Abstract
Background
The filamentous fungi Paecilomyces variotii and Penicillium roqueforti are prevalent food spoilers and are of interest as potential future cell factories. A functional CRISPR/Cas9 genome editing system would be beneficial for biotechnological advances as well as future (genetic) research in P. variotii and P. roqueforti.
Results
Here we describe the successful implementation of an efficient AMA1-based CRISPR/Cas9 genome editing system developed for Aspergillus niger in P. variotii and P. roqueforti in order to create melanin deficient strains. Additionally, kusA− mutant strains with a disrupted non-homologous end-joining repair mechanism were created to further optimize and facilitate efficient genome editing in these species. The effect of melanin on the resistance of conidia against the food preservation stressors heat and UV-C radiation was assessed by comparing wild-type and melanin deficient mutant conidia.
Conclusions
Our findings show the successful use of CRISPR/Cas9 genome editing and its high efficiency in P. variotii and P. roqueforti in both wild-type strains as well as kusA− mutant background strains. Additionally, we observed that melanin deficient conidia of three food spoiling fungi were not altered in their heat resistance. However, melanin deficient conidia had increased sensitivity towards UV-C radiation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40694-021-00111-w.
Keywords: CRISPR/Cas9, Cell factory, Melanin, Food spoilage, Food spoiling fungi, Polyketide synthase, Conidia, Aspergillus niger, Penicillium roqueforti, Paecilomyces variotii
Introduction
The genome editing system by clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) has proven to be a powerful tool in filamentous fungi, providing new insights and opportunities within food, agricultural, clinical and biotechnological research [1–4]. Currently, the CRISPR/Cas9 gene editing tool has been introduced in over 40 species of filamentous fungi and oomycetes to date [5]. In this paper, we describe a functional CRISPR/Cas9 genome editing protocol for two food spoilage fungi Paecilomyces variotii and Penicillium roqueforti.
The CRISPR/Cas9 genome editing system introduces a double stranded break (DSB) on a specific genomic DNA site. Fungi have two main DNA repair mechanisms that can restore the DSB created by CRISPR/Cas9, namely the non-homologous end-joining repair mechanism (NHEJ) and the homology directed repair mechanism (HDR). For genome editing purposes, many studies rely on the HDR mechanism in order to control genomic editing (e.g. gene replacement studies), by providing the fungus with homologous DNA created in vitro [6]. This allows for precise DNA insertion, replacement or removal in the genome. However, many filamentous fungi prefer repair via NHEJ over HDR, which complicates this precise genome editing. In order to promote DNA repair by HDR in fungi, genes involved in the NHEJ repair mechanism can be deleted. A mutant fungus with a deleted kusA gene is defective in the NHEJ repair mechanism, therefore a DSB can only be repaired by HDR as shown in several filamentous fungi such including e.g. Neurospora crassa [7], Aspergillus niger [8].
The thermotolerant nature of P. variotii spores makes this fungus a relevant food spoiler [9–12]. P. variotii is a known spoiler of fruit juices, sauce, canned products and non-carbonized sodas [13, 14]. Additionally, P. variotii strains have been reported to produce industrially interesting, often thermostable, enzymes such as tannases, amylases, β-glucosidases and an alcohol oxidase [15–21]. Recently, a genome of P. variotii has been published [22] in which the first method on targeted gene disruptions in this fungus using Agrobacterium tumefaciens is described. Although A. tumefaciens mediated transformations are shown to be efficient and beneficial over other transformation methods in certain cases [23], it does require optimization of multiple factors and can be tedious compared to the relatively quick and easy to use PEG-mediated transformations [24].
The filamentous fungus P. roqueforti is best known as the ‘blue cheese’ fungus for its use in blue cheese production [25]. However, P. roqueforti is also a known food spoiler that can produce mycotoxins such as PR-toxin and roquefortine-C, which form potential health risks for humans [26–29]. As such, P. roqueforti has been intensively studied for its secondary metabolite production and specifically its mycotoxin production [30–33]. Additionally, P. roqueforti has biotechnological potential as a cell factory, as it produces proteolytic enzymes of interest to the cheese-making industry and high-value secondary metabolites such as mycophenolic acid [33–37]. A CRISPR/Cas9 genome editing system has been described for Penicillium chrysogenum, a closely related species to P. roqueforti, using a similar approach as has been used for Aspergillus species by providing a CRISPR/Cas9 plasmid during PEG-mediated transformation [38]. This has led to the possibility of large scale genome re-engineering making P. chrysogenum a useful platform organism as cell factory for production of natural products [39]. Taken together, a functional CRISPR/Cas9 targeted genome editing protocol based on PEG-mediated transformations of CRISPR/Cas9 plasmids would be beneficial for future research on food spoilage capabilities and potential biotechnological advances in both P. variotii and P. roqueforti.
Many food spoiling fungi, such as P. variotii and P. roqueforti, produce asexual derived spores (conidia) that can withstand commonly used preservation treatments such as UV radiation or heat [40, 41]. Recently, the conidia of P. variotii have been reported to survive 60 °C for 20 min, being the most heat resistant of this type of asexual spores [10]. Additionally, conidia of food spoilage fungi are able to survive UV radiation levels used for decontamination by food industry [42–45]. It is yet unclear if pigmentation provides stress resistance against these preservation techniques in food spoiling fungi. In many ascomycetes, disruption of a specific polyketide synthase (PKS) gene results in loss of conidial pigmentation. As a consequence, these transformants produce lighter or white conidia [46–49]. Comparing the conidia of these mutants with their parental strain will lead to new insights into the potential roles of melanin in preservation stress resistance of conidia.
In this research, a functional CRISPR/Cas9 genome editing system for P. variotii and P. roqueforti is implemented to create melanin deficient mutants of both fungi, and subsequently comparing these mutants to their wild-type parental strains, using a recently described CRISPR/Cas9 deletion system developed for A. niger [50] with minor adaptations. This CRISPR/Cas9 genome editing system developed for A. niger is based on the expression of Cas9 driven from the tef1 promoter [51]. The Cas9 expression cassette, together with the guide RNA expression cassette and the hygromycin selection marker are located on a plasmid that also contains the AMA1 sequence which enables autonomous replication in Aspergillus species, thereby making integration of the vector into the genome less likely [52]. This AMA1-based CRISPR/Cas9 genome editing system allows for the temporal presence of the CRISPR/Cas9 plasmid and therefore marker-free genome editing [50]. The CRISPR/Cas9 genome editing method is considerably faster than the already established marker-free genome editing method which relies on recyclable markers. The CRISPR/Cas9 based genome editing method allows for the creation of multiple mutations in a single transformation experiment, as demonstrated in A. niger [50], whereas the recycling method requires deletions to be performed one at a time.
Understanding the resistance mechanisms of conidia from food spoiling fungi will help us in designing novel targeted preservation techniques able to inactivate conidia without altering food flavor profiles. In order to investigate this, a working CRISPR/Cas9 genome editing system has been developed for P. variotii and P. roqueforti. These genome editing systems could enhance future research and provide a stepping stone towards creating novel biotechnologically relevant cell factories.
Results
Construction of melanin deficient mutants in P. variotii and P. roqueforti using CRISPR/Cas9 based genome editing.
In order to investigate the impact of melanin on stress resistance in P. roqueforti and P. variotii, melanin deficient strains in these species were made using a recently described CRISPR/Cas9 deletion system [50]. Polyketide synthase (PKS) homologues from P. roqueforti and P. variotii were identified as best bi-directional BLASTp hits with both the FwnA protein (gene ID: An09g05730) from A. niger [47] and the Pks17 protein (gene ID: Pc21g16000) from P. chrysogenum DS68530 [38]. The BLASTp searches identified Pro_LCP9604111_2|g6432.t1 as the bi-directional best hit in P. roqueforti. The P. variotii protein ID456077, recently described as PvpP in [49], was identified as the bi-directional best hit in P. variotii. The P. roqueforti protein Pro_LCP9604111_2|g6432.t1 is 68% identical to FwnA from A. niger and 92% identical to Pks17 from P. chrysogenum. The P. variotii protein ID456077 (PvpP) is 67% identical to FwnA and 65% identical to Pks17.
Optimal guide RNAs for CRISPR/Cas9 genome editing for these genes were chosen based on CHOPCHOP predictors [53] (Additional file 1: Table S1). The guide RNAs were chosen to target an exon near the start codon of the open reading frame (ORF). The guide RNAs targeting the PKS homologous were cloned into the autonomously replicating vector pFC332, expressing the Cas9 nuclease, creating the CRISPR/Cas9 plasmids pPT13.1 and pPT9.3 to use for transformation of P. variotii and P. roqueforti respectively (Additional file 1: Table S1). The CRISPR/Cas9 plasmids were subsequently transformed to protoplasts of P. variotii CBS101075 and P. roqueforti LCP9604111. Transformants were obtained that produced white conidia, indicative of a disrupted melanin biosynthesis and subsequent loss of pigmentation, on the primary transformation plates for both fungi (Fig. 1). The efficiency of obtaining transformants with white conidia on the primary transformation plates in P. variotii and P. roqueforti was 83% (728 colonies with white conidia from a total of 876 transformants) and 97% (56 colonies with white conidia from a total of 58 transformants) respectively (Table 1).
Fig. 1.
