Significance
Although plenty of studies have identified the RNA polymerase II (Pol II) pausing events in eukaryotes and demonstrated the regulatory roles of such events in gene expression and RNA processing, there was no study that systematically modeled the global landscape of Pol II pausing in the whole genome. Here, we have developed a deep learning framework that can accurately predict the Pol II pausing events from the contextual DNA sequences. Through applying our powerful computational approach to predict the pausing tendencies on interested regions in the human genome, we provided useful insights into understanding the relations between Pol II pausing events and alternative splicing, transcription factors, histone modifications, and DNA methylation.
Keywords: Pol II pausing, deep learning, alternative splicing
Abstract
RNA polymerase II (Pol II) generally pauses at certain positions along gene bodies, thereby interrupting the transcription elongation process, which is often coupled with various important biological functions, such as precursor mRNA splicing and gene expression regulation. Characterizing the transcriptional elongation dynamics can thus help us understand many essential biological processes in eukaryotic cells. However, experimentally measuring Pol II elongation rates is generally time and resource consuming. We developed PEPMAN (polymerase II elongation pausing modeling through attention-based deep neural network), a deep learning-based model that accurately predicts Pol II pausing sites based on the native elongating transcript sequencing (NET-seq) data. Through fully taking advantage of the attention mechanism, PEPMAN is able to decipher important sequence features underlying Pol II pausing. More importantly, we demonstrated that the analyses of the PEPMAN-predicted results around various types of alternative splicing sites can provide useful clues into understanding the cotranscriptional splicing events. In addition, associating the PEPMAN prediction results with different epigenetic features can help reveal important factors related to the transcription elongation process. All these results demonstrated that PEPMAN can provide a useful and effective tool for modeling transcription elongation and understanding the related biological factors from available high-throughput sequencing data.
Plenty of studies have discovered that the eukaryotic transcription elongation is not a stand-alone process. Instead, it is often coupled with many cotranscriptional RNA processing events, such as precursor mRNA (pre-mRNA) capping, splicing, and cleavage (1). During these processes, RNA polymerase II (Pol II) does not read out the DNA sequence at even speeds. In fact, the transcription elongation rate is dynamic and interplays with many cotranscriptional regulatory factors (2, 3). Recently, accumulating evidence has revealed that RNA Pol II complexes are unevenly distributed along gene bodies (4, 5) and can pause in specific regions in nearly 40% of genes (6, 7), often associating with the kinetic competition and coordination between transcription elongation and cotranscriptional events (4, 8). For example, it has been reported that Pol II frequently pauses in promoter-proximal regions as well as 20 to 40 nucleotides downstream from the transcription start sites of hsp70 in Drosophila and c-fos in mammals (9–11), which regulate the gene expression and RNA processing events (e.g., 5′-end capping and 3′-end processing). In addition, the transcription elongation rates have been shown to be crucially involved in the regulation of alternative splicing outcomes (12–14). For instance, it has been observed that Pol II pauses on a strong splice site and enables a weaker splice site upstream to be recognized by the spliceosome, thus resulting in the inclusion of the corresponding weak exon into the final mature RNA (15). Although these studies have demonstrated the regulatory roles of Pol II pausing in cotranscriptional processes, they were mainly based on the statistical analyses of Pol II densities from low-resolution sequencing data in a limited number of genes and genetic regions. They were mainly dependent on qPCR and high-throughput chromatin immunoprecipitation (ChIP-seq) techniques to provide snapshots of the relative abundance of Pol II along gene bodies (16, 17), which were generally limited in resolution (>200 base pairs [bp]) and strand specificity that are generally important for our deep understanding of the regulatory mechanisms underlying Pol II pausing and cotranscription processes (5).
Recently, the native elongating transcript sequencing (NET-seq) technique was able to provide a genome-wide, single-nucleotide resolution and strand-specific quantification of Pol II abundance in vivo (7, 18). Based on the high-quality quantitative measurement of Pol II density, Pol II pausing events can be identified at single-nucleotide resolution (18). Despite the advent of this high-throughput DNA sequencing technique in characterizing Pol II distributions, the underlying contextual DNA patterns related to Pol II pausing and transcription elongation and the corresponding associations with cotranscriptional processes are still not fully understood. Thus, accurately modeling the transcription elongation process at nucleotide resolution in genome-wide scale and systematically extracting the sequence features underlying Pol II pausing can greatly advance our understanding of the regulatory mechanisms of gene transcription.
In recent years, deep learning frameworks have been widely applied in numerous genomic data analysis tasks, such as prediction of antigen presentation by major histocompatibility complex (19), modeling of translation initiation and elongation (20, 21), and identification of nucleic acid–protein binding sites (22). Here, we developed PEPMAN (polymerase II elongation pausing modeling through attention-based deep neural network) to address the RNA Pol II pausing prediction problem using contextual sequences surrounding the target loci. Our PEPMAN framework is a machine learning-based method that systematically models Pol II pausing events. By fully taking advantage of the superior predictive capacity of deep neural networks (22) and the interpretability of attention mechanism (23), PEPMAN can accurately predict Pol II pausing events in human genome and capture the important contextual sequence features around Pol II pausing sites. In addition, the PEPMAN prediction results enable us to systematically investigate the underlying relations between Pol II pausing and other essential cotranscriptional processes, such as alternative splicing, transcription factor binding, histone modification, and DNA methylation. All these results demonstrate that PEPMAN can provide a powerful and useful tool to study the transcription elongation process from high-throughput sequencing data.
Results
The PEPMAN Framework.
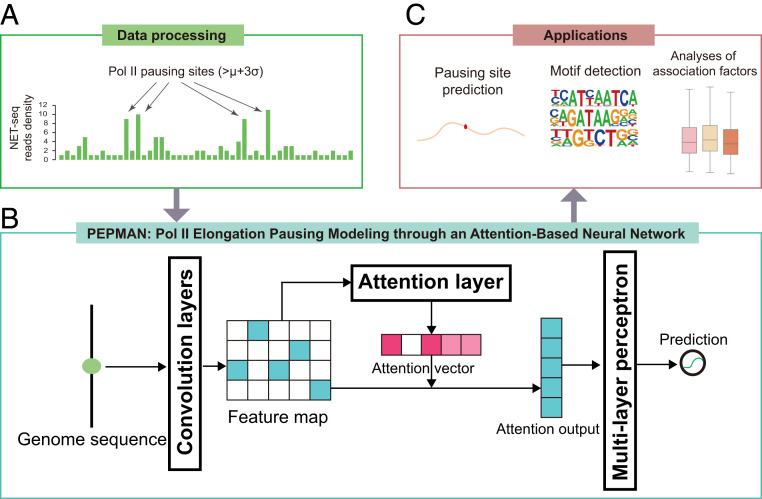
For modeling the Pol II pausing events in human genome, the PEPMAN framework formalized this problem as a classification task, in which the output scores of the employed deep neural network quantified the probabilities of Pol II pausing in genome loci (Fig. 1). In our study, a Pol II pausing event was said to occur at a given genome locus (which is also defined as a Pol II pausing site) if its read count derived from NET-seq is larger than four and higher than three standard deviations above the mean of surrounding 200 nucleotides (Fig. 1A), following similar criteria as in the previous research (24). Our sensitivity analyses demonstrated that such criteria were guaranteed to yield a high precision set of positive samples with a good genomic coverage (SI Appendix). In addition, we randomly sampled other genome loci as background from gene bodies, which were then combined with Pol II pausing sites to train a prediction model (Materials and Methods).
