Significance
RORγt is a nuclear receptor associated with several diseases. Various synthetic ligands have been developed that target the canonical orthosteric or a second, allosteric pocket of RORγt. We show that orthosteric and allosteric ligands can simultaneously bind to RORγt and that their potency is positively influenced by the other ligand, a phenomenon called cooperative dual ligand binding. The mechanism behind cooperative binding in proteins is poorly understood, primarily due to the lack of structural data. We solved 12 crystal structures of RORγt, simultaneously bound to various orthosteric and allosteric ligands. In combination with molecular dynamics, we reveal a mechanism responsible for the cooperative binding behavior. Our comprehensive structural studies provide unique insights into how cooperative binding occurs in proteins.
Keywords: nuclear receptors, RORγt, allosteric modulators, structure elucidation, drug discovery
Abstract
Cooperative ligand binding is an important phenomenon in biological systems where ligand binding influences the binding of another ligand at an alternative site of the protein via an intramolecular network of interactions. The underlying mechanisms behind cooperative binding remain poorly understood, primarily due to the lack of structural data of these ternary complexes. Using time-resolved fluorescence resonance energy transfer (TR-FRET) studies, we show that cooperative ligand binding occurs for RORγt, a nuclear receptor associated with the pathogenesis of autoimmune diseases. To provide the crucial structural insights, we solved 12 crystal structures of RORγt simultaneously bound to various orthosteric and allosteric ligands. The presence of the orthosteric ligand induces a clamping motion of the allosteric pocket via helices 4 to 5. Additional molecular dynamics simulations revealed the unusual mechanism behind this clamping motion, with Ala355 shifting between helix 4 and 5. The orthosteric RORγt agonists regulate the conformation of Ala355, thereby stabilizing the conformation of the allosteric pocket and cooperatively enhancing the affinity of the allosteric inverse agonists.
Allosteric ligands bind to pockets on proteins that typically do not overlap with the canonical, orthosteric binding pockets that are usually targeted by endogenous ligands (1, 2). Thus, allosteric ligands exert their effects via different structural modes of action (1–3). This can convey advantages over orthosteric ligands in terms of potency, because competition with endogenous ligands is removed, and selectivity, because allosteric sites are less conserved across protein families (1). Molecules that target allosteric pockets are therefore of high interest for drug development. Such ligands have been identified for several important protein classes, like G protein coupled receptors (GPCRs) and kinases (4–6), with some of those compounds developed into marketed drugs (7, 8).
Simultaneous binding of an endogenous, orthosteric ligand and an allosteric drug at different binding sites (dual ligand binding) is a fascinating pharmacological concept since this can modulate the overall physiological effect of the drug. Of particular significance are cooperative dual ligand binding events where binding of one ligand enhances that of the other (9–11), as observed for GPCR ligands in particular (12, 13). However, detailed structural insights into the mechanics of cooperative ligand binding remain scarce (14). This, in part, results from the absence of high-resolution structural data, required to visualize the effects of dual ligand binding. Better structural understanding of cooperativity in dual ligand binding is therefore required to accelerate the development of new allosteric drugs.
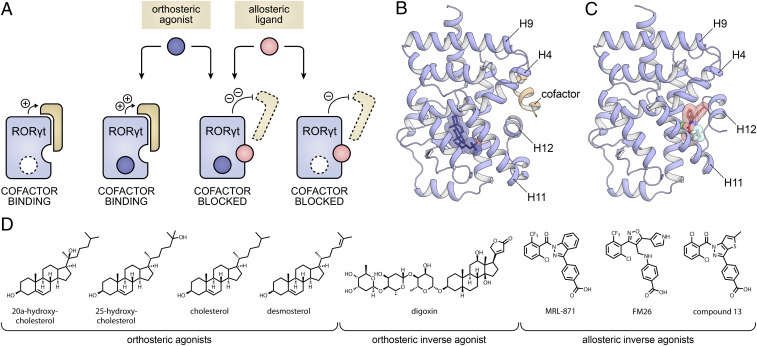
Dual ligand binding has occasionally been observed for nuclear receptors (NRs), but there is no clear mechanistic understanding of connected cooperative effects (15, 16). NRs are a class of transcription factors that can be modulated by endogenous and synthetic small molecules (17) and constitute attractive drug targets, with 16% of all drugs targeting this protein class (18). Mechanistical understanding and exploiting of cooperative dual ligand binding in NRs harbors great potential for drug development. An interesting NR in this context is the retinoic acid receptor-related orphan receptor γ t (RORγt) that plays an essential role in the differentiation of T helper 17 (Th17) cells, associated with the pathogenesis of autoimmune diseases (19–21). Inhibition of RORγt with small molecules, in order to disrupt the Th17/IL-17 pathway, is a promising strategy toward reducing the inflammatory response (20, 22–30). The RORγt ligand binding domain (LBD) has been shown to contain both a clearly defined canonical, orthosteric binding site (Fig. 1 A and B), accessible for both endogenous and synthetic compounds (24, 31–33), and a high-affinity second binding site, termed allosteric pocket, formed by helices 3, 4, and 11 and reoriented helix 12 (H12), to which allosteric inverse agonists can bind (Fig. 1C) (34–41). The crystal structures of the RORγt LBD bound to either an orthosteric or allosteric modulator indicate the possibility for dual ligand binding in this NR (Fig. 1 B and C) (38, 40).
Fig. 1.
(A) Conceptual representation of the RORγt LBD which is intrinsically active in the apo state, inducing cofactor binding. In the presence of an orthosteric agonist (blue), cofactor binding is further increased, while in the presence of an allosteric inverse agonist (pink), cofactor binding is blocked. In the presence of both an orthosteric and an allosteric ligand, the cofactor blockage is most efficient because of cooperative dual ligand binding. (B) Crystal structure of the RORγt LBD with the agonist 25-OH bound to the orthosteric site (blue sticks; PDB entry 3L0L). (C) Crystal structure of the RORγt LBD with inverse agonist MRL-871 bound to the allosteric site (red sticks; PDB entry 5C4O). The structures show that the orthosteric and allosteric binding sites do not overlap and highlight the prominently different orientation of H12. (D) Chemical structures of RORγt orthosteric agonists (20-OH, 25-OH, CHL, and DSM), orthosteric inverse agonist (digoxin), and allosteric inverse agonists (MRL-871, FM26, and compound 13) used in this study.
Here we reveal the biochemical and structural proof of RORγt dual ligand binding in a variety of orthosteric and allosteric ligand combinations. Our study also provides a detailed mechanistic explanation of cooperativity between the two binding sites in this NR. Extensive dual ligand binding studies combined with dual ligand protein cocrystallography and molecular dynamics (MD) simulations highlight the cooperative binding events and shed light on the underlying molecular mechanism controlling protein conformation and enhanced dual ligand affinity (Fig. 1A).