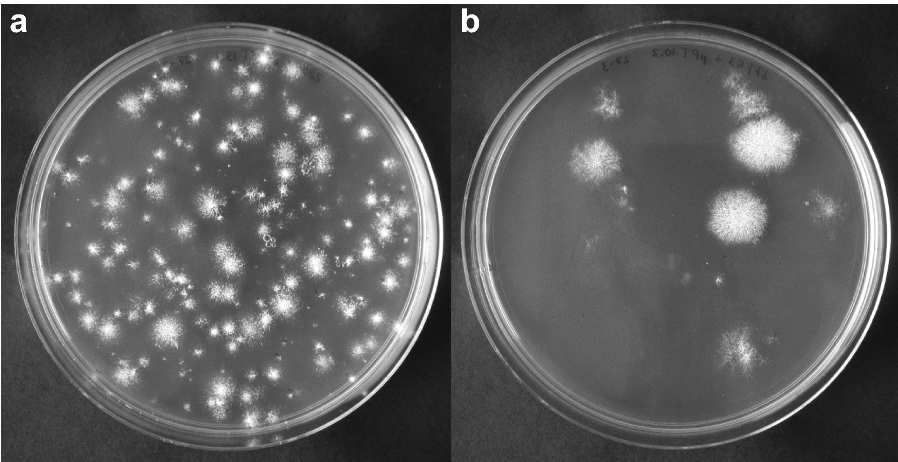
Transformation plates obtained when transforming P. variotii and P. roqueforti with PKS gene targeting CRISPR/Cas9 plasmids. a Transformation plate of P. variotii CBS101075 transformed with AMA1-based CRISPR/Cas9 plasmid pPT13.1 targeting gene pvpP needed for melanin biosynthesis. Typically around 100 transformants are obtained when adding 2 µg of plasmid depending on the amount of protoplasts obtained. b Transformation plate of P. roqueforti LCP9604111 transformed with AMA1-based CRISPR/Cas9 plasmid pPT9.3 targeting gene pksA. Typically around 10 transformants are obtained when adding 2 µg of plasmid depending on the amount of protoplasts obtained
Table 1.
Gene editing efficiencies of P. variotii and P. roqueforti using CRISPR/Cas9
| P. variotii | P. roqueforti | |||
|---|---|---|---|---|
| Brown conidia | White conidia | Green conidia | White conidia | |
| Phenotype obtained on first transformation plate | 148/876 (17%) | 728/876 (83%) | 2/58 (3%) | 56/58 (97%) |
| Hygromycin resistance loss after one round of non-selective growth | 32/40 (80%) | 9/40 (23%) | 1/2 (50%) | 16/56 (29%) |
Numbers represent transformants and were calculated over multiple transformation experiments. The average amount of transformants obtained per transformation using 2 µg of CRISPR/Cas9 plasmid was ± 100 colonies for P. variotii and ± 10 colonies for P. roqueforti depending on amounts of obtained protoplasts
The efficiency of a white-coloured mutant losing hygromycin resistance in P. variotii and P. roqueforti was 23% (9 out of 40) and 29% (15 out of 56), respectively. White sporulating mutants that lost their hygromycin resistance under non-selective growth conditions were purified further and subsequently checked for mutations in the PKS genes by performing diagnostic PCRs and DNA sequencing. Melanin deficient mutants P. roqueforti (PT34.2) and P. variotii (PT32.5) were chosen for further analysis (Fig. 2). Both strains contain a 14 bps deletion in the pksA and pvpP gene respectively causing a frameshift and thus a probable genetic loss of function (Additional file 2: Figure S1). These results show that CRISPR/Cas9 genome editing using the AMA1-based expression vectors are effectively disrupting target genes in both species.
Fig. 2.
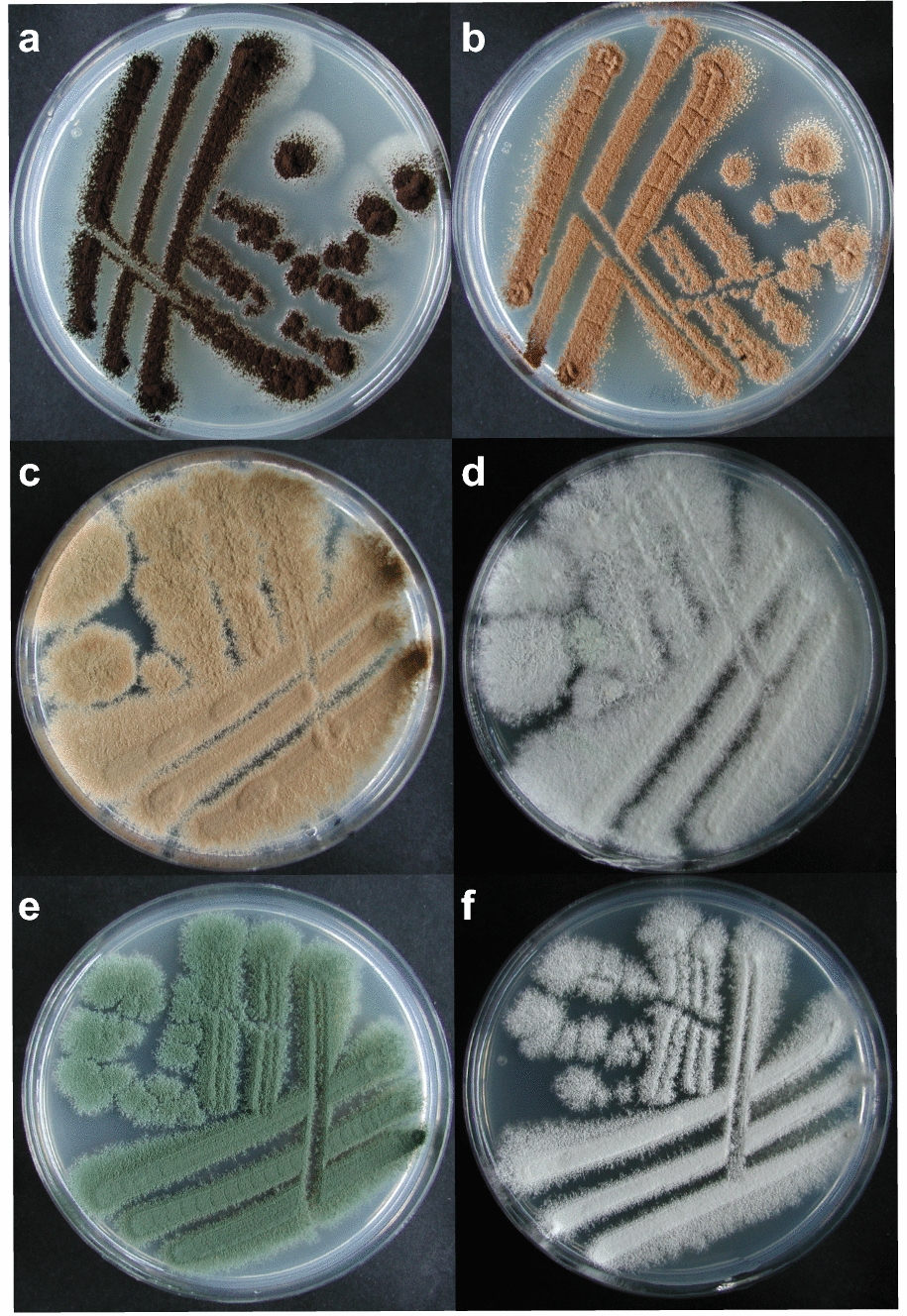
Phenotypes of parental strains and PKS mutants. a Phenotype A. niger N402, it has black conidia. b Phenotype A. niger MA93.1, it has fawn coloured / light brown conidia. c Phenotype P. variotii CBS101075 strain, it has fawn coloured conidia. d Phenotype P. variotii pksA− strain PT32.5, it has white conidia. e Phenotype P. roqueforti LCP9604111, it has green conidia. f Phenotype P. roqueforti pksA− strain PT34.2, it has white conidia
The low efficiency of hygromycin loss after one round of non-selective growth in P. variotii transformants
The low efficiency of hygromycin loss in white-coloured transformants of 23% (9 out of 40) is lower compared to the efficiency of hygromycin loss in brown transformants of 80% (32 out of 40) in P. variotii. To investigate the low efficiency of hygromycin loss and its link with the phenotype, eleven white-coloured (of which two showed hygromcin loss) and six brown-coloured (of which four showed hygromycin loss) transformants of P. variotii CBS101075 were analysed by diagnostic PCR and sequencing. When analyzing the pvpP locus of the eleven white-coloured P. variotii transformants, we discovered that only 3 out of 11 (27%) mutants had the expected small indel mutation of which 1 lost the hygromycin resistant phenotype after one round of non-selective growth (Table 2). Also, 4 out of the 11 (36%) transformants analysed had part of the pPT13.1 plasmid integrated in the pvpP locus at the site where the double stranded break (DSB) took place, of which one lost the hygromcin resistant phenotype. The sequence and size of parts of the integrated pPT13.1 plasmid were variable. We were unable to obtain PCR products for the remaining 4 out of 11 (36%) transformants, indicating a large insertion or deletion that hampered the PCR.
Table 2.
Plasmid integration in P. variotii CBS101075 transformants that did not lose hygromycin resistance
| Transformants with small indel | Transformants containing part of pPT13.1 plasmid at DBS site | No proper PCR product obtained | |
|---|---|---|---|
| White conidia | 3/11 (27%) | 4/11 (36%) | 4/11 (36%) |
| Brown conidia | 6/6 (100%) | 0/6 (0%) | 0/6 (0%) |
The six brown P. variotii transformants analysed, of which four showed hygromycin resistance loss after one round of non-selective growth, all contained an in-frame deletion in the pvpP locus at the DSB site (3 bps, 6 bps, 6 bps, 9 bps, 12 bps and 15 bps respectively). This indicates that the brown P. variotii transformants were modified by CRISPR/Cas9 gene editing, but had a functional PvpP enzyme despite the DSB and subsequent indel caused by CRISPR/Cas9.
