Significance
Coping with UV-B is crucial for plant survival in sunlight. The UV-B photoreceptor UVR8 regulates gene expression associated with photomorphogenesis, acclimation, and UV-B stress tolerance. UV-B photon reception by UVR8 homodimers results in monomerization, followed by interaction with the key signaling protein COP1. We have discovered a UV-B hypersensitive UVR8 photoreceptor that confers strongly enhanced UV-B tolerance and generated a UVR8 variant based on the underlying mutation that shows extremely enhanced constitutive signaling activity. Our findings provide key mechanistic insight into how plants respond and acclimate to UV-B radiation.
Keywords: UV-B, photoreceptor, acclimation, signal transduction, abiotic stress
Abstract
The plant ultraviolet-B (UV-B) photoreceptor UVR8 plays an important role in UV-B acclimation and survival. UV-B absorption by homodimeric UVR8 induces its monomerization and interaction with the E3 ubiquitin ligase COP1, leading ultimately to gene expression changes. UVR8 is inactivated through redimerization, facilitated by RUP1 and RUP2. Here, we describe a semidominant, hyperactive allele, namely uvr8-17D, that harbors a glycine-101 to serine mutation. UVR8G101S overexpression led to weak constitutive photomorphogenesis and extreme UV-B responsiveness. UVR8G101S was observed to be predominantly monomeric in vivo and, once activated by UV-B, was not efficiently inactivated. Analysis of a UVR8 crystal structure containing the G101S mutation revealed the distortion of a loop region normally involved in stabilization of the UVR8 homodimer. Plants expressing a UVR8 variant combining G101S with the previously described W285A mutation exhibited robust constitutive photomorphogenesis. This work provides further insight into UVR8 activation and inactivation mechanisms and describes a genetic tool for the manipulation of photomorphogenic responses.
Plant growth and development rely on appropriate responses to the light environment. Plants have evolved different photoreceptor families that respond to photons of specific wavelengths: red/far-red light-sensing phytochromes (phyA–E); blue light-sensing cryptochromes (cry1 and cry2), phototropins (phot1 and phot2), and Zeitlupe family proteins (ztl, fkf1, and lkp1); and the ultraviolet (UV)-B-sensing UV RESISTANCE LOCUS 8 (UVR8) (1–4). Photoreceptors shape plant development throughout the entire life cycle. At the seedling stage, photoreceptor-mediated light perception initiates processes such as inhibition of hypocotyl elongation, cotyledon expansion, biogenesis of the photosynthetic machinery, and synthesis of photoprotective pigments (5–7). Notably, phytochrome, cryptochrome, and UVR8 pathways converge on the CONSTITUTIVELY PHOTOMORPHOGENIC 1–SUPPRESSOR OF PHYA-105 (COP1–SPA) E3 ubiquitin ligase complex, which is responsible for degradation of photomorphogenesis-promoting factors and is inactivated by photoreceptors under light (8, 9). Perception of UV-B and blue light through UVR8 and cry1, respectively, is crucial for UV-B protection and survival in the field (10).
UVR8 is a β-propeller protein that exists in a homodimeric ground state held together by an intricate network of salt-bridge interactions (4, 11–13). Absorption of UV-B photons by specific tryptophan residues that provide chromophore function, of which W285 plays a prime role, disrupts the electrostatic interactions stabilizing the homodimer, resulting in UVR8 monomerization. The active UVR8 monomer interacts with the WD40 domain of COP1 using a cooperative binding mechanism involving the core β-propeller domain of UVR8 together with its disordered C terminus, which harbors a so-called valine-proline (VP) motif (14–17). Interaction of UVR8 with COP1 inhibits COP1 activity by direct competition with COP1 substrates and remodeling of COP1 E3 ligase complex composition (6, 17, 18). The COP1 substrate ELONGATED HYPOCOTYL 5 (HY5), a crucial positive regulator of light signaling (19), is induced transcriptionally and stabilized posttranscriptionally and is necessary for the regulation of most UVR8-induced genes (14, 20–24). Additionally, UVR8 regulates the stability or DNA-binding activity of a set of transcription factors affecting UV-B responses (25–32).
REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2 are among early UV-B-induced genes and encode components of a negative feedback loop that down-regulates UVR8 signaling (33). RUP1 and RUP2 physically interact with UVR8 and promote UVR8 redimerization back to its inactive state (33, 34). Additionally, a role for RUP1 and RUP2 as components of an E3 ligase complex mediating HY5 degradation has recently been suggested (35).
To dissect UVR8 function, several studies have made use of targeted mutagenesis to assess the role of individual tryptophan residues in photoreception and the importance of electrostatic interactions in mediating dimerization (4, 11, 12, 36–39). For example, UVR8W285F is locked in its inactive homodimeric form and is thus unresponsive to UV-B (4, 11, 12, 37). Several other mutant forms, including UVR8W285A and UVR8D96N,D107N, display a constitutive interaction with COP1, whereas their downstream effects differ (37–39). Notably, UVR8W285A is a weak UVR8 dimer that does not respond to UV-B activation (4, 11, 37). Overexpression of UVR8W285A leads to constitutive photomorphogenesis, likely through inhibition of COP1 (37). On the other hand, UVR8D96N,D107N is constitutively monomeric; however, despite its constitutive interaction with COP1, UV-B exposure is required to induce physiological responses (11, 39).
We present here uvr8-17D, a semidominant, hypersensitive uvr8 mutant allele that harbors the UVR8G101S protein variant. We found that UVR8G101S is predominantly monomeric in vivo, requires UV-B for activation, and confers an exaggerated UV-B response when expressed at wild-type levels, which is attributed to impaired redimerization and sustained UVR8 activity. Moreover, we describe the engineered UVR8G101S,W285A variant with amazingly strong constitutive activity in vivo.
Results
uvr8-17D Exhibits Enhanced UV-B-Induced Photomorphogenesis.
To identify regulators of the UVR8 pathway, we screened an ethyl methanesulfonate (EMS)-mutagenized Arabidopsis population (Col-0 accession) for mutants with an aberrant hypocotyl phenotype when grown for 4 d under weak white light supplemented with UV-B. Several mutants with long hypocotyls contained mutations in the UVR8 coding sequence; we thus named these alleles uvr8-16 and uvr8-18 to uvr8-30 (SI Appendix, Fig. S1A). Conversely, several of the mutants displaying short hypocotyls in the screen conditions contained mutations in RUP2; we named these alleles rup2-3 to rup2-10 (SI Appendix, Fig. S1B). However, one mutant with enhanced UV-B responsiveness did not carry a RUP2 mutation and showed a semidominant hypocotyl phenotype under UV-B (SI Appendix, Fig. S2A). Interestingly, in this mutant we found a G-to-A transition in the UVR8 locus that results in a glycine-101 to serine (G101S) amino acid change (Fig. 1A); we thus named this allele uvr8-17D.
