Significance
All mitochondria import most of their proteins from the cytosol. Even though the targeting signals of imported proteins are well conserved within eukaryotes, this is not the case for the mitochondrial outer membrane receptors that recognize these signals. Here we compare the substrate preferences of protein import receptors from the parasitic protozoans Trypanosoma brucei and Trichomonas vaginalis, as well as from yeast. Using biochemical and proteomic analysis, combined with complementation experiments, we show that evolutionarily unrelated receptors can share the same substrate preferences. Moreover, we provide evidence that receptors sharing the same domain structure and topology can have different substrate specificity. In summary, our study illustrates how determinism and contingencies have shaped the evolution of mitochondrial import receptors.
Keywords: mitochondria, protein import, receptors, Trypanosoma, Trichomonas
Abstract
Mitochondrial protein import requires outer membrane receptors that evolved independently in different lineages. Here we used quantitative proteomics and in vitro binding assays to investigate the substrate preferences of ATOM46 and ATOM69, the two mitochondrial import receptors of Trypanosoma brucei. The results show that ATOM46 prefers presequence-containing, hydrophilic proteins that lack transmembrane domains (TMDs), whereas ATOM69 prefers presequence-lacking, hydrophobic substrates that have TMDs. Thus, the ATOM46/yeast Tom20 and the ATOM69/yeast Tom70 pairs have similar substrate preferences. However, ATOM46 mainly uses electrostatic, and Tom20 hydrophobic, interactions for substrate binding. In vivo replacement of T. brucei ATOM46 by yeast Tom20 did not restore import. However, replacement of ATOM69 by the recently discovered Tom36 receptor of Trichomonas hydrogenosomes, while not allowing for growth, restored import of a large subset of trypanosomal proteins that lack TMDs. Thus, even though ATOM69 and Tom36 share the same domain structure and topology, they have different substrate preferences. The study establishes complementation experiments, combined with quantitative proteomics, as a highly versatile and sensitive method to compare in vivo preferences of protein import receptors. Moreover, it illustrates the role determinism and contingencies played in the evolution of mitochondrial protein import receptors.
Intracellular endosymbionts lack protein import systems, whereas such systems are a defining feature of mitochondria and plastids, both of which evolved from bacterial endosymbionts (1–3). Today, more than 95% of all mitochondrial proteins are imported from the cytosol, which makes mitochondrial protein import a key process required for mitochondrial biogenesis (4–6). The question of how mitochondrial protein import evolved is therefore central to understand how the endosymbiotic bacterial ancestor of mitochondria converted into an organelle that is genetically integrated into the host cell (7–9).
Proteins are targeted to mitochondria by internal or external import signals, the most frequent one of which is the N-terminal presequence found in 60 to 70% of all imported proteins (10, 11). Interestingly, the various mitochondrial import signals are conserved even between highly diverged eukaryotes (6). The import signals are decoded by receptors, which are integral mitochondrial outer membrane (OM) proteins that are associated with the heterooligomeric protein translocase of the OM (TOM complex) (6, 12). Contrary to the core components of the TOM complex (Tom40, Tom22, and Tom7), which are highly conserved in essentially all eukaryotes, these receptors evolved independently in different eukaryotic lineages, even though they recognize the same conserved import signals (6).
The best studied prototypical import receptors are Tom20 and Tom70 of yeast, orthologs of which are found in all members of the eukaryotic supergroup of the opisthokonts (13). Tom20 is an N-terminally anchored OM membrane protein, and its cytosolic domain contains a single tetratricopeptide repeat (TPR). Tom20 preferentially recognizes precursor proteins that have N-terminal presequences. It binds to the hydrophobic surface of the presequence and transfers the precursors to the highly conserved Tom22 that functions as a secondary receptor (14–17). Tom70 is the primary receptor for proteins that have multiple membrane spanning domains, such as mitochondrial carrier proteins, but also binds to hydrophobic precursor proteins that have presequences (18–20). Moreover, it has been shown that binding of Tom70 to the mitochondrial presequence-like stretches that are present in the mature part of many precursor proteins increases the import efficiency (21). Tom70 is N-terminally anchored in the membrane. Its large cytosolic domain consists of 11 TPR motifs. The three TPR motifs proximal to the membrane interact with cytosolic Hsp70 or Hsp90, from which Tom70 can receive precursor proteins (22, 23). The remaining eight TPR motifs directly recognize substrate proteins (24, 25). In yeast, Tom20 and Tom70 have partially redundant functions. Tom70 is not essential for growth and respiration. Loss of Tom20 causes a stronger phenotype; it abolishes respiration but is not lethal. Finally, even the deletion of Tom70 and Tom20 does not kill the cells, provided that the secondary receptor Tom22 is still present (15, 26–29).
A single import receptor, termed Tom20, is associated with the TOM complex of plant mitochondria. Yeast and plant Tom20 (30) are superficially similar: both have a single transmembrane domain (TMD) and a soluble domain containing one (in yeast) and two TPR motifs (in plants). Furthermore, both proteins have the same domain organization provided that they are aligned in an antiparallel way. Thus, whereas yeast Tom20 is N-terminally anchored, plant Tom20 is a C-terminally anchored protein. This strongly suggests that yeast and plant Tom20, while both being import receptors, have different evolutionary origins (31, 32). Moreover, plants have another TPR domain-containing OM protein, termed OM64, that is not associated with the TOM complex, but implicated in protein import (31, 33).
ATOM46 and ATOM69 are the two receptor subunits of the atypical translocase of the OM (ATOM) of trypanosomatids (34). ATOM69 is superficially similar to yeast Tom70. Both have the same molecular mass and multiple TPR-like motifs. ATOM69, in addition, has an N-terminal CS/Hsp20-like domain, which potentially can bind to cytosolic chaperones. Analogous to plant Tom20, ATOM69 is C-terminally membrane-anchored, whereas yeast Tom70 has an N-terminal TMD. ATOM46 also has an N-terminal membrane anchor and a cytosolic armadillo (ARM) repeat domain, a protein–protein interaction module specific for eukaryotes. The cytosolic domains of ATOM69 and ATOM46 were shown to bind a number of different precursor proteins and are essential for normal growth (34). ATOM69 and ATOM46 have been found in all kinetoplastids as well as in euglenoids (35). Except for the TPR domain in ATOM69, the two import receptors of trypanosomes do not resemble the TOM subunits of other species, indicating that they evolved independently from both the yeast and the plant receptors.
Recently, an analysis of the TOM complex in Trichomonas vaginalis hydrogenosomes, which are mitochondria-derived hydrogen-producing organelles that lack their own genome (36), identified Tom36 and Tom46 (37). The two proteins are paralogues and consist of an N-terminal CS/Hsp20-like domain, three TPR-like sequences, and a C-terminal membrane anchor, which is reminiscent of trypanosomal ATOM69, although the mass of both hydrogenosomal proteins is much lower than that of ATOM69. Moreover, HHpred analysis, using Tom36 as a query, retrieved ATOM69 as the first hit (37). The cytosolic domains of Tom36 and Tom46 were able to bind hydrogenosomal precursor proteins, suggesting they may function as protein import receptors. However, despite the similarities between ATOM69 and Trichomonas Tom36/Tom46, phylogenetic analysis suggests that they evolved independently of each other, and therefore reflect yet another example of convergent evolution, although a diversification of a common ancestor cannot be ruled out (37).
Here, we have investigated the substrate specificity of the trypanosomal import receptors ATOM46 and ATOM69 using inducible RNA interference (RNAi) cell lines and biochemical methods. We could correlate the observed receptor preference with specific features of the recognized substrate proteins, such as the presence of a predicted presequence, average hydrophobicity, and presence of TMDs. Moreover, we devised a method that allows for identification of which trypanosomal precursor proteins can be recognized by heterologous import receptors. Using this method, the mitochondrial proteomes are quantitatively compared between Trypanosoma brucei cell lines lacking either ATOM46 or ATOM69 and with T. brucei cell lines in which ATOM46 or ATOM69 were replaced by either Tom20 from yeast or Tom36 from Trichomonas.