Fig. 1.
Schematic overview of the PEPMAN pipeline. (A) Data preprocessing. The NET-seq read counts that are larger than three standard deviations above the mean are defined as Pol II pausing sites. (B) The PEPMAN architecture. The contextual sequence surrounding a target site is first one-hot encoded and then passed through a two-layer CNN. The encoded feature map is then fed into an attention layer to calculate the attention vector, which stores the importance scores of individual nucleotide positions to the final prediction. Next, the attention vector is combined with the original feature map and then passed into an MLP to predict the pausing probability of the input target site. (C) Downstream applications of PEPMAN. The text has more details.
We assumed that the prediction of Pol II tendencies is determined by the contextual sequences around individual Pol II pausing sites. Thus, we also extended each pausing site 100 nucleotides both upstream and downstream to obtain a 201-bp-long sequence as an input to the convolutional neural network (CNN) employed in PEPMAN to learn the underlying sequence features (Fig. 1B and SI Appendix, Fig. S2A). The trained PEPMAN model can be used to model the Pol II pausing tendency of any given genomic locus (Fig. 1C). Furthermore, the attention mechanism employed in our model can highlight the important locations of the contextual sequence when predicting Pol II pausing tendencies and thus, can be used to identify the sequence motifs around individual Pol II pausing sites. More importantly, we can associate the prediction results of PEPMAN with the alternative splicing events, which may offer useful hints into understanding the interplay between Pol II pausing and alternative splicing. Moreover, the analyses of the prediction results of PEPMAN enabled us to investigate the relations between Pol II pausing and other transcription-related factors, such as transcription factor binding sites, histone modification, and DNA methylation, which thus may provide insights into the cotranscriptional process (Fig. 1C).
PEPMAN Accurately Predicts the Pol II Pausing Events.
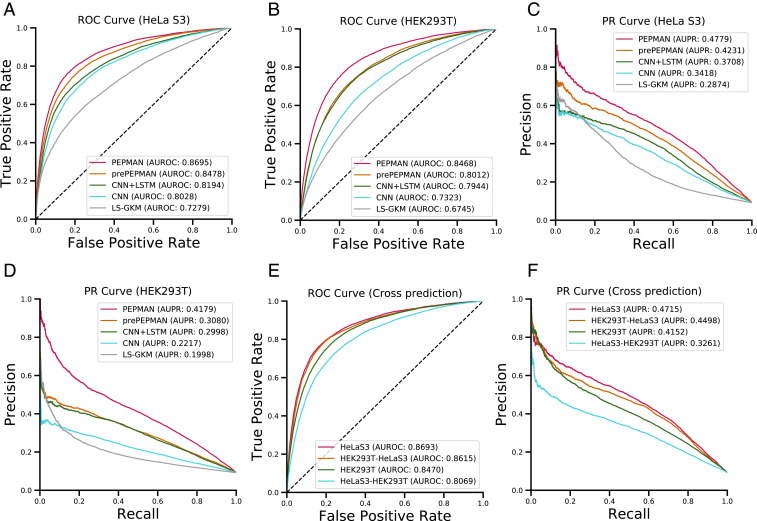
We evaluated the prediction performance of PEPMAN on the high-throughput NET-seq data derived from the human HeLa S3 and human embryonic kidney 293T (HEK293T) cell lines (5). Because our method is a method for modeling Pol II pausing events, we also implemented several machine learning or deep learning-based methods (22, 25) that were previously used in other biological data analysis tasks and regarded them as the baselines for comparison. In particular, we first compared the performance of PEPMAN with that of a conventional machine learning-based method large-scale gapped k-mer (LS-GKM) (25), which was an updated version of gapped k-mer support vector machine (gkm-SVM) (26) that was originally used for predicting the regulatory elements in DNA sequences. LS-GKM first transfers an input DNA sequence into a gapped -mer frequency feature vector space and then builds an SVM classifier to distinguish important sequence features from background signals (25). Our comparison showed that PEPMAN can achieve a superior prediction performance over LS-GKM, with increases of the area under the receiver-operating characteristic curve (AUROC) of 14.2 and 17.3% and the area under the precision recall curve (AUPR) of 18.4 and 21.5% in HeLa S3 and HEK293T cells, respectively (Fig. 2 A–D). In addition, to compare our model with other deep neural network-based methods in biological data analysis tasks, we also implemented a CNN-based model similar to the DeepBind architecture (22), which was originally used for predicting the sequence specificities of DNA and RNA binding proteins, and a hybrid convolutional and bidirectional long short-term memory (LSTM) recurrent neural network (CNN + LSTM) model similar to DanQ (27), which was previously applied in predicting the mutational effects of DNA sequences. As in PEPMAN, these two methods first embed input sequences into one hot-encoded feature representations, which are then fed into deep neural networks for prediction. The comparison results showed that PEPMAN still can achieve better performance than both CNN-based and hybrid frameworks in the HeLa S3 cell line, with increases of 6.8 and 4.9% in AUROC, respectively, and 13.3 and 9.0% in AUPR, respectively (Fig. 2 A and C). In the HEK293T cell line, the increases were larger, reaching to 11.5 and 5.3% in AUROC, respectively, and 19.2 and 10.9% in AUPR, respectively (Fig. 2 B and D). Notably, we also found that the performance of the PEPMAN model without attention mechanism decreased by 2.1 and 4.7% in AUROC and 5.0 and 10.7% in AUPR in HeLa S3 and HEK293T cell lines, respectively (Fig. 2 A–D), which thus verified the important role of the attention layer employed in our model. All these results demonstrated that PEPMAN is a general framework that can be applied to different cell lines and that can accurately predict Pol II pausing events and greatly outperform the baseline methods.
Fig. 2.
Performance evaluation of PEPMAN on the test data (chromosomes 17 to 22 and X; the ratio of positive to negative samples was 1:10). (A and B) Receiver-operating characteristic (ROC) curves and corresponding area under receiver-operating characteristic curve (AUROC) scores of PEPMAN and different baselines in HeLa S3 and HEK293T cell line, respectively. (C and D) Precision recall (PR) curves and the corresponding area under precision recall (AUPR) scores of PEPMAN and different baselines in HeLa S3 and HEK293T cell lines, respectively. Pre-PEPMAN denotes the PEPMAN model without attention mechanism, CNN + LSTM denotes a reimplementation of DanQ (27), CNN denotes a reimplementation of DeepBind (22), and LS-GKM stands for a conventional SVM-based method (25). (E and F) ROC curves/AUROC scores and PR curves/AUPR scores of cross-cell line prediction of different models between HEK293T and HeLa S3 cells, respectively. HeLa S3 and HEK293T denote the performance of PEPMAN on the original test datasets. HEK293T–HeLa S3 and HeLa S3–HEK293T denote the cross-cell line performance of the models that were trained on data from the former and tested on data from the latter. Detailed AUROC and AUPR scores of PEPMAN and baselines in HeLa S3 and HEK293T cell lines over 10 repeats are shown in SI Appendix, Table S1.