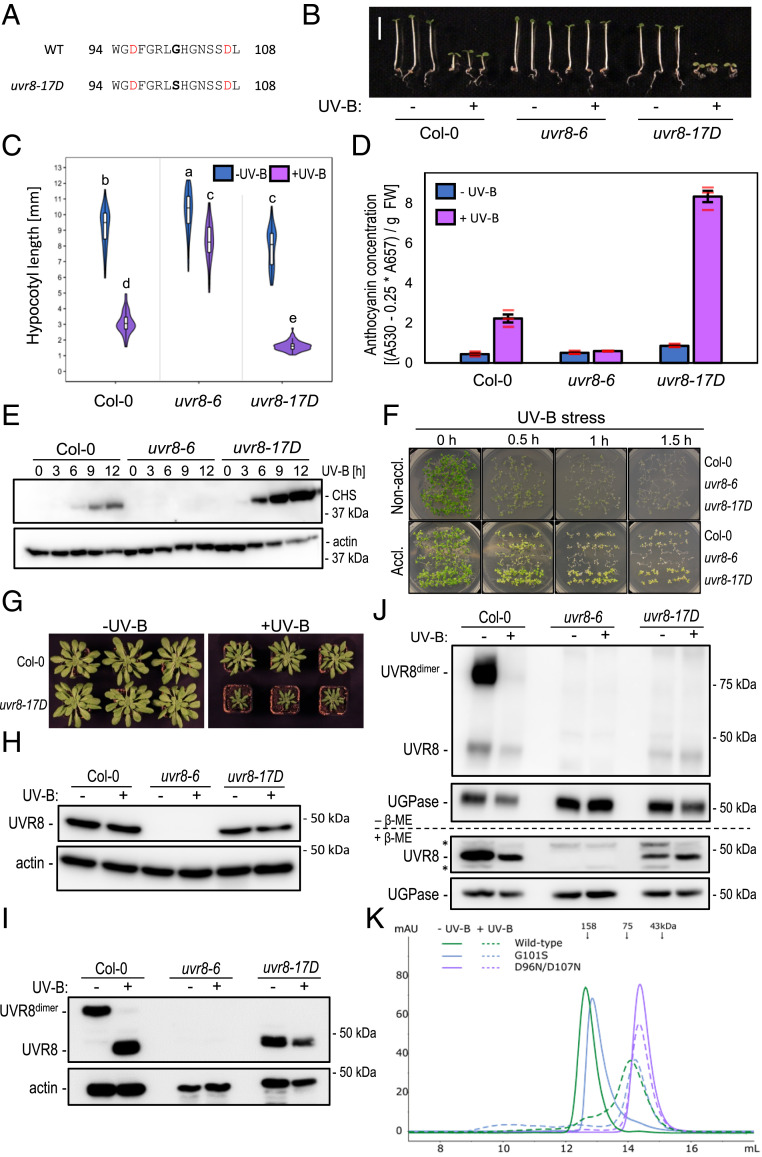
Fig. 1.
Enhanced UV-B photomorphogenesis in plants expressing constitutively monomeric UVR8G101S. (A) UVR8 amino acid sequence of residues 94 to 108 in wild type (WT) and uvr8-17D. The G101S mutation in uvr8-17D is indicated in bold. Asp-96 and -107 involved in dimer interaction are indicated in red. (B) Representative images of wild-type (Col-0), uvr8-6 null mutant, and uvr8-17D seedlings grown in white light or white light supplemented with UV-B. (Scale bar, 5 mm.) (C) Quantification of hypocotyl length (n > 60); shared letters indicate no statistically significant difference in the means (P > 0.05). (D) Anthocyanin concentration; values of independent measurements (red bars), means, and SEM are shown (n = 3). (E) Immunoblot analysis of CHS and actin (loading control) protein levels in Col-0, uvr8-6, and uvr8-17D grown in white light for 4 d and then white light supplemented with UV-B for 0 to 12 h. (F) Survival of Col-0, uvr8-6, and uvr8-17D seedlings after UV-B stress. Seedlings were grown for 7 d in white light (nonacclimated) or white light supplemented with UV-B (acclimated), then exposed to varying durations (0 to 1.5 h) of broadband UV-B stress. Pictures were taken after a 7-d recovery period. (G) Rosette phenotype of Col-0 and uvr8-17D grown for 56 d under short-day conditions in white light or white light supplemented with UV-B. (H) Immunoblot analysis of UVR8 and actin (loading control) protein levels in 7-d-old Col-0, uvr8-6, and uvr8-17D seedlings. (I) UVR8 dimer/monomer status in Col-0 and uvr8-17D. Protein samples were extracted from dark-grown seedlings either exposed or not exposed to 15 min of saturating UV-B and analyzed using immunoblot analysis of samples separated through SDS-PAGE without prior heat denaturation. Actin is shown as loading control. (J) Immunoblot analysis of UVR8 in DSP cross-linked extracts (Top, –β-ME) of dark-grown Col-0, uvr8-6, and uvr8-17D seedlings either exposed or not exposed to 15 min of saturating UV-B. Cross-linking was reversed by treating with 5% β-ME (Bottom, +β-ME). UGPase is shown as loading control. β-ME, β-mercaptoethanol. (K) Size-exclusion chromatography of recombinant UVR8, UVR8G101S, and UVR8D96N,D107N proteins purified from Sf9 insect cells, with and without UV-B treatment.
uvr8-17D seedlings exhibited shorter hypocotyls and elevated anthocyanin levels under UV-B compared with wild type, whereas under white light there were only minor phenotypic differences (Fig. 1 B–D). Hypocotyl lengths were indistinguishable in dark-grown seedlings, suggesting that uvr8-17D does not exhibit significant constitutive activity (SI Appendix, Fig. S2 B and C). Consistent with the UV-B-hypersensitive phenotype, CHALCONE SYNTHASE (CHS) and HY5 accumulated to higher levels in uvr8-17D than in wild type (Fig. 1E and SI Appendix, Fig. S2D). uvr8-17D seedlings also showed enhanced UV-B acclimation and stress tolerance (Fig. 1F). Compared to wild type, uvr8-17D plants showed greater dwarfing at rosette stage under UV-B, whereas they grew normally in the absence of UV-B (Fig. 1G). Altogether, the uvr8-17D phenotype and UV-B responsiveness resembled that of UVR8 overexpression (UVR8-OX) lines (SI Appendix, Fig. S3 A–C) (14); however, UVR8G101S levels in uvr8-17D were comparable to endogenous UVR8 levels in wild type (Fig. 1H and SI Appendix, Fig. S3C).
