Posttranslational modifications of proteins are often key to understanding their biological function, localization, and fate. In particular, the covalent attachment of ubiquitin, a small 76-amino acid polypeptide, to substrates has attracted recent attention and is being exploited to generate novel drugs capable of removing pathogenic targets in a selective fashion (proteolysis-targeting chimeras) (1). Conjugation of ubiquitin can be reversed by deubiquitinating enzymes (DUBs), reflecting additional regulation (2). Control for reversing protein ubiquitylation has been the subject of Satpal Virdee and coworkers (3), who have developed a DUB amino acid profiling assay that led to the discovery of a class of ubiquitin esterases, classically assigned as ubiquitin specific proteases. As reported in PNAS, the authors show, for a subset of DUBs from the Machado−Josephin Domain (MJD) family, previously with unknown function, their ability to cleave ubiquitin not only from lysine-based “classical” isopeptide bonds but also from ubiquitin moieties linked to serine and threonine side chains via ester linkages (3).
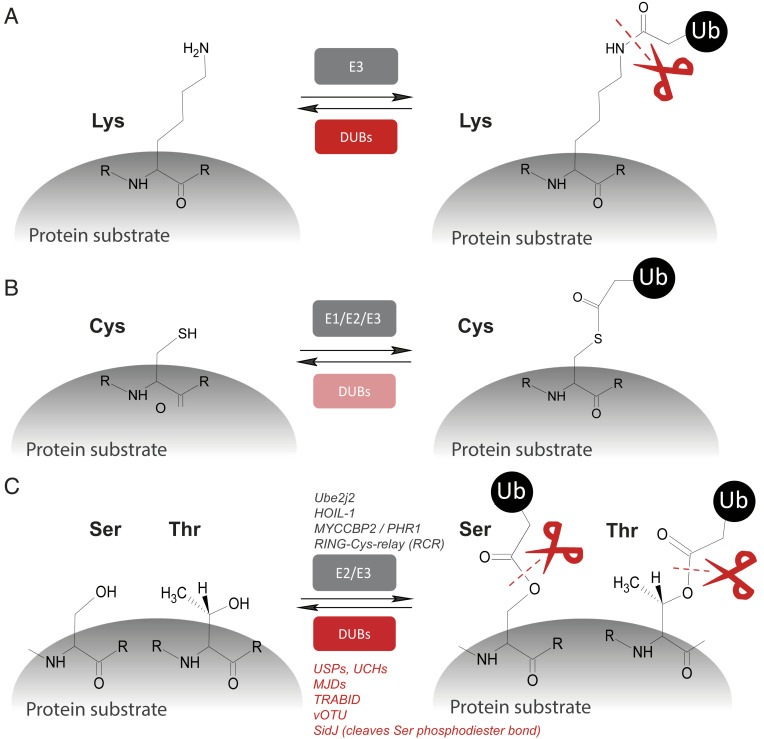
Protein ubiquitination seems predominantly restricted to the attachment of ubiquitin C termini to lysine (Lys) residues, forming “classical” isopeptide and, in the case of linear ubiquitin chains, peptide bonds (Fig. 1A). This is, in part, due to the unique length of the hydrocarbon chain of Lys that appears to be most optimal for E2/E3 enzymes’ reaction and transition of ubiquitin molecules to substrates (4). At the same time, there has been a precedence for ubiquitin covalent linkages other than to lysine epsilon amine side chains representing “canonical ubiquitylation.” For instance, in the context of ubiquitin conjugation to substrate proteins, the ubiquitin C terminus is transiently reacting with cysteine (Cys) thiol groups to form ubiquitin thioesters, in particular, with E2 and E3 enzymes (Fig. 1B) (5). At present, there is little evidence of DUBs reversing this reaction, although ubiquitin thioester formation may regulate DUB function itself, as proposed for USP4 and USP15 (6). Evidence for noncanonical ubiquitylation on alternative amino acid residues was suggested by several observations. First, the discovery of enzymatically catalyzed ubiquitin adenosine 5′-diphosphate (ADP) ribosylation was made, leading to the attachment to protein substrate’s serine (Ser) residues via phosphodiester bonds (ubiquitin attachment via arginine through the ribosyl group), mediated by bacterial pathogen-derived effectors (7). Ubiquitin ADP ribosyl Ser phosphodiesters can be hydrolyzed by the esterase activity of SidJ (8). Second, protein ubiquitination via esterification of Thr and Ser residues was demonstrated to occur via the E2 enzyme Ube2j2 (9) and the E3 ligase HOIL-1 (10). Also, Satpal Virdee’s team initially discovered MYCCBP2, an E3 ubiquitin ligase of the RING-Cys relay family, capable of performing this reaction (11, 12) (Fig. 1C). However, it was previously unknown whether mammalian DUBs might target these unusual sites for deubiquitination. To address this, the same group has now extended this line of research and developed