We also performed cross-cell line prediction between HeLa S3 and HEK293T and found that our model can still obtain decent performance when trained on the data from one cell line and tested on that from the other cell line (Fig. 2 E and F). These results indicated that PEPMAN can learn cross-cell line features of Pol II pausing sites, and both HeLa S3 and HEK293T cell lines may share a number of similar sequence features contributing to the prediction of Pol II pausing sites. To further investigate the similarities between the Pol II pausing sites of HeLa S3 and HEK293T, we checked the overlap between the two gold standard datasets (i.e., HeLa S3 and HEK293T). We found that between the 36,429 and 548,35 positive samples from HeLa S3 and HEK293T cell lines, respectively, there were 22,110 samples overlapping within a 201-bp window. In other words, over 60% of positive samples of HeLa S3 cell line overlapped with those of HEK293T cell line. This result probably could explain the high accuracy of our model in cross-cell line prediction. In addition, to understand why HEK293T–HeLa S3 displayed competitive accuracy with HeLa S3 and better performance than HeLa S3–HEK293T (Fig. 2 E and F), we also checked the sequencing depth of NET-seq data of both HeLa S3 and HEK293T cell lines. We found that the sequencing depth of HEK293T was larger than that of HeLa S3 (555 vs. 360 million uniquely aligned reads for HEK293T and HeLa S3, respectively). This result indicated that a number of pausing sites may not be detected in the HeLa S3 cell line possibly due to the low sequence depth. For example, among the expressed genes that did not contain pausing sites in the HeLa S3 cell line, over 30% of them recalled pausing sites in the HEK293T cell line. Meanwhile, we observed that the expression levels of these genes were not tissue specifically expressed between HeLa S3 and HEK293T cell lines. Thus, the pausing sites from HEK293T may be able to complement those pausing sites that were not detected in the HeLa S3 cell line because of the low sequencing depth. These results could provide a possible reason behind the competitive accuracy of HEK293T–HeLa S3 with HeLa S3 and also explain why HEK293T–HeLa S3 displayed better performance than HeLa S3–HEK293T.
PEPMAN Highlights Important Sequence Motifs Associated with Pol II Pausing.
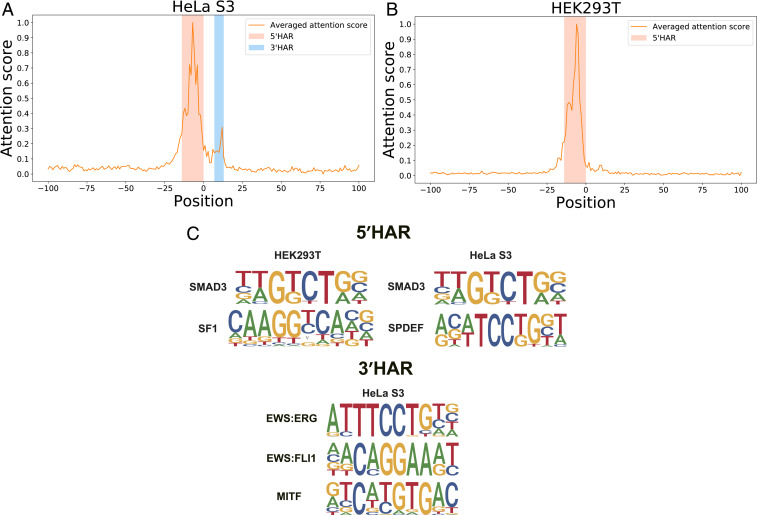
A major advantage of PEPMAN over other deep learning-based frameworks is that PEPMAN further incorporates the attention mechanism, thus enabling one to capture the important sequence features through examining the attention vectors of samples. Here, we first examined the distribution of attention scores through averaging the attention vectors over all samples in the test dataset. As shown in Fig. 3A, in the HeLa S3 cell line, two high-attention regions (HARs) appeared in the contextual sequences of the predicted Pol II pausing sites (i.e., a relatively higher peak on the 5′-end direction [around positions −14 to 0] and a relatively lower peak on the 3′-end direction [around positions 7 to 12], which were defined as 5′HAR and 3′HAR, respectively). This observation indicated that the contextual sequences around a 10- to 20-bp window on both sides of the target sites are essential for Pol II pausing prediction in HeLa S3 cells. Similarly, HEK293T cells also displayed an obvious 5′HAR on the upstream of Pol II pausing sites (Fig. 3B), which was quite similar to the result in HeLa S3 cells (Fig. 3A). On the other hand, unlike the result of HeLa S3 cells, we did not observe a 3′HAR on the downstream of Pol II pausing sites for HEK293T cells (Fig. 3B). To demonstrate the contribution of HARs in predicting Pol II pausing sites, we masked out the regions in samples with low-attention scores and retrained the model. As shown in SI Appendix, Table S2, comparing with the original PEPMAN model, the model (PEPMAN) that was trained on the masked samples only displayed a minor decrease in performance. Moreover, we also masked out those positions with high-attention scores and retrained the model. As expected, the performance of this model (PEPMAN) decreased significantly compared with the original PEPMAN model. These results revealed that the attention mechanism module employed in our framework can accurately detect the important positions for predicting Pol II pausing sites.
Fig. 3.
The high-attention regions (HARs) indicating the important contextual sequence features in predicting Pol II pausing events. (A and B) The distributions of attention scores of the contextual sequences over all samples in the test data in HeLa S3 and HEK293T cell lines, respectively. For the HeLa S3 cell line (A), two HARs were highlighted (one on the 5′-end direction, denoted by 5′HAR, and the other on the 3′-end direction, denoted by 3′HAR), while for the HEK293T cell line (B), only one HAR (i.e., 5′ HAR) was highlighted. (C) The known sequence motifs from the database TRANSFAC (28) enriched in the 5′HARs of HeLa S3 and HEK293T cell lines and the 3′HAR of the HeLa S3 cell line, which were determined by the motif-calling program HOMER (29), with Benjamini q values .
To further demonstrate the superiority of our feature attribution strategy, we also compared our attention mechanism approach with other methods, including a random selection scheme and an integrated gradient-based approach (30). In particular, we first selected the important positions indicated by our scheme and baseline methods and then trained a conventional neural network using only these chosen important features as input. Intuitively, a better feature attribution method should lead to superior prediction performance in this setting. As shown in SI Appendix, Fig. S3, we observed that both our attention mechanism and integrated gradients yielded better performance than the random selection method. In addition, our attention mechanism scheme exhibited greater improvement over integrated gradients. All these results demonstrated that the attention mechanism module employed in our framework plays an important role in capturing important features and enhancing the prediction of Pol II pausing sites.