Results
RORγt-mediated transcriptional activity is correlated with binding of cofactor proteins to its LBD (Fig. 1A). This interaction is controlled by the conformation of RORγt H12, also called the activation-function 2 (AF-2) helix. H12 can adopt a stabilized agonistic conformation that promotes coactivator binding or a destabilized inverse agonistic state that inhibits intrinsic cofactor recruitment by RORγt. Even though the biomolecular, structural effects of agonistic or inverse agonistic ligands on coactivator binding do not necessarily translate to the cellular activation or inactivation of target genes under RORγt control for a variety of reasons, they do provide the crucial starting points for mechanistic understanding. A diverse set of RORγt ligands was selected to evaluate the apparent dual ligand binding behavior (Fig. 1D). Although there is no singularly defined endogenous ligand, cholesterol (CHL) and its derivatives including 20α-hydroxycholesterol (20-OH), 25-hydroxycholesterol (25-OH), and desmosterol (DSM) are known to be orthosteric agonists that promote coactivator binding by stabilizing H12 in an agonistic conformation (Fig. 1 A and B) (31, 42). Digoxin is an exemplary orthosteric inverse agonist that destabilizes the folding of H12, thereby inhibiting intrinsic coactivator recruitment (33). MRL-871 (34, 35), FM26 (38), and Glenmark’s compound 13 (39, 40, 43) are all allosteric inverse agonists, with varying potencies, that reposition H12 into a distinct conformation that prevents coactivator binding (Fig. 1 A and C) (35).
Dual Ligand Binding Enhances the Stability of the RORγt LBD.
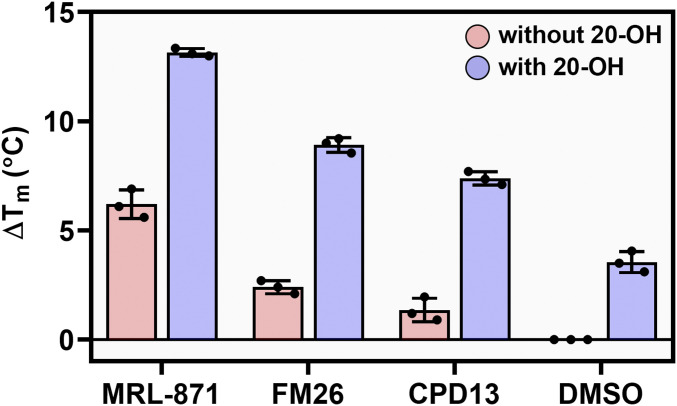
Ligand binding typically improves the thermal stability of NRs via structural and dynamic changes to the protein fold (44, 45). Thermal RORγt protein denaturation assays were performed to investigate the effect of single and dual ligand binding, as indicated by the RORγt melting temperature, Tm (45, 46). In the presence of the 20-OH, the Tm of RORγt increased by 3.6 °C, relative to the DMSO control, indicating enhanced thermal stability of the protein upon orthosteric agonist binding (Fig. 2, DMSO, blue bar). Similarly, the allosteric ligands each increased the RORγt Tm between 1 and 7 °C (Fig. 2, pink bars). When the allosteric ligands were used in combination with the orthosteric agonist 20-OH, the Tm values increased between 7 and 14 °C, strongly exceeding the individual and additive effects of the two types of ligands. The significant synergistic enhancement of the thermal stability of RORγt upon dual ligand binding is a strong indication for a cooperative behavior between the two binding sites.
Fig. 2.
Thermal stability of RORγt (5 µM) using a TSA format in presence of allosteric ligands MRL-871 (15 µM), FM26 (60 µM), and compound 13 (20 µM) (lowest concentrations giving a maximal ΔTm so maximal binding) as well as DMSO, in the absence (blue bars) and presence (pink bars) of 20-OH (60 µM). Data are recorded in triplicate from three independent experiments. Data are presented as mean ΔTm (normalized to DMSO) ± SD.
Orthosteric RORγt Ligands Enhance the Potency of Allosteric RORγt Ligands.
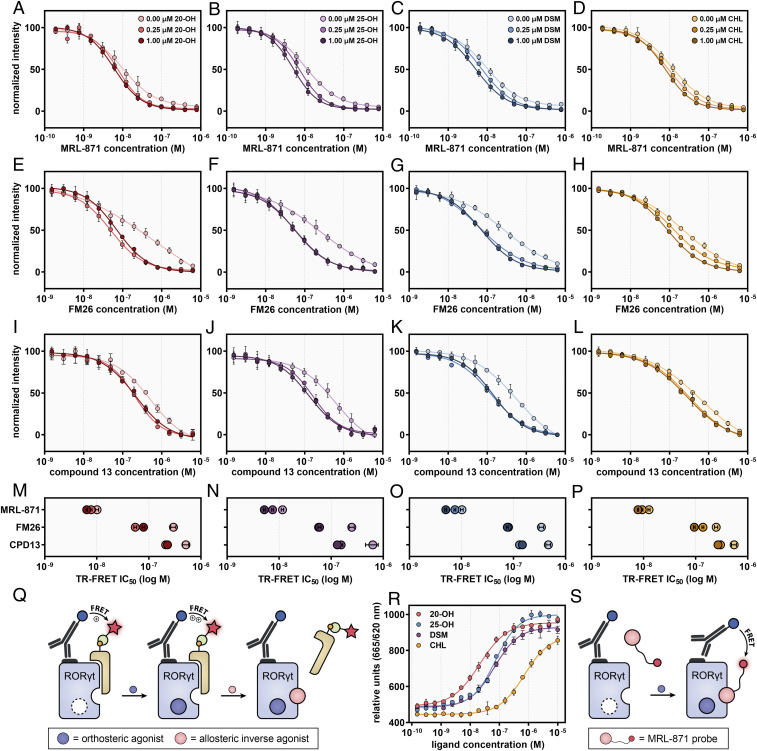
An established time-resolved fluorescence resonance energy transfer (TR-FRET) coactivator binding assay (47) was used to investigate the impact of dual ligand binding on the cofactor displacement potency of allosteric RORγt inverse agonists. Fluorescence emission occurs when the cofactor binds to the RORγt LBD, by FRET pairing from an anti-His terbium cryptate donor to a d2-labeled cofactor (Fig. 3Q). The orthosteric ligands CHL, 20-OH, and DSM all showed dose-dependent agonistic behavior as expected (dose–response curves and EC50 [half maximal effective concentration] /IC50 (half maximal inhibitory concentration) values in SI Appendix, Fig. S1 and Table S1) (21, 42). Surprisingly, 25-OH was found to be a partial inverse agonist, contrary to previous reports that showed it would be a RORγt agonist (21, 42). As expected, digoxin was found to be an orthosteric inverse agonist (33). The IC50 value for digoxin was significantly increased when the compound was titrated into assay mixtures containing fixed concentrations of the orthosteric agonist CHL (SI Appendix, Fig. S2 and Table S2). This confirmed that digoxin and CHL compete for binding in the same pocket (38).
Fig. 3.