We tested whether UVR8G101S is affected in its homodimeric ground state or in UV-B-activated monomerization by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis without sample heat denaturation, as previously described (4). In this assay, UVR8G101S migrated as a constitutive monomer, whereas wild-type UVR8 derived from seedlings grown in the absence of UV-B or irradiated with saturating UV-B migrated as a homodimer or a monomer, respectively (Fig. 1I). Moreover, in contrast to wild-type UVR8, no UVR8G101S homodimers could be detected in vivo by dithiobis(succinimidyl propionate) (DSP) cross-linking, even in conditions without UV-B (Fig. 1J). However, when assessing the oligomeric state of recombinant UVR8G101S in vitro, we found through analytical size-exclusion chromatography experiments that the UVR8G101S mutant protein purified from insect cells behaved similarly to wild-type UVR8, in that it eluted as a dimer under −UV-B conditions and as a monomer after UV-B treatment (Fig. 1K). We thus conclude that UVR8G101S is a hypersensitive UVR8 variant, which is very likely associated with its monomeric state in vivo.
UVR8G101S Overexpression Leads to Extreme UV-B Photomorphogenesis.
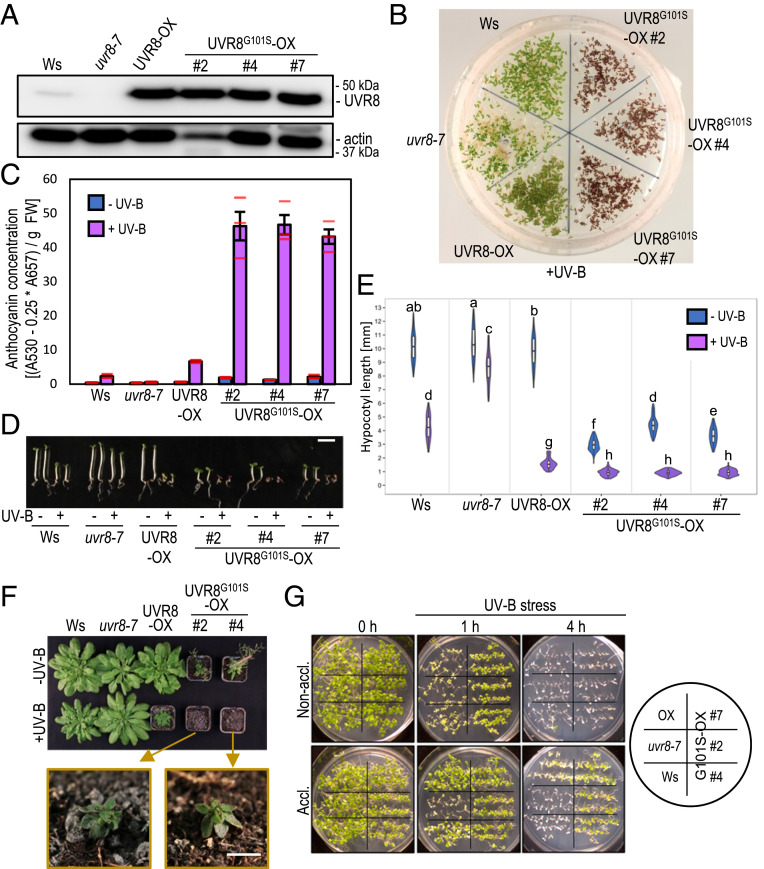
UVR8G101S expression at levels comparable to a previously described UVR8-OX line (14) (Fig. 2A) resulted in strikingly enhanced anthocyanin accumulation under UV-B (Fig. 2 B and C). Interestingly, hypocotyl elongation was strongly reduced in UVR8G101S-OX lines when compared with wild type and UVR8-OX, even in the absence of UV-B (Fig. 2 D and E). To determine whether the latter was due to the extreme sensitivity of UVR8G101S to very low levels of UV-B from the fluorescent tubes in this light field (37) or to the intrinsically constitutive activity of overexpressed UVR8G101S, we measured the hypocotyl length of seedlings grown in the dark or in monochromatic red or blue light. Although UVR8G101S-OX seedlings displayed only a slightly altered hypocotyl-growth phenotype in darkness, some of the lines showed open cotyledons, suggesting that UVR8G101S-OX induces weak constitutive photomorphogenesis (SI Appendix, Fig. S4 A and B). More strikingly, comparable to their phenotype described above (Fig. 2 D and E), UVR8G101S-OX seedlings were indeed shorter in continuous red or blue conditions (SI Appendix, Fig. S4 C–F), suggesting that UVR8G101S broadly sensitizes photomorphogenesis. Consistently, mature UVR8G101S-OX plants exhibited dwarfing and early flowering even in the absence of UV-B, with dwarfing further exaggerated under UV-B (Fig. 2F). In agreement with this enhanced photomorphogenesis, UVR8G101S-OX lines showed a constitutively enhanced UV-B tolerance that was even further enhanced upon UV-B acclimation (Fig. 2G).
Fig. 2.
Overexpression of UVR8G101S results in strongly enhanced UV-B photomorphogenesis. (A) Immunoblot analysis of UVR8 and actin (loading control) protein levels in wild type (Ws), uvr8-7, uvr8-7/Pro35S:UVR8 (UVR8-OX), and three independent uvr8-7/Pro35S:UVR8G101S (UVR8G101S-OX #2, #4, and #7) lines. (B) Representative image of plate-grown seedlings grown under UV-B. (C) Anthocyanin concentration of seedlings depicted in B. Values of independent measurements (red bars), means, and SEM are shown (n = 3). (D) Representative images of seedlings grown in white light or white light supplemented with UV-B. (Scale bar, 5 mm.) (E) Quantification of hypocotyl length of seedlings depicted in D (n > 60). Shared letters indicate no statistically significant difference in the means (P > 0.05). (F) Rosette phenotype of representative plants grown for 56 d under short-day conditions in white light or white light supplemented with UV-B. The closeups show UV-B-grown UVR8G101S-OX plants. (Scale bar, 5 mm.) (G) Survival of seedlings grown for 7 d in white light (nonacclimated) or white light supplemented with UV-B (acclimated) then exposed for varying durations (0 to 4 h) to UV-B stress. Pictures were taken after a 7-d recovery period.
UVR8G101S Shows No Increased Binding Affinity for COP1 under UV-B.