Next, we examined the enriched transcription factor binding motifs in the HARs. More specifically, we extracted those positions with the highest 5% attention scores in each HAR and then calculated the statistically enriched sequence motifs (with negative samples as background) using the motif-calling program Hypergeometric Optimization of Motif EnRichment (HOMER) (29) (Materials and Methods). We found that a number of the enriched sequence motifs were significantly related to the transcription regulation process (Fig. 3C). For example, we found that both HeLa S3 and HEK293T shared a common motif of SMAD3, which is a protein that has been shown to regulate the alternative splicing of a cancer stem cell marker CD44 through colocalizing with PCBP1 in a variable exon region to inhibit spliceosome assembly (31). For the tissue-specific motifs of transcription factors such as SF1, a previous study had shown that in the HEK293T cell line, SF1 can bind to CA150 and repress Pol II transcription by inhibiting the transcription elongation process (32). Other examples are EWS:ERG and EWS:FLI1 (in the 3′HAR of HeLa S3), two known fusion proteins of EWS with ERG and FLI1, respectively, which are both members of the ETS family (33). More specifically, EWS is a protein containing an RNA binding domain, which interacts with TFIID and Pol II using its C-terminal half and plays an important role in Pol II transcription regulation (34). In particular, the fusion of EWS and FLI1 interacts with the seventh largest subunit of human RNA Pol II (hsRPB7) to influence the promoter selectivity (35). Among the enriched motifs in the 3′HAR of HeLa S3, microphthalmia-associated transcription factor (MITF) is another transcription factor containing the basic helix–loop–helix (bHLH) structure that is involved in controlling gene activities through recognizing a short DNA sequence CACGTG called E box, which is located in the promoter regions and has been found to play an important role in the regulation of gene expression (36). All of the above results revealed that although the two cell lines shared common sequence patterns, they also owned certain unique features and that our attention mechanism can detect the positions representing the cell line-specific features that can be supported by the previous known evidence in the literature.
PEPMAN Provides Useful Clues for Understanding the Mechanisms of Cotranscriptional Splicing.
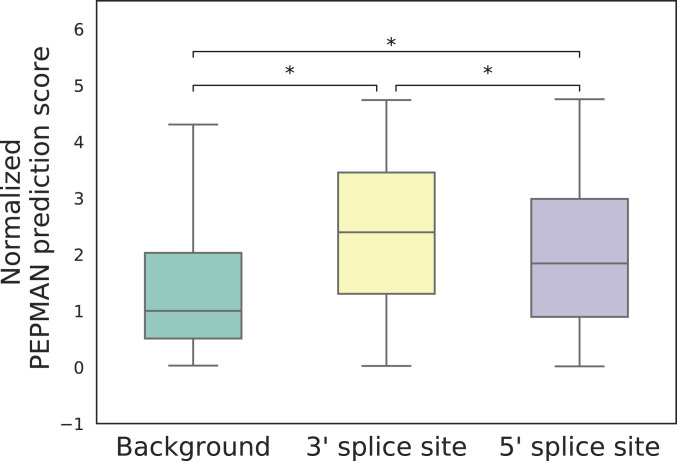
Cotranscriptional splicing was first reported by Beyer and Osheim (12) in Drosophila and was found predominant in Saccharomyces cerevisiae (13). In this process, the assembly of the spliceosome and its regulatory factors competes with transcription elongation, thus resulting in a biased distribution of Pol II complexes around splice sites (2). For example, Pol II tends to accumulate at the 3′ ends of introns in yeast (37) and also displays higher density around the 3′ splice sites (3′SSes) compared with the 5′ splice sites (5′SSes) in humans (5). In addition, it was found that the transcription elongation rates can regulate the outcomes of alternative splicing, and various molecular mechanisms had been proposed to illustrate the related phenomena (8, 38, 39). For instance, the potential regulation mechanism of exon skipping was illustrated by a kinetic model, in which Pol II pauses at the 3′SSes to enable the inclusion of upstream exons (15, 39, 40). Here, we investigated the relation between Pol II pausing and cotranscriptional splicing based on the PEPMAN prediction scores around the exon–intron boundaries. More specifically, we first extracted all exons in the chromosomes in the test dataset and then calculated the PEPMAN prediction scores in their 3′SSes and 5′SSes. We also compared these scores with those of 5,000 background loci that were randomly sampled from the genome. We found that the PEPMAN prediction scores of both 3′SSes and 5′SSes were significantly higher than those of background loci (Fig. 4), which was consistent with the previous finding that Pol II tends to accumulate on alternative splicing sites (2). In addition, 3′SSes generally had much higher predicted pausing potentials than 5′SSes (Fig. 4), which also agreed well with the previous result that Pol II complexes accumulated more on 3′SSes than on 5′SSes (5). All these results indicated that the PEPMAN prediction scores are consistently correlated with the previously known patterns of RNA Pol II distributions on splice sites.
Fig. 4.
Comparative analyses of the PEPMAN prediction scores between splice sites and splice sites. Background: 5,000 randomly sampled positions from the genome. The PEPMAN prediction scores of splice sites are normalized by the median of the prediction results in background. *, two-sided Wilcoxon rank-sum test.
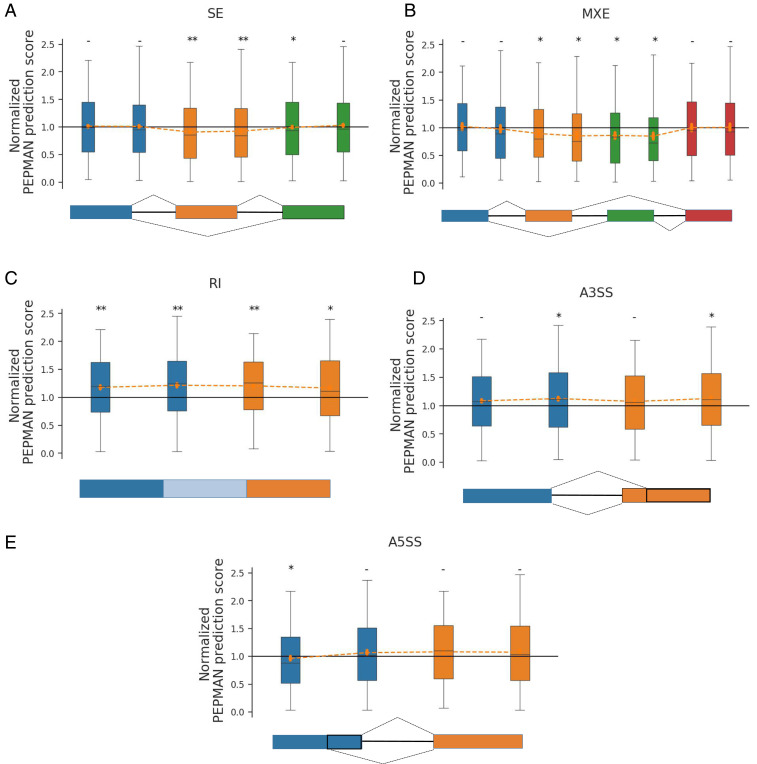
To further investigate the relations between Pol II pausing and individual types of alternative splicing, we first extracted all of the previously well-studied alternative splicing events from the ASpedia database (41) and then analyzed the corresponding PEPMAN prediction scores. The alternative splicing events are basically classified into five categories (i.e., skipping exons, mutual exclusive exons [MXEs], retained introns [RIs], alternative 3′ splice sites [A3SSes], and alternative 5′ splice sites [A5SSes]). Here, we compared the predicted tendencies of Pol II pausing for individual types of alternative splicing events with those of constitutive exons (i.e., those exons that are not engaged in alternative splicing) in our test data. In particular, we calculated the PEPMAN prediction scores for both 3′SSes and 5′SSes of exons for each type of alternative splicing and then normalized them with the median scores of the corresponding boundaries of constitutive exons. Our results showed that the predicted tendencies of Pol II pausing were relatively lower on skipping exons than on constitutive exons (Fig. 5A), which can also be supported by the previous finding that the Pol II density is generally smaller on skipping exons (5). We also found that MXEs had a lower predicted pausing tendency similar to skipping exons (Fig. 5B), indicating that these two types of alternative splicing may share common cotranscriptional mechanisms. On the other hand, RIs showed an opposite pattern in which the RIs preferred a stronger pausing tendency (Fig. 5C), which was consistent with the previous finding that the slow transcription elongation rates can enhance intron retention more efficiently (42). Taken together, our analyses indicated that higher pausing tendencies and slow elongation rates may promote intron retention and inhibit exon skipping. On the contrary, the PEPMAN prediction scores on the exon–intron boundaries of A3SSes and A5SSes were quite close to those of constitutive exons (Fig. 5 D and E). Previous studies had revealed that the sequences on exon–intron boundaries of A3SSes and A5SSes are similar to those of constitutive exons, although the sequences on splice sites in alternative splicing exons of A3SSes and A5SSes resemble those on skipping exons (43). This result was consistent with our finding that the transcription elongation rates on exon–intron boundaries of A3SSes and A5SSes may display similar patterns to those on constitutive exons. In summary, PEPMAN can provide a useful tool to investigate the relations between Pol II pausing and specific alternative splicing events and thus, offers useful hints to better understand the cotranscriptional splicing process.