(A–L) Dose–response curves of competitive TR-FRET coactivator recruitment assays by titration of allosteric ligands MRL-871 (A–D), FM26 (E–H), and compound 13 (I–L) to RORγt in the presence of fixed concentrations of 20-OH (A, E, and I), 25-OH (B, F, and J), DSM (C, G, and K), and CHL (D, H, and J) (0.00, 0.25, and 1.00 µM). The data were normalized with regards to plateau levels (nonnormalized data are provided in SI Appendix, Fig. S3). The data for FM26 were adapted from Meijer et al. (38). (M–P) Overview of the IC50 values for MRL-871, FM26, and compound 13, at different fixed concentrations of the orthosteric ligands. The IC50 values decreased as the concentration of orthosteric ligand increased. (Q) Schematic representation of the TR-FRET coactivator recruitment assay. When RORγt is in its apo or agonist-bound state, the cofactor binds to the LBD, resulting in FRET pairing from an anti-His terbium cryptate donor to the D2-labeled streptavidin (binding to biotin-labeled cofactor). (R) Dose–response curves of a TR-FRET assay with an AlexaFluor647-MRL-871 allosteric probe, by titration of the orthosteric ligands 20-OH, 25-OH, DSM, and CHL to a fixed concentration of RORγt (20 nM) and allosteric MRL-871 probe (100 nM). (S) Schematic representation of the TR-FRET assay using the AlexaFluor647-labeled MRL-871 probe. When the probe binds to the RORγt LBD, fluorescence emission occurs via FRET pairing between an anti-His terbium cryptate donor and the probe. Data are recorded in triplicate from three independent experiments (one representative dataset shown). Error bars represent the SD of the mean.
Next, dose–response curves for the allosteric ligands were obtained in presence and absence of orthosteric ligands (Fig. 3 A–L). In isolation these allosteric compounds exhibit dose-dependent inverse agonistic behavior, as has been reported previously (35, 38, 40). Crucially, when the allosteric ligands were titrated to assay mixtures containing different fixed concentrations of orthosteric ligands, their IC50 values decreased (i.e., they became more potent) as the concentration of orthosteric ligand increased (Fig. 3 M–P and SI Appendix, Table S3). This synergistic effect is demonstrated by the dose–response curves shifting to the left and increasing Hill slopes (Fig. 3 A–L). Although 25-OH was found to be an inverse agonist, the enhancing effect on the potency of the allosteric ligands was still observed. These data are compelling evidence that there is cooperative dual ligand binding to RORγt that enhances the potency of the allosteric inverse agonists.
Differences between the cooperative responses were observed for the orthosteric ligands. 25-OH and DSM had the most profound effect on allosteric inverse agonist IC50 values compared to 20-OH and CHL (Fig. 3 M–P). Furthermore, 25-OH and DSM appear to show a maximum cooperative response at the lowest concentration of 0.25 μM, compared to CHL and 20-OH which show a greater effect at higher concentration (1.00 μM) (Fig. 3 M–P). These differences are likely to be a result of a combination of slightly differing agonist binding modes and affinities to the orthosteric pocket.
Orthosteric RORγt Ligands Increase the Binding Affinity of a Fluorescently Labeled Allosteric RORγt Ligand.
An orthogonal TR-FRET assay format (Fig. 3S) was used to evaluate the effect of orthosteric ligands on the binding affinity of an AlexaFluor647-labeled MRL-871 probe (SI Appendix, Fig. S4) to the allosteric pocket of RORγt LBD. Fluorescence emission from the probe occurs upon binding to the RORγt LBD as a result of a FRET pairing with an anti-His terbium cryptate donor bound to the protein. The assay thus directly measures allosteric ligand binding, as opposed to coactivator recruitment to the LBD as described above. Titration of all four orthosteric ligands to a fixed concentration of RORγt and the MRL-871 probe increased fluorescence emission in a dose-dependent manner (Fig. 3R). This shows that orthosteric ligand binding increases the affinity of the allosteric probe and thus provides further compelling and consistent evidence for cooperative dual ligand binding to RORγt.
As seen in the coactivator recruitment assay, the orthosteric ligands increased the binding affinity of the allosteric probe by different extents. Here 20-OH had the most profound effect followed by 25-OH and DSM. CHL was least effective at inducing allosteric probe binding. The pattern of activity was not entirely consistent with that observed in the coactivator binding assay where 20-OH had a lesser effect compared to 25-OH and DSM. Coactivator binding and allosteric ligand binding are discrete events, and thus, it is possible that subtle structural differences have a greater impact on one binding event compared to the other.
Cocrystal Structures Give Molecular Insights in the Simultaneous Binding of Orthosteric and Allosteric RORγt Ligands.
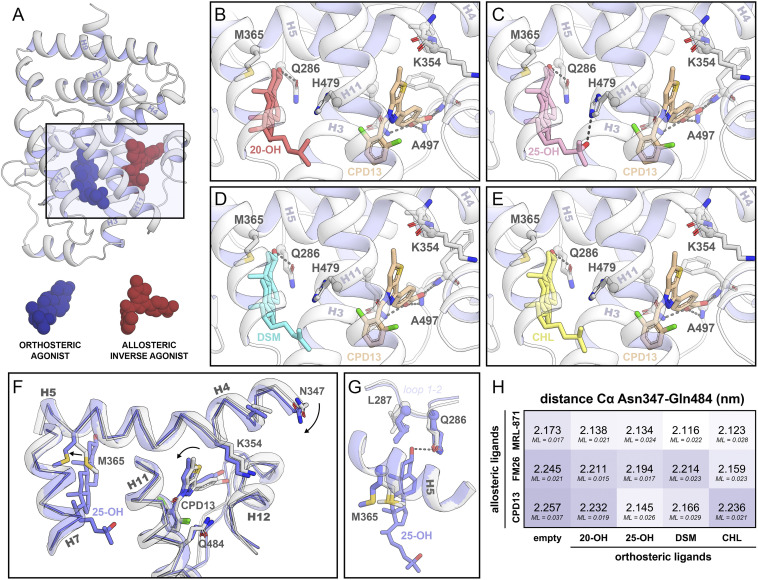
X-ray protein crystallography was used to examine the impact of cooperative dual ligand binding on protein flexibility, protein folding, and ligand binding modes. Crystal packing and buffer additives can have a significant impact on the overall fold and flexibility of parts of the protein. Therefore, conditions were screened to establish identical crystal packing. This allowed for comparison of the crystal structures and minimized the possibility of crystallization artifacts (SI Appendix, Fig. S5). Using this approach, the first ternary complexes of RORγt bound to both an orthosteric and allosteric ligand were crystallized. In total, 12 high-resolution crystal structures were solved, including all combinations of the four orthosteric (20-OH, 25-OH, DSM, and CHL) and three allosteric ligands (MRL-871, FM26, and compound 13) (Fig. 4 B–E and SI Appendix, Fig. S6 and Tables S5–S7).
Fig. 4.