Construction of NHEJ repair disrupted mutants in P. variotii and P. roqueforti using CRISPR/Cas9 genome editing
In order to facilitate future genome editing in P. variotii and P. roqueforti, a kusA− strain was made for both P. variotii and P. roqueforti. When performing transformations of a kusA− mutant parental strain, the double stranded break caused by CRISPR/Cas9 cannot be repaired by non-homologous end-joining creating indels and instead relies on a homologous repair DNA fragment, which could be donor DNA provided by the user. This will enable targeted and complete gene knock-out or gene replacement studies. Gene deletion mutants in the kusA gene of P. variotii DTO217-A2 and P. roqueforti DTO013-F5 backgrounds were made, as these strains produce heat resistant conidia [10], a phenotype of interest for future food spoilage studies. The kusA homologous of P. variotii and P. roqueforti were identified by BLASTp analysis (best bi-directional hits) using the KusA protein from A. niger (An15g02700) as a query. Only a single homologous protein with significant identity score was found in P. variotii DTO217-A2: Pva_DTO217A2_1|g5897.t1 (66% identity). Similarly, a single homologous protein was found in P. roqueforti: Pro_LCP9604111_2|g3395.t1 (67% identity). Plasmids pPT23.1 and pPT22.4, containing specific guide RNA and the Cas9 expression cassettes, were made for creating kusA− strains in P. variotii and P. roqueforti respectively (Additional file 1: Table S1). The guide RNAs were chosen to target an exon near the start codon of the ORFs. The creation of the kusA− mutants of P. variotii and P. roqueforti relied on the creation of indels caused by NHEJ repair to disrupt the kusA homologous genes of these species. From 26 P. variotii transformants obtained only a single transformant lost the hygromycin resistant phenotype after one round of non-selective growth. In the case of P. roqueforti only 1 transformant was obtained and this single transformant lost the hygromycin resistance phenotype after one round of non-selective growth. Sequencing the kusA locus of transformants of strains P. variotii PT39.26 and P. roqueforti PT43.1 showed small indels in the kusA locus (7 bp deletion in the kusA gene of P. variotii and 22 bp deletion in P. roqueforti) resulting in frameshifts and thus potentially a disrupted kusA gene (Additional file 3: Figure S2).
The impact of a disrupted NHEJ repair mechanism on the genome editing efficiency in P. variotii (kusA−)
The P. variotii PT39.26 (kusA−) strain was tested for its genome editing efficiency by transformation with the previously used pPT13.1 plasmid, which contains the pvpP specific guide RNA to create a double stranded break in the pvpP gene. A transformation of the PT39.26 strain with the pPT13.1 plasmid without providing a homologous repair DNA fragment did not give any transformants on the primary transformation plate, as expected. This indicates that indeed the NHEJ repair mechanism has been impaired in this strain and thus the strain cannot repair its double stranded break without the help of a homologous piece of DNA. Next, transformations were performed with the addition of donor DNA to allow the repair of the DSB created by the guide via homologous recombination. When donor DNA was added, putative transformants were obtained on the transformation plates. The donor DNA was a fused PCR product of both 5′ and 3′ untranslated flanks of the pvpP gene, which would theoretically result in the removal of the entire translated region of pvpP (6677 bps). In this transformation a total of fifteen transformants were obtained which all had the white-coloured phenotype. The transformants were purified from the first transformation plate and 14 out of the 15 (93%) purified transformants lost their hygromycin resistant phenotype after one round of non-selective conditions. These efficiencies are similar to those observed in A. niger and a major improvement over the original ratio of 1 out of 26 (3.8%) for creating a kusA− strain observed in P. variotii DTO217-A2 or the 9 out of 40 (23%) ratio observed in P. variotii CBS101075. Genomic DNA was isolated for eight of these transformants. Diagnostic PCR revealed that all eight transformants were repaired using the homologous piece of DNA provided, making a full knock-out of the 6677 bps gene pvpP (Additional file 4: Figure S3). The PCR products of two transformants were excised from gel and subsequently send for sequencing. This confirmed repair using the HDR mechanism, replacing the original pvpP gene with the provided donor DNA fragment that only contained the fused flanks. In this way, we obtained the pvpP knock-out strain P. variotii PT42.1 (kusA−, ΔpvpP). All kusA− mutant strains described in this study have no visible alteration in morphology and no visible change in colony diameter or radial growth rate when compared to their parental strains (Additional file 5: Figure S4).
Heat resistance of conidia from food spoiling fungi not affected in melanin deficient mutants
Heat inactivation assays were performed to determine the heat resistance of conidia from the P. variotii PT32.5 (pvpP−) and P. roqueforti PT34.2 (pksA−) deletion strains when compared to their parental strains. Additionally, we included the previously made ∆fwnA strain from A. niger (MA93.1) and its parental strain (N402) [47]. Note that the strains with intact kusA genes have been used for phenotyping, as the NHEJ disruption in the kusA− strains could potentially impact resistance against DNA damage caused by either UV radiation or heat. In order to observe at least a two log reduction in microbial load within 30 min, heating temperatures had to be adjusted per species. Conidia from P. variotii are more heat resistant than their A. niger and P. roqueforti counterparts [10, 54] and thus heat inactivation was done in a 60 °C water bath for P. variotii conidia instead of a 56 °C water bath for P. roqueforti and A. niger. Heat inactivation curves of wild-type and melanin deficient conidia from A. niger, P. variotii and P. roqueforti are shown in Fig. 3A, 3B and 3C respectively. Decimal reduction values were calculated based on these graphs and given in Table 3.
Fig. 3.
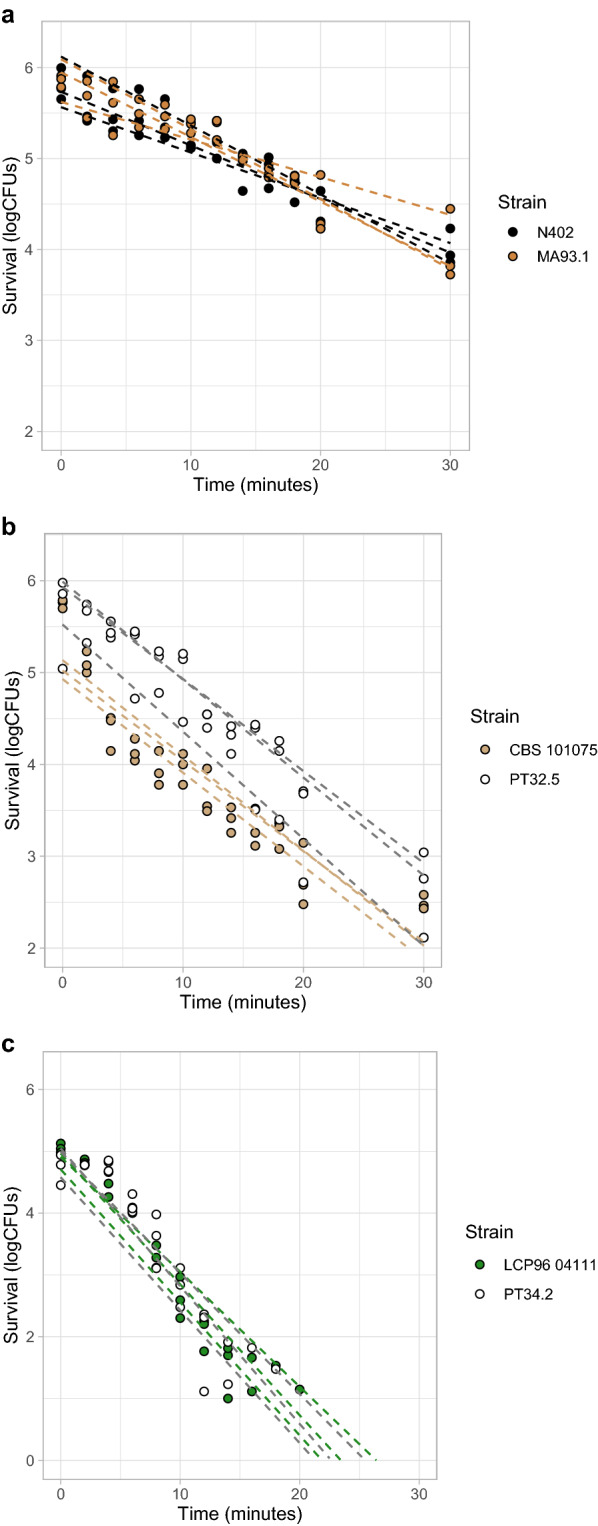
Heat resistance of three food spoiling fungi and their melanin deficient mutants. Colours used correspond with the phenotype of the conidia, see Fig. 2a The heat inactivation curves of A. niger N402 wild-type conidia (black lines) and A. niger MA93.1 melanin deficient mutant conidia (brown lines). The A. niger strains were heat treated in a 56 °C water bath. Heat inactivation shows only a 2-log reduction in microbial load for wild-type and mutant when treated for 30 min. b The heat inactivation curves of P. variotii CBS101075 wild-type conidia (light brown lines) and P. variotii PT32.5 melanin deficient mutant conidia (grey lines). The P. variotii strains were heat treated in a 60 °C water bath. Heat inactivation shows a 3-log reduction in microbial load for wild-type and mutant when treated for 30 min. c The heat inactivation curves of P. roqueforti LCP9604111 wild-type conidia (green lines) and P. roqueforti PT34.2 melanin deficient mutant conidia (grey lines). The P. roqueforti strains were heat treated in a 56 °C water bath. Heat inactivation shows at least a 5-log reduction in microbial load for wild-type and mutant when treated for 30 min. Three biological triplicates were measured. Inactivation curves were drawn based on the linear regression model
Table 3.