Fig. 5.
Analyses of the tendencies of Pol II pausing predicted by PEPMAN on different alternative splicing (AS) events, including (A) skipping exons (SEs), (B) mutual exclusive exons (MXEs), (C) retained introns (RIs), (D) alternative splice sites (A3SSes), and (E) alternative splice sites (A5SSes). In each panel, the lower indicates the splicing types, while the upper shows the PEPMAN prediction scores on the corresponding splice sites. Each color represents a single exon, and each box plot represents the normalized prediction scores on the corresponding exon–intron boundary. Introns are represented by lines, and the RI in C is represented by a light blue box. The PEPMAN prediction scores of 3′SSes and 5′SSes in individual AS events are normalized by the median of the prediction results of the corresponding splice sites from 5,000 randomly selected constitutive exons. *: , two-sided Wilcoxon rank-sum test; **: , two-sided Wilcoxon rank-sum test; -: , two-sided Wilcoxon rank-sum test.
PEPMAN Associates Transcription Elongation with Epigenetic Features.
The cotranscriptional splicing process had been reported to be modulated by different epigenetic factors such as histone modification, transcription factor binding, and DNA methylation (44). In particular, several previous studies revealed that histone modification may regulate the transcription elongation rates and further influence the alternative splicing events. For instance, it was found that exon skipping can be induced by the high elongation rates in the context of hyperacetylation of H3K9 and increased levels of H3K36me3 (2, 45). In addition, it had been found that inhibition of histone deacetylases can lead to the increased processivity of Pol II along an alternative splicing element, thus changing the altered splicing outcome (46). Moreover, several transcription factors had been reported to govern the transcription elongation rates and thus, affect the Pol II processivity (47, 48). It had also been reported that DNA methylation can regulate Pol II pausing and alternative splicing through mediating the binding of methyl-sensitive DNA binding proteins such as MeCP2 and CTCF (49, 50). Therefore, it is generally necessary to gain comprehensive understanding of the relations between Pol II pausing and various types of epigenetic features. To achieve this goal, we first collected the ChIP-seq data of 10 histone modifications and nine transcription factors as well as information about methylated and unmethylated –C–phosphate–G– (CpG) sites for all of the chromosomes in the test dataset. For each histone modification or transcription factor, we also randomly sampled a nonbinding genomic region of equal length from the same gene for each binding area as negative control. For DNA methylation, we also randomly sampled 5,000 genomic loci from the same gene as background.
As shown in Fig. 6A, 9 of 10 histone modifications displayed significant changes in the PEPMAN prediction scores compared with negative control (i.e., H3K27ac, H3K27me3, H3K36me3, H3K4me1, H3K4me2, H3K4me3, H3K79me2, H3K9ac, and H4K20me1: , and , respectively; two-sided Wilcoxon rank-sum test). Among these histone modifications with significant change, H3K79me2 had been previously verified to activate transcription and regulate transcription elongation through the methyltransferase DOT1L of H3K79 (51, 52). H3K4me3 had been shown to promote transcription initiation, and H3K9ac has been reported to release the Pol II pausing by recruiting the super elongation complex to chromatin (53). In addition, we observed that the transcription factor binding motifs were also highly associated with the PEPMAN prediction scores (Fig. 6B), indicating that transcription factors may also act as strong or direct regulators of Pol II pausing. For example, NELFA, the most significant factor, is one of the subunits of the negative elongation factor (NELF) that had been known to directly interact with the RNA Pol II complex and promote Pol II pausing in the promoter-proximal regions (3). Also, DDX5 had been previously reported to affect the three-dimensional chromatin structure through influencing the CTCF/Cohesin binding in the targeted exons, which may indirectly interrupt the functions of CTCF to produce Pol II pausing (54). In addition, the DRB sensitivity-inducing factor (DSIF) had been reported to inhibit transcription elongation rates together with NELF, which can be rescued by the positive transcription elongation factor b (P-TEFb), a kinase that phosphorylates the C-terminal domain of the RPB1 subunit of Pol II (55). Moreover, previous studies indicated that DNA methylation can also play a certain role in transcription regulation. For example, DNA methylation can inhibit CTCF binding, which leads to Pol II pausing, resulting in the weakened inclusion of exon 5 of CD45 (49). Being consistent with this finding, our analyses showed that the unmethylated CpG sites tended to have a higher potential of Pol II pausing than the methylated CpG sites (Fig. 6C). Overall, the above analyses demonstrated that most of the epigenetic factors that were previously known to associate with transcription elongation were also highly related to PEPMAN prediction scores, which suggested that PEPMAN can provide a reasonably accurate tool for identifying the important regulators of the transcription elongation process.
Fig. 6.
Analyses of the relations between PEPMAN prediction scores and different regulatory factors, including (A) histone modifications, (B) transcription factor binding sites, and (C) DNA methylation. In A and B, negative control was generated by randomly sampling the genomic regions from the same genes, each of which did not overlap with any binding area of the corresponding epigenetic factor and had the same length of an epigenetic binding peak. The sums of PEPMAN prediction scores of individual sites within peak regions were compared. In C, 5,000 randomly selected positions in the test data in which the guanine–cytosine content distributions were consistent with the DNA methylation sites were used as background. The PEPMAN prediction scores of the DNA methylation sites were normalized by the median of the prediction results of background. *: , two-sided Wilcoxon rank-sum test; **: , two-sided Wilcoxon rank-sum test; +: , two-sided Wilcoxon rank-sum test; -: , two-sided Wilcoxon rank-sum test.
Discussion
In our work, we provided a useful deep learning framework to accurately predict the pausing tendencies of Pol II pausing in single-nucleotide resolution. We found that Pol II pausing was strongly associated with alternative splicing, histone modifications, transcription factors, and DNA methylations based on PEPMAN prediction results. A number of associations can be supported by the previous studies in the literature. Meanwhile, six of nine histone modifications and five of nine transcription factors (TFs) that exhibited significant associations with Pol II pausing in our analyses (Fig. 6) had not been studied before (to our best knowledge). These findings can provide useful insights into understanding the regulation and functions of transcription elongation dynamics.
