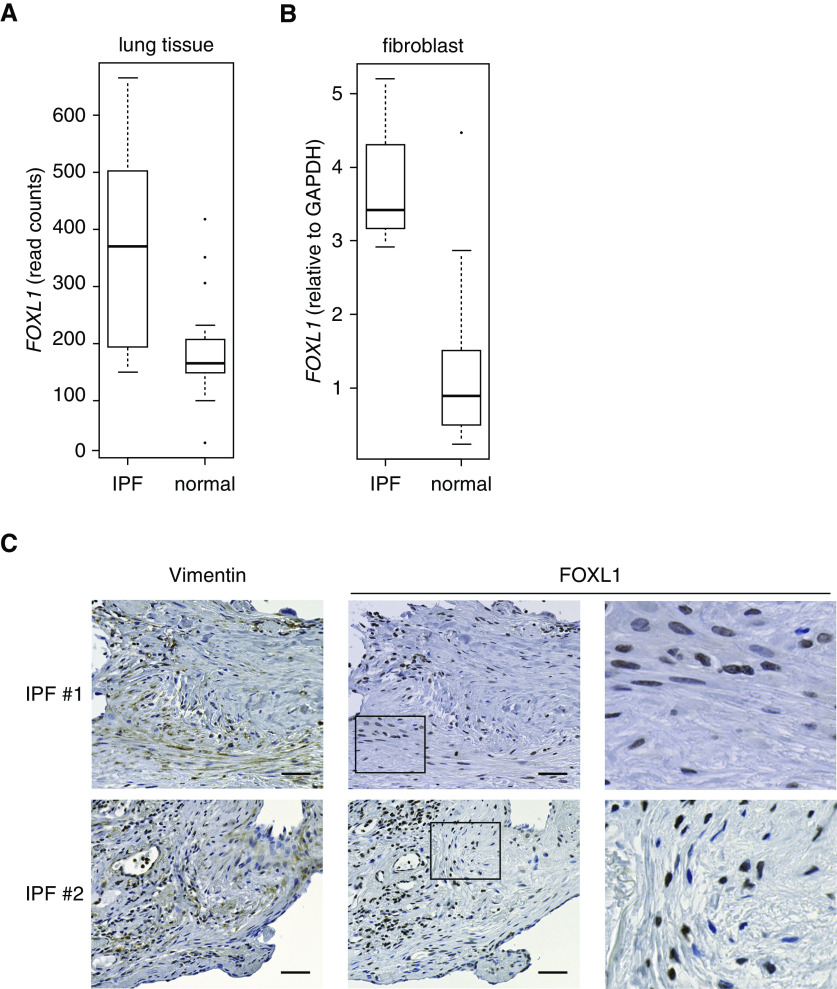
It is widely accepted that fibroblasts are key effector cells responsible for tissue fibrosis. Clusters of fibroblasts distributed in IPF lung tissue are presumed to be the sites of fibrotic reaction (23). To explore the clinical significance of FOXL1 expression in the lungs, we compared the transcript levels of FOXL1 between normal and IPF lung tissues using the GSE92592 dataset (42) and found higher FOXL1 expression levels in IPF lung tissue (Figure 7A).
Figure 7.
FOXL1 is expressed in the fibroblasts of idiopathic pulmonary fibrosis (IPF) lung tissue. (A) Transcript levels of FOXL1 in normal (n = 19) and IPF (n = 20) lung tissues. RNA-seq data were obtained from the GSE92592 dataset. (B) Transcript levels of FOXL1 in lung fibroblast lines derived from normal subjects (n = 14) and lung fibroblast lines derived from patients with IPF (n = 3). Quantitative RT-PCR was performed, and relative expression levels were normalized to those of GAPDH. (C) IHC for vimentin and FOXL1 in IPF lung tissue. Two different IPF cases are presented. High-magnification views of rectangular area of FOXL1 immunostaining are shown to the right. Note that nuclear FOXL1 staining is observed in the fibroblasts positive for vimentin. Scale bars, 50 µm.
Previous studies demonstrated that GREM1 and FSTL1 are increased in IPF lung tissue and are involved in IPF pathogenesis (43, 44). Because fibroblasts are the major source of these molecules, we compared the transcript levels of FOXL1 with those of GREM1 and FSTL1 in normal and IPF lung tissues. In agreement with the previous reports (43, 44), GREM1 and FSTL1 expression levels were higher in IPF lung tissue (Figure E5B). Moreover, they showed positive correlations with FOXL1 (r = 0.54 for GREM1 and r = 0.69 for FSTL1), supporting the notion that GREM1 and FSTL1 are positively regulated by FOXL1 (Table E7 and Figure E5A). These observations also suggested that FOXL1 might be functionally active to regulate a subset of target genes in IPF lung tissue.