D-values of heat inactivated conidia from three food spoiling fungi and their melanin mutants
| Species and strain names | D-value ± standard deviation in time (minutes) | |
|---|---|---|
| A. niger | N402 | 16.8 ± 3.4 |
| A. niger | MA93.1 | 17.1 ± 6.2 |
| P. variotii | CBS101075 | 9.3 ± 0.7 |
| P. variotii | PT32.5 | 9.9 ± 0.3 |
| P. roqueforti | LCP9604111 | 4.9 ± 0.4 |
| P. roqueforti | PT34.2 | 4.7 ± 0.3 |
D-values are measured at 60 °C for P. variotii and 56 °C for A. niger and P. roqueforti
There were no significant differences in D-values based on Student’s t-tests between wild-type and mutant (all p-values were p > 0.05). Therefore, no significant difference in heat resistance between wild-type conidia and their melanin deficient mutant conidia was observed.
UV-C radiation resistance of conidia from food spoiling fungi is affected in melanin deficient mutants
A UV-C radiation assay was performed to determine the UV resistance of the melanin deficient mutants A. niger MA93.1 (ΔfwnA), P. variotii PT32.5 (pvpP−) and P. roqueforti PT34.2 (pksA−). UV inactivation curves of conidia from A. niger, P. variotii and P. roqueforti are shown in Fig. 4A, 4B and 4C respectively. The results show that conidia from all three food spoiling species have reduced UV resistance when melanin biosynthesis is disrupted. Decimal reduction values were calculated to quantify this difference and are listed in Table 4.
Fig. 4.
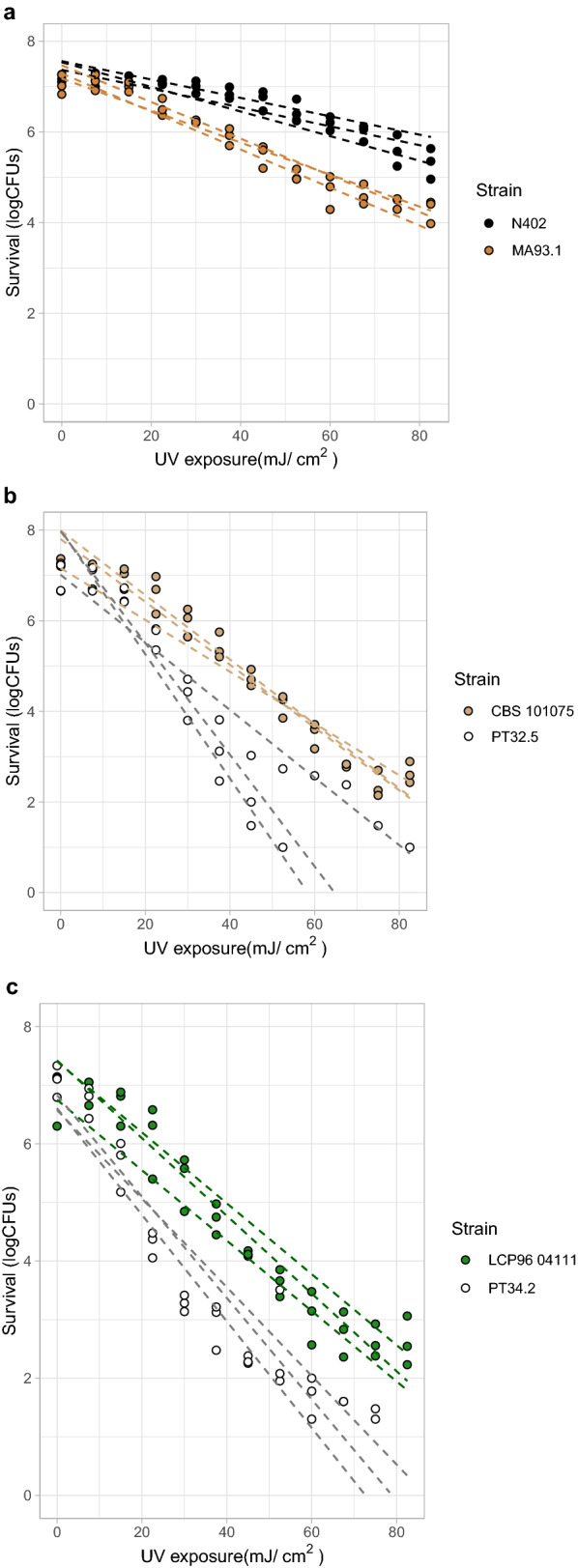
UV-C radiation resistance of melanin deficient mutants from food spoilage fungi. Colours used correspond with the phenotype of the conidia, see Fig. 2. a The UV inactivation curves of A. niger N402 wild-type conidia (black lines) and A. niger MA93.1 melanin deficient mutant conidia (brown lines). A maximum of 3-log reduction in microbial load was observed in the mutant after UV treatment. b The UV inactivation curves of P. variotii CBS101075 wild-type conidia (light brown lines) and P. variotii PT32.5 melanin deficient mutant conidia (grey lines). A maximum of 7-log reduction in microbial load was observed in the mutant after UV treatment. c The UV inactivation curves of P. roqueforti LCP9604111 wild-type conidia (green lines) and P. roqueforti PT34.2 melanin deficient mutant conidia (grey lines). A maximum of 7-log reduction in microbial load was observed in the mutant after UV treatment. Three biological triplicates were measured. Inactivation curves were drawn based on linear regression model
Table 4.
D-values of UV inactivated conidia from three food spoiling fungi and their melanin mutants
| Species and strain names | D-value ± standard deviation in dose (mJ/cm2) | |
|---|---|---|
| A. niger | N402 | 44.7 ± 5.6 |
| A. niger | MA93.1 | 25.6 ± 1.9 |
| P. variotii | CBS101075 | 15.3 ± 1.5 |
| P. variotii | PT32.5 | 9.6 ± 2.7 |
| P. roqueforti | LCP9604111 | 16.1 ± 0.7 |
| P. roqueforti | PT34.2 | 11.9 ± 0.9 |
Student’s t-tests revealed significant UV radiation reductions when wild-type is compared to the melanin mutant in A. niger (p = 0.01), P. roqueforti (p = 0.01), but not for P. variotii (p = 0.06) although a similar trend is visible. The A. niger wild-type conidia are more resistant to UV radiation than the wild-type conidia from both P. variotii and P. roqueforti (p < 0.00 and p < 0.00 respectively). Interestingly, even the fawn coloured A. niger melanin mutant MA93.1 is significantly more resistant to UV than the P. variotii and P. roqueforti wild-type conidia (p < 0.00 and p < 0.00 respectively). This suggests that other melanin types or pigments in the MA93.1 strain, produced independently from the FwnA enzyme, contribute significantly to the UV radiation resistance of A. niger conidia. Overall, the pks− mutants were more sensitive to UV radiation than their parental strains, indicating that melanin contribute to UV resistance of conidia from food spoiling fungi.
Discussion
CRISPR/Cas9 genome editing protocol for P. variotii and P. roqueforti
In this article a previously described CRISPR/Cas9 genome editing protocol for A. niger [50] was successfully implemented to perform genome editing in P. roqueforti and P. variotii. The efficiency of obtaining white-coloured transformants was 728 out of 876 (83%) in P. variotii CBS101075 and 56 out of 58 (97%) in P. roqueforti LCP9604111 (Table 1). This efficiency is comparable with previous findings in A. niger [50]. The total amount of transformants obtained per transformation experiment was consistently lower in P. roqueforti (n = 10 per plate) when compared to P. variotii (n = 100 per plate), which is probably due to less efficient protoplasting of P. roqueforti using adjusted A. niger protocols. As such, the protoplasting protocol for P. roqueforti should be further optimized.
The low efficiency of hygromycin loss after one round of non-selective growth in P. variotii transformants
When analyzing 40 P. variotii CBS101075 pvpP− transformants with white conidia, only 9 out of 40 (23%) lost the hygromycin resistance phenotype after one round of non-selective growth. This is in contrast to 40 P. variotii transformants with the wild-type brown conidia, where 32 out of 40 (80%) lost the hygromycin resistance phenotype after one round of non-selective growth. Also, when transforming P. variotii CBS101075 with empty vector pFC332 a similar 17 out of 24 (71%) ratio of transformants that lose their hygromycin resistant phenotype after one round of non-selective growth is obtained. In A. niger the reported hygromycin loss efficiency is also around 80% [50]. This suggests that having a white-coloured phenotype in P. variotii transformants is tied to retaining the hygromycin resistant phenotype, which could be explained by integration of the hygromycin resistance marker on the pvpP locus itself where the double stranded break occurs. Indeed, further investigation of P. variotii transformants revealed that a significant portion of at least 4 out of 11 (36%) transformants with white conidia contained pieces of the pPT13.1 plasmid at the site where the double stranded break had occurred (Table 2). This observation suggests that the double stranded break in the genomic DNA of P. variotii is somehow repaired using the CRISPR/Cas9 containing vector itself. The integration of the AMA1-based vector containing the hygromycin selection marker explains the stability of the hygromycin marker even under non-selective conditions in P. variotii. Since the hygromycin loss efficiency in P. roqueforti is also low (29%, see Table 1), we speculate that the same plasmid integration is happening in P.roqueforti transformations.