Results
Import Receptors Are Essential and Affect Mitochondrial Proteins Differentially.
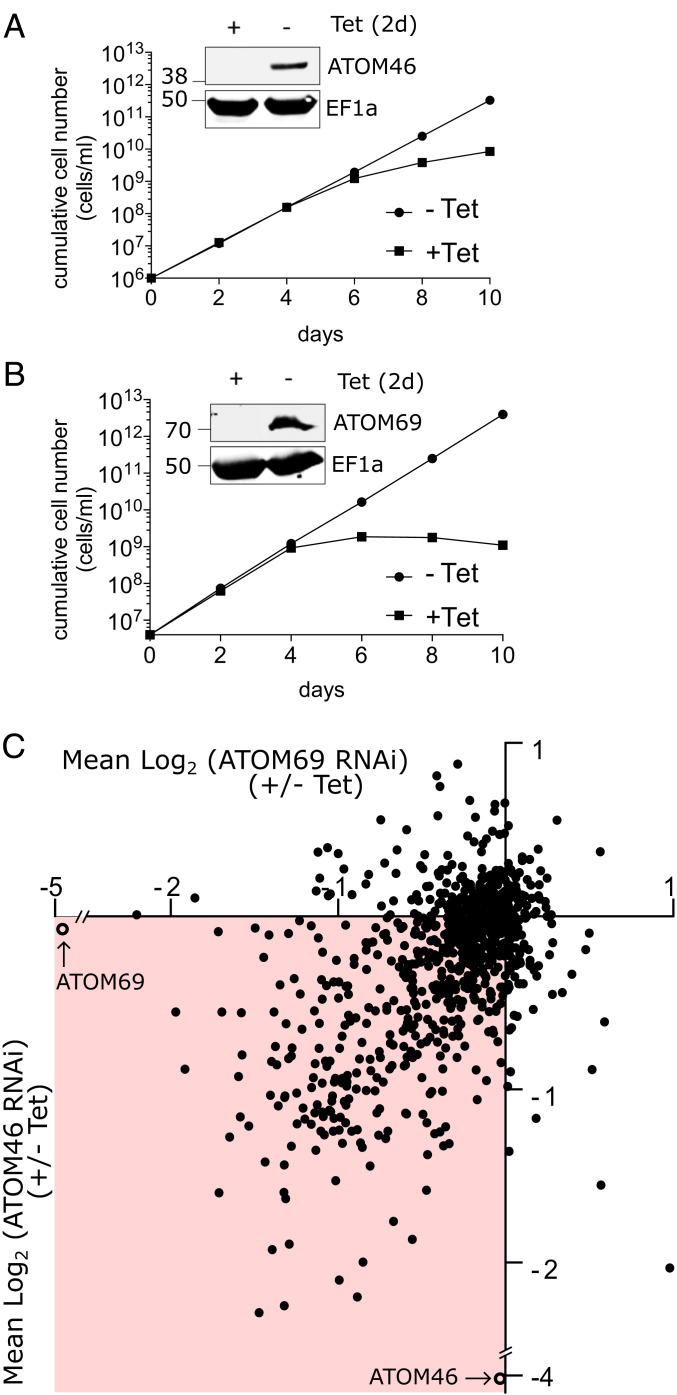
In order to investigate the substrate preference of the two mitochondrial protein import receptors ATOM46 and ATOM69, we used transgenic procyclic T. brucei cell lines, which allow their tetracycline (Tet)-inducible ablation by RNAi. We have previously reported that RNAi targeting the open reading frame (ORF) of ATOM46, while not affecting growth at 27 °C, results in a growth arrest at 33 °C (34). For the present study, we generated an additional RNAi cell line that targets the 3′-UTR of the ATOM46 mRNA. In this cell line, ATOM46 is more efficiently down-regulated, and growth slows down after 6 d of induction at 27 °C (Fig. 1A). Induction of RNAi against the ORF of ATOM69 results in efficient down-regulation of ATOM69 and a growth arrest after 4 d (Fig. 1B) (34). Thus, both protein import receptors are essential for normal growth of procyclic trypanosomes.
Fig. 1.
Import receptors are essential and affect mitochondrial proteins differentially. (A and B) Growth curves of uninduced and induced ATOM46 3′-UTR RNAi and the ATOM69 ORF RNAi cell lines. (Inset) Immunoblots probed for ATOM46 and ATOM69, respectively. Elongation factor 1a (EF1a) serves as loading control. (C) The two RNAi cell lines were analyzed by SILAC combined with quantitative proteomics. Scatter plot depicting the change in abundance of 856 mitochondrial proteins after ablation of ATOM69 (x-axis) and ATOM46 (y-axis), respectively. Both targets of RNAi are efficiently down-regulated (open circles).
Abolishing mitochondrial protein import by ablation of ATOM40, the protein import pore of the OM, results in a reduction of the steady-state levels of mitochondrial proteins (38, 39). Thus, quantitative proteomic comparison of mitochondria-enriched fractions from induced (+Tet) versus uninduced (−Tet) RNAi cells ablated for either of the two receptors should allow the delineation of the substrate preferences of ATOM46 and ATOM69, respectively. Procyclic T. brucei was grown in heavy and light stable isotope labeling by amino acids in cell culture (SILAC) media. Subsequently, the noninduced and induced (+/−Tet) SILAC cultures were mixed, and a crude mitochondrial fraction was purified using mild digitonin treatment and centrifugation. The resulting pellet was subjected to quantitative mass spectrometry (MS)-based proteomics analysis to determine the fold change in the abundance of proteins after Tet induction for each of the receptors. Nonmitochondrial contaminants were removed in silico by filtering with the previously determined mitochondrial importome (39). This resulted in a total of 856 proteins, corresponding to 76.4% of the mitochondrial proteome, that were detected and quantified in both datasets. Subsequently, the results for both receptors were combined in a scatter plot, which depicts on the x and y axes the fold change in the abundance of each detected mitochondrial protein after ablation of ATOM69 and ATOM46, respectively. The plot shows that the targets for the RNAi were efficiently down-regulated (ATOM46, 16-fold; ATOM69, 32-fold) and that ablation of either receptor does not affect the steady-state levels of the other. The steady-state levels of most mitochondrial proteins were decreased upon ablation of both receptors but to various extents, indicating that these substrates require both receptors for efficient import (Fig. 1C, highlighted in red). However, there is a minority of proteins that were found to be up-regulated upon ablation of either or both of the receptors (Fig. 1C, nonhighlighted quadrants).
ATOM46 and ATOM69 Bind Their Substrates by Different Mechanisms.
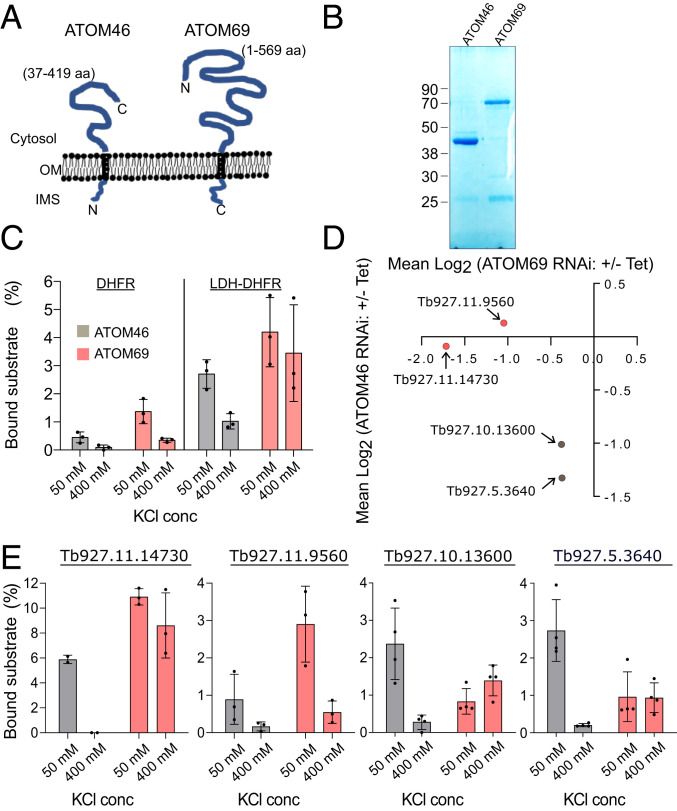
A subset of mitochondrial proteins is more extensively down-regulated after ablation of only one receptor, suggesting that it has a receptor preference (Fig. 1C). In order to find out whether this preference reflects receptor binding, we performed in vitro binding assays using the cytosolically exposed soluble receptor domains of ATOM46 and ATOM69 (Fig. 2 A and B) that were bound to agarose beads as described before (34).