We further compared FOXL1 expression levels in lung fibroblasts derived from normal and IPF lung tissues. Consistent with the results in lung tissue samples, FOXL1 expression was increased in lung fibroblasts derived from IPF lung tissue at the transcript level (Figure 7B).
Finally, we investigated FOXL1 protein expression in IPF lung tissue and found FOXL1 immunoreactivity in the nuclei of fibroblasts, which were also positive for vimentin (Figure 7C). Positive staining of FOXL1 in fibroblasts was commonly observed among the lung specimens derived from three different patients with IPF, suggesting its role in IPF pathogenesis.
Discussion
Fibroblasts are heterogeneous populations, and previous transcriptome analyses have demonstrated organ-specific gene signatures that represent “positional memory” of cultured fibroblasts. In search of a lung-specific fibroblast signature, our unbiased screening using the FANTOM5 database, including 45 different fibroblast lines, identified FOXL1 as a transcription factor highly activated in fibroblasts derived from the lungs. This was further supported by the findings based on the ENCODE database that the FOXL1 gene in lung fibroblasts is selectively hypomethylated and associated with super-enhancers.
Recent studies demonstrated that Foxl1-positive mesenchymal cells in the murine intestine, which are also positive for Pdgfra, are crucial for the intestinal stem cell niche (12, 13). It is also suggested that mesenchymal cells marked by Foxl1 and Pdgfra expression are so-called “telocytes” (45–47), which constitute a morphologically defined cell type found in various organs, including the lungs (48). It is known that Pdgfra+ alveolar fibroblasts contribute to alveolar epithelial self-renewal in mice (15, 17), and it was noteworthy that Foxl1 expression was found in Pdgfra+ murine lung mesenchymal cells by reanalysis of single-cell RNA-seq data (Figure E2B) (17).
It should be noted that the 45 fibroblast lines analyzed in this study do not include fibroblasts of gut origin. Considering that both the lungs and the gastrointestinal tract are endoderm-derived organs, it would be interesting to identify similarities and differences between lung fibroblasts and gastrointestinal fibroblasts.
In the present study, we explored genes possibly regulated by FOXL1 and investigated functional roles of FOXL1 in cultured human lung fibroblasts. Our RNA-seq analysis suggested that FOXL1 modulates key signaling pathways, including CTGF, BMP, and PDGF, through transcriptional regulation of their ligands, antagonists, or receptors. Moreover, cell culture studies revealed that FOXL1 could regulate key cellular responses such as cell growth and collagen gel contraction, all of which are important for the processes of wound healing and fibrogenesis.
Recent evidence suggests that fibroblast- or mesenchyme-derived Wnt and BMP signals are crucial for alveolar epithelial cell proliferation and differentiation (38, 49). Although we did not find a clear decrease in Wnt ligands after FOXL1 knockdown, our results indicate that FOXL1 could positively regulate BMP ligands (BMP2 and BMP4) and antagonists (GREM1 and FSTL1). Indeed, quantitative RT-PCR experiments showed positive correlations of expression levels between FOXL1 and these BMP ligands/antagonists (38). FOXL1-mediated modulation of BMP signaling and its pathophysiological role in the context of epithelial–mesenchymal interactions are worthy of further investigation.
In IPF lung tissue, pathologically activated lung fibroblasts are responsible for excessive ECM deposition and fibrotic responses (22). Furthermore, alveolar epithelial cell proliferation and differentiation are dysregulated in IPF lung tissue (50) at least partly because of the influence of activated fibroblasts. Taken together with our finding that FOXL1 is expressed in fibroblasts of IPF lung tissue, FOXL1 may be an important player in the pathological processes of IPF.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Sayaka Arakawa and Sayaka Igarashi for their assistance for primary cultured lung fibroblasts obtained at Tokyo National Hospital. They also thank Isao Asari for his excellent technical assistance with immunohistochemistry. Human lung specimens were provided by the University of North Carolina Marsico Lung Institute Tissue Procurement and Cell Culture Core, which is supported in part by the Cystic Fibrosis Foundation (grant BOUCHE19R0) and the National Institutes of Health (grant DK065988). The authors also thank Rodney C. Gilmore for performing RNA ISH. They also thank the members of the Department of Respiratory Medicine of the Graduate School of Medicine at the University of Tokyo for their support and useful discussion.
Footnotes
Supported by JSPS (Japanese Society for the Promotion of Science) KAKENHI grants 19K23981 (N.M.), 18K08170 (A.S.), and 16H02653 (T.N.). The RNA in situ hybridization experiment was supported in part by the Cystic Fibrosis Foundation (grant OKUDA19I0) and a research grant from Cystic Fibrosis Research Incorporation.