Obtaining a transformant with a mutation in the targeted gene is highly efficient, thus this methodology suffices when a single gene disrupted mutant is made. However, the high stability of the hygromycin marker limits the possibility for sequential rounds of transformations. Additionally, this methodology does not allow for efficiently obtaining full knock-out mutants, nor the efficient targeted integration of new pieces of DNA (for example to produce heterologous proteins) as the NHEJ repair mechanism is the preferred DNA repair mechanism over HDR in most filamentous fungi [6]. Disrupting the NHEJ repair mechanism is therefore beneficial as it could potentially circumvents all these drawbacks. Therefore, the created kusA− mutants that are incapable of performing NHEJ are valuable tools for future genome editing in both P. variotii and P. roqueforti.
The impact of a disrupted NHEJ repair mechanism on the genome editing efficiency in P. variotii (kusA−)
It was noticed that in the process of obtaining the kusA− mutant in P. variotii DTO217-A2, only 1 out of 26 (3%) of transformants lost the hygromycin resistance phenotype after one round of non-selective growth. This again stresses the inefficiency of obtaining a marker free mutant after one round of growth on non-selective media when transforming a kusA+ wild-type P. variotii strain with a AMA1-based CRISPR/Cas9 plasmid as previously seen when isolating melanin mutants (Table 1). After obtaining this single P. variotii PT39.26 (kusA−) strain, we investigated its genome editing efficiency. P. variotii PT39.26 (kusA−) was transformed using the pPT13.1 plasmid (pFC332 containing a pvpP targeting sgRNA) together with the addition of donor DNA consisting of the fused flanks of the pvpP gene. Obtaining white-coloured transformants in this transformation was highly efficient as 15 out of the 15 transformants produced white spores. The percentage of these transformants that lost their hygromycin resistant phenotype after one round of non-selective growth was also highly efficient, 14 out of 15 (93%). This is a major improvement over the 1 out of 26 (3%) ratio of hygromycin resistance loss mentioned earlier when creating the kusA− strain. Further analysis of eight of these white-coloured P. variotii transformants revealed that all eight transformants lost the complete pvpP gene (6677 bps) as checked by diagnostic PCR (Additional file 4: Figure S3), indicating that repair by HDR had taken place. This was further confirmed by sequencing 2 out of the 8 transformants which showed the knock-out of the complete 6677 bps as expected. Thus, eight knock-out mutants were obtained where no chunks of pPT13.1 plasmid were integrated into the pvpP locus. Taken together, the increased efficiency in hygromycin resistance loss and the absence of pPT13.1 chunks on the pvpP locus in transformants obtained from the P. variotii PT39.26 kusA− strain indicate that by disrupting the NHEJ repair mechanism in P. variotii DTO217-A2 the integration of the AMA1 containing vector into the genome is prevented or severely reduced. Therefore, we conclude that the high degree of plasmid integration into the target site where DSB took place was due to repair facilitated by the NHEJ mechanism. Increasing overall efficiency of obtaining genetic alterations by HDR will open up future research for generating full knock-out mutants as well as targeted integration of DNA (e.g. fluorescent proteins or non-native enzymes). The kusA− strains can be safely used for strain development as genome stability is not severely altered in NHEJ disrupted filamentous fungi [55].
In future research, it could be preferred to restore the kusA locus after complete knock-out of the target genes has been confirmed. For example, conidia of the ΔkusA mutant of A. niger show increased sensitivity to UV radiation resistance [8]. Thus, in order to investigate the impact of pvpP in the UV-C radiation resistance of conidia obtained from P. variotii PT42.1 (kusA−, ΔpvpP) it would be desirable to restore the kusA gene back to wild-type. This can be done by performing the same CRISPR/Cas9 genome editing system described for the creation of the ΔpvpP, but changing the target sequence to target the disrupted kusA locus and provide donor DNA with the intact wild-type kusA gene and its flanks. However, the restored wild-type kusA gene should not be recognized by the guide RNA targeting the disrupted kusA locus. This can be achieved by either targeting the indel itself, or incorporating silent point mutations in the donor DNA.
The contribution of melanin on the heat resistance and UV radiation resistance of conidia in three food spoiling fungi
There have been reports on yeast species in which melanization correlates to heat resistance [56, 57]. However, no altered heat resistance was observed in the conidia of the three food spoiling filamentous fungi with disrupted melanin biosynthesis. It has been previously shown that the conidia of A. niger strain MA93.1 have no altered heat inactivation resistance when compared to the parental strain N402 [58]. Here we similarly show no heat inactivation alterations for the mutant strains P. roqueforti PT34.2 and P. variotii PT32.5 when compared to their parental strains. These results suggests that melanin does not play a significant role in conidial heat resistance in these food spoiling fungi. In contrast, conidia disrupted in their melanin production showed increased susceptibility to UV-C radiation in all three food spoiling fungi. The UV-C radiation resistance of A. niger MA93.1 is lowered compared to the parental strain, as was previously shown [58]. Additionally, here we show that a gene disruption in the pksA homologue of P. roqueforti LCP9604111 and the pvpP gene in P. variotii CBS101075 also reduce UV-C radiation resistance of the conidia from these mutants. Interestingly, this result is in contrast to the polyketide synthase (alb1) knockdown mutant of Penicillium marneffei, which did not show altered UV-C radiation resistance [48]. It is apparent that the melanin deficient A. niger MA93.1 strain is more UV-C radiation resistant than either P. roqueforti or P. variotii wild-type strains (p < 0.00 student’s t-test). This suggests that the remaining pigmentation in the A. niger MA93.1 strain is a significant factor in UV-C radiation resistance, explaining the significant differences in resistance between A. niger and both the P. roqueforti and P. variotii strains (Fig. 4). The A. niger conidia seem to have an additional type of pigmentation that does not require the functionality of the FwnA enzyme. Which type of pigmentation still remains in A. niger MA93.1 is currently unknown. There are reports of several Aspergillus species producing other melanin types besides DHN-melanin, such as DOPA-melanin or pyomelanin [59, 60], which would make likely candidates. A total of eight different melanin types have been described for fungi alone, each with their own distinct biosynthesis pathway [61]. The relation between the type of fungal melanin and subsequent UV radiation and heat resistance is currently unknown. Additionally, we noted that UV radiation resistance of the three melanin deficient mutants is about two-thirds of their wild-type parental strains in each species. This finding suggests that the relative contribution of the PKS produced pigmentation to the UV radiation resistance of conidia is similar in each species.
Conclusions
We have shown the successful implementation of the AMA1-based CRISPR/Cas9 genome editing system in P. variotii and P. roqueforti, which is capable of creating indels in the targeted gene. However, we observed a large amount of plasmid integration events when using the AMA1-based plasmids in this way, resulting in mutant strains which are often no longer marker-free. We demonstrate that a kusA− background can be used to prevent or otherwise severely reduce plasmid integration, which allows for efficient marker-free genome editing and additionally facilitates the creation of complete knock-outs by relying on the homology directed DNA repair mechanism.
We have used the AMA1-based CRISPR/Cas9 plasmids to create melanin deficient mutants of P. variotii and P. roqueforti, in order to analyse their preservation stress resistance. We show that the melanin-lacking conidia of food spoilers P. variotii, P. roqueforti and A. niger are not altered in their heat resistance compared to their parental strains. In contrast, mutant conidia of food spoilers P. variotii, P. roqueforti and A. niger have increased sensitivity towards UV-C radiation. As such, the presence of DHN-melanin in conidia of three food spoiling fungi does not contribute to their heat resistance, but does contribute to their UV-C radiation resistance.
Materials and methods
Strains, growth conditions, spore harvesting, media and molecular techniques
The A. niger, P. roqueforti and P. variotii strains used in this study are listed in Table 5.
Table 5.
Strains used in this study
| Strain name | Genotype | Reference |
|---|---|---|
| Aspergillus niger N402 | cspA1 | [62] |
| Aspergillus niger MA93.1 | cspA1, fwnA::hygB in N402 | [47] |
| Paecilomyces variotii CBS101075 | wild-type | [22] |
| Paecilomyces variotii PT32.5 | pvpP− in CBS101075 | This study |
| Paecilomyces variotii DTO217-A2 | wild-type | [10] |
| Paecilomyces variotii PT39.26 | kusA− in DTO217-A2 | This study |
| Paecilomyces variotii PT42.1 | kusA−, ΔpvpP in DTO217-A2 | This study |
| Penicillium roqueforti LCP9604111 | wild-type | [54] |
| Penicillium roqueforti PT34.2 | pksA− in LCP9604111 | This study |
| Penicillium roqueforti DTO013-F5 | wild-type | Westerdijk Fungal Biodiversity Institute, CBS collection |
| Penicillium roqueforti PT43.1 | kusA− in DTO013-F5 | This study |
The Escherichia coli strain DH5α was used for cloning purposes. Fungal strains were grown for 7 days at 25 °C on malt extract agar (MEA) unless noted otherwise. All media used and spore harvesting methods are described by Arentshorst et al. [63]. Standard PCR and E. coli cloning techniques were used according to Sambrook et al. [64]. Spore suspensions were made using physiological salt buffer (0.9% NaCl + 0.02% Tween 80 in demiwater) unless noted otherwise. The P. roqueforti strains were harvested and washed in ACES buffer (10 mM N‐(2‐acetamido)‐2‐aminoethanesulfonic acid, 0.02% Tween 80, pH 6.8) according to van den Brule et al. [10].