Fig. 2.
ATOM46 and ATOM69 bind their substrate by different mechanisms. (A) Topology of ATOM46 and ATOM69, the cytosolic domains expressed in E. coli, are indicated. (B) SDS-PAGE of purified cytosolic receptor domains of ATOM46 and ATOM69 after expression in E. coli. (C) Binding of the artificial substrates DHFR and LDH-DHFR at low (50 mM) and high (500 mM) KCl concentrations to the indicated isolated cytosolic receptor domains. (D) Scatter plot as in Fig. 1C depicting the substrates that were selected for in vitro transcription and translation and subsequent binding assays. Substrates Tb927.11.14730 and Tb927.11.9560 show a preference for ATOM69 (red) and substrates Tb927.10.13600 and Tb927.5.3640 for ATOM46 (gray). (E) Binding assays using the four selected substrates Tb927.11.14730, Tb927.11.9560, Tb927.10.13600, and Tb927.5.3640 that show receptor preference in the RNAi cell lines. SEs are indicated.
The specificity of the binding assay was determined using cytosolic dihydrofolate reductase (DHFR) from mouse or a derivative thereof that was fused to the N-terminal 14 amino acids of trypanosomal dihydrolipoyl dehydrogenase, which comprises the mitochondrial presequence (LDH-DHFR; Fig. 2C). Moreover, in order to gain insight into the nature of receptor substrate interactions, all binding assays were done at low (50 mM) and high (400 mM) KCl concentrations, respectively. Should receptor binding mainly be guided by electrostatic interactions, it should be sensitive to high salt concentrations, whereas, if it is mainly based on hydrophobic interactions, the presence of salt will not influence or even could increase the binding. As shown in Fig. 2C, LDH-DHFR has a slight preference for ATOM69 and binds more efficiently to the cytosolic domains of the receptors than DHFR alone, as might be expected. Binding of LDH-DHFR to ATOM46 seems to be mainly electrostatic, whereas binding to ATOM69 is not reduced by high salt, suggesting this to be a hydrophobic interaction.
Next, we tested receptor binding of four trypanosomal mitochondrial proteins, two of which are predicted to have a preference for ATOM69 (substrates Tb927.11.14730 and Tb927.11.9560) and two which are predicted to have a preference for ATOM46 (substrates Tb927.10.13600 and Tb927.5.3640; Fig. 2D). The choice was based on the fact that the SILAC data suggest a strong receptor preference for these substrates, and because radiolabeled versions of the proteins could efficiently be produced by in vitro transcription translation reactions using a rabbit reticulocyte lysate. Fig. 2E shows that, at low salt concentrations, substrates Tb927.11.14730 and Tb927.11.9560 were more efficiently bound to ATOM69, whereas substrates Tb927.10.13600 and Tb927.5.3640 preferentially interacted with ATOM46. These results (Fig. 2E) are congruent with the SILAC RNAi analysis (Fig. 2D) and suggest that the reduction in the mitochondrial steady-state levels of substrate proteins observed in the RNAi cell lines might indeed be caused by a lack of receptor substrate interactions.
The results furthermore show that binding of all substrates (including LDH-DHFR) to ATOM46 is salt-sensitive, suggesting a dominant role for electrostatic interactions. The situation is different for ATOM69, where binding of substrate proteins is generally not affected (Tb927.11.14730, Tb927.5.3640, Tb927.10.13600, and LDH-DHFR) by high salt concentrations, indicating a major role for hydrophobic interactions. Substrate Tb927.11.9560 is different, as its binding to ATOM69 is salt-sensitive and therefore likely electrostatic. In summary, these results suggest that ATOM46 binds its substrates mainly by electrostatic interactions, whereas ATOM69 can interact with its substrates by both hydrophobic and electrostatic interactions.
Receptor Preference Correlates with Physicochemical Features of the Substrates.
The only extensively characterized mitochondrial protein import receptors are Tom20 and Tom70 from yeast. While they have partially redundant functions and overlapping substrate specificities, Tom20 preferentially recognizes presequence-containing proteins (14–17), while Tom70 binds precursor proteins with multiple TMDs such as mitochondrial carrier proteins (18–20). In order to determine whether the trypanosomal ATOM46 and ATOM69 behave in a similar way, we investigated whether physicochemical features of the substrates can be correlated with their apparent receptor preference.
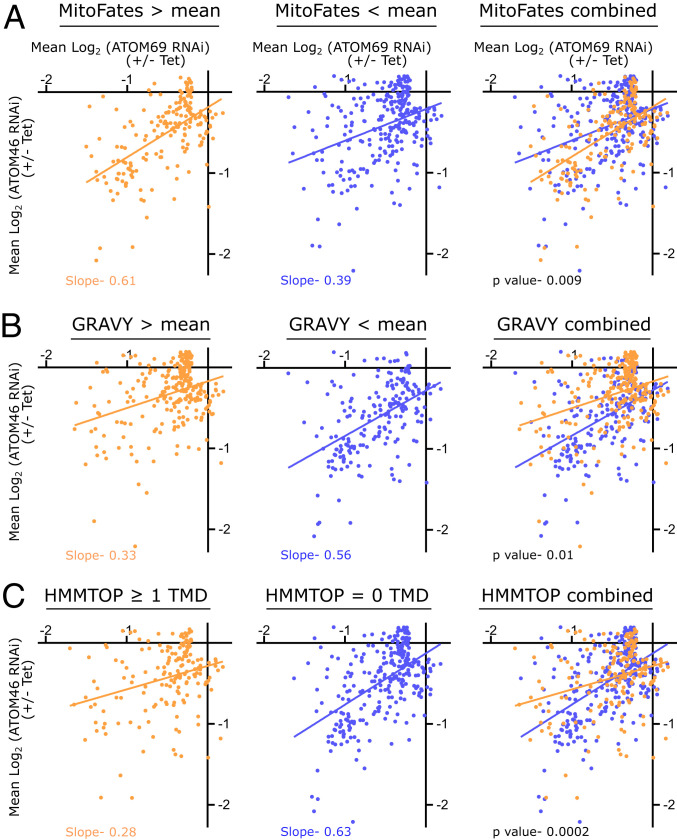
Before we started with this analysis, we filtered the set of 854 mitochondrial proteins detected in both RNAi cell lines for strong substrate candidates for either of the two import receptors. For this, we first removed 80 proteins that were elevated in abundance in either or both RNAi cell lines, applying a threshold of 1.15 fold (0.2 log2). As receptor substrates should be decreased in abundance, these 80 proteins were not considered as binding partners of ATOM46 or ATOM69. Second, we filtered for proteins whose abundance was only slightly decreased, applying a threshold of 0.87 fold (−0.2 log2). These 320 proteins were little affected by the depletion of the import receptors and thus do not represent strong substrate candidates. A possible reason for this is that these proteins might have a slow turnover and are therefore less affected in their steady-state level by the RNAi under the experimental conditions employed. The remaining set of 454 mitochondrial proteins was analyzed for three parameters: 1) the presence of a mitochondrial presequence, 2) overall hydrophobicity [grand average of hydropathy (GRAVY) index], and 3) the presence of predicted TMDs. We used the MitoFates algorithm to predict N-terminal mitochondrial presequences with a probability score of zero (unlikely to contain a presequence) to one (likely to contain a presequence) (40). The mean MitoFates value calculated for all 454 mitochondrial proteins was 0.364. This value was used to divide the proteins into two groups, the first one consisting of 184 proteins with MitoFates values above the mean and the second one consisting of 270 proteins with MitoFates values below the mean (depicted in orange and blue, respectively, in Fig. 3A). Linear regression shows that the proteins with a higher presequence probability (MitoFates value >0.364) cluster closer to the y-axis (Fig. 3A, orange line) compared to the second group of proteins, and thus are, on average, more strongly down-regulated in cells lacking ATOM46. In contrast, the proteins with a lower presequence probability cluster closer to the x-axis (Fig. 3A, blue line), indicating that they are more affected by the loss of ATOM69 (Fig. 3A). These findings suggest that, in trypanosomes, presequence-containing proteins have a preference for ATOM46, and the ones lacking it, for ATOM69.