Although it is convenient to analyze the associations between different genomic features and Pol II pausing using only NET-seq data (SI Appendix, Figs. S4 and S5), some of the associations can only be found using PEPMAN prediction scores. For example, the analyses based on the PEPMAN predictions showed that for the MXE event, the middle exons displayed significantly lower Pol II pausing tendencies (Fig. 5B), while the results from NET-seq data cannot reveal this relation (SI Appendix, Fig. S4B). In addition, CTCF and DDX5 displayed stronger statistical significance in the analysis results from PEPMAN prediction scores than those from NET-seq data (Fig. 6B and SI Appendix, Fig. S5B). Furthermore, although NET-seq data can provide a useful source of information for studying transcription elongation, they are often noisy and of low coverage in certain genomic regions (18, 24). In such regions, it is generally hard to analyze the Pol II pausing activities using only NET-seq data. To further demonstrate this point in detail, we also particularly investigated the associations with TFs in those low-coverage regions (i.e., with bottom 30% coverage of NET-seq data). As shown in SI Appendix, Fig. S6, in such regions with low coverage, the analysis with NET-seq data failed to identify almost all of the known associations that had been supported by the previous evidence in the literature, including CTCF (54), DDX5 (54), and DSIF (55). Only the association with NELFA appeared in the analysis results from NET-seq data. On the other hand, the PEPMAN prediction scores can still reveal almost all these associations between Pol II pausing and TFs (except SWI), even in the low-coverage regions (SI Appendix, Fig. S6). All these results demonstrated that PEPMAN can learn the underlying sequence features determining Pol II pausing, which are not subject to the influence of sequencing depth and bias in experimental data.
It would also be interesting to understand the relations between Pol II pausing and expression levels of genes. To investigate this point, we first predicted the pausing tendencies using our trained model along the gene bodies and then calculated average pausing score for each gene in the test dataset. Next, we checked the correlation between these prediction scores and the corresponding RNA sequencing (RNA-seq) read count densities of individual genes, which were the RNA-seq read count densities obtained from the same paper as in the NET-seq data (5). As shown in SI Appendix, Fig. S7A, the correlation between RNA-seq read count densities and predicted Pol II pausing scores was 0.41 (Spearman correlation), which indicated that Pol II pausing displayed a moderate correlation with the expression of the corresponding genes. Meanwhile, our analyses indicated that some lowly expressed genes contained a number of high prediction scores, indicating the false position predictions in them. We looked into several specific genes with low expression levels (i.e., logarithm of RNA-seq densities <−5) and high prediction scores (i.e., logarithm of averaged PEPMAN scores >−0.75). For example, for gene KISS1R, we found that there were a number of high peaks along the prediction scores on the gene body, although there were only five NET-seq read counts on this gene (SI Appendix, Fig. S8). This result indicated that there were some specific sequence features on these high-prediction regions, which may result in false-positive predictions. To further investigate this problem, we extracted the sequences of these high prediction scores on the genes with low expression levels and then called the enriched sequence motifs using the program HOMER (29). Intriguingly, we found that these regions were enriched with the CAGCTG-like core sequence, which is a binding motif of Group A bHLH proteins (SI Appendix, Fig. S9). In addition, we previously found that there was a Group B binding motif (MITF) that was also enriched with the sequence CACGTG on the HARs of PEPMAN. Thus, it was possible that PEPMAN had learned the enriched bHLH sequence motifs from the Pol II pausing sites and predicted high scores on those lowly expressed genes that were enriched with the bHLH binding motifs.
Note that although the gene expression levels displayed a relatively higher correlation with the NET-seq read counts (SI Appendix, Fig. S7B), such a quantitative analysis may be biased by the abundance of the expressed transcripts. According to the NET-seq protocol (5), the sequenced read counts are inevitably biased toward those highly expressed genes. Thus, when defining the Pol II pausing events, we should not simply compare the NET-seq read counts at a genome-wide level (24). Instead, it would be better to consider the local NET-seq read density, as such a strategy has already taken into account the abundance of the expressed nascent RNA within the same gene. In our study, we followed the same rigorous scheme as in ref. 24 to define the Pol II pausing events from NET-seq data. Such a definition of pausing events had been shown to be an effective scheme to study the transcription elongation activities from NET-seq data (24).
To further investigate the biological functions of the Pol II pausing-associated genes, we also applied a gene ontology (GO) term analysis (56) on those genes with average PEPMAN scores larger than 0.5. As shown in SI Appendix, Fig. S10, the Pol II pausing-associated genes were highly enriched in the functions and processes related to DNA binding and transcription regulation. Such GO analysis results indicated that Pol II pausing may play an important regulatory role in controlling transcription activities.
Conclusion
Deciphering the regulatory codes of Pol II pausing can provide useful and insightful understanding on how the transcription elongation rates affect gene expression and messenger RNA (mRNA) splicing. PEPMAN is an attempt to apply a machine learning-based framework to systematically model the Pol II pausing events from high-throughput sequencing data. The comparative analyses showed that the deep learning architecture employed in PEPMAN can achieve superior prediction performance over other baseline prediction methods. Through interpreting our prediction model via attention mechanism, we also discovered possible important regulatory regions and their corresponding sequence motifs in the neighborhood of Pol II pausing sites.
We also demonstrated that PEPMAN can accurately predict the tendencies of Pol II pausing around the alternative splicing sites. Our analysis results indicated that there may exist different regulatory cotranscriptional mechanisms behind different types of alternative splicing. In addition, the analyses of the relations between the PEPMAN prediction scores and different epigenetic factors also indicated that PEPMAN can learn the underlying important regulatory features of transcription elongation. Through the comprehensive analyses between PEPMAN prediction results and histone modifications, transcription factor binding motifs, and DNA methylation, we provided a thorough overview about the relations between Pol II pausing and individual types of epigenetic features. Since our framework can be applied to almost all species and cell lines with available NET-seq data, we expect that it can be used to gain a better understanding of Pol II pausing among different cell lineages and organisms. In summary, we believe that our PEPMAN framework can offer a powerful and useful tool in understanding the regulation of transcription elongation.
Materials and Methods
NET-seq and RNA-seq Data.
NET-seq and RNA-seq data of HeLa S3 and HEK293T cell lines were acquired from previous research (5). According to the statistics information summarized in ref. 5, in total 768 million and 1,203 billion reads were sequenced and 360 million and 555 million reads were uniquely aligned for HeLa S3 and HEK293T cell lines, respectively. In addition, most genes contained 100 to 1,000 reads in both HeLa S3 and HEK293T cell lines (figures 1D and S1H in ref. 5). Thus, the NET-seq datasets used in our study should provide a sufficient set of high-quality Pol II pausing data to train our model. To only consider the Pol II pausing events in the expressed genes, we excluded those genes in which no RNA-seq reads were mapped onto their bodies. We followed the same principle as in ref. 24 and extracted the positive and negative samples according to the following criteria: 1) the read density was at least three standard deviations above the mean over a surrounding 201-bp window that did not contain any other Pol II pausing event; 2) if the genomic distance of two positive samples is less than 201 bp, we only kept the one with a higher read density; and 3) the read count was larger than four regardless of sequencing coverage. Those with at least one read coverage that did not satisfy the above criteria were defined as negative samples. In summary, we obtained 36,429 positive samples and 1,699,724 negative samples for the HeLa S3 cell line and 54,835 positive samples and 1,568,693 negative samples for the HEK293T cell line.