Author Contributions: Conception and design: N.M., M.H., H.I.S., and A.S. Analysis and interpretation: N.M., M.H., H.I.S., M. Saito, Y.M., K.O., M. Suzukawa, and A.S. Drafting the manuscript: N.M. and A.S. Supervision: R.C.B., A.H., and T.N.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2019-0396OC on September 18, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Micke P, Ostman A. Tumour-stroma interaction: cancer-associated fibroblasts as novel targets in anti-cancer therapy? Lung Cancer. 2004;45:S163–S175. doi: 10.1016/j.lungcan.2004.07.977. [DOI] [PubMed] [Google Scholar]
- 2.Peyser R, MacDonnell S, Gao Y, Cheng L, Kim Y, Kaplan T, et al. Defining the activated fibroblast population in lung fibrosis using single-cell sequencing. Am J Respir Cell Mol Biol. 2019;61:74–85. doi: 10.1165/rcmb.2018-0313OC. [DOI] [PubMed] [Google Scholar]
- 3.Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci USA. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horie M, Yamaguchi Y, Saito A, Nagase T, Lizio M, Itoh M, et al. Transcriptome analysis of periodontitis-associated fibroblasts by CAGE sequencing identified DLX5 and RUNX2 long variant as novel regulators involved in periodontitis. Sci Rep. 2016;6:33666. doi: 10.1038/srep33666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horie M, Miyashita N, Mikami Y, Noguchi S, Yamauchi Y, Suzukawa M, et al. TBX4 is involved in the super-enhancer-driven transcriptional programs underlying features specific to lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2018;314:L177–L191. doi: 10.1152/ajplung.00193.2017. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki HI, Young RA, Sharp PA. Super-enhancer-mediated RNA processing revealed by integrative microRNA network analysis. Cell. 2017;168:1000–1014, e15. doi: 10.1016/j.cell.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox’s in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- 10.Kaestner KH, Bleckmann SC, Monaghan AP, Schlöndorff J, Mincheva A, Lichter P, et al. Clustered arrangement of winged helix genes fkh-6 and MFH-1: possible implications for mesoderm development. Development. 1996;122:1751–1758. doi: 10.1242/dev.122.6.1751. [DOI] [PubMed] [Google Scholar]
- 11.Kaestner KH, Silberg DG, Traber PG, Schütz G. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- 12.Aoki R, Shoshkes-Carmel M, Gao N, Shin S, May CL, Golson ML, et al. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol Gastroenterol Hepatol. 2016;2:175–188. doi: 10.1016/j.jcmgh.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Tóth B, Kondo A, Massasa EE, et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557:242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics: tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 15.Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boström H, Gritli-Linde A, Betsholtz C. PDGF-A/PDGF alpha-receptor signaling is required for lung growth and the formation of alveoli but not for early lung branching morphogenesis. Dev Dyn. 2002;223:155–162. doi: 10.1002/dvdy.1225. [DOI] [PubMed] [Google Scholar]
- 17.Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148, e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkarni T, O’Reilly P, Antony VB, Gaggar A, Thannickal VJ. Matrix remodeling in pulmonary fibrosis and emphysema. Am J Respir Cell Mol Biol. 2016;54:751–760. doi: 10.1165/rcmb.2015-0166PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders YY, Liu H, Scruggs AM, Duncan SR, Huang SK, Thannickal VJ. Epigenetic regulation of caveolin-1 gene expression in lung fibroblasts. Am J Respir Cell Mol Biol. 2017;56:50–61. doi: 10.1165/rcmb.2016-0034OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato K, Logsdon NJ, Shin YJ, Palumbo S, Knox A, Irish JD, et al. Impaired myofibroblast dedifferentiation contributes to nonresolving fibrosis in aging. Am J Respir Cell Mol Biol. 2020;62:633–644. doi: 10.1165/rcmb.2019-0092OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet. 2017;389:1941–1952. doi: 10.1016/S0140-6736(17)30866-8. [DOI] [PubMed] [Google Scholar]
- 22.Thannickal VJ, Zhou Y, Gaggar A, Duncan SR. Fibrosis: ultimate and proximate causes. J Clin Invest. 2014;124:4673–4677. doi: 10.1172/JCI74368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinz B, Phan SH, Thannickal VJ, Prunotto M, Desmoulière A, Varga J, et al. Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am J Pathol. 2012;180:1340–1355. doi: 10.1016/j.ajpath.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito A, Nagase T. Hippo and TGF-β interplay in the lung field. Am J Physiol Lung Cell Mol Physiol. 2015;309:L756–L767. doi: 10.1152/ajplung.00238.2015. [DOI] [PubMed] [Google Scholar]