Fig. 3.
Receptor preference correlates with physicochemical features of the substrates. (A) Mitochondrial presequence prediction values for 454 mitochondrial proteins that were down-regulated (greater than 0.15 fold, −0.2 log2) in at least one of the two RNAi cell lines were determined using MitoFates. The mean of these values was calculated, and the dataset was divided into two groups: proteins with a predicted presequence value above (orange) or below (blue) the mean. Linear regressions of both groups are indicated by the orange and blue lines, respectively. (B) As in A, but GRAVY scores are analyzed. Proteins with a GRAVY score above the mean are indicated in orange, whereas proteins below the mean are depicted in blue. (C) As in A and B, but proteins were analyzed for predicted TMDs using HMMTOP. The dataset was divided into orange (≥1 TMDs) and blue (0 TMD) proteins. Slopes for regression lines and P value are indicated.
A similar analysis was done using the GRAVY score. The mean GRAVY score of all 454 analyzed proteins was −0.371. Then, two groups were defined, the first one consisting of 250 proteins that are more hydrophobic than the mean (Fig. 3B, orange) and the second one consisting of 204 proteins that are more hydrophilic than the mean (Fig. 3B, blue). The linear regressions (Fig. 3B, orange and blue lines) show that the more hydrophobic proteins cluster more toward the x-axis and thus are, on average, more strongly down-regulated in cells lacking ATOM69 than the more hydrophilic proteins. This suggests that mitochondrial import of the more hydrophobic proteins depends more strongly on ATOM69, whereas the more hydrophilic proteins preferentially bind to the ATOM46 protein import receptor. Finally, in Fig. 3C, the 454 mitochondrial proteins were analyzed using HMMTOP, which predicts their TMDs. As a result, the proteins were divided in two groups: the first containing proteins that contain one or more predicted TMDs (169 proteins; Fig. 3C, orange) and the second encompassing proteins that lack predicted TMDs (285 proteins; Fig. 3C, blue). In line with the GRAVY score analysis, proteins lacking TMDs were more severely affected in the absence of ATOM46, whereas TMD-containing proteins were more efficiently decreased in abundance in cells lacking ATOM69.
In summary these results indicate that the preferred substrates for ATOM46 are presequence-containing, more hydrophilic proteins that lack TMDs, whereas ATOM69 prefers substrates without a presequence that are more hydrophobic and contain at least one TMD. However, these are only preferences, and many proteins appear to depend to similar extents on both receptors.
The CS/Hsp20-Like Domain of ATOM69 Is Essential for Growth at 33 °C.
ATOM69 has a large N-terminal cytosolic region and also a domain with TPR-like motifs (SI Appendix, Fig. S1A), which is adjacent to the C-terminal membrane anchor. Additionally, the receptor has a CS/Hsp20-like domain close to its N terminus. It has been shown for other proteins that this domain can bind to Hsp90 and possibly Hsp70. In order to determine whether the CS/Hsp20-like domain of ATOM69 is essential for its function, we performed complementation experiments. An RNAi cell line targeting the 3′-UTR of the ATOM69 mRNA was used as a host to express either full-length ATOM69 or variants truncated either just at the N terminus to remove the CS/Hsp20-like domain alone (ΔN103-ATOM69) or the CS/Hsp20-like domain plus some adjacent sequences (ΔN187-ATOM69). All three ATOM69 versions were expressed carrying a C-terminal c-Myc tag in order to distinguish them from the endogenous ATOM69. Immunoblots show that, in all three complemented cell lines, the endogenous ATOM69 was replaced by the ectopically expressed c-Myc–tagged version of ATOM69 (SI Appendix, Fig. S1B). It should be noted that the ATOM69 antiserum recognizes the untagged, endogenous, full-length protein and the ΔN103 c-Myc–tagged ectopically expressed ATOM69. Cell fractionations and immunofluorescence microscopy analyses indicate that all ATOM69 versions behaved like the integral mitochondrial OM protein ATOM40 (SI Appendix, Fig. S2). Moreover, blue native polyacrylamide gel electrophoresis (BN-PAGE) analyses indicate that all three versions of ATOM69 associate with the ATOM complex (SI Appendix, Fig. S1C).
Subsequently, we tested growth of all complemented cell lines at 27 °C, the standard growth temperature for procyclic T. brucei cells (SI Appendix, Fig. S1 D, Top). Normal growth was restored when full-length ATOM69 was expressed. Deletion of the CS/Hsp20-like domain slowed down growth for 2 d only, and normal growth resumed later. However, when the two cell lines expressing either full-length ATOM69 or ΔN103-ATOM69 were grown at 33 °C, a different result was obtained. Complementation with full-length ATOM69 still largely restored growth (SI Appendix, Fig. S1 D, Bottom Left), whereas complementation with ΔN103-ATOM69 did not (SI Appendix, Fig. S1D, Bottom Right). These results suggest that, at elevated temperature, the CS/Hsp20-like domain becomes essential for receptor function.
Finally, expression of the most extensively deleted ATOM69 version lacking the 187 N-terminal amino acids including the CS/Hsp20-like domain (ΔN187-ATOM69) could not restore growth.
Chimeric Trichomonas Tom36 Import Receptor Integrates into the ATOM Complex.