CRISPR/Cas9 plasmids construction
All plasmids and primers used in this study are listed in the supplementary data (Additional file 1: Table S1, Additional file 6: Table S2). In silico work was performed on FASTA files obtained from JGI [65]. Plasmid construction was based on earlier work performed in A. niger [50]. A detailed protocol on the CRISPR/Cas9 plasmid construction and subsequent transformations in P. variotii and P. roqueforti can be found in the supplementary data (Additional file 7: Protocol S1). Briefly, the plasmids pTLL108.1 and pTLL109.2 were used as templates for creating the 5′ flank and 3′ flank of the sgRNA respectively. After fusion PCR using the pTE1_for and pTE1_rv primers, a PacI (Fermentas, Thermo Scientific™) digestion on the purified PCR product was performed O/N. The PacI digested sgRNA was ligated into a PacI digested and dephosphorylated pFC332 plasmid and subsequently cloned into E. coli DH5α. The ampicillin resistant colonies were grown under selective pressure overnight and miniprepped (GeneJET Plasmid Miniprep Kit, Thermo Scientific™), after which restriction analysis was done with SacII (Fermentas, Thermo Scientific™) to check for correct insertion of the sgRNA. Lastly, sequencing was performed as a final check to ensure correct sgRNA is present in the newly constructed plasmid.
Transformation protocol and DNA isolation
Fungal transformations were performed according to van Leeuwe et al. [50] with a few adaptations. Hygromycin concentrations used for selection during transformation were adjusted per species, chosen based on the lowest concentration still preventing background growth. As such, the final concentrations used were 100 µg/ml hygromycin for P. roqueforti transformations and 200 µg/ml hygromycin for P. variotii transformations. Since these wild-type strains were kusA+, gene disruptions relied on non-homologous end joining (NHEJ) for repair resulting in the creation of indels, see van Leeuwe et al. [50] for more information. In contrast, after the P. variotii PT39.26 (kusA−) strain was obtained transformation with pPT13.1 was done with the addition of a repair DNA fragment to obtain P. variotii PT42.1 (kusA−, ΔpvpP), see Results section. The mycelium for protoplasting and subsequent transformation of P. roqueforti was pre-grown in CM for 2 days at 25 °C instead of 1 day at 30 °C for A. niger and P. variotii. Protoplasting was done in SMC medium with Lysing enzymes (Sigma) essentially as described previously [63]. Protoplast formation was checked by light microscopy every 15 min for both P. roqueforti and P. variotii. The protoplasting process was commonly stopped after 45 min, when protoplasts were visually present. Genomic DNA isolations were done according to Arentshorst et al. [63].
Heat resistance assay
The heat inactivation assay was based on van den Brule et al. [10] with few exceptions. At t = 0 a total of 200 µl spore suspension of 1*10^8 conidia/ml was added to pre-heated 19.8 mL ACES buffer (P. roqueforti) or 19.8 mL physiological salt buffer (P. variotii and A. niger). The temperatures of the water bath were adjusted per species (56 °C A. niger, 56 °C P. roqueforti, 60 °C P. variotii). The P. variotii and A. niger conidia were treated in a static water bath with magnetic stirring (Julabo Corio C-BT19) at 180 rpm inside 50 ml Erlenmeyers. The P. roqueforti conidia were treated in a shaking water bath (Grant OLS200) at 100 rpm. Samples were taken every 2 min until t = 20 min. Additionally t = 30 min was taken as a final sample. Heat inactivation curves and standard deviations were made based on three biological replicates. Dilutions were made in either ACES buffer (P. roqueforti) or physiological salt buffer (P. variotii and A. niger) corresponding with their heating menstruum. Spore suspensions were plated on MEA plates for colony counting. The colony forming units (CFUs) were counted after 7 days of growth at 25 °C. Decimal reduction values (D-values) were calculated using the linear regression model.
UV-C radiation resistance assay
The UV-C radiation resistance assay was done in a UV crosslinker (Hoefer UVC 500 Ultraviolet Croslinker). A total of 2*10^7 conidia per mL were UV exposed inside open Petri dishes (total starting volume = 25 mL). After each UV dose, 1 mL of spore suspension was taken and subsequently serially diluted and plated on MEA plates. Survival was measured by CFUs scored after 7 days. The lowest dose applied was 7.5 mJ/cm2 and then increased by 7.5 mJ/cm2 in a stepwise manner with a maximum dose of 82.5 mJ/cm2. The UV radiation resistance assays were performed in biological triplicates. Decimal reduction values (D-values) were calculated based on the linear regression model. Significance was tested using an unpaired Student’s t-test (significant if p < 0.05).
Supplementary Information
Additional file 1: Table S1. Plasmids used in this study.
Additional file 2: Figure S1. Indels found in P. variotii PT32.5 and P. roqueforti PT34.2. Sequencing results of pksA genes after mutation caused by CRISPR/Cas9 double stranded break and subsequently repair with the NHEJ repair machinery.a Indel of 14 bps in the pvpP gene in P. variotii causing a frameshift soon after the start codon. This frameshift likely disrupts gene function. b Indel of 14 bps in the pksA gene of P. roqueforti causing a frameshift soon after the start codon. This frameshift likely disrupts gene function. Pictures were made using Benchling [Biology Software] (2020). Retrieved from https://benchling.com.
Additional file 3: Figure S2. Indels found in kusA− strains P. variotii PT39.26 and P. roqueforti PT43.1.Sequencing results of kusA genes after mutation caused by CRISPR/Cas9 double stranded break and subsequently repair with the NHEJ repair machinery. a Indel of 7 bps in the kusA gene in P. variotii PT39.26 causing a frameshift soon after the start codon. This frameshift likely disrupts gene function. b Indel of 22 bps in the kusA gene of P. roqueforti PT43.1 causing a frameshift soon after the start codon. This frameshift likely disrupts gene function. Pictures were made using Benchling [Biology Software] (2020). Retrieved from https://benchling.com.
Additional file 4: Figure S3. Diagnostic PCR on eight transformants in P. variotii PT39.26 missing the pvpP gene. a Diagnostic PCR to investigate the presence of the pvpP gene by amplifying outside the used flanks. If the gene is absent a band size of 2619 bps is expected. If the gene pvpP is still present, a band size of 9344 bps is present. The eight transformants all have lost the pvpP gene. The PCR fragments loaded on #6 and #7 were purified and subsequently send for sequencing. b Diagnostic PCR to investigate the presence of the pvpP gene by amplifying inside the gene. If the gene is absent, no band is expected. In wild-type situation, a PCR fragment of 1095 bps is expected. No transformants show the presence of pvpP gene. Taken together, these results show that 8/8 transformants had a full knock-out of the pvpP gene. Both contained the expected sequence for the repair DNA fragment, indicating repair by HDR.
Additional file 5: Figure S4. Growth rate of P. variotii and P. roqueforti strains. Colony diameters were measured of P. variotii and P. roqueforti strains growing 2-5 days at 25°C point-inoculated on MEA plates. a Colony diameters of strains plotted against time. The average growth rate in millimeters was estimated by performing a best fit using linear regression. The slope represents the average millimeter increase in diameter per day. On average, all P. variotii strains increased 18-19 mm in diameter per day and all P. roqueforti strains increased 10-11 mm in diameter per day. No differences were observed between mutant strains and their parental strains. b Morphology of P. roqueforti strains growing on MEA plates. No differences are visible between mutant strains and their parental strains except for the expected change in spore coloration. c Morphology of P. variotii strains growing on MEA plates. No differences are visible between mutant strains and their parental strains except for the expected change in spore coloration
Additional file 6: Table S2. Primers used in this study.
Additional file 7. Protocol S1. A detailed protocol for CRISPR/Cas9 mediated gene knock-out in Penicillium roqueforti and Paecilomyces variotii.
Acknowledgements
We would like to thank Prof Dr. Han de Winde (Leiden University) for his contributions during work discussions and his comments on the drafted manuscript.
Authors' contributions
SJS and PPPT designed and performed most of the experiments. MP and HABW designed and performed the heat resistance assays of P. roqueforti and provided expertise on P. roqueforti where needed. Similarly, TvdB, JH and JD designed and performed the heat resistance assays of P. variotii and provided expertise on P. variotii where needed. All authors (SJS, PPPT, MP, TvdB, JH, JD, HABW and AFJR) contributed to data interpretation and writing of the final version of the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the TIFN project “Heterogeneity in spores of food spoilage fungi” (AF-15507).