Alternative Splicing Sites.
In our analyses, the constitutive 3′SSes and 5′SSes were extracted from human genome (57). Five types of alternative splicing events were obtained from the ASpedia database (41), from which both alternative splicing sites and their adjacent exon–intron boundaries were extracted. We also extended each splice site 100 nucleotides both upstream and downstream to obtain a 201-bp contextual sequence, as in the previous construction procedure of training samples. Note that here we only analyzed those splice sites in the expressed genes of chromosomes in the test dataset (i.e., chromosomes 17 to 22 and chromosome X). Overall, we obtained 3,488 skipping exons; 340 MXEs; 1,199 RIs; 1,645 A3SSes; and 899 A5SSes.
Histone Modifications, Transcription Factor Binding Sites, and DNA Methylation Sites.
We collected the ChIP-seq data of nine transcription factors (i.e., CTCF, CHD2, DDX5, DSIF, ELL2, NELFA, NELFE, SWI, and TFIIF) and 10 histone modifications (i.e., H3K27ac, H3K36me3, H3K4me1, H3K4me3, H3K27me3, H3K4me2, H3K79me2, H3K9ac, H3K9me3, and H4K20me1) for the HeLa S3 cell line from ENCODE (58) and the ChIP-Atlas database (59). The CpG DNA methylation sites were obtained from the HAIB Methyl 450K Bead Arrays data in ENCODE (58). The experimentally measured strength of CpG methylation was defined between 0 and 1,000, and we chose the CpG sites with methylation scores greater than 600 as methylated and those with scores less than 200 as unmethylated, following the original setting provided in ENCODE (58). We also extended each ChIP-seq peak 100 nucleotides both upstream and downstream to obtain a 201-bp contextual sequence. In our analyses, we mainly focused on those loci in chromosomes in the test dataset (i.e., chromosomes 17 to 22 and X).
Feature Extraction by a CNN.
For a 201-bp contextual sequence, we first convert it into a one hot-encoded feature matrix (also called feature map), which is then used as an input to PEPMAN. We then apply a two-layer CNN over this input feature map (SI Appendix, Fig. S2A). More specifically, given an input sequence , we first apply the zero padding scheme on its corresponding one hot-encoded feature map (i.e., padding both ends of the feature map with zero values) and then perform a convolution operation on the padded matrix : that is,
| [1] |
where ; (here, represents the kernel number); stands for kernel size; and stand for the elements of and , respectively; represents the learnable weight matrix; and stands for an element of the output matrix . After the convolution operation, the parametric rectified linear activation function (PReLU) is used to imitate the neuron activation: that is,
| [2] |
where stands for a single unit in each layer and stands for a trainable parameter representing the negative slope coefficient. In the end, we obtain the output feature (denoted by ) of the first convolution layer: that is,
| [3] |
The second layer of convolution operation is implemented similarly. After the two layers of convolution operation, we obtain a feature matrix , where stands for the learned feature of the th position and and represent the length and the dimension of the feature matrix, respectively.
Pol II Pausing Site Prediction with the Attention Mechanism.
To capture the important contextual sequence features involved in the determinants of RNA Pol II pausing, we incorporate an attention layer in our model, using a similar strategy to the previous works (23, 60). As shown in SI Appendix, Fig. S2B, given the feature matrix , we compute the attention score of the th position in the attention vector by
| [4] |
| [5] |
where stands for a weight matrix ( is a hyperparameter that needs to be determined; is the dimension of the feature matrix) and represents a weight vector. Next, we multiply the attention vector with the original feature matrix and then feed the result into a two-layer multilayer perception (MLP) network, followed by a sigmoid activation function. Overall, the procedure of predicting the Pol II pausing probability of an input sequence based on the above operations can be defined by the following formula:
| [6] |
where represents the sigmoid activation and stands for the two-layer MLP network.
In our study, we mainly followed the same principle as in the previous study (24) to use the 201-bp window to define the Pol II pausing events. To check how this window size can affect the prediction results, we also tested different window sizes (i.e., 151, 101, and 51 bp). As shown in SI Appendix, Table S3, the window size parameter used for defining the Pol II pausing sites did not significantly change the prediction performance, and our model yielded relatively robust prediction results for different window sizes. Note that according to our test results shown in Fig. 3, the most important contextual sequence features (i.e., the HARs) are typically located in positions −14 to 12 bp around the pausing sites. Thus, it would not be surprising to observe the stable performance of the different window sizes tested.
Model Training.
In order to determine the hyperparameters in PEPMAN, such as kernel size, kernel number, number of hidden units in the attention layer, learning rate, batch size, and number of hidden units, we first divided our data into three independent sets, including training, validation, and test datasets. More specifically, samples from chromosomes 1 to 13 were used as training data, those from chromosomes 14 to 16 as validation data, and the rest as test data (excluding chromosome Y). Overall, we obtained 25,297, 3,988, and 7,116 positive samples for training, validation, and test, respectively, in the HeLa S3 cell line and 38,992, 5,484, and 10,331 positive samples for training, validation, and test, respectively, in the HEK293T cell line. We next used a grid search strategy according to the performance on the validation set to obtain the best hyperparameters settings (SI Appendix, Table S4). During training, to address the issue arising from the imbalanced positive and negative samples and cover the negative samples as much as possible, for each epoch, we also resampled a new group of negative samples with a specific ratio to positive samples (i.e., 10:1) and combined them with positive samples as training data. For the objective function, we used a binary cross-entropy loss to train our model: that is,
| [7] |
where represents the th input sequence, represents its corresponding true label, and stands for the total number of training samples. The Adam optimizer (61) was used to minimize our training loss. In addition, we reduced the learning rate by multiplying a factor 0.8 for every five epochs. The early-stopping strategy (62) and the dropout method (63) were applied to overcome the overfitting problem. Batch normalization was used to alleviate the gradient vanishing and gradient exploding problem (64). We accelerated the training process with NVIDIA GTX 1080Ti GPU, in which PEPMAN costs about 100 s for an epoch with 278,267 training samples.
Motif Discovery.
To identify the regulatory elements that are specifically enriched in the HARs identified by PEPMAN, for each positive sample in the test dataset, we first selected those regions whose attention scores were among the top 5% list within the input contextual sequence. We also extracted the same number of sequences from negative samples and used them as background. We then ran the findMotifs.pl Perl script provided by the program HOMER (29) to extract the known sequence motifs that were significantly enriched in positive samples from the TRANSFAC database (28).
Supplementary Material
Acknowledgments
We thank Dr. Hailin Hu, Mr. Tingzhong Tian, and Ms. Yipin Lei for helpful discussions about this work. This work was supported in part by National Natural Science Foundation of China Grants (61872216, 81630103, 31900862); the Turing Artificial Intelligence Institute of Nanjing; and the Zhongguancun Haihua Institute for Frontier Information Technology.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007450118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or SI Appendix. The code is available in Github at https://github.com/fpy94/PEPMAN.