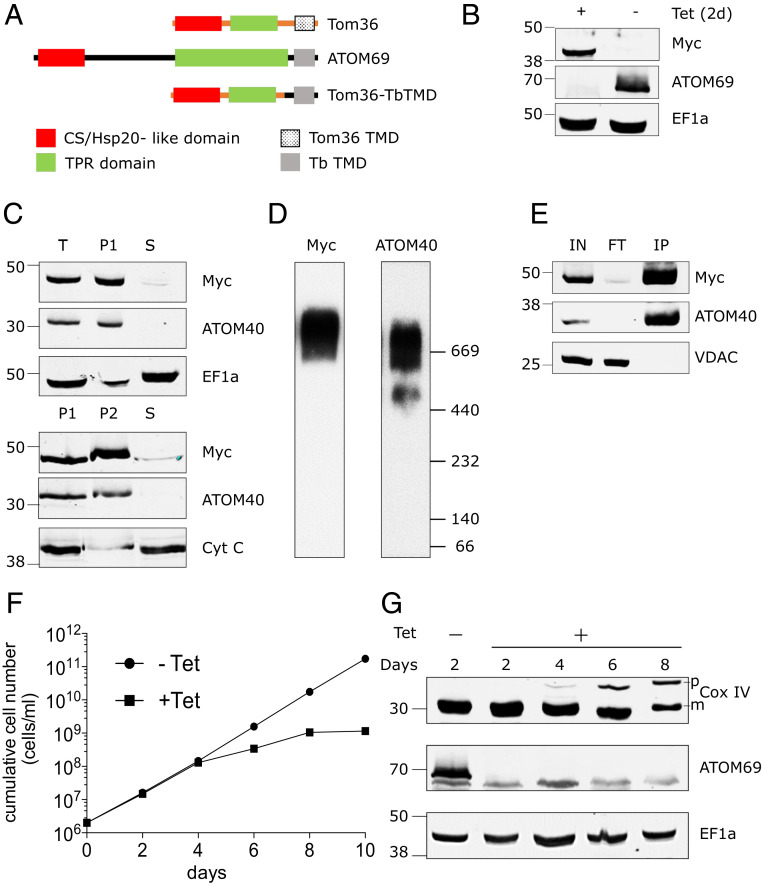
There is substantial evidence that the various targeting signals for mitochondrial proteins are functionally conserved in all eukaryotes (6). This contrasts to the mitochondrial protein import receptors, which evolved independently from each other in at least three eukaryotic lineages (34). Complementation experiments of the type discussed above can also be used to determine whether import receptors of various origins can functionally replace ATOM69 and/or ATOM46 when expressed in trypanosomes. An interesting candidate for such an analysis is Tom36 from T. vaginalis, which appears to function as an import receptor in hydrogenosomes (37). Tom36 has a similar domain architecture as ATOM69: both proteins have an N-terminal CS/Hsp20-like domain that is followed by a region of the protein that contains TPR-like repeats and a C-terminal TMD (Fig. 4A). However, the Trichomonas protein is only half the size of ATOM69, and phylogenetic analysis suggests that the two proteins have independent evolutionary origins (37). Thus, we expressed an N-terminally c-Myc–tagged version of Tom36, in which the endogenous TMD was replaced by the ATOM69 membrane anchor, in the background of the ATOM69 RNAi cell line. Upon addition of Tet, ATOM69 is ablated and the chimeric c-Myc–tagged Tom36 is expressed (Fig. 4B). Crude cell fractionation indicates that the expressed Tom36 chimera behaves like ATOM40, an integral mitochondrial membrane protein (Fig. 4C). Moreover, BN-PAGE and immunoprecipitation of the tagged chimeric Tom36 demonstrate that the heterologous receptor is efficiently incorporated into the trypanosomal ATOM complex (Fig. 4 D and E). However, Tom36 cannot complement the growth arrest caused by the lack of ATOM69 (Fig. 4F), even though the growth phenotype is less severe than for the noncomplemented ATOM69 RNAi cell line (Fig. 1B and SI Appendix, Fig. S3A). In line with this, accumulation of Cox IV precursor proteins is still observed in the complemented cell line (Fig. 4G). Since Tom36 is integrated into the ATOM complex, the absence of growth complementation indicates Tom36 incompatibility in fully assuming ATOM69 function.
Fig. 4.
Chimeric Trichomonas Tom36 import receptor integrates into the ATOM complex. (A) Domain structure of T. vaginalis Tom36, ATOM69, and its chimera (Tom36-TbTMD). Tom36-TbTMD is expressed in the background of ATOM69 3′-UTR RNAi. (B) Immunoblots probed with antisera against c-Myc (Myc) and ATOM69 show the efficient replacement of endogenous ATOM69 by the chimeric Tom36-TbTMD. Elongation factor 1a (EF1a) serves as a loading control. (C) Immunoblot analysis of total-cell (“T”), digitonin-extracted mitochondria-enriched pellet (P1), and soluble (“S”) fractions of cells expressing c-Myc–tagged chimeric protein (Top). ATOM40 and EF1a serve as mitochondrial and cytosolic markers, respectively. The digitonin-extracted mitochondria-enriched pellet (P1) was subjected to carbonate extraction. Immunoblot analysis of total (P1), pellet (P2), and soluble fraction (“S”) after carbonate extraction (Bottom). ATOM40 and cytochrome C (Cyt C) serve as integral and peripheral membrane markers, respectively. (D) Crude mitochondrial fractions of the tagged Tom36-TbTMD–expressing cell line were analyzed by BN-PAGE. The corresponding immunoblots were probed using antisera against c-Myc (Myc) and ATOM40. (E) Coimmunoprecipitation using tagged Tom36-TbTMD (Myc) as the bait. Voltage-dependent anion channel (VDAC) serves as a negative control. (F) Growth curves of uninduced (−Tet) and induced (+Tet) ATOM69 3′-UTR RNAi cell line ectopically expressing the tagged Tom36-TbTMD. (G) Immunoblots showing steady-state levels of CoxIV and ATOM69 in whole-cell extracts of the same cell line as in F collected at the indicated time points after Tet induction. EF1a serves as loading control.
Tom36 Restores Import of a Subpopulation of Proteins.
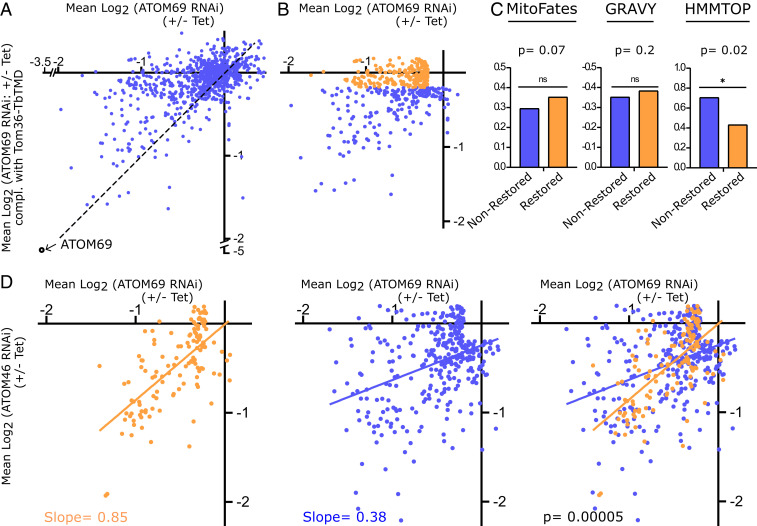
Expression of Trichomonas Tom36 does not restore growth of ATOM69-lacking trypanosomes (SI Appendix, Fig. S3A). However, it is possible that the chimeric receptor recognizes and restores mitochondrial import of a subset of proteins. We compared mitochondria-enriched fractions of the induced ATOM69 RNAi cell line with its counterpart, in which ATOM69 was replaced by the Tom36 chimera, using SILAC-based proteomics. The scatter plot in Fig. 5A depicts on the x- and y-axes the mean log2 SILAC ratios of mitochondrial proteins after ablation of ATOM69 and after its replacement by the Tom36 chimera, respectively. Proteins that are not recognized by Tom36 should cluster around the diagonal of the plot (Fig. 5A, dotted line). This is true for many proteins, but, surprisingly, a sizeable group clusters along the x-axis, indicating that, for these substrates, replacement of ATOM69 by the Tom36 chimera, to a large part, restored import.
Fig. 5.
Tom36 restores import of a subpopulation of proteins. (A) Scatter plot depicting fold change in abundance of 895 mitochondrial proteins upon ablation of ATOM69 (x-axis) and after ATOM69 has been replaced by Tom36-TbTMD (y-axis). (B) Orange dots depict the 222 proteins, the import of which is efficiently restored, with a down-regulation between +0.2 and −0.2 log2. Blue dots depict the remaining 211 proteins, the import of which is not efficiently restored. For both groups, 115 significantly up-regulated proteins whose levels were above 0.2 log2 and 346 proteins whose levels changed less than ±0.2 log2 in both the ATOM69 RNAi as well as the Tom36 complemented cell lines were subtracted. (C) Comparison of the mean of MitoFates, GRAVY index, and TMD predictions between nonrestored and restored groups. (D) The 127 restored proteins that are also detected in the receptor preference dataset of Fig. 3 are highlighted in orange. Orange and blue lines show the linear regressions of the 127 detected restored (orange) and the 327 remaining proteins (blue). Slopes for regression lines and P values are indicated.