Crystal structures of RORγt in complex with orthosteric and allosteric ligands. (A) Cartoon representation of RORγt in complex with an orthosteric (blue spheres) and an allosteric ligand (red spheres). The rectangle indicates the location of the zoomed-in orthosteric and allosteric LBP. (B–E) The orthosteric and allosteric LBP of RORγt in the presence of various orthosteric ligands (20-OH in red, 25-OH in pink, DSM in blue, and CHL in yellow) and the allosteric ligand compound 13 (brown). (F) Comparison of the crystal structures of RORγt in the presence (blue; PDB entry 6T50) or absence (white; PDB entry 6TLM) of an orthosteric modulator. The presence of the orthosteric modulator shifts helix 4 toward the allosteric pocket, thereby clamping the allosteric ligand. (G) Focused view of the orthosteric LBP. Side chains of Gln286, Leu287, and Met365 are shown for all crystal structures containing ligands in both pockets (12 structures in blue) as well as in absence of an orthosteric modulator (three structures in white; PDB entries 5C4O, 6SAL, and 6TLM). The presence of the orthosteric ligand locks Met365 into a defined state which is conserved for all 12 crystal structures containing orthosteric ligands. (H) Distance (in nm) between the α-carbons of Asn347 (helix 4) and Gln484 (helix 11) in the crystal structures. The maximum-likelihood coordinate error (ML; in nm) is provided for every structure.
All 12 ternary RORγt crystal structures reveal the protein folded into a conformation where H12 is positioned over the allosteric ligand and thus physically preventing potential coactivator binding. This is consistent with the binary crystal structures of RORγt in complex with an allosteric modulator only (35, 38, 40). Previously, we reported that allosteric ligands FM26 and compound 13 introduce more bulk toward helix 4 of RORγt compared to MRL-871, shifting helix 4 toward helix 9 (SI Appendix, Fig. S7) (38, 40). Interestingly, the additional binding of an orthosteric ligand is seen to reverse this process in all the ternary crystal structures containing an orthosteric ligand and FM26 or compound 13 (Fig. 4 F and H and SI Appendix, Fig. S8). For the structures containing MRL-871, a similar but less pronounced effect is observed.
Superposition of the crystal structures showed that the binding modes of the CHL derivatives are comparable to those seen in earlier binary structures of RORγt in complex with the orthosteric ligands 20α- and 25-OH (Protein Data Bank [PDB] entries 3KYT and 3L0L, respectively) (42). Comparison of the orthosteric pockets shows that the conformation of surrounding residues is predominantly unaffected by binding of the CHL derivatives, which is the same for the allosteric pocket. However, an altered conformation was observed for Met365 which, in the absence of an orthosteric ligand, was oriented toward the center of the orthosteric pocket. The CHL derivatives occupy this part of the pocket, thereby locking Met365 in a distinctly repositioned conformation (Fig. 4G). This Met365 repositioning is correlated with a restriction of the movement of helix 5 and leads to conformational change of helix 4 toward the allosteric ligand, a clamping effect that is seen in all the crystal structures (Fig. 4 F and H and Movie S1 capturing the helix movement). The distance between α-carbons of Asn347 on helix 4 and Gln484 on helix 11 is used as a measure for the clamping motion (Fig. 4H). Although the movement of helix 4 is restricted by the allosteric ligand, the presence of the orthosteric ligands can reduce this distance by 0.1 nm. To accommodate for the altered conformation of helix 4, the allosteric ligands slightly twist, following the motion of helix 4 (Fig. 4F and SI Appendix, Fig. S8). A larger movement of helix 4 toward the allosteric ligand is correlated with a larger twist of the allosteric ligand (SI Appendix, Fig. S8). Consequently, the conformation of the loop between helix 11 and 12 changes due to the polar interactions of the conserved carboxylic acid of the allosteric ligands with the protein backbone. Altogether, the structural data reveal a molecular mechanism of how the presence of an orthosteric ligand influences the binding behavior of the allosteric ligand.
Orthosteric Ligands Restrict the Conformational Flexibility of RORγt Met365 and Alter the Conformation of Helices 7 and 11.
MD simulations were performed to investigate the interplay between orthosteric and allosteric ligand binding. For this, cocrystal structures of RORγt in complex with both ligands were compared to the respective structure with only the allosteric modulator bound (Fig. 5A). To improve the reliability of the simulations, five independent simulations were performed per complex, each starting from a random initial velocity distribution.
Fig. 5.
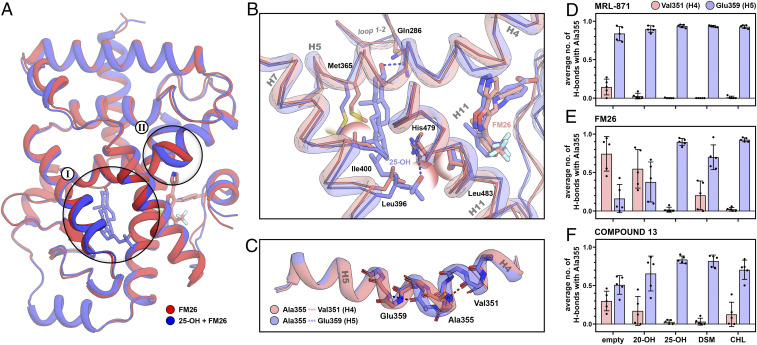
Comparison of the RORγt complexes bound to an allosteric ligand in the presence (blue) or absence (red) of an orthosteric ligand using MD. (A) Superposition of the average structure of FM26 with and without 25-OH. The orthosteric LBP (I) and the transition between helices 4 and 5 of RORγt (II) are highlighted. (B) Focused view of the orthosteric LBP of RORγt. Polar interactions are shown as dashed lines, and steric clashes are shown as semitransparent spheres. (C) Isolated helices 4 and 5 showing the conformational switch of Ala355 from helix 4 to 5 upon orthosteric ligand binding. (D–F) The average number of hydrogen bonds of Ala355 with Val351 (helix 4; red) or Glu359 (helix 5; blue) in the presence of different orthosteric and allosteric modulators over the course of the simulation. Bars represent the average value over five independent simulations with the individual values represented as black spheres and the error bar showing the SD.
For all simulations, no large conformational changes in the tertiary structure of the protein or ligand conformation were observed (SI Appendix, Figs. S10–S12). The presence of any orthosteric ligand significantly reduced the overall flexibility of the complete protein backbone (SI Appendix, Fig. S13). Consistent with the crystal structures, Met365 of RORγt showed limited conformational freedom in the orthosteric pocket due to steric hindrance with the C ring of the CHL derivatives. The specific conformation of Met365 leads to a repositioning of Ile400 on helix 7, thereby shifting this helix away from helix 5 (Fig. 5B). The aliphatic tail of the CHL derivatives is oriented toward Leu483 on helix 11, causing this helix to move toward the allosteric ligand, restricting the overall mobility of both the allosteric ligand and helix 11 (Fig. 5B and SI Appendix, Fig. S13).
Orthosteric Ligands Influence the Helix Participation of Ala355 Resulting in Clamping of the Allosteric Ligand Binding Pocket.