In agreement with the constitutive activity of UVR8G101S when overexpressed, yeast two-hybrid (Y2H) analysis showed that UVR8G101S interacts weakly with COP1 in the absence of UV-B (SI Appendix, Fig. S5A). UV-B irradiation of the yeast strongly enhanced the UVR8G101S–COP1 interaction to a level comparable to the UVR8–COP1 interaction under UV-B, which, however, is strictly UV-B dependent (SI Appendix, Fig. S5A) (4). Similarly, in comparison to UVR8, recombinant UVR8G101S showed enhanced in vitro binding to the COP1 WD40 domain in the absence of UV-B as shown through quantitative grating-coupled interferometry (GCI)-binding assays (SI Appendix, Fig. S5B). UV-B enhanced the affinity of both UVR8 and UVR8G101S for COP1, but the effect was much stronger for wild-type UVR8 (SI Appendix, Fig. S5B). Importantly, the G101S mutation did not increase the affinity of UV-B-exposed UVR8G101S for COP1 in vitro. This suggests that intrinsically increased affinity of UVR8G101S for COP1 does not underlie the enhanced activity of UVR8G101S in planta. However, it should be noted that in addition to the intrinsic binding affinity, the in planta interaction of UVR8 with COP1 is affected by negative regulation through the activities of RUP1 and RUP2 facilitating UVR8 redimerization (34), which may be impaired in the constitutively monomeric UVR8G101S. Indeed, coimmunoprecipitation of COP1 with YFP-UVR8 and YFP-UVR8G101S in transgenic lines not only confirmed the weak constitutive interaction of UVR8G101S with COP1, but also revealed an enhanced association of UVR8G101S with COP1 under UV-B (SI Appendix, Fig. S5C). In agreement with increased COP1 interaction (40), nuclear accumulation of UVR8G101S was enhanced compared to UVR8 (SI Appendix, Figs. S5 and S6). Mechanistically, COP1 activity under UV-B is mainly inhibited through the C-terminal VP motif of UVR8 (15–17), and, in agreement, we observed that CRISPR/Cas9-generated mutations causing truncation of the UVR8 C terminus suppressed the UV-B-hypersensitive phenotype of uvr8-17D (SI Appendix, Fig. S7 A–D).
UVR8G101S,W285A Shows Strong Constitutive Photomorphogenesis.
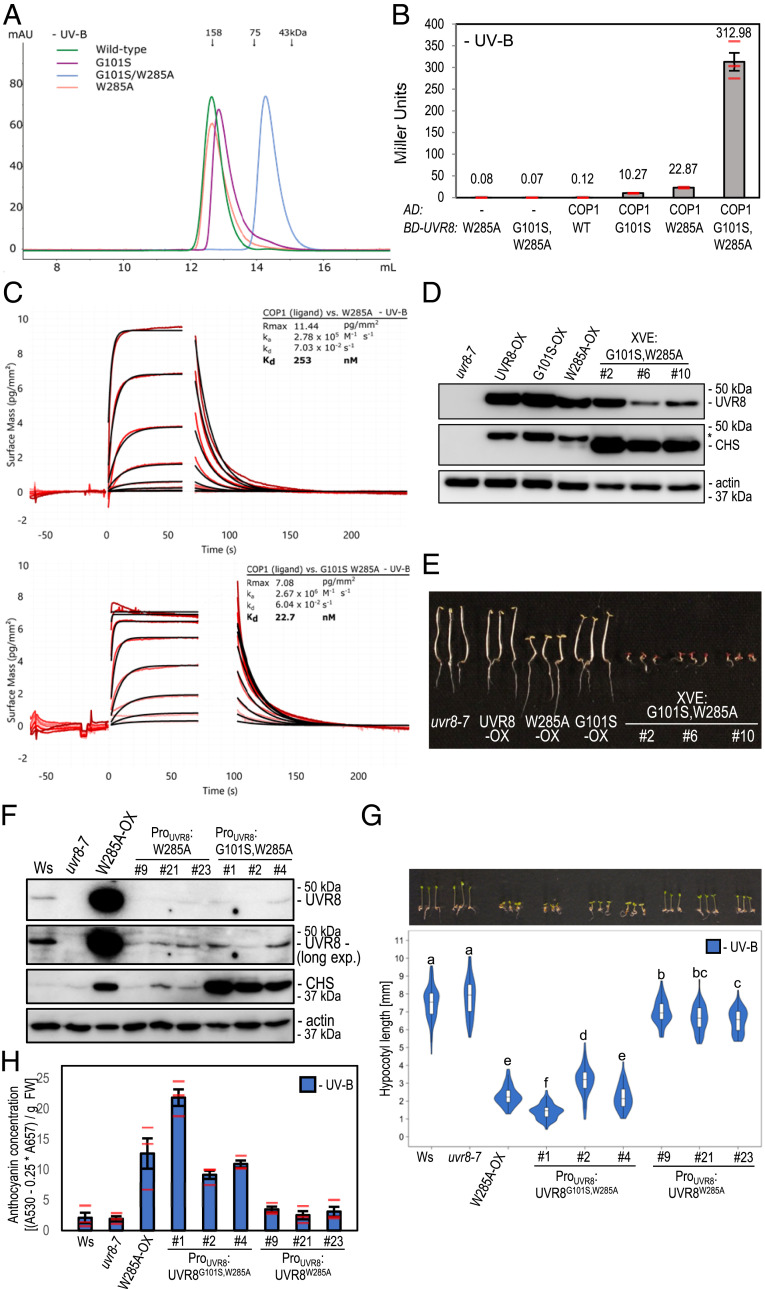
We hypothesized that combining the G101S mutation conferring UV-B hypersensitivity with the W285A mutation conferring weak constitutive activity (37) would create a strong constitutive UVR8 variant. Thus, we created UVR8G101S,W285A, which, in contrast to UVR8W285A and UVR8G101S that were still able to form homodimers in vitro, appeared fully monomeric in vitro as well as in the cross-linking assay in vivo (Fig. 3A and SI Appendix, Fig. S8A). We observed a strongly elevated constitutive interaction of UVR8G101S,W285A with COP1 compared to the COP1–UVR8G101S or COP1–UVR8W285A interaction in the Y2H system (Fig. 3B) as well as in GCI assays (Fig. 3C and see also SI Appendix, Fig. S5B). When we attempted to generate transgenic UVR8G101S,W285A-OX lines (Pro35S:UVR8G101S,W285A), we noticed seedling lethality in the T1 generation, strongly resembling cop1 null alleles (41). We thus generated lines expressing UVR8G101S,W285A under control of the estradiol-inducible XVE system or the UVR8 promoter. Indeed, upon estradiol treatment, XVE:UVR8G101S,W285A lines showed an extreme photomorphogenic phenotype in darkness (Fig. 3 D and E). In lines with weak expression of UVR8W285A and UVR8G101S,W285A under the native UVR8 promoter (Fig. 3F), the former (ProUVR8:UVR8W285A) showed only a very minor phenotype in the absence of UV-B, similar to previous reports (36, 37), whereas the latter (ProUVR8:UVR8G101S,W285A) showed a striking constitutive photomorphogenic phenotype resembling that associated with UVR8W285A-OX (Fig. 3 F–H and SI Appendix, Fig. S8 B and C). Of note, this constitutive photomorphogenesis was despite UVR8G101S,W285A protein levels being lower than endogenous UVR8 in wild type (Fig. 3F). We conclude that the combination of G101S and W285A in UVR8 yields a fully monomeric variant with very strong constitutive activity in vivo.