representative model substrates and screen 53 DUBs for nonlysine activity, thereby providing important insights into DUB function (3). Satpal Virdee’s team found that, generally, ubiquitin-specific protease (USP) and ubiquitin C-terminal hydrolase (UCH) class DUBs exert both isopeptidase and esterase activity with comparable kinetics. In contrast, ovarian tumor domain (OTU) DUBs had little esterase activity, with the exception of TRABID and virally encoded vOTU. Notably, Satpal Virdee’s team (3) uncovered that a poorly studied class of DUBs of the MJD family (13) has potent and highly selective threonine esterase activity (Fig. 1C). These findings suggest that nonlysine ubiquitination appears to be common and coexisting with its conventional counterpart, possibly serving distinct biochemical purposes (3).
Fig. 1.
DUB esterases as part of the ubiquitin code arsenal. Protein substrate (de)-ubiquitylation on different amino acid side chains. (A) Classical ubiquitin (Ub) conjugation and deubiquitylation of lysine “isopeptide bonds.” (B) Cys-ubiquitylation yielding Ub-thioesters, mostly prevalent in transthioylation reactions involving E1, E2, and E3 enzymes. (C) Ser/Thr ubiquitylation and cleavage via DUBs (in red) from the USP, UCH, MJD, and OTU subfamilies. Scissors indicate DUB esterase activities (in red).
These intriguing observations made by the Virdee team open up a plethora of routes for ubiquitin biology, but there remain unanswered questions. For instance, from a thermodynamic point of view, thioester bonds are less stable as compared to (iso)-peptide bonds under aqueous conditions, such as aqueous, neutral pH and physiological temperature (37 °C) (14, 15). This is, at least in part, a possible reason why analytical detection of nonlysine ubiquitylation on proteins has been a challenge, as their chemically labile nature may lead to premature hydrolysis during sample preparation and/or measurement in biochemical or mass spectrometry-based assays. On the other hand, such modifications make biological sense, for example, as short-lived reaction intermediates, which is the case for ubiquitin thioesters during transthioylation and trans(iso)-peptide reactions, until formation of the final product, typically a Lys-Ub protein substrate, which is chemically more stable. Considering all these variations of ubiquitin-linked chemistry, perhaps it does not completely come as a surprise that nonlysine-linked ubiquitin also exists beyond Cys residues, such as ubiquitin esters with Ser/Thr residues of targeted proteins as described by Virginia De Cesare et al. (3).
It remains to be determined, in future studies, what exactly the stability and lifetime of protein adducts are that contain intracellular esters that are expected to be less stable as compared to thioesters and, in particular, (iso)-peptide bonds. The authors argue that, under certain circumstances, this may not necessarily be the case (3). As stated above, ester bonds are much more prone to hydrolysis than are peptide bonds, which suggests that ubiquitin chains based on ester linkages, if proven to widely exist in vivo, may play a more transient role than ubiquitin chains based on conventional (iso)-peptide linkages.