References
- 1.Saldi T., Cortazar M. A., Sheridan R. M., Bentley D. L., Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol. 428, 2623–2635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oesterreich F. C., Bieberstein N., Neugebauer K. M., Pause locally, splice globally. Trends Cell Biol. 21, 328–335 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Narita T., et al. , Human transcription elongation factor NELF: Identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 23, 1863–1873 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adelman K., Lis J. T., Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreas M., et al. , Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A., A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer A., Churchman L. S., Genome-wide profiling of RNA polymerase transcription at nucleotide resolution in human cells with native elongating transcript sequencing. Nat. Protoc. 11, 813–833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dujardin G., et al. , Transcriptional elongation and alternative splicing. Biochim. Biophys. Acta 1829, 134–140 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Li J., Gilmour D. S., Promoter proximal pausing and the control of gene expression. Curr. Opin. Genet. Dev. 21, 231–235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rougvie A. E., Lis J. T., The RNA polymerase II molecule at the 5 end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54, 795–804 (1988). [DOI] [PubMed] [Google Scholar]
- 11.Strobl L. J., Eick D., Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-MYC in vivo. EMBO J. 11, 3307–3314 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beyer A. L., Osheim Y. N., Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes. Dev. 2, 754–765 (1988). [DOI] [PubMed] [Google Scholar]
- 13.Oesterreich F. C., Preibisch S., Neugebauer K. M., Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell 40, 571–581 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Kadener S., Fededa J. P., Rosbash M., Kornblihtt A. R., Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. U.S.A. 99, 8185–8190 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dujardin G., et al. , How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 54, 683–690 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Kim T. H., et al. , A high-resolution map of active promoters in the human genome. Nature 436, 876–880 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raha D., Hong M., Snyder M., ChIP-seq: A method for global identification of regulatory elements in the genome. Curr. Protoc. Mol. Biol. 21, 21.19.1–21.19.14 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Churchman L. S., Weissman J. S., Native elongating transcript sequencing (NET-seq). Curr. Protoc. Mol. Biol. 4, 4.14.1–4.14.17 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Hu Y., et al. , ACME: Pan-specific peptide–MHC class I binding prediction through attention-based deep neural networks. Bioinformatics 35, 4946–4954 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Zhang S., et al. , Analysis of ribosome stalling and translation elongation dynamics by deep learning. Cell. Syst. 5, 212–220 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Zhang S., Hu H., Jiang T., Zhang L., Zeng J., TITER: Predicting translation initiation sites by deep learning. Bioinformatics 33, i234–i242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alipanahi B., Delong A., Weirauch M. T., Frey B. J., Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Hu H., et al. , DeepHINT: Understanding HIV-1 integration via deep learning with attention. Bioinformatics 15, 1660–1667 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Churchman L. S., Weissman J. S., Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D., LS-GKM: A new gkm-SVM for large-scale datasets. Bioinformatics 32, 2196–2198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Setty M., Leslie C. S., SeqGL identifies context-dependent binding signals in genome-wide regulatory element maps. PLoS Comput. Biol. 11, e1004271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quang D., Xie X., DanQ: A hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 44, e107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matys V., et al. , Transfac® and its module Transcompel®: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundararajan M., Taly A., Yan Q., Axiomatic attribution for deep networks. arXiv:1703.01365 (4 March 2017). [Google Scholar]
- 31.Tripathi V., et al. , Direct regulation of alternative splicing by SMAD3 through PCBP1 is essential to the tumor-promoting role of TGF-. Mol. Cell 64, 549–564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldstrohm A. C., Albrecht T. R., Suñé C., Bedford M. T., Garcia-Blanco M. A., The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21, 7617–7628 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wasylyk B., Hahn S. L., Giovane A., The ETS family of transcription factors. Eur. J. Biochem. 211, 7–18 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Bertolotti A., et al. , EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: Interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 18, 1489–1497 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petermann R., et al. , Oncogenic EWS-FLI1 interacts with HSRPB7, a subunit of human RNA polymerase II. Oncogene 17, 603–610 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Massari M. E., Murre C., Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429–440 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexander R. D., Innocente S. A., Barrass J. D., Beggs J. D., Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell 40, 582–593 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C., Effects of RNA secondary structure on alternative splicing of pre-mRNA: Is folding limited to a region behind the transcribing RNA polymerase? Cell 54, 393–401 (1988). [DOI] [PubMed] [Google Scholar]
- 39.Roberts G. C., Gooding C., Mak H. Y., Smith C. W., Proudfoot N. J., Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26, 5568–5572 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Mata M., et al. , A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12, 525–532 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Daejin H., Jihyun K., Young C. S., Charny P., Aspedia: A comprehensive encyclopedia of human alternative splicing. Nucleic Acids Res. 46, D58–D63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fong N., et al. , Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes. Dev. 28, 2663–2676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koren E., Lev-Maor G., Ast G., The emergence of alternative 3 and 5 splice site exons from constitutive exons. PLoS Comput. Biol. 3, e97 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B., Carey M., Workman J. L., The role of chromatin during transcription. Cell 128, 707–719 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Schor I. E., Rascovan N., Pelisch F., Alló M., Kornblihtt A. R., Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 106, 4325–4330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hnilicová J., et al. , Histone deacetylase activity modulates alternative splicing. PLoS One 6, e16727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J., Ha K. S., La Porta A., Landick R., Block S. M., Applied force provides insight into transcriptional pausing and its modulation by transcription factor NUSA. Mol. Cell 44, 635–646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wada T., et al. , DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human SPT4 and SPT5 homologs. Genes. Dev. 12, 343–356 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shukla S., et al. , CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maunakea A. K., Chepelev I., Cui K., Zhao K., Intragenic DNA methylation modulates alternative splicing by recruiting MECP2 to promote exon recognition. Cell Res. 23, 1256–1269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen A. T., Zhang Y., The diverse functions of DOT1 and H3K79 methylation. Genes. Dev. 25, 1345–1358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood K., Tellier M., Murphy S., DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules 8, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gates L. A., et al. , Acetylation on histone h3 lysine 9 mediates a switch from transcription initiation to elongation. J. Biol. Chem. 292, 14456–14472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giraud G., Terrone S., Bourgeois C. F., Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Rep. 51, 613–622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peterlin B. M., Price D. H., Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., Panther version 14: More genomes, a new panther go-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cunningham F., et al. , Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis C. A., et al. , The encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 46, D794–D801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oki S., et al. , ChIP-atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 19, e46255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin Z., et al. , A structured self-attentive sentence embedding. arXiv:1703.03130 (9 March 2017).
- 61.Kingma D. P., Ba J., Adam: A method for stochastic optimization. arXiv:1412.6980 (22 December 2014).
- 62.Prechelt L., “Early stopping—But when?” in Neural Networks: Tricks of the Trade, Montavon G., G. Orr, Müller K.-R., Eds. (Lecture Notes in Computer Science, Springer, Berlin, Germany, 1998), pp. 55–69. [Google Scholar]
- 63.Hinton G. E., Srivastava N., Krizhevsky A., Sutskever I., Salakhutdinov R. R., Improving neural networks by preventing co-adaptation of feature detectors. arXiv:1207.0580 (3 July 2012).
- 64.Ioffe S., Szegedy C., Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv:1502.03167 (11 February 2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. The code is available in Github at https://github.com/fpy94/PEPMAN.