For further analysis, we defined two groups of substrates. The first one consists of proteins that, upon replacement of ATOM69 by Tom36, were up- and down-regulated less than 1.15 fold and not more than 0.87 fold (+0.2 to −0.2 log2). It represents 222 substrates whose import is efficiently restored by the Tom36 chimera (Fig. 5B, orange dots) and was termed the restored group. The second group, termed the nonrestored group, consists of the remaining 211 substrates. It represents all proteins whose import was not or was less efficiently restored by Tom36 (Fig. 5B, blue dots). Next, we compared the means for: 1) mitochondrial presequence probability, 2) overall hydrophobicity, and 3) the number of predicted TMDs between the restored and the nonrestored groups. The results show that the restored group has slight but nonsignificant preference for proteins that carry a presequence, whereas there was no difference for overall hydrophobicity between the two groups. Strikingly, however, the restored group was significantly enriched for proteins that lack TMDs (Fig. 5C). Thus, the physicochemical features of the restored group are different than the ones preferred by ATOM69. This is surprising and indicates that the shared domain architecture and topology of ATOM69 and Tom36 does not result in the two receptors having the same preferred substrates.
These results prompted us to analyze whether the restored group shows a preference for either ATOM46 or ATOM69, respectively. To that end, we used the same dataset that was used to investigate receptor preference (Fig. 3). It contained data points for 127 of 222 substrates of the restored group (Fig. 5D). The 127 restored substrates (Fig. 5D, orange) showed a strong and highly significant preference for ATOM46 when compared to all other remaining (Fig. 5D, blue) proteins. This is counterintuitive, as the complemented cell line is depleted for ATOM69 but has wild type levels of ATOM46. Why should expression of Tom36 chimera restore the import of proteins that prefer ATOM46?
Tom36 and ATOM69 prefer different substrates but share an CS/Hsp20-like domain, which allows interaction with cytosolic chaperones. Thus, it might be this domain, normally provided by ATOM69, that allows complementation of import of the restored group of proteins.
It has recently been shown that the main function of yeast Tom70 is to recruit chaperones to the OM and that import of many soluble matrix proteins also depends on Tom70 (41). The restored group is significantly enriched for mitoribosomal proteins but depleted for inner membrane proteins and subunits of the oxidative phosphorylation complexes (SI Appendix, Fig. S3 B–D). The latter two groups contain many TMDs and are therefore not the preferred substrate of Tom36. The enrichment of mitoribosomal proteins, on the contrary, is surprising, since Trichomonas does not have organellar ribosomes. However, we know that the mitoribosomal proteins of the protein-dominated mitoribosome of T. brucei form an extensive and intricate interaction network (42). Mitoribosomal proteins are, in principle, soluble, and thus preferred substrates for ATOM46. However, they may nevertheless require cytosolic chaperones for efficient import, since, without them, they may aggregate before they can be stably integrated into the mitoribosome. Tom36 appears to have a similar substrate preference than ATOM46, but it can also recruit cytosolic chaperones. It might therefore be the CS/Hsp20 domain of Tom36, normally provided by ATOM69, that restores import of mitoribosomal proteins in the absence of ATOM69.
Chimeric Yeast Tom20 Integrates into the ATOM Complex but Does Not Restore Import.
ATOM46 and yeast Tom20 both prefer presequence-containing substrates. We therefore wanted to test whether Tom20 is able to compensate for the loss of ATOM46. To that end, we expressed a C-terminally c-Myc–tagged version of Tom20, in which the endogenous N-terminal TMD was replaced by the ATOM46 membrane anchor, in the background of ATOM46 RNAi (SI Appendix, Fig. S4A). Upon addition of Tet, ATOM46 is ablated and the chimeric c-Myc–tagged Tom20 is expressed (SI Appendix, Fig. S4B). Crude cell fractionation indicates that the heterologously expressed Tom20 version behaves like the integral mitochondrial OM protein ATOM40 (SI Appendix, Fig. S4C). Moreover, BN-PAGE and immunoprecipitation of the chimeric Tom20 demonstrate that it is efficiently incorporated into the trypanosomal ATOM complex (SI Appendix, Fig. S4 D and E). However, Tom20 cannot complement the growth arrest caused by the lack of ATOM46 (SI Appendix, Fig. S4F). In line with this, accumulation of Cox IV precursor proteins is still observed (SI Appendix, Fig. S4G).
To find out whether import of at least a few proteins was restored in the complemented cell line, we prepared mitochondria-enriched fractions of the induced ATOM46 RNAi cell line. These fractions were compared with the cell line in which ATOM46 was replaced by the chimeric Tom20 using SILAC-based proteomics. The growth curve of the two cell lines in SILAC medium is depicted in SI Appendix, Fig. S5A. The 950 mitochondrial proteins that were detected in both datasets are presented in the scatter plot (SI Appendix, Fig. S5B). The result shows that most proteins cluster around the diagonal (SI Appendix, Fig. S5B, dotted line), indicating they are down-regulated to a very similar extent in both cell lines. Thus, unlike in the case of the Trichomonas receptor, we do not find a significant subpopulation of proteins that had their import restored. This indicates that, even though the chimeric Tom20 integrates into the ATOM complex, it does not productively recognize trypanosomal proteins.
Discussion
Mitochondria are monophyletic. Thus, the last eukaryotic common ancestor already had mitochondria, including the core subunits of the main mitochondrial protein import systems (1). However, the mitochondrial protein import receptors in the OM were likely not present, but evolved later, after a first divergence of eukaryotes (6). This explains why we find evolutionarily unrelated receptor pairs in yeast (Tom20/Tom70), plants (Tom20/OM64), and trypanosomes (ATOM46/ATOM69). We also expect that there are more such examples in other lineages (37). The mitochondrial protein import machinery mediates import of ∼1,500 different proteins into this organelle. Moreover, the targeting signals on the imported proteins are often functionally exchangeable between different systems (6, 43). Consequently, the receptor pairs in the different systems have, in principle, the exact same function. They represent examples of parallel evolution of the same molecular function over great phylogenetic distances. This comes close to “replaying the tape of life” as originally envisioned in a thought experiment by Gould (44). A comparative analysis of import receptors should therefore allow the elucidation of the role contingency and determinism have played in their evolution (45). However, while the yeast import receptors, Tom20 and Tom70, have been studied in great detail, we do not have the same depth of information about trypanosomal ATOM46 and ATOM69, even though they represent the second best characterized receptor pair.
We devised an in vivo method to determine the substrate preference of the two trypanosomal import receptors. It quantifies the changes in the mitochondrial proteome after inducible ablation of either of the two receptors using SILAC-based quantitative proteomics and provides a global picture of their substrate preferences. In an extension of this approach, we combined it with complementation studies. Cell lines were produced in which either of the two receptors was physically replaced by a heterologous receptor adapted to be integrated into the ATOM complex. Subsequently, the mitochondrial proteomes of the uncomplemented and complemented cell lines were quantitatively compared using SILAC-based proteomics. The advantage of the approach is its high resolution: the restoration of import of only a small number of proteins that is not sufficient to complement growth can easily be detected. Moreover, it can, in principle, be applied to any putative receptor variant, including heterologous receptors from species representing interesting branches of the eukaryotic evolutionary tree that, by themselves, are not experimentally accessible. A caveat in the latter case is that receptor function is tested against the proteome of a heterologous organism, in our case T. brucei. In the present study, we tested Tom36 from Trichomonas or Tom20 from yeast in an ATOM69 or an ATOM46 RNAi cell line, respectively. Complementation with orthologous receptors from related trypanosomatids was not tried.
Using the method described above, in combination with biochemistry, we showed that most proteins require both receptors for efficient import. Moreover, we demonstrated that the import receptor ATOM46 prefers substrate proteins that have an N-terminal presequence, are more hydrophilic, and lack TMDs, which is very similar to the substrate preference of the structurally distinct yeast Tom20. Our data furthermore show that substrate binding to ATOM46 is mainly based on electrostatic interactions, whereas, for yeast Tom20, it is based on hydrophobic interactions (17, 46). Thus, ATOM46 and Tom20 mediate import of the same type of substrate proteins, but they appear to use different mechanisms to do so. Interestingly, and possibly due to this different mechanism, Tom20 cannot replace ATOM46 when expressed in T. brucei.