We investigated in detail the helix 4 shift which was observed in our crystallographic data upon binding of the orthosteric ligand. The MD simulations show an unusual mechanism by which this helix movement takes place. In the crystal structures, Ala355 is located at the end of helix 4, but during our simulations, Ala355 showed the ability to exchange its participation between helices 4 and 5. The RORγt structures containing both ligands significantly bias Ala355 toward the helix 5 conformation compared to the complexes with only the allosteric ligand present (Fig. 5C). This altered equilibrium of conformations is more distinct for complexes with the bulkier allosteric ligands FM26 and compound 13 since these ligands promote the helix 4 conformation for Ala355 in absence of an orthosteric ligand. The participation in helix 5 by Ala355 induces a shift of helix 4 toward the allosteric ligand, moving the ligand deeper into the allosteric binding pocket (SI Appendix, Fig. S9). In agreement with the crystal structures, an apparent clamping motion of helix 4 can be observed for all structures containing an orthosteric ligand, illustrated by the distance between the α-carbons of Asn347 and Gln484 (SI Appendix, Fig. S14).
The effect of different orthosteric ligands on the conformation of Ala355 was determined by measuring the average number of H-bonds of Ala355 with the backbone of Val351 (helix 4) and Glu359 (helix 5) over the course of the simulation (Fig. 5 C–F). The structures containing MRL-871 showed that together with binding of any of the orthosteric ligands, Ala355 is almost exclusively in the helix 5 conformation (Fig. 5D). For the complex of RORγt with FM26 alone, Ala355 is primarily in the helix 4 conformation, but in the presence of an orthosteric ligand, the equilibrium completely shifts toward helix 5 (Fig. 5E). A similar behavior is observed for compound 13 (Fig. 5F). For all structures containing 25-OH, Ala355 showed to be almost exclusively in the helix 5 conformation, independent of which allosteric ligand is bound. In contrast, 20-OH only had a minor effect on the conformational equilibrium. It appears that the flexibility of helix 5 plays a key role in defining the conformation of Ala355. Without an orthosteric ligand present, helix 5 acts as a spring, allowing movement of helices 4 to 5 and allowing Ala355 to sample both conformations. Upon binding of an orthosteric ligand, the conformation of Met365 is locked making the spring more rigid, promoting the helix 5 conformation. This provides an explanation of why orthosteric ligands with less conformational freedom in the ligand binding pocket (LBP), due to additional polar interactions or more rigid alkene bonds for 25-OH and DSM respectively, more effectively induce the helix 5 conformation. The absolute conformation of Ala355 is directly correlated to the binding mode of the allosteric ligand. Therefore, the extent by which the orthosteric ligand adjusts the conformational equilibrium of this residue will define the cooperative binding behavior.
Discussion
Cooperative dual ligand binding is a relevant but poorly understood concept in drug discovery. Instead of competing with an endogenous ligand, the endogenous ligand and an allosteric ligand can collaborate to produce a modulated pharmacological response. It is difficult to rationally design ligands that show a predictable cooperative binding behavior, which is primarily caused by a lack of structural understanding of the underlying cooperativity. In this work, we used a combination of biochemical data, protein crystallography, and MD simulations to produce a mechanistic explanation of how cooperative dual ligand binding occurs for the NR RORγt. The thermal shift data indicated cooperative stabilization of RORγt folding by dual ligand binding. The TR-FRET cofactor recruitment assays demonstrated the functional effect of the cooperative binding by an enhanced inhibitory potency of the allosteric ligands on cofactor binding, in the presence of an orthosteric ligand. Although all orthosteric ligands show cooperative behavior in these TR-FRET data, they all do this to different extents. DSM and 25-OH give the highest cooperative response, showing a significant decrease in IC50 values of the allosteric ligands, whereas CHL and 20-OH show only minor effects. In an orthogonal TR-FRET assay format we showed that orthosteric ligand binding directly increased the binding affinity of an allosteric probe, which provided additional evidence for cooperative dual ligand binding.
The determination of the ternary crystal structures of RORγt with all combinations of orthosteric and allosteric ligands allowed for the elucidation of a mechanistic explanation for the cooperative dual ligand binding behavior. Orthosteric ligands lock Met365 in a distinct conformation that leads to a conformational change of helices 4 to 5 which results in a clamping effect of the allosteric binding pocket. This results in a modest conformational change of the allosteric ligand. Frequently, these small differences between crystal structures go unnoticed because of a lack of proper reference structures, but they can be critical to explain protein functioning (48). The generation of the 12 closely related ternary structures allowed for the clear delineation of these differences.
MD simulations further confirmed the clamping behavior of the orthosteric pocket upon orthosteric ligand binding. The clamping motion was achieved by a yet, to our knowledge, unknown characteristic of Ala355 that showed the ability to transition between the end of helix 4 and the beginning of helix 5, which plays a significant role in the clamping effect. Restricting the conformational flexibility of Met365 with an orthosteric ligand limits the movement of helices 4 to 5, thereby promoting the helix 5 conformation of Ala355, resulting in a conformational change of helix 4 toward the allosteric ligand. DSM and 25-OH more effectively directed the absolute conformation of Ala355 to helix 5 compared to CHL and 20-OH. A similar trend was observed for these compounds in the TR-FRET data where DSM and 25-OH also showed the largest effect on the increase in potency for the allosteric ligands. This is likely to be caused by the reduced flexibility of these ligands in the orthosteric LBP and, therefore, more effective locking of Met365. Ultimately, the conformation of Ala355 is directly correlated to the binding mode and, as a result, the binding affinity of the allosteric ligand. Considering the TR-FRET data, the helix 5 conformation of Ala355, and the associated clamping motion of helix 4, shows a positive effect on the binding affinity of the allosteric ligand and provides an explanation for the cooperative binding behavior.
Kojetin et al. already demonstrated the essential role of helices 4 to 5 in the allosteric regulation of dimerization and the AF-2 site of RXR (48). The bent conformation of helices 4 to 5 is a common characteristic within the NR family. Like RORγt, most of the NR family members contain a conformationally flexible residue at the transition between these two helices (49). Therefore, it is likely that orthosteric ligand binding also has a significant effect on the dynamics and conformation of helices 4 to 5 of other NRs (48). In addition to H12, also helix 4 is essential for the recruitment of cofactors (17). An altered behavior of helix 4 as a result of orthosteric ligand binding could therefore potentially influence the cofactor binding behavior across all NRs.
In summary, our data provide a mechanistic explanation for cooperative dual ligand binding in NRs, via a mechanism in RORγt that operates via an internal conformational change of the LBD. The specific RORγt cooperativity data in this study let us speculate that similar mechanistic concepts can also be found to govern other NRs and protein classes containing two binding sites. These mechanistic insights bring the pharmacological concept of cooperative dual ligand binding for NRs a step closer to implementation in NR drug discovery. The potential to further enhance the pharmacological effects of allosteric ligands by an interplay with the endogenous orthosteric NR ligands provides a highly attractive entry for a novel NR pharmacology.
Materials and Methods
RORγt-LBD Expression and Purification (Used for Thermal Shift and TR-FRET Assays).