Availability of data and materials
All datasets supporting the results of this article are included within the article or in the additional files. Plasmids and strains are available upon request. Strains have been deposited at the KNAW strain archive.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shi TQ, Liu GN, Ji RY, Shi K, Song P, Ren LJ, et al. CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl Microbiol Biotechnol. 2017;101:7435–7443. doi: 10.1007/s00253-017-8497-9. [DOI] [PubMed] [Google Scholar]
- 2.Song R, Zhai Q, Sun L, Huang E, Zhang Y, Zhu Y, et al. CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Appl Microbiol Biotechnol. 2019;103:6919–6932. doi: 10.1007/s00253-019-10007-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ouedraogo JP, Tsang A. CRISPR_Cas systems for fungal research. Fungal Biol Rev. 2020;34:189–201. doi: 10.1016/j.fbr.2020.10.002. [DOI] [Google Scholar]
- 4.Muñoz IV, Sarrocco S, Malfatti L, Baroncelli R, Vannacci G. CRISPR-CAS for fungal genome editing: a new tool for the management of plant diseases. Front Plant Sci. 2019;10:135. doi: 10.3389/fpls.2019.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster M, Kahmann R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet Biol. 2019;130:43–53. doi: 10.1016/j.fgb.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Krappmann S. Gene targeting in filamentous fungi: the benefits of impaired repair. Fungal Biol Rev. 2007;21:25–29. doi: 10.1016/j.fbr.2007.02.004. [DOI] [Google Scholar]
- 7.Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci U S A. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer V, Arentshorst M, El-Ghezal A, Drews AC, Kooistra R, van den Hondel CAMJJ, et al. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J Biotechnol. 2007;128:770–775. doi: 10.1016/j.jbiotec.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Houbraken J, Samson RA, Frisvad JC. Byssochlamys: significance of heat resistance and mycotoxin production. Adv Exp Med Biol. 2006;571:211–224. doi: 10.1007/0-387-28391-9_14. [DOI] [PubMed] [Google Scholar]
- 10.van den Brule T, Punt M, Teertstra W, Houbraken J, Wösten H, Dijksterhuis J. The most heat-resistant conidia observed to date are formed by distinct strains of Paecilomyces variotii. Environ Microbiol. 2019;22:986–999. doi: 10.1111/1462-2920.14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houbraken J, Varga J, Rico-Munoz E, Johnson S, Samson RA. Sexual reproduction as the cause of heat resistance in the food spoilage fungus Byssochlamys spectabilis (anamorph Paecilomyces variotii) Appl Environ Microbiol. 2008;74:1613–1619. doi: 10.1128/AEM.01761-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dantigny P, Guilmart A, Radoi F, Bensoussan M, Zwietering M. Modelling the effect of ethanol on growth rate of food spoilage moulds. Int J Food Microbiol. 2005;98:261–269. doi: 10.1016/j.ijfoodmicro.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Piecková E, Samson RA. Heat resistance of Paecilomyces variotii in sauce and juice. J Ind Microbiol Biotechnol. 2000;24:227–230. doi: 10.1038/sj.jim.2900794. [DOI] [Google Scholar]
- 14.Pitt JI, Hocking AD. Fungi and food spoilage. 3. New York: Springer, US; 2009. [Google Scholar]
- 15.Guimarães LHS, Peixoto-Nogueira SC, Michelin M, Rizzatti ACS, Sandrim VC, Zanoelo FF, et al. Screening of filamentous fungi for production of enzymes of biotechnological interest. Brazilian J Microbiol. 2006;37:474–480. doi: 10.1590/S1517-83822006000400014. [DOI] [Google Scholar]
- 16.Battestin V, Macedo GA. Tannase production by Paecilomyces variotii. Bioresour Technol. 2007;98:1832–1837. doi: 10.1016/j.biortech.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 17.Kondo T, Morikawa Y, Hayashi N. Purification and characterization of alcohol oxidase from Paecilomyces variotii isolated as a formaldehyde-resistant fungus. Appl Microbiol Biotechnol. 2008;77:995–1002. doi: 10.1007/s00253-007-1237-9. [DOI] [PubMed] [Google Scholar]
- 18.Job J, Sukumaran RK, Jayachandran K. Production of a highly glucose tolerant β-glucosidase by Paecilomyces variotii MG3: optimization of fermentation conditions using Plackett-Burman and Box-Behnken experimental designs. World J Microbiol Biotechnol. 2010;26:1385–1391. doi: 10.1007/s11274-010-0311-0. [DOI] [Google Scholar]
- 19.Michelin M, Silva TM, Benassi VM, Peixoto-Nogueira SC, Moraes LAB, Leão JM, et al. Purification and characterization of a thermostable α-amylase produced by the fungus Paecilomyces variotii. Carbohydr Res. 2010;345:2348–2353. doi: 10.1016/j.carres.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 20.de Herrera Bravo Laguna I, Toledo Marante FJ, Mioso R. Enzymes and bioproducts produced by the ascomycete fungus Paecilomyces variotii. J Appl Microbiol. 2015;119:1455–1466. doi: 10.1111/jam.12934. [DOI] [PubMed] [Google Scholar]
- 21.Battestin V, Macedo GA, De Freitas VAP. Hydrolysis of epigallocatechin gallate using a tannase from Paecilomyces variotii. Food Chem. 2008;108:228–233. doi: 10.1016/j.foodchem.2007.10.068. [DOI] [Google Scholar]
- 22.Urquhart AS, Mondo SJ, Mäkelä MR, Hane JK, Wiebenga A, He G, et al. Genomic and genetic insights into a cosmopolitan fungus, Paecilomyces variotii (Eurotiales) Front Microbiol. 2018;9:3058. doi: 10.3389/fmicb.2018.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idnurm A, Bailey AM, Cairns TC, Elliott CE, Foster GD, Ianiri G, et al. A silver bullet in a golden age of functional genomics: the impact of agrobacterium-mediated transformation of fungi. Fungal Biol Biotechnol. 2017;4:1–28. doi: 10.1186/s40694-016-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D, Tang Y, Lin J, Cai W. Methods for genetic transformation of filamentous fungi. Microb Cell Fact. 2017;16:1–13. doi: 10.1186/s12934-016-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martín JF, Coton M. Blue Cheese: Microbiota and Fungal Metabolites. In: Fermented Foods Heal Dis Prev. 1st ed. Elsevier; 2017. 10.1016/B978-0-12-802309-9.00012-1
- 26.Dubey MK, Aamir M, Kaushik MS, Khare S, Meena M, Singh S, et al. PR Toxin - biosynthesis, genetic regulation, toxicological potential, prevention and control measures: Overview and challenges. Front Pharmacol. 2018;9:288. doi: 10.3389/fphar.2018.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallo A, Giuberti G, Bertuzzi T, Moschini M, Masoero F. Study of the effects of PR toxin, mycophenolic acid and roquefortine C on in vitro gas production parameters and their stability in the rumen environment. J Agric Sci. 2015;153:163–176. doi: 10.1017/S0021859614000343. [DOI] [Google Scholar]
- 28.Fernández-Bodega MA, Mauriz E, Gómez A, Martín JF. Proteolytic activity, mycotoxins and andrastin A in Penicillium roqueforti strains isolated from Cabrales, Valdeón and Bejes-Tresviso local varieties of blue-veined cheeses. Int J Food Microbiol. 2009;136:18–25. doi: 10.1016/j.ijfoodmicro.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Finoli C, Vecchio A, Galli A, Dragoni I. Roquefortine C occurrence in blue cheese. J Food Prot. 2001;64:246–251. doi: 10.4315/0362-028X-64.2.246. [DOI] [PubMed] [Google Scholar]
- 30.Larsen TO, Gareis M, Frisvad JC. Cell cytotoxicity and mycotoxin and secondary metabolite production by common penicillia on cheese agar. J Agric Food Chem. 2002;50:6148–6152. doi: 10.1021/jf020453i. [DOI] [PubMed] [Google Scholar]
- 31.Ohmomo S, Sato T, Utagawa T, Abe M. Isolation of Festuclavine and Three New Indole Alkaloids, Roquefortine A, B and C from the Cultures of Penicillium Roqueforti. Nippon Någeikagaku Kaishi. 1975;39:1333–1334. [Google Scholar]
- 32.Proctor RH, Hohn TM. Aristolochene synthase. Isolation, characterization, and bacterial expression of a sesquiterpenoid biosynthetic gene (Ari1) from Penicillium roqueforti. J Biol Chem. 1993;268:4543–4548. doi: 10.1016/S0021-9258(18)53644-9. [DOI] [PubMed] [Google Scholar]
- 33.Kinsella JE, Hwang D. Biosynthesis of flavors by Penicillium roqueforti. Biotechnol Bioeng. 1976;18:927–938. doi: 10.1002/bit.260180706. [DOI] [Google Scholar]
- 34.Mioso R, Toledo Marante FJ. Herrera Bravo de Laguna I. Penicillium roqueforti: a multifunctional cell factory of high value-added molecules. J Appl Microbiol. 2015;118:781–791. doi: 10.1111/jam.12706. [DOI] [PubMed] [Google Scholar]
- 35.Ismaiel AA, Ahmed AS, El-Sayed ESR. Optimization of submerged fermentation conditions for immunosuppressant mycophenolic acid production by Penicillium roqueforti isolated from blue-molded cheeses: enhanced production by ultraviolet and gamma irradiation. World J Microbiol Biotechnol. 2014;30:2625–2638. doi: 10.1007/s11274-014-1685-1. [DOI] [PubMed] [Google Scholar]
- 36.García-Estrada C, Martín JF. Biosynthetic gene clusters for relevant secondary metabolites produced by Penicillium roqueforti in blue cheeses. Appl Microbiol Biotechnol. 2016;100:8303–8313. doi: 10.1007/s00253-016-7788-x. [DOI] [PubMed] [Google Scholar]
- 37.Coton E, Coton M, Hymery N, Mounier J, Jany JL. Penicillium roqueforti: an overview of its genetics, physiology, metabolism and biotechnological applications. Fungal Biol Rev. 2020;34:59–73. doi: 10.1016/j.fbr.2020.03.001. [DOI] [Google Scholar]
- 38.Pohl C, Kiel JAKW, Driessen AJM, Bovenberg RAL, Nygård Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth Biol. 2016;5:754–764. doi: 10.1021/acssynbio.6b00082. [DOI] [PubMed] [Google Scholar]
- 39.Guzmán-Chávez F, Zwahlen RD, Bovenberg RAL, Driessen AJM. Engineering of the filamentous fungus Penicillium chrysogenum as cell factory for natural products. Front Microbiol. 2018;9:2768. doi: 10.3389/fmicb.2018.02768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerrero-Beltrán JA, Barbosa-Cánovas GV. Review: advantages and limitations on processing foods by UV light. Food Sci Technol Int. 2004;10:137–147. doi: 10.1177/1082013204044359. [DOI] [Google Scholar]