ATOM69 prefers more hydrophobic proteins that lack a presequence and have TMDs. Its substrate preference is therefore complementary to ATOM46 and similar to yeast Tom70. Biochemical analysis shows that binding of substrates to yeast Tom70 is not sensitive to high salt concentrations (47), whereas, in the case of ATOM69, this was the case, but not for all substrates. This indicates that receptor–substrate binding for Tom70 is dominated by hydrophobic interactions, whereas ATOM69 uses both electrostatic and/or hydrophobic interactions, depending on which substrate is being recognized.
In a complementation experiment, we replaced ATOM69 with the hydrogenosomal import receptor Tom36 from Trichomonas (37). Expression of Tom36 restored import of a fairly large set of substrates in cells lacking ATOM69. This may have been expected because Tom36 and ATOM69 look superficially very similar: both have an N-terminal CS/Hsp20 domain followed by TPR repeats and a C-terminal TMD, resulting in the same domain architecture and topology.
However, surprisingly, the substrates, the import of which was efficiently restored by Tom36, were enriched for proteins that lack TMDs and that are the preferred substrates of ATOM46 (Fig. 5). This suggests that the restored import of these proteins might be due to the CS/Hsp20 box of Tom36, which allows it to recruit cytosolic chaperones and is absent from ATOM46. Thus, it appears that Tom36 evolved to preferentially recognize proteins that lack TMDs, but whose import might nevertheless require cytosolic chaperones such as mitoribosomal proteins. Due to the lack of the respiratory complexes, the fraction of integral membrane proteins might be smaller in hydrogenosomes than in bona fide mitochondria, which might explain the substrate preference of Tom36. Alternatively, it is also possible that Tom46, a paralogue of Tom36 that is only loosely associated with the TOM complex but capable of binding to hydrogenosomal precursor proteins (37), preferentially recognizes hydrophobic proteins.
Using the results presented in this study, we can begin to analyze the roles determinism and contingencies played in the evolution of the primary mitochondrial protein import receptors. It seems that different paths of evolution repeatedly resulted in import receptor pairs, where one member has a preference for presequence-containing proteins that are generally hydrophilic and the other member prefers more hydrophobic substrates with multiple TMDs. Examples for this are found in yeast and kinetoplastids, as well as probably in plants (6, 8). Moreover, the receptor showing a preference for hydrophobic proteins needs to interact with cytosolic chaperones, either through specialized TPR domains (23–25) or via a CS/Hsp20-like domain. Indeed, we could show that, for ATOM69, the presence of the CS/Hsp20-like domain is required for normal growth at elevated temperature. This is in line with recent results in yeast, which demonstrated that the most important activity of Tom70 is to recruit cytosolic chaperones to the mitochondrial OM. In fact, tethering of an unrelated chaperone binding domain to the OM complemented most of the defects caused by Tom70 deletion (41). Whether hydrogenosomes also have two receptors with different substrate specificities, or whether Tom36 and Tom46 are functionally equivalent, is unclear at present. Finally, it appears that essentially all receptors have acquired similar protein–protein interaction motifs such as TPR or ARM domains, both of which form alpha-solenoid structures (48). The convergent features described above are likely caused by the deterministic force of natural selection. This illustrates that there are limits to the ways the cell can solve the problem of importing ∼1,500 different proteins into mitochondria. These convergent features may therefore ultimately reflect constraints imposed by the laws of physics.
However, while replaying the tape of life in the case of import receptors results in many shared features, it is also clear that contingency plays an important role. Thus, the mechanism of substrate–receptor recognition, even in the case of receptors that have similar substrate preferences, does not need to be identical. The best examples for this are ATOM46 and yeast Tom20. Both prefer presequence-containing, more hydrophilic proteins, but, while ATOM46 mainly relies on electrostatic interactions to recognize its substrates, yeast Tom20 uses hydrophobic interactions to do so. Moreover, the topology of functionally similar receptors is not “conserved.” Yeast Tom20 is N-terminally membrane-anchored, whereas the membrane anchor of plant Tom20 is at the C terminus. A similar situation is found for Tom70, with its TMD is at the N terminus, and ATOM69 or Trichomonas Tom36, which have C-terminal TMDs. There is a remarkable flexibility in how evolution can shape receptors. ATOM69 and Tom36 have distinct substrate preferences. However, their evolution is based on the same toolkit, namely: 1) a CS/Hsp20-domain, 2) a segment containing multiple TPRs, and 3) a C-terminal TMD, which form two functionally distinct receptors that, however, exhibit the same domain structure and topology.
In summary, when we zoom in on the molecular details of the various receptors, we see the influence of randomness in that there are multiple molecular solutions to achieve the same task, which ultimately is to import ∼1,500 different proteins into mitochondria. Looking at the big picture, on the contrary, we see that natural selection independently produced receptors that share many common features. Identifying such universally “conserved” features more accurately requires input from functional studies using as many independently evolved import receptors as possible. To obtain such information is a challenging but worthwhile endeavor. It will help to identify the essential immutable features all import receptors share, not due to common descent, but due to the same physical constraints acting on them. This, in turn, may lead to a deeper level of understanding of the fundamental biological process of mitochondrial protein import.
Materials and Methods
Transgenic Cell Lines.
Transgenic procyclic T. brucei cell lines were generated using strain 29–13 (49) (described in detail in SI Appendix). ATOM46 and ATOM69 RNAi cell lines (Fig. 1 A and B) were generated using a pLew100-derived stem-loop plasmid. For the complementation experiments shown in SI Appendix, Figs. S1 and S2, DNA fragments corresponding to the full-length and truncated ATOM69 ORF variants were cloned into modified pLew100 vectors and transfected into the ATOM69 RNAi cell line (50). For the complementation experiments shown in Figs. 4 and 5, Tom36-TbTMD, encoding the soluble domain of T. vaginalis Tom36 and the C-terminal membrane anchor of ATOM69, was cloned into a modified pLew100 vector that allows the addition of a C- or N-terminal c-Myc epitope.
Purification of Cytosolic Receptor Domains by Ni-NTA Affinity Chromatography.
The soluble domain of ATOM46 and ATOM69 were fused with a hexa-histidine tag and expressed in Escherichia coli. The tagged proteins were purified using Ni-NTA affinity chromatography. Ni-NTA beads that were not exposed to His-tag proteins served as bed volume control for the background in the binding assays. The procedures are described in detail in the SI Appendix.
Receptor Binding Assays.
[35S]-Met–labeled substrate proteins were synthesized using an in vivo transcription translation system as previously described (34). For binding assays, beads loaded with receptor domains and equal volumes of control beads were used. After washing, the beads were resuspended in one bed volume of assay buffer containing 3 µL each of [35S]-Met–labeled in vitro translated precursor proteins. The beads were incubated for 1 h and washed. Subsequently, bound proteins were eluted and analyzed on a 13% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by digital autoradiography. The procedures are described in detail in the SI Appendix.
Immunoprecipitations.
Digitonin-extracted mitochondria-enriched pellet fractions were solubilized in lysis buffer. After centrifugation, the lysate [input (IN)] was incubated with 40 µL of anti–c-Myc beads. Bound proteins were eluted by boiling in SDS-PAGE sample buffer. The IN, flow-through, and eluate fractions were subjected to SDS-PAGE and immunoblotting. The procedures are described in detail in the SI Appendix.
Quantitative Proteomics of SILAC RNAi Experiments.