Fig. 3.
Low expression of UVR8G101S,W285A is sufficient for strong constitutive photomorphogenesis. (A) Size-exclusion chromatography of recombinant UVR8 (wild-type), UVR8G101S (G101S), UVR8W285A (W285A), and UVR8G101S,W285A (G101S/W285A) proteins purified from Sf9 insect cells. (B) Quantitative Y2H analysis of the interaction between COP1 and UVR8 (WT), UVR8G101S, UVR8W285A, and UVR8G101S,W285A in the absence of UV-B. AD, activation domain; BD, DNA-binding domain. (C) Binding kinetics of the full-length UVR8W285A and UVR8G101S,W285A versus the COP1 WD40 domain obtained by GCI experiments. Sensorgrams of protein injected are shown in red, with their respective heterogenous ligand binding model fits in black. The following amounts were typically used: ligand – COP1 (2,000 pg/mm2); analyte – UVR8 (2 μM highest concentration). ka, association rate constant; kd, dissociation rate constant; Kd, dissociation constant. (D) Immunoblot analysis of UVR8, CHS, and actin (loading control) protein levels in seedlings of uvr8-7, uvr8-7/Pro35S:UVR8 (UVR8-OX), uvr8-7/Pro35S:UVR8G101S #2 (G101S-OX), uvr8-7/Pro35S:UVR8W285A (W285A-OX), and three independent lines of uvr8-7/XVE:UVR8G101S,W285A (XVE:G101S,W285A #2, #6, and #10) grown on 5 μM estradiol. Asterisk shows residual UVR8 signal after stripping of the polyvinylidene difluoride (PVDF) membrane. (E) Representative image of seedlings described in D grown for 4 d in darkness on plates supplemented with 5 μM estradiol. (Scale bar, 5 mm.) (F) Immunoblot analysis of UVR8, CHS, and actin (loading control) protein levels in wild type (Ws), uvr8-7, uvr8-7/Pro35S:UVR8W285A, and three independent lines of each of uvr8-7/ProUVR8:UVR8W285A (#9, #21, and #23) and uvr8-7/ProUVR8:UVR8G101S,W285A (#1, #2, and #4). (G) Representative seedling images and quantification of hypocotyl length (n > 60) for seedlings described in F. Shared letters indicate no statistically significant difference in the means (P > 0.05). (Scale bar, 5 mm.) (H) Anthocyanin concentration of seedlings described in F. Values of independent measurements (red bars), means, and SEM are shown (n = 3).
The Combination of G101S and W285F Confirms a Basal Constitutive Activity for UVR8G101S.
We further generated and characterized a UVR8G101S,W285F variant. The W285F mutation is known to abolish all UV-B responsiveness (4, 37), whereas, as described above for UVR8G101S-OX (Fig. 2 C–E), the G101S mutation results in weak constitutive activity in conditions devoid of UV-B and a constitutively monomeric conformation in vivo. Indeed, integrating the G101S mutation into the UVR8W285F variant caused strongly reduced dimer stability in UVR8G101S,W285F, mimicking the conformation of UVR8G101S (SI Appendix, Fig. S8A). As further predicted, UVR8G101S,W285F retained a basal interaction with COP1 in the Y2H system, similar to the COP1–UVR8G101S interaction in the absence of UV-B; however, by contrast, the COP1–UVR8G101S,W285F interaction was not further enhanced under UV-B (SI Appendix, Fig. S9A). UVR8G101S,W285F-OX lines exhibited a phenotype in both the absence and presence of UV-B resembling the phenotype of UVR8G101S-OX lines in −UV-B conditions (SI Appendix, Fig. S9 B–E), confirming that the constitutive phenotype induced by UVR8G101S overexpression in the absence of UV-B is not linked to W285-mediated photoactivation.
The G101S Mutation Distorts a Critical Loop Interaction in the UVR8 Dimer.
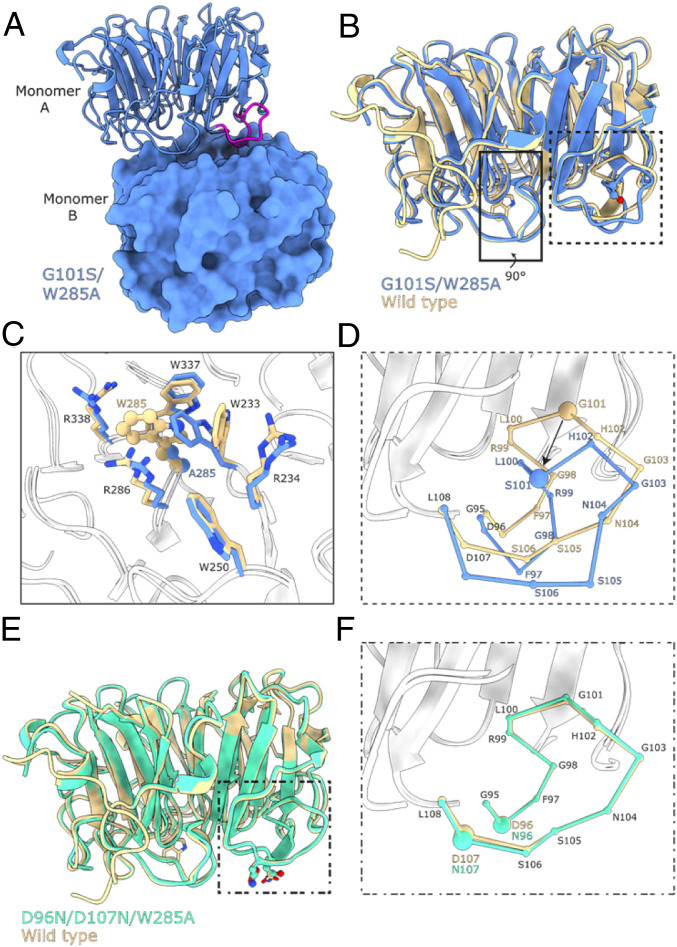
We next investigated the effects of the G101S mutation in structural detail. No crystals could be obtained for the UVR8G101S protein, yet crystals developed for UVR8G101S,W285A, diffracting to 1.75-Å resolution (SI Appendix, Table S1). The structure revealed a nonsymmetric dimeric arrangement of UVR8G101S,W285A in the crystal lattice, as the relative position of the monomers are rotated ∼10° with respect to the previously determined symmetric UVR8 wild-type dimer (SI Appendix, Figs. S10 and S11) (11). The altered dimeric assembly of UVR8G101S,W285A contains a unique salt-bridge/hydrogen bond network between the two monomers with a reduced number of interactions compared to the wild-type dimer (SI Appendix, Fig. S11 and Table S2). The G101S mutation maps to a loop that forms part of the crystallographic UVR8 dimer interface (Fig. 4A and SI Appendix, Fig. S12). A structural superposition with wild-type UVR8 revealed that the position of the loop in UVR8G101S,W285A is distorted by the G101S mutation (Fig. 4 B–D). Importantly, the slightly larger Ser side chain cannot be accommodated within the loop. This forces a movement of the loop that affects the position of Asp-96 and -107, the mutation of which to Asn has been previously shown to impair UVR8 homodimerization (11, 39). The W285A mutation results in rearrangements of adjacent Trp side chains reminiscent of the movements observed in the UVR8W285A crystal structure (12), highlighting the potential changes upon UV-B irradiation (Fig. 4C). We also obtained crystal structures of UVR8D96N,D107N and UVR8D96N,D107N,W285A variants that revealed no large structural rearrangements in the loop (Fig. 4 E and F, and SI Appendix, Fig. S13). Together, our structural analysis reveals that the G101S mutation affects the conformation of a loop region involved in UVR8 homodimer stabilization.