A pET15b expression vector encoding the human RORγt LBD (residues 265 to 518) with an N-terminal His6-tag was transformed by heat shock into BL21(DE3) Escherichia coli cells. Single colonies were used to inoculate precultures of 8 mL LB-media containing 100 μg/mL ampicillin. After overnight incubation at 37 °C each preculture was transferred to 1L TB media supplemented with ampicillin (100 μg/mL) and incubated at 37 °C until an OD600 nm = 1.0 was reached. Protein expression was then induced with 0.5 mM isopropyl-b-d-thiogalactoside (IPTG), and cultures were incubated for 16 h at 18 °C. The cells were collected by centrifugation and suspended in lysis buffer (300 mM NaCl, 20 mM Tris HCl, pH 8.0, 20 mM imidazole, 1 mM TCEP, 10% vol/vol glycerol, cOmplete, EDTA-free Protease Inhibitor Mixture tablets (1 tablet/50 mL lysate) and benzonase [0.1 μL/1 mL]). After lysis using a C3 Emulsiflex-C3 homogenizer (Avestin) the cell lysate was cleared by centrifugation at 4 °C, and the protein was purified via Ni2+ affinity column chromatography. Fractions containing the protein of interest were combined and dialyzed to 150 mM NaCl, 20 mM Tris (pH 8.0), 5 mM DTT, and 10% vol/vol glycerol.
Thermal Shift Assay.
Thermal shift assays (TSAs) were performed using 40-μL samples containing 5 μM RORγt-LBD, allosteric ligand (15 μM MRL-871, 60 μM FM26, 20 μM compound 13 [lowest concentrations giving a maximal ΔTm]) or compound combination (combining the previous allosteric compound concentrations with 60 μM 20α-OH-CHL) and 2.5× SYPRO Orange (Sigma) in buffer containing 150 mM NaCl, 10 mM Hepes, 1% DMSO, and 1% ethanol. The samples were heated from 35 to 75 °C at a rate of 0.3 °C per 15 s in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Excitation (575/30 nm) and emission (630/40 nm) filters were used and the reported melting values were calculated as the minimum in the negative derivative of the resulting melting curve. ΔTm values are determined as mean ± SD from three independent experiments performed in triplicate and normalized to DMSO.
TR-FRET Coactivator Recruitment Assay.
Assays were conducted using 100 nM N-terminal biotinylated SRC-1 box2 peptide (Biotin-N-PSSHSSLTARHKILHRLLQEGSPSD-CONH2) and 20 nM His6-RORγt-LBD in buffer containing 10 mM Hepes, 150 mM NaCl, 5 mM DTT, 0.1% BSA (wt/vol), and 0.1 mM CHAPS, pH 7.5. A terbium labeled anti-His antibody (CisBio Bioassays, 61HISTLA) and D2-labeled streptavidin (CisBio Bioassays, 610SADLA) were used at the concentrations recommended by the supplier. Compounds (dissolved in DMSO) were titrated using a 2× dilution series in Corning white low-volume, low-binding 384-well plates at a final volume of 10 μL. The final DMSO concentration was 2% vol/vol throughout. The plate was incubated at room temperature for 30 min and centrifuged before reading (excitation = 340 nm; emission = 665 and 620 nm) on a Tecan infinite F500 plate reader using the parameters recommended by CisBio Bioassays. The data were analyzed with Origin Software. The dose–response curve was fitted represented by
where y is the FRET ratio ([acceptor/donor] * 1,000), A1 is the bottom asymptote, A2 is the top asymptote, p is the Hill slope, x is the ligand concentration in μM, and x0 is the IC50 value in μM. Where dose–response curves did not reach a bottom asymptote, this was fixed at the value of the negative control. The data were normalized with regard to plateau levels (positive and negative control values). Data were recorded in triplicate from three independent experiments. Error bars represent the SD of the mean.
Competition TR-FRET Coactivator Recruitment Assay.
Competition assays were performed in an analogous fashion to that described above only in the presence of fixed concentrations of CHL/DSM/20-OH/25-OH: 0 μM (DMSO), 0.25 μM, 1.0 μM such that the final concentration of DMSO remained at 1.2% vol/vol.
Ligand Binding TR-FRET Assay.
Assays were conducted using 100 nM Alexa647-labeled MRL-871 and 20 nM His6-RORγt-LBD in buffer as described above. A terbium-labeled anti-His antibody (CisBio Bioassays, 61HISTLA) was used at the concentrations recommended by the supplier.
RORγt-LBD Expression and Purification (Used for Crystallography).
The plasmid used for crystallography was ordered from Genscript. The pET15b vector incorporated human RORγt LBD (AA265-507) with a C455H mutation to enhance crystallization. Using heat shock, the plasmid was transformed into E. coli BL21 (DE3) cells. A single colony was used to culture overnight at 37 °C in 25 mL of LB medium supplied with 100 μg/mL ampicillin. These cultures were transferred to 2 L of 2× YT medium supplied with 0.05% antifoam SE-15 (Sigma Aldrich) with 100 μg/mL ampicillin. After an OD600 of 0.6 is reached, protein expression was induced by adding 0.25 mM IPTG. The protein expression continued overnight at 15 °C. Using centrifugation at 10,000 RCF for 10 min at 4 °C, the cell pellet was collected and thereafter dissolved in lysis buffer (20 mM Tris, 500 mM NaCl, 2 mM TCEP, 0.1% Tween20, 10% glycerol, 10 cOmplete Protease Inhibitor Mixture tablets [Roche], and 25 U/mL Bezonase Nuclease [Millipore], adjusted to pH 8.0). The cells were lysed using an Emulsiflex-C3 homogenizer (Avestin), and the crude protein solution was obtained by centrifugation at 40,000 RCF for 40 min at 4 °C. This solution was loaded on a 5-mL Ni-NTA Superflow cartridge (QIAGEN) which was equilibrated with buffer A (20 mM Tris, 500 mM NaCl, 20 mM imidazole, 2 mM TCEP, 0.1% Tween20, and 10% glycerol). The column was washed with 10 column volumes (CVs) of buffer A and 10 CVs of buffer A with 50 mM imidazole to eliminate unspecific binding of proteins to the resin. The product was eluted from the column using 8 CVs of buffer A with 200 mM imidazole. The elution fraction was dialyzed overnight in buffer A without imidazole. In addition, 1.2 U/mg restriction-grade thrombin was added to the purified protein sample to remove the purification tag. The purified sample was then concentrated using an Amicon Ultra centrifugal filter with a 10-kDa cutoff (Millipore) and loaded on a Superdex 75 pg 16/60 size-exclusion column (GE Life Sciences) using 20 mM Tris, 100 mM NaCl, and 5 mM DTT (adjusted to pH 8.0) as running buffer. Fractions of 3 mL were collected and analyzed using a quadrupole time-of-flight liquid chromatography-mass spectrometry (LC/MS) to only combine fractions where the correct mass of RORγtC455H was found. These fractions were concentrated to a final concentration of 11.1 mg/mL, aliquoted, and stored at −80 °C.
X-Ray Crystallography.