SILAC RNAi experiments were primarily done as described previously (39). T. brucei RNAi cell lines for ATOM46 (Fig. 1A) and ATOM69 (Fig. 1B), as well as the corresponding derived ATOM69 RNAi cell lines, expressing either Tom36-TbTMD (Fig. 5A) or TbTMD-Tom20 (SI Appendix, Fig. S4A), were induced with Tet for 6, 5, 4, and 9 d, respectively. The induced and the noninduced cultures were mixed in a 1:1 ratio, and a crude mitochondria-enriched pellet fraction of the mixed cells was generated by digitonin treatment. Proteins of mitochondria-enriched fractions were either separated by SDS-PAGE or digested with trypsin and processed for liquid chromatography-MS analysis as described before (39). Further quantification was done as described before (51, 52). Datasets S1–S4 provide lists of all proteins identified in the individual datasets. All SILAC RNAi experiments were done in three biological replicates including a label switch. The procedures are described in detail in the SI Appendix.
Miscellaneous.
Digitonin extractions were done as described before (50). Transgenic cell lines expressing epitope-tagged proteins were harvested, washed, and lysed with digitonin, followed by differential centrifugation that yielded an organelle enriched (pellet) and a cytosolic (soluble) fraction. For the carbonate extractions, organelle-enriched pellet fractions were resuspended in 100 mM Na2CO3 and centrifuged, yielding a soluble (peripheral protein) and a pellet (integral protein) fraction. BN-PAGE was done as described before (53). A list of antibodies and procedure details are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Philip Stettler for help with the statistical analysis. Work in the laboratory of A.S. is supported by the National Centre of Competence in Research (NCCR) “RNA & Disease” and Grant 175563, both funded by the Swiss National Science Foundation. A.M. is supported by Grant Agency of Charles University (GAUK) 250937, and J.T. is supported by European Regional Development (ERD) funds [Centre for research of pathogenicity and virulence of parasites (CePaViP) CZ.02.1.01/0.0/0.0/16_019/0000759]. Work in the laboratory of B.W. was supported by European Research Council Consolidator Grant 648235 and Deutsche Forschungsgemeinschaft (German Research Foundation) Project 403222702/SFB 1381 and Germany’s Excellence Strategy [Centre for Integrative Biological Signalling Studies-Excellence Cluster (CIBSS–EXC)-2189–Project 390939984].
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.W.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017774118/-/DCSupplemental.
Data Availability.
All study data are available in the text or the SI Appendix
References
- 1.Roger A. J., Muñoz-Gómez S. A., Kamikawa R., The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Archibald J. M., Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 25, R911–R921 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Dacks J. B., et al., The changing view of eukaryogenesis–Fossils, cells, lineages and how they all come together. J. Cell Sci. 129, 3695–3703 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Hansen K. G., Herrmann J. M., Transport of proteins into mitochondria. Protein J. 38, 330–342 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Pfanner N., Warscheid B., Wiedemann N., Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 20, 267–284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mani J., Meisinger C., Schneider A., Peeping at TOMs-diverse entry gates to mitochondria provide insights into the evolution of eukaryotes. Mol. Biol. Evol. 33, 337–351 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Dolezal P., Likic V., Tachezy J., Lithgow T., Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Schneider A., Evolution of mitochondrial protein import–Lessons from trypanosomes. Biol. Chem. 401, 663–676 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Fukasawa Y., Oda T., Tomii K., Imai K., Origin and evolutionary alteration of the mitochondrial import system in eukaryotic lineages. Mol. Biol. Evol. 34, 1574–1586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mossmann D., Meisinger C., Vögtle F. N., Processing of mitochondrial presequences. Biochim. Biophys. Acta 1819, 1098–1106 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Vögtle F. N., et al., Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Perry A. J., et al., Structure, topology and function of the translocase of the outer membrane of mitochondria. Plant Physiol. Biochem. 46, 265–274 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Endo T., Kohda D., Functions of outer membrane receptors in mitochondrial protein import. Biochim. Biophys. Acta 1592, 3–14 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Söllner T., Griffiths G., Pfaller R., Pfanner N., Neupert W., MOM19, an import receptor for mitochondrial precursor proteins. Cell 59, 1061–1070 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Ramage L., Junne T., Hahne K., Lithgow T., Schatz G., Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 12, 4115–4123 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitoh T., et al., Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 26, 4777–4787 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe Y., et al., Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100, 551–560 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Hines V., Schatz G., Precursor binding to yeast mitochondria. A general role for the outer membrane protein Mas70p. J. Biol. Chem. 268, 449–454 (1993). [PubMed] [Google Scholar]
- 19.Hines V., et al., Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 9, 3191–3200 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steger H. F., et al., Import of ADP/ATP carrier into mitochondria: Two receptors act in parallel. J. Cell Biol. 111, 2353–2363 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Backes S., et al., Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 217, 1369–1382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hachiya N., et al., Reconstitution of the initial steps of mitochondrial protein import. Nature 376, 705–709 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Young J. C., Hoogenraad N. J., Hartl F. U., Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Chan N. C., Likić V. A., Waller R. F., Mulhern T. D., Lithgow T., The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J. Mol. Biol. 358, 1010–1022 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Wu Y., Sha B., Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 13, 589–593 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Harkness T. A., Nargang F. E., van der Klei I., Neupert W., Lill R., A crucial role of the mitochondrial protein import receptor MOM19 for the biogenesis of mitochondria. J. Cell Biol. 124, 637–648 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moczko M., et al., Identification of the mitochondrial receptor complex in Saccharomyces cerevisiae. FEBS Lett. 310, 265–268 (1992). [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto H., et al., Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 284, 31635–31646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lithgow T., Junne T., Wachter C., Schatz G., Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J. Biol. Chem. 269, 15325–15330 (1994). [PubMed] [Google Scholar]
- 30.Heins L., Schmitz U. K., A receptor for protein import into potato mitochondria. Plant J. 9, 829–839 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Lister R., et al., Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19, 3739–3759 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry A. J., Hulett J. M., Likić V. A., Lithgow T., Gooley P. R., Convergent evolution of receptors for protein import into mitochondria. Curr. Biol. 16, 221–229 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Chew O., et al., A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett. 557, 109–114 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Mani J., et al., Mitochondrial protein import receptors in Kinetoplastids reveal convergent evolution over large phylogenetic distances. Nat. Commun. 6, 6646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammond M. J., et al., A uniquely complex mitochondrial proteome from Euglena gracilis. Mol. Biol. Evol. 37, 2173–2191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller M., The hydrogenosome. J. Gen. Microbiol. 139, 2879–2889 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Makki A., et al., Triplet-pore structure of a highly divergent TOM complex of hydrogenosomes in Trichomonas vaginalis. PLoS Biol. 17, e3000098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pusnik M., et al., Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr. Biol. 21, 1738–1743 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Peikert C. D., et al., Charting organellar importomes by quantitative mass spectrometry. Nat. Commun. 8, 15272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukasawa Y., et al., MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics 14, 1113–1126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Backes S., et al. , The mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. bioRxiv [preprint] (2020). https://www.biorxiv.org/content/10.1101/2020.09.14.296194v1. Accessed 20 November 2020. [DOI] [PMC free article] [PubMed]
- 42.Ramrath D. J. F., et al., Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes. Science 362, eaau7735 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Long S., Jirku M., Ayala F. J., Lukes J., Mitochondrial localization of human frataxin is necessary but processing is not for rescuing frataxin deficiency in Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 105, 13468–13473 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gould S. J., Wonderful Life: The Burgess Shale and the Nature of History (W.W. Norton & Co., New York, 1990). [Google Scholar]
- 45.Blount Z. D., Lenski R. E., Losos J. B., Contingency and determinism in evolution: Replaying life’s tape. Science 362, eaam5979 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Brix J., Rüdiger S., Bukau B., Schneider-Mergener J., Pfanner N., Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 274, 16522–16530 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Brix J., Dietmeier K., Pfanner N., Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 272, 20730–20735 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Groves M. R., Barford D., Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9, 383–389 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Wirtz E., Leal S., Ochatt C., Cross G. A., A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Bochud-Allemann N., Schneider A., Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 277, 32849–32854 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Cox J., et al., Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Schägger H., von Jagow G., Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are available in the text or the SI Appendix