Fig. 4.
UVR8G101S,W285A distorts a critical interaction loop at the dimer interface. (A) Ribbon and surface representations of the UVR8G101S,W285A dimer. Highlighted in magenta is the critical loop containing the G101S mutation. (B) Superposition of UVR8G101S,W285A (blue) with a wild-type UVR8 (PDB:4D9S, yellow) in ribbon representation. The site of mutation, residue 101, is represented in a ball-and-stick manner and colored by their atomic identity. Residue 285 is represented by sticks. (C) Zoomed-in view of the site of the W285A mutation. The site of mutation is represented in a ball-and-stick manner and the surrounding residues are shown as sticks. (D) Zoomed-in view of the loop containing the G101S mutation. The loop is represented with each ball corresponding to a Cα carbon to highlight the structural rearrangements of the loop. (E) Superposition of UVR8D96N,D107N,W285A (green) with UVR8 represented as in B. (F) Zoomed-in view of the loop containing G101 represented as in D.
Enhanced Activity of UVR8G101S Is Caused by Impaired Inhibition through RUP1 and RUP2.
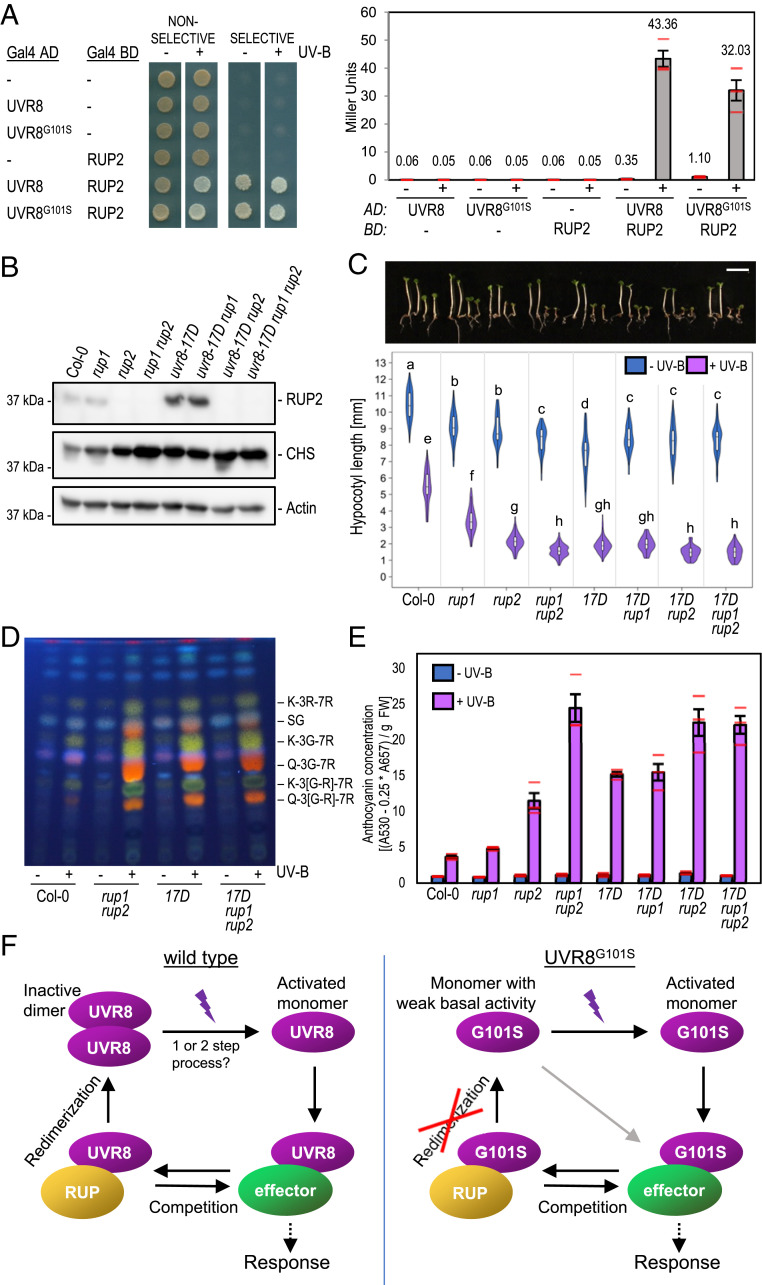
Mutations in RUP1 and RUP2 enhance the UV-B photomorphogenic phenotype, as active monomeric UVR8 is more prevalent in rup1 rup2 (33, 34). We hypothesized that monomeric UVR8G101S enhances UV-B signaling due to its inability to dimerize, thus mimicking a rup1 rup2 mutant after UV-B exposure. We first tested the interaction of UVR8 and UVR8G101S with RUP2 in the Y2H system. Overall, the UVR8–RUP2 interaction was only slightly affected in yeast strains containing UVR8G101S compared to UVR8 (Fig. 5A).
Fig. 5.