Allosteric ligands MRL-871, FM26, and compound 13 were dissolved in DMSO to final concentrations of 40, 30, and 20 mM, respectively. The CHL derivatives were poorly soluble in DMSO and were therefore dissolved in EtOH to a final concentration of 40 mM. All ligands were aliquoted to prevent freeze–thaw cycles and to prevent evaporation of the ligand solution. For both ligands, 1.7 to 2.5 equivalents were added to the RORγtC455H solution (11.1 mg/mL), and the mixture was incubated on ice. After 1 h, the sample was centrifuged at 20,000 RCF for 20 min at 4 °C to eliminate ligand and protein precipitate. All crystals were produced using a sitting drop crystallization method. MRC-2 well (Hampton Research) plates were prepared using a Mosquito pipetting robot (TTP Labtech) and stored at room temperature. Dependent on the ligand combination used, different crystallization and cryoprotection conditions were used (summarized in SI Appendix, Table S4). In general, crystals grew to their final size overnight and nucleated at the bottom of the well, thereby attaching to the plastic surface. An Ultra Micro-Needle (HR4-849, Hampton Research) was used to dent the plastic right next to the crystal to release the crystal. Diffraction data of the crystals containing 20-OH were collected at the P11 beamline of the Positron Electron Tandem Ring Anlage III (PETRA III) facility at Deutsches Elektronen-Synchrotron (DESY, Hamburg, Germany) while the other crystals were measured at the i03 beamline of the Diamond Light Source (Oxford, United Kingdom). All crystals were measured at 100 K using a wavelength of 1 Å. Initial data processing was performed using the CCP4i2 suite (version 7.0.077) (50). Diffraction Integration for Advanced Light Sources (DIALS) was used to integrate the data, and Aimless was used for scaling (51, 52). Using the RORγt crystal structure in complex with allosteric ligand FM26 (PDB entry 6SAL) as a search model for molecular replacement, Phaser was used to phase the data, and ligand restrains were generated using AceDRG (53, 54). REFMAC and Crystallographic Object-Oriented Toolkit (COOT) were used for sequential refinement and model building (55, 56). Final refinement was performed using phenix.refine from the Phenix software suite (version 1.16_3459) (stereo images are available in SI Appendix, Figs. S15–S26) (57). For all structures, no Ramachandran outliers were observed, except for one in 6TLT. The Ramachandran statistics showed that 98 to 99% of the residues are in the preferred conformation, and 1 to 2% are in the allowed conformation (Ramachandran statistics per dataset are available in SI Appendix, Tables S5–S7). Figs. 1, 4, and 5 were made with PyMOL (version 2.2.3, Schrödinger) (58).
MD Studies.
The GROMACS 2019.3 MD package was used to perform the simulations (59). X-ray structures of RORγt in complex with an allosteric ligand (PDB entries 5C4O, 6SAL, and 6TLM) and both the orthosteric and allosteric ligand (PDB entries 6T4G, 6T4I, 6T4J, 6T4K, 6T4T, 6T4U, 6T4W, 6T4X, 6T4Y, 6T50, 6TLQ, and 6TLT) were used. Whenever necessary, the protein was N-terminally truncated to Thr268 in order to use the same protein sequence for all simulations. The FF14SB force field was used to parameterize the protein (60). Ligands were parameterized separately using the General Amber Force Field (GAFF) (61). The complex was immersed in a cubic box with ∼22,500 TIP3P waters extending 20 Å away from the protein surface (62). The system charge was neutralized using one Cl− ion. The system was first energy minimized using the steepest decent minimization algorithm using a maximum number of 50,000 steps. Next, the system was progressively equilibrated by performing three heavy-atom restrained in the isothermal-isovolumetric (NVT) simulations for 100 ps at 100, 200, and 300 K consecutively (Velocity-rescale thermostat) with a time (coupling) constant of 0.1 ns (63). The final step of equilibration was performed for 100 ps in the isothermal-isobaric (NPT) ensemble at 300 K (Parrinello–Rahman barostat) with a time (coupling) constant of 2.0 ns (64). During all stages, the maximum force on the protein and ligand atoms was set to 1,000 kJ mol−1 nm−2, and the bonds were restrained using the Linear Constraint Solver (LINCS) algorithm (65). The long-range electrostatics were calculated using the Particle Mesh Ewald method with a short range cutoff of 1.0 nm and a grid spacing of 0.16 nm (66). Five independent simulation runs of 100 ns were performed for each system, with every run starting from a random initial velocity distribution.
Supplementary Material
Acknowledgments
We thank Pim J. de Vink for discussions and feedback. We also thank the tutors of the DLS-CCP4 Data Collection and Structure Solution Workshop 2017 at Diamond Light Source (Oxfordshire, United Kingdom). This work was supported by the Netherlands Organization for Scientific Research through Gravity program 024.001.035 and Vici grant 016.150.366 and the European Union through a Marie Skłodowska-Curie Actions (MSCA) Individual Fellowship (R.G.D., H2020-MSCA-IEF-2016, grant no. 705188). This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative.
Footnotes
Competing interest statement: L.B. is scientific cofounder of Ambagon Therapeutics, a 14-3-3 drug discovery company. F.A.M., R.G.D., and L.B. are coinventors of patent WO2020149740: Substituted heterocyclic compounds and their use as retinoid-related orphan receptor (ROR) gamma-t inhibitors.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021287118/-/DCSupplemental.
Data Availability.
Coordinates and structure factors for the RORγt complexes have been deposited in the PDB at Research Collaboratory for Structural Bioinformatics (RCSB) under accession codes 6T4G, 6T4I, 6T4J, 6T4K, 6T4T, 6T4U, 6T4W, 6T4X, 6T50, 6TLQ, and 6TLT.