- 41.Mañas P, Pagán R. Microbial inactivation by new technologies of food preservation. J Appl Microbiol. 2005;98:1387–1399. doi: 10.1111/j.1365-2672.2005.02561.x. [DOI] [PubMed] [Google Scholar]
- 42.Can FO, Demirci A, Puri V, Gourama H. Decontamination of hard cheeses by pulsed UV-light. Am Soc Agric Biol Eng Annu Int Meet. 2014;77:1723–1731. doi: 10.4315/0362-028X.JFP-13-559. [DOI] [PubMed] [Google Scholar]
- 43.Keklik NM, Krishnamurthy K, Demirci A. Microbial decontamination of food by ultraviolet (UV) and pulsed UV light. Microbial decontamination in the food industry, 1st edition. London: Elsevier Ltd; 2012. p. 344–69. 10.1533/9780857095756.2.344
- 44.Begum M, Hocking AD, Miskelly D. Inactivation of food spoilage fungi by ultra violet (UVC) irradiation. Int J Food Microbiol. 2009;129:74–77. doi: 10.1016/j.ijfoodmicro.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 45.Put HMC, De JJ. The heat resistance of ascospores of four Saccharomyces spp. isolated from spoiled heat-processed soft drinks and fruit products. J Appl Bacteriol. 1982;52:235–243. doi: 10.1111/j.1365-2672.1982.tb04845.x. [DOI] [PubMed] [Google Scholar]
- 46.Tsai HF, Wheeler MH, Chang YC, Kwon-Chung KJ. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J Bacteriol. 1999;181:6469–6477. doi: 10.1128/JB.181.20.6469-6477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jørgensen TR, Park J, Arentshorst M, van Welzen AM, Lamers G, VanKuyk PA, et al. The molecular and genetic basis of conidial pigmentation in Aspergillus niger. Fungal Genet Biol. 2011;48:544–553. doi: 10.1016/j.fgb.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Woo PCY, Tam EWT, Chong KTK, Cai JJ, Tung ETK, Ngan AHY, et al. High diversity of polyketide synthase genes and the melanin biosynthesis gene cluster in Penicillium marneffei. FEBS J. 2010;277:3750–3758. doi: 10.1111/j.1742-4658.2010.07776.x. [DOI] [PubMed] [Google Scholar]
- 49.Urquhart AS, Hu J, Chooi YH, Idnurm A. The fungal gene cluster for biosynthesis of the antibacterial agent viriditoxin. Fungal Biol Biotechnol. 2019;6:1–13. doi: 10.1186/s40694-019-0072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Leeuwe TM, Arentshorst M, Ernst T, Alazi E, Punt PJ, Ram AFJ. Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi. Fungal Biol Biotechnol. 2019;6:1–13. doi: 10.1186/s40694-018-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nødvig CS, Nielsen JB, Kogle ME, Mortensen UH. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE. 2015;10:e0133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gems D, Johnstone IL, Clutterbuck AJ. An autonomously replicating plasmid transforms Aspergillus nidulans at high frequency. Gene. 1991;98:61–67. doi: 10.1016/0378-1119(91)90104-J. [DOI] [PubMed] [Google Scholar]
- 53.Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:171–174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Punt M, van den Brule T, Teertstra WR, Dijksterhuis J, den Besten HMW, Ohm RA, et al. Impact of maturation and growth temperature on cell-size distribution, heat-resistance, compatible solute composition and transcription profiles of Penicillium roqueforti conidia. Food Res Int. 2020;136:109287. doi: 10.1016/j.foodres.2020.109287. [DOI] [PubMed] [Google Scholar]
- 55.Álvarez-Escribano I, Sasse C, Bok JW, Na H, Amirebrahimi M, Lipzen A, et al. Genome sequencing of evolved aspergilli populations reveals robust genomes, transversions in A. flavus, and sexual aberrancy in non-homologous end-joining mutants. BMC Biol. 2019;17:1–17. doi: 10.1186/s12915-019-0702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosas ÁL, Casadevall A. Melanization affects susceptibility of Cryptococcus neoformans to heat and cold. FEMS Microbiol Lett. 1997;153:265–272. doi: 10.1016/S0378-1097(97)00239-5. [DOI] [PubMed] [Google Scholar]
- 57.Jiang H, Liu NN, Liu GL, Chi Z, Wang JM, Zhang LL, et al. Melanin production by a yeast strain XJ5-1 of Aureobasidium melanogenum isolated from the Taklimakan desert and its role in the yeast survival in stress environments. Extremophiles. 2016;20:567–577. doi: 10.1007/s00792-016-0843-9. [DOI] [PubMed] [Google Scholar]
- 58.Esbelin J, Mallea S, Ram AFJ, Carlin F. Role of pigmentation in protecting Aspergillus niger conidiospores against pulsed light radiation. Photochem Photobiol. 2013;89:758–761. doi: 10.1111/php.12037. [DOI] [PubMed] [Google Scholar]
- 59.Schmaler-Ripcke J, Sugareva V, Gebhardt P, Winkler R, Kniemeyer O, Heinekamp T, et al. Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus. Appl Environ Microbiol. 2009;75:493–503. doi: 10.1128/AEM.02077-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonçalves RCR, Lisboa HCF, Pombeiro-Sponchiado SR. Characterization of melanin pigment produced by Aspergillus nidulans. World J Microbiol Biotechnol. 2012;28:1467–1474. doi: 10.1007/s11274-011-0948-3. [DOI] [PubMed] [Google Scholar]
- 61.Toledo AV, Franco MEE, Yanil Lopez SM, Troncozo MI, Saparrat MCN, Balatti PA. Melanins in fungi: Types, localization and putative biological roles. Physiol Mol Plant Pathol. 2017;99:2–6. doi: 10.1016/j.pmpp.2017.04.004. [DOI] [Google Scholar]
- 62.Bos CJ, Debets AJM, Swart K, Huybers A, Kobus G, Slakhorst SM. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr Genet. 1988;14:437–443. doi: 10.1007/BF00521266. [DOI] [PubMed] [Google Scholar]
- 63.Arentshorst M, Ram AFJ, Meyer V. Using non-homologous end-joining-deficient strains for functional gene analyses in filamentous fungi. Methods Mol Biol. 2012;835:133–150. doi: 10.1007/978-1-61779-501-5_9. [DOI] [PubMed] [Google Scholar]
- 64.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. New York: CSHL Press; 2001. [Google Scholar]
- 65.Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014;42:26–31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Plasmids used in this study.
Additional file 2: Figure S1. Indels found in P. variotii PT32.5 and P. roqueforti PT34.2. Sequencing results of pksA genes after mutation caused by CRISPR/Cas9 double stranded break and subsequently repair with the NHEJ repair machinery.a Indel of 14 bps in the pvpP gene in P. variotii causing a frameshift soon after the start codon. This frameshift likely disrupts gene function. b Indel of 14 bps in the pksA gene of P. roqueforti causing a frameshift soon after the start codon. This frameshift likely disrupts gene function. Pictures were made using Benchling [Biology Software] (2020). Retrieved from https://benchling.com.
Additional file 3: Figure S2. Indels found in kusA− strains P. variotii PT39.26 and P. roqueforti PT43.1.Sequencing results of kusA genes after mutation caused by CRISPR/Cas9 double stranded break and subsequently repair with the NHEJ repair machinery. a Indel of 7 bps in the kusA gene in P. variotii PT39.26 causing a frameshift soon after the start codon. This frameshift likely disrupts gene function. b Indel of 22 bps in the kusA gene of P. roqueforti PT43.1 causing a frameshift soon after the start codon. This frameshift likely disrupts gene function. Pictures were made using Benchling [Biology Software] (2020). Retrieved from https://benchling.com.
Additional file 4: Figure S3. Diagnostic PCR on eight transformants in P. variotii PT39.26 missing the pvpP gene. a Diagnostic PCR to investigate the presence of the pvpP gene by amplifying outside the used flanks. If the gene is absent a band size of 2619 bps is expected. If the gene pvpP is still present, a band size of 9344 bps is present. The eight transformants all have lost the pvpP gene. The PCR fragments loaded on #6 and #7 were purified and subsequently send for sequencing. b Diagnostic PCR to investigate the presence of the pvpP gene by amplifying inside the gene. If the gene is absent, no band is expected. In wild-type situation, a PCR fragment of 1095 bps is expected. No transformants show the presence of pvpP gene. Taken together, these results show that 8/8 transformants had a full knock-out of the pvpP gene. Both contained the expected sequence for the repair DNA fragment, indicating repair by HDR.
Additional file 5: Figure S4. Growth rate of P. variotii and P. roqueforti strains. Colony diameters were measured of P. variotii and P. roqueforti strains growing 2-5 days at 25°C point-inoculated on MEA plates. a Colony diameters of strains plotted against time. The average growth rate in millimeters was estimated by performing a best fit using linear regression. The slope represents the average millimeter increase in diameter per day. On average, all P. variotii strains increased 18-19 mm in diameter per day and all P. roqueforti strains increased 10-11 mm in diameter per day. No differences were observed between mutant strains and their parental strains. b Morphology of P. roqueforti strains growing on MEA plates. No differences are visible between mutant strains and their parental strains except for the expected change in spore coloration. c Morphology of P. variotii strains growing on MEA plates. No differences are visible between mutant strains and their parental strains except for the expected change in spore coloration
Additional file 6: Table S2. Primers used in this study.
Additional file 7. Protocol S1. A detailed protocol for CRISPR/Cas9 mediated gene knock-out in Penicillium roqueforti and Paecilomyces variotii.
Data Availability Statement
All datasets supporting the results of this article are included within the article or in the additional files. Plasmids and strains are available upon request. Strains have been deposited at the KNAW strain archive.