The enhanced activity of UVR8G101S depends on the presence of RUP1 and RUP2. (A) Y2H analyses of the interactions of RUP2 with UVR8 and UVR8G101S in the presence or absence of UV-B. Left: growth assay on selective SD/−Trp/−Leu/−His medium. Right: quantitative β-galactosidase assay. AD, activation domain; BD, DNA-binding domain. (B) Immunoblot analysis of RUP2, CHS, and actin (loading control) protein levels in seedlings of wild type (Col-0), rup1, rup2, rup1 rup2, uvr8-17D, uvr8-17D rup1, uvr8-17D rup2, and uvr8-17D rup1 rup2 grown for 4 d in weak white light supplemented with UV-B. (C) Representative images of seedlings described in B (17D, uvr8-17D) and quantification of hypocotyl length (n > 60). Shared letters indicate no statistically significant difference in the means (P > 0.05). (Scale bar, 5 mm.) (D) High-performance thin layer chromatography (HPTLC) analysis of the flavonol glycoside levels in 4-d-old seedlings of wild type (Col-0), rup1 rup2, uvr8-17D, and uvr8-17D rup1 rup2 grown in white light or white light supplemented with UV-B. Identified compounds include: K-3R-7R, kaempferol-3-O-rhamnoside-7-O-rhamnoside; SG, sinapoyl glucose; K-3G-7R, kaempferol-3-O-glucoside-7-O-rhamnoside; Q-3G-7R, quercetin-3-O-glucoside-7-O-rhamnoside; K-3[G-R]-7R, kaempferol 3-O-[rhamnosyl-glucoside]-7-O-rhamnoside; Q-3[G-R]-7R, quercetin 3-O-[rhamnosyl-glucoside]-7-O-rhamnoside. (E) Anthocyanin concentration of seedlings described in B. Values of independent measurements (red bars), means, and SEM are shown (n = 3). (F) Working model of the UVR8 photocycle in the case of wild type (Left) and G101S-mutated UVR8 (Right). In the wild type, UV-B induces monomerization of dimeric UVR8 in a one- or two-step photon absorption process. The activated monomer then interacts with effector proteins such as COP1 and transcription factors to induce a photomorphogenic response. RUP1/RUP2 (RUP) proteins compete with these effectors to abrogate signaling activity. Afterward, RUP-bound UVR8 undergoes redimerization and is brought back to its initial inactive state. UVR8G101S exists as a monomer in vivo and exhibits weak constitutive activity in the absence of UV-B, as seen in overexpression lines. UV-B absorption fully activates the UVR8G101S monomer and RUP proteins still compete with other UVR8 signaling effectors. However, the full cycle cannot be completed because redimerization is intrinsically impossible. This results in sustained UVR8 signaling, leading to enhanced UV-B photomorphogenesis.
We then constructed higher-order mutant combinations between uvr8-17D, rup1, and rup2. Under UV-B, RUP2 was detected at an elevated level in uvr8-17D and uvr8-17D rup1 when compared to that in wild type and rup1, respectively (Fig. 5B). Interestingly, uvr8-17D was similar to rup1 rup2 in terms of hypocotyl length and flavonol accumulation (Fig. 5 C and D), whereas uvr8-17D seedlings displayed an intermediate phenotype between a rup2 and a rup1 rup2 mutant regarding anthocyanin accumulation (Fig. 5E). Together with the observation that the anthocyanin phenotypes of uvr8-17D rup2 and uvr8-17D rup1 rup2 are stronger than that of the single uvr8-17D mutant (Fig. 5E), this suggests that RUP2 is still able to partially inhibit UVR8G101S.
Interestingly, in the presence of functional RUP1 and RUP2, the UVR8G101S mutation significantly enhances the UV-B phenotype (uvr8-17D versus wild type; Fig. 5 C and E); however, in their absence, UVR8G101S does not induce stronger UV-B signaling compared to UVR8 (uvr8-17D rup1 rup2 versus rup1 rup2; Fig. 5 C and E). Together, this demonstrates that, whereas RUP1 and RUP2 proteins retain some ability to negatively regulate UVR8G101S, the enhanced activity of this variant is only evident in a background where RUP1 and RUP2 are present. We conclude that the enhanced activity of UVR8G101S under UV-B is due to impaired redimerization by RUP1 and RUP2, as UVR8G101S shows a strongly reduced ability to dimerize in vivo.
Discussion
uvr8-17D is a UV-B-hypersensitive UVR8 photoreceptor allele that is linked with a single glycine-101 to serine amino acid change. The UV-B hypersensitivity of uvr8-17D is associated with its monomeric conformation in vivo, suggesting that redimerization facilitated by RUP1 and RUP2 is impaired and that UVR8G101S therefore remains active much longer than wild-type UVR8 (Fig. 5F). Our work suggests that UVR8 dimerization is not absolutely required to keep UVR8 inactive, but it is important to make UVR8 inactivatable after activation by UV-B. In agreement, uvr8-17D phenotypes under UV-B resemble those of rup1 rup2 double mutants, further supporting the prime importance of UVR8 redimerization to optimally balance UV-B-induced photomorphogenesis with plant growth and development (33, 34, 42–44).
Intriguingly, UVR8G101S appeared monomeric in vivo, whereas in vitro it appeared monomeric in a gel-based assay and dimeric in a size-exclusion chromatography experiment (Fig. 1K and SI Appendix, Fig. S14). The SDS-PAGE assay is not sensitive enough to recognize weak dimeric variants such as UVR8W285A, which appears monomeric (4, 11, 12, 37). However, in cross-linking and size-exclusion chromatography assays, UVR8W285A is indeed dimeric, whereas UVR8G101S cross-linked as a dimer only poorly and only when overexpressed. UVR8G101S was however dimeric in size-exclusion chromatography assays, which could be linked to the different concentrations of UVR8 protein used and the different chemical environments between the in vitro assay and the in vivo cellular environment.
The monomeric nature of UVR8G101S in vivo is reminiscent of the UVR8D96N,D107N variant previously generated based on structural information (SI Appendix, Fig. S15A) (11, 39). Similar to UVR8G101S, UVR8D96N,D107N also shows weak constitutive COP1 binding (SI Appendix, Fig. S15 B–E) and requires UV-B for functional activation (39). By contrast though, no UV-B hypersensitivity was previously reported for transgenic lines expressing UVR8D96N,D107N (39). However, in our phenotypic assays, transgenic UVR8D96N,D107N-OX lines phenocopied the UV-B-hypersensitive phenotype of UVR8G101S-OX (SI Appendix, Fig. S15 F and G), suggesting that UVR8D96N,D107N and UVR8G101S indeed have similar enhancing effects on UV-B responsiveness in vivo, consistent with our conclusion that the uvr8-17D phenotype is directly linked to its monomeric conformation in vivo. In support, our atomic structure of UVR8G101S,W285A suggests a direct mechanistic link between the mutation of G101 and the misorientation of the D96 and D107 residues, likely underlying the impaired homodimer formation and weak constitutive interaction with COP1. In addition, the constitutive interaction of UVR8G101S and UVR8D96N,D107N with COP1 may sensitize other photomorphogenic pathways by sequestering COP1, although this effect is only apparent when these UVR8 variants are strongly overexpressed.