References
- 1.Meijer F. A., Leijten-van de Gevel I. A., de Vries R. M. J. M., Brunsveld L., Allosteric small molecule modulators of nuclear receptors. Mol. Cell. Endocrinol. 485, 20–34 (2019). [DOI] [PubMed] [Google Scholar]
- 2.van Westen G. J. P., Gaulton A., Overington J. P., Chemical, target, and bioactive properties of allosteric modulation. PLoS Comput. Biol. 10, e1003559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Changeux J. P., Christopoulos A., Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell 166, 1084–1102 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Thal D. M., Glukhova A., Sexton P. M., Christopoulos A., Structural insights into G-protein-coupled receptor allostery. Nature 559, 45–53 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Wu P., Clausen M. H., Nielsen T. E., Allosteric small-molecule kinase inhibitors. Pharmacol. Ther. 156, 59–68 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Lewis J. A., Lebois E. P., Lindsley C. W., Allosteric modulation of kinases and GPCRs: Design principles and structural diversity. Curr. Opin. Chem. Biol. 12, 269–280 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Jacobsen S. E., Gether U., Bräuner-Osborne H., Investigating the molecular mechanism of positive and negative allosteric modulators in the calcium-sensing receptor dimer. Sci. Rep. 7, 46355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capdeville R., Buchdunger E., Zimmermann J., Matter A., Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discov. 1, 493–502 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Whitty A., Cooperativity and biological complexity. Nat. Chem. Biol. 4, 435–439 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Stefan M. I., Le Novère N., Cooperative binding. PLoS Comput. Biol. 9, e1003106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cattoni D. I., Chara O., Kaufman S. B., González Flecha F. L., Cooperativity in binding processes: New insights from phenomenological modeling. PLoS One 10, e0146043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wootten D., Christopoulos A., Sexton P. M., Emerging paradigms in GPCR allostery: Implications for drug discovery. Nat. Rev. Drug Discov. 12, 630–644 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Lu J., et al., Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat. Struct. Mol. Biol. 24, 570–577 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Cui Q., Karplus M., Allostery and cooperativity revisited. Protein Sci. 17, 1295–1307 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang J., et al., Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPARγ. eLife 7, e43320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Putcha B.-D. K., Wright E., Brunzelle J. S., Fernandez E. J., Structural basis for negative cooperativity within agonist-bound TR:RXR heterodimers. Proc. Natl. Acad. Sci. U.S.A. 109, 6084–6087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darimont B. D., et al., Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12, 3343–3356 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos R., et al., A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov I. I., et al., The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Fauber B. P., Magnuson S., Modulators of the nuclear receptor retinoic acid receptor-related orphan receptor-γ (RORγ or RORc). J. Med. Chem. 57, 5871–5892 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Solt L. A., Burris T. P., Action of RORs and their ligands in (patho)physiology. Trends Endocrinol. Metab. 23, 619–627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bronner S. M., Zbieg J. R., Crawford J. J., RORγ antagonists and inverse agonists: A patent review. Expert Opin. Ther. Pat. 27, 101–112 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Pandya V. B., Kumar S., Sachchidanand R., Sharma R., Desai R. C., Combating autoimmune diseases with retinoic acid receptor-related orphan receptor-γ (RORγ or RORc) inhibitors: Hits and misses. J. Med. Chem. 61, 10976–10995 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Cyr P., Bronner S. M., Crawford J. J., Recent progress on nuclear receptor RORγ modulators. Bioorg. Med. Chem. Lett. 26, 4387–4393 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Huh J. R., et al., Identification of potent and selective diphenylpropanamide RORγ inhibitors. ACS Med. Chem. Lett. 4, 79–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanis V. M., et al., 3-substituted quinolines as RORγt inverse agonists. Bioorg. Med. Chem. Lett. 29, 1463–1470 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Imura C., et al., A novel RORγt inhibitor is a potential therapeutic agent for the topical treatment of psoriasis with low risk of thymic aberrations. J. Dermatol. Sci. 93, 176–185 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Tian J., et al., Discovery of N-indanyl benzamides as potent RORγt inverse agonists. Eur. J. Med. Chem. 167, 37–48 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Kotoku M., et al., Discovery of second generation RORγ inhibitors composed of an azole scaffold. J. Med. Chem. 62, 2837–2842 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Duan J. J.-W., et al., Structure-based discovery of phenyl (3-phenylpyrrolidin-3-yl)sulfones as selective, orally active RORγt inverse agonists. ACS Med. Chem. Lett. 10, 367–373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X., et al., Sterol metabolism controls T(H)17 differentiation by generating endogenous RORγ agonists. Nat. Chem. Biol. 11, 141–147 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Kumar N., et al., The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol. Pharmacol. 77, 228–236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huh J. R., et al., Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORγt activity. Nature 472, 486–490 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karstens W. F. J., et al. , “RORgammaT Inhibitors”. PCT Int. Appl. WO2012/106995 (2012).
- 35.Scheepstra M., et al., Identification of an allosteric binding site for RORγt inhibition. Nat. Commun. 6, 8833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouvry G., et al., Discovery of phenoxyindazoles and phenylthioindazoles as RORγ inverse agonists. Bioorg. Med. Chem. Lett. 26, 5802–5808 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Fauber B. P., et al., Discovery of imidazo[1,5-a]pyridines and -pyrimidines as potent and selective RORc inverse agonists. Bioorg. Med. Chem. Lett. 25, 2907–2912 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Meijer F. A., et al., Ligand-based design of allosteric retinoic acid receptor-related orphan receptor γt (RORγt) inverse agonists. J. Med. Chem. 63, 241–259 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaudari S. S., et al., “Bicyclic Heterocyclic Compounds as ROR gamma Modulators” PCT Int. Appl. WO2015/008234 (2015).
- 40.de Vries R. M. J. M., Doveston R. G., Meijer F. A., Brunsveld L., Elucidation of an allosteric mode of action for a thienopyrazole RORγt inverse agonist. ChemMedChem 15, 561–565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang X., et al., A novel series of cysteine-dependent, allosteric inverse agonists of the nuclear receptor RORγt. Bioorg. Med. Chem. Lett. 30, 126967 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Jin L., et al., Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol. Endocrinol. 24, 923–929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gege C., Retinoid-related orphan receptor γ t modulators: Comparison of Glenmark’s me-too patent application (WO2015008234) with the originator application from Merck Sharp and Dohme (WO2012106995). Expert Opin. Ther. Pat. 25, 1215–1221 (2015). [DOI] [PubMed] [Google Scholar]
- 44.DeSantis K. A., Reinking J. L., Use of differential scanning fluorimetry to identify nuclear receptor ligands. Methods Mol. Biol. 1443, 21–30 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Niesen F. H., Berglund H., Vedadi M., The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Gao K., Oerlemans R., Groves M. R., Theory and applications of differential scanning fluorimetry in early-stage drug discovery. Biophys. Rev. 12, 85–104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Degorce F., et al., HTRF: A technology tailored for drug discovery—A review of theoretical aspects and recent applications. Curr. Chem. Genomics 3, 22–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kojetin D. J., et al., Structural mechanism for signal transduction in RXR nuclear receptor heterodimers. Nat. Commun. 6, 8013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leijten-van de Gevel I. A., Brunsveld L., Delineation of the molecular determinants of the unique allosteric binding site of the orphan nuclear receptor RORγt. J. Biol. Chem. 295, 9183–9191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Potterton L., et al., CCP4i2: The new graphical user interface to the CCP4 program suite. Acta Crystallogr. D Struct. Biol. 74, 68–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clabbers M. T. B., et al., Electron diffraction data processing with DIALS. Acta Crystallogr. D Struct. Biol. 74, 506–518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy A. J., Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long F., et al., AceDRG: A stereochemical description generator for ligands. Acta Crystallogr. D Struct. Biol. 73, 112–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murshudov G. N., et al., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afonine P. V., et al., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The PyMOL Molecular Graphics System , Version 2.2.3 (Schrödinger LLC).
- 59.Abraham M. J., et al., Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 60.Maier J. A., et al., ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A., Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 63.Bussi G., Donadio D., Parrinello M., Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 65.Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M., LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997). [Google Scholar]
- 66.Holden Z. C., Richard R. M., Herbert J. M., Periodic boundary conditions for QM/MM calculations: Ewald summation for extended Gaussian basis sets. J. Chem. Phys. 139, 244108 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the RORγt complexes have been deposited in the PDB at Research Collaboratory for Structural Bioinformatics (RCSB) under accession codes 6T4G, 6T4I, 6T4J, 6T4K, 6T4T, 6T4U, 6T4W, 6T4X, 6T50, 6TLQ, and 6TLT.