- 25.Noguchi S, Saito A, Mikami Y, Urushiyama H, Horie M, Matsuzaki H, et al. TAZ contributes to pulmonary fibrosis by activating profibrotic functions of lung fibroblasts. Sci Rep. 2017;7:42595. doi: 10.1038/srep42595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, III, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902, e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horie M, Saito A, Mikami Y, Ohshima M, Morishita Y, Nakajima J, et al. Characterization of human lung cancer-associated fibroblasts in three-dimensional in vitro co-culture model. Biochem Biophys Res Commun. 2012;423:158–163. doi: 10.1016/j.bbrc.2012.05.104. [DOI] [PubMed] [Google Scholar]
- 30.Horie M, Saito A, Yamauchi Y, Mikami Y, Sakamoto M, Jo T, et al. Histamine induces human lung fibroblast-mediated collagen gel contraction via histamine H1 receptor. Exp Lung Res. 2014;40:222–236. doi: 10.3109/01902148.2014.900155. [DOI] [PubMed] [Google Scholar]
- 31.Okuda K, Chen G, Subramani DB, Wolf M, Gilmore RC, Kato T, et al. Localization of secretory mucins MUC5AC and MUC5B in normal/healthy human airways. Am J Respir Crit Care Med. 2019;199:715–727. doi: 10.1164/rccm.201804-0734OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, et al. FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hrycaj SM, Marty-Santos L, Cebrian C, Rasky AJ, Ptaschinski C, Lukacs NW, et al. Hox5 genes direct elastin network formation during alveologenesis by regulating myofibroblast adhesion. Proc Natl Acad Sci USA. 2018;115:E10605–E10614. doi: 10.1073/pnas.1807067115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44:D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 37.Tadokoro T, Gao X, Hong CC, Hotten D, Hogan BL. BMP signaling and cellular dynamics during regeneration of airway epithelium from basal progenitors. Development. 2016;143:764–773. doi: 10.1242/dev.126656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung MI, Bujnis M, Barkauskas CE, Kobayashi Y, Hogan BLM. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 2018;145:dev163014. doi: 10.1242/dev.163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green J, Endale M, Auer H, Perl AK. Diversity of interstitial lung fibroblasts is regulated by platelet-derived growth factor receptor α kinase activity. Am J Respir Cell Mol Biol. 2016;54:532–545. doi: 10.1165/rcmb.2015-0095OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noskovičová N, Petřek M, Eickelberg O, Heinzelmann K. Platelet-derived growth factor signaling in the lung: from lung development and disease to clinical studies. Am J Respir Cell Mol Biol. 2015;52:263–284. doi: 10.1165/rcmb.2014-0294TR. [DOI] [PubMed] [Google Scholar]
- 41.Ostman A, Heldin CH. PDGF receptors as targets in tumor treatment. Adv Cancer Res. 2007;97:247–274. doi: 10.1016/S0065-230X(06)97011-0. [DOI] [PubMed] [Google Scholar]
- 42.Schafer MJ, White TA, Iijima K, Haak AJ, Ligresti G, Atkinson EJ, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun. 2017;8:14532. doi: 10.1038/ncomms14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong Y, Geng Y, Li L, Li X, Yan X, Fang Y, et al. Blocking follistatin-like 1 attenuates bleomycin-induced pulmonary fibrosis in mice. J Exp Med. 2015;212:235–252. doi: 10.1084/jem.20121878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koli K, Myllärniemi M, Vuorinen K, Salmenkivi K, Ryynänen MJ, Kinnula VL, et al. Bone morphogenetic protein-4 inhibitor gremlin is overexpressed in idiopathic pulmonary fibrosis. Am J Pathol. 2006;169:61–71. doi: 10.2353/ajpath.2006.051263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondo A, Kaestner KH. Emerging diverse roles of telocytes. Development. 2019;146:dev175018. doi: 10.1242/dev.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popescu LM, Gherghiceanu M, Suciu LC, Manole CG, Hinescu ME. Telocytes and putative stem cells in the lungs: electron microscopy, electron tomography and laser scanning microscopy. Cell Tissue Res. 2011;345:391–403. doi: 10.1007/s00441-011-1229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaestner KH. The intestinal stem cell niche: a central role for Foxl1-expressing subepithelial telocytes. Cell Mol Gastroenterol Hepatol. 2019;8:111–117. doi: 10.1016/j.jcmgh.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hussein MM, Mokhtar DM. The roles of telocytes in lung development and angiogenesis: an immunohistochemical, ultrastructural, scanning electron microscopy and morphometrical study. Dev Biol. 2018;443:137–152. doi: 10.1016/j.ydbio.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359:1118–1123. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulkarni T, de Andrade J, Zhou Y, Luckhardt T, Thannickal VJ. Alveolar epithelial disintegrity in pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2016;311:L185–L191. doi: 10.1152/ajplung.00115.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.