This is relevant, as the entire study has been performed predominantly with recombinant enzymes and artificial substrates, so the biological significance as well as how widespread ubiquitin esterification of protein substrates is remains to be demonstrated (3). There is precedence in the literature describing how ubiquitination of Ser, Thr, or Lys residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3 (16). Also, Ser-Thr ubiquitination mediates down-regulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu (17), and ester bonds were observed between ubiquitin and components of the MYD88 Myddosome (18). However, the role of DUBs—in particular, how selective OTUs and the MJDs, such as JosD1, with apparent selective esterase activity, may act as ubiquitin-specific carboxylic ester hydrolases (CEHs) in this cellular context—is not yet fully understood (19). According to the De Cesare study, many DUBs have apparent cysteine protease and esterase activities, and structural features discriminating between these two enzymatic activities are at the beginning of being unraveled. For instance, CEHs are predominantly serine hydrolases and classified as clans A-D (20), whereas a large subset of DUB cysteine proteases belong to the papain-like family of the clan CA (21). Dissecting this will open up another layer of regulation that controls ubiquitin-driven biological processes.
In summary, the discovery made in the study by Virginia De Cesare et al. (3) expands the biochemical concept of protein ubiquitylation, suggesting Ser/Thr ubiquitin conjugates may be widespread and intertwined with classical ubiquitin modifications. This has potentially wide implications for our general understanding of the ubiquitin system and associated biology in the normal, but also a pathophysiological, context.
Acknowledgments
This work was funded by the Engineering and Physical Sciences Research Council EPSRC grant (EP/N034295/1) and by the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Science (CIFMS), China (Grant 2018-I2M-2–002, to B.M.K.).
Footnotes
The author declares no competing interest.
See companion article, “Deubiquitinating enzyme amino acid profiling reveals a class of ubiquitin esterases,” 10.1073/pnas.2006947118.
References
- 1.Burslem G. M., Crews C. M., Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 181, 102–114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clague M. J., Urbé S., Komander D., Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20, 338–352 (2019). [DOI] [PubMed] [Google Scholar]
- 3.De Cesare V., et al., Deubiquitinating enzyme amino acid profiling reveals a class of ubiquitin esterases. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.2006947118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liwocha J., et al., Linkage-specific ubiquitin chain formation depends on a lysine hydrocarbon ruler. Nat. Chem. Biol., 10.1038/s41589-020-00696-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheffner M., Nuber U., Huibregtse J. M., Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373, 81–83 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Wijnhoven P., et al., USP4 auto-deubiquitylation promotes homologous recombination. Mol. Cell 60, 362–373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhogaraju S., et al., Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167, 1636–1649.e13 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Ronau J. A., Hochstrasser M., The DUB blade goes snicker-snack: Novel ubiquitin cleavage by a Legionella effector protein. Cell Res. 27, 845–846 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., et al., Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J. Cell Biol. 187, 655–668 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelsall I. R., Zhang J., Knebel A., Arthur J. S. C., Cohen P., The E3 ligase HOIL-1 catalyses ester bond formation between ubiquitin and components of the Myddosome in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 116, 13293–13298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pao K. C., et al., Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature 556, 381–385 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Mabbitt P. D., et al., Structural basis for RING-Cys-Relay E3 ligase activity and its role in axon integrity. Nat. Chem. Biol. 16, 1227–1236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seki T., et al., JosD1, a membrane-targeted deubiquitinating enzyme, is activated by ubiquitination and regulates membrane dynamics, cell motility, and endocytosis. J. Biol. Chem. 288, 17145–17155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foley J. F., Serine ubiquitylation. Sci. Signal. 9, ec303 (2016). [Google Scholar]
- 15.McDowell G. S., Philpott A., Non-canonical ubiquitylation: Mechanisms and consequences. Int. J. Biochem. Cell Biol. 45, 1833–1842 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Wang X., et al., Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177, 613–624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokarev A. A., Munguia J., Guatelli J. C., Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu. J. Virol. 85, 51–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen P., Kelsall I. R., Nanda S. K., Zhang J., HOIL-1, an atypical E3 ligase that controls MyD88 signalling by forming ester bonds between ubiquitin and components of the Myddosome. Adv. Biol. Regul. 75, 100666 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClellan A. J., Laugesen S. H., Ellgaard L., Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol. 9, 190147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Black D. S., Reilly P. J., Carboxylic ester hydrolases: Classification and database derived from their primary, secondary, and tertiary structures. Protein Sci. 25, 1942–1953 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawlings N. D., et al., The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]