The effects of UVR8G101S and UVR8D96N,D107N thus suggest that monomeric UVR8 variants require UV-B to trigger the UV-B signaling pathway, confirming that engineered constitutive UVR8 monomers are not sufficient for strong constitutive UVR8 activity. This confirms that monomerization and activation of UVR8 by UV-B can clearly be uncoupled, at least for mutated UVR8 variants (39). However, in these cases, the mutant variants are simply monomeric versions that have not been UV-B activated. Whether wild-type UVR8 proteins that monomerize in response to UV-B—a process associated with broader structural changes that likely also affect the VP-containing C terminus (13, 16, 17, 45, 46)—can or even need to be similarly UV-B activated remains to be shown. Nonetheless, it may be that the activation of wild-type UVR8 is a two-step process, with photon absorption events required both for chromophore excitation that leads to monomerization and for functional activation. An engineered monomeric UVR8 mutant may be primed for activation by bypassing the need for monomerization prior to activation. Alternatively, one photon absorption event may be sufficient for both monomerization and activation. In this case, monomerization and activation by UV-B would be intrinsically linked in wild-type UVR8. Another possibility is that monomeric wild-type UVR8 absorbs additional UV-B photons as an efficient way to keep the photoreceptor active, without going through redimerization and reactivation by monomerization. Conclusive evidence to address these possibilities will require structural determination of full-length UV-B-activated monomer versus an engineered monomeric UVR8.
Despite remaining monomeric in vivo, UVR8G101S retains an interaction with and remains weakly negatively regulated by RUP1 and RUP2. The UVR8–COP1 interaction may be inhibited by direct interaction of RUP proteins with the VP-containing C terminus of UVR8, which is the same domain that UVR8 uses to bind both COP1 and direct transcription factor targets (15–17, 25, 26, 47). This implies that UVR8 redimerization may proceed via two steps: 1) RUPs outcompete COP1 and other VP-domain interactors, separating them from UVR8; and 2) RUPs facilitate UVR8 redimerization. As redimerization occurs at a normal rate in a cop1 background (34), competitive removal of COP1 alone is evidently not sufficient to induce redimerization. In uvr8-17D, step 1 could explain the remaining partial activity of RUPs on UVR8G101S, whereas step 2 is abolished.
Importantly, the uvr8-17D rup1 rup2 triple mutant did not show phenotypic differences compared with rup1 rup2, suggesting the mechanisms underlying UVR8G101S hyperactivity are linked to RUP1/RUP2 activity. As a result of the more prevalent active UVR8 monomer, the increased signaling induced by UVR8G101S originates early and upstream in the pathway and all subsequent UVR8 signaling mechanisms, including UVR8 nuclear accumulation, interaction with COP1, inhibition of its E3 ligase activity, and inhibition of transcription factor activity, are likely to be indirectly promoted by the relatively increased activity of UVR8G101S.
UVR8 is widely conserved across the green lineage, and several studies have confirmed its role in mediating UV-B responses in a diverse range of species (48–56). G101 is conserved in UVR8 orthologs, as are residues involved in photoreception and dimer stability (4, 52, 57) (SI Appendix, Fig. S16). The phenotype of uvr8-17D holds the exciting possibility that a homologous glycine-to-serine mutation leads to enhanced UV-B responses in other species as well. This may eventuate as a convenient way to generate UV-B-hypersensitive phenotypes, including enhanced acclimation and UV-B tolerance, made possible by a single nucleotide substitution. Additionally, expressing UVR8G101S,W285A at very low levels is sufficient to produce a striking constitutive photomorphogenic phenotype. Notwithstanding the use of UVR8G101S in other plant species, both for fundamental research and potential biotechnological applications, uvr8-17D may serve as a genetic tool to further investigate UVR8 activity regulation and the effects of enhanced UVR8 signaling in Arabidopsis.
Methods
Hypocotyl length measurements, extraction, and analysis of anthocyanins and flavonoids, immunoblot analysis, protein cross-linking, Y2H analysis, protein purification from Sf9 cell cultures, analytical size-exclusion chromatography, in vitro methylation, protein crystallization and data collection, crystallographic structure solution and refinement, and GCI are described in SI Appendix, Materials and Methods.
Plant Material.
Arabidopsis thaliana mutants uvr8-6 (14), cop1-4 (58), hy5-215 (59), rup1-1, rup2-1, and rup1-1 rup2-1 (33) are all in the Columbia (Col) accession. uvr8-7, uvr8-7/Pro35S:UVR8 line #2 (UVR8-OX) (14), uvr8-7/Pro35S:UVR8W285A line #3, and uvr8-7/Pro35S:UVR8W285F line #1 (37) are in the Wassilewskija (Ws) accession. uvr8 and rup2 alleles identified in this work stem from an EMS-mutagenized population in the Col accession.
Combinatorial mutants uvr8-17D rup1-1, uvr8-17D rup2-1, and uvr8-17D rup1-1 rup2-1 were generated by genetic crossing. Genotyping of rup1-1 and rup2-1 alleles was performed as described (33), and uvr8-17D genotyping was through PCR amplification of a genomic fragment with a forward (5ʹ-TCG GGA TGA GAT GAT GAC-3ʹ) and a reverse primer (5ʹ-TAG ACC CAA CAT TGA CCC-3ʹ) followed by digestion with HaeIII (NEB) yielding diagnostic fragments of 344/173/145 bp for wild type and 489/173 bp for uvr8-17D.
To generate transgenic lines expressing UVR8 variants, mutations corresponding to G101S, W285A, W285F, and D96N/D107N were introduced into the UVR8 coding sequence in pDONR207 (Thermo Fisher Scientific) by site-directed mutagenesis. UVR8G101S, UVR8G101S,W285A, UVR8G101S,W285F, and UVR8D96N,D107N sequences were inserted into the Gateway-compatible binary vector pB2GW7 (60). UVR8G101S,W285A was also inserted into the Gateway-compatible binary vector pMDC7 allowing estradiol-inducible expression (61). To generate lines expressing YFP-tagged constructs, UVR8 and UVR8G101S were inserted into the Gateway-compatible binary vector pB7WGY2 (60). The control line expressing Pro35S:StrepII-3xHA-YFP in a Col background has been previously described (62).
To generate transgenic lines expressing UVR8 under its own promoter, the UVR8 promoter (1,809 bp upstream of the translational start ATG) was respectively cloned upstream of the UVR8W285A and UVR8G101S,W285A coding sequences and introduced into the Gateway-compatible binary vector pGWB501 (63).
The CRISPR/Cas9 system was used to delete the UVR8 C terminus in wild type and uvr8-17D. An sgRNA directed against the UVR8 sequence was inserted into the pHEE401E vector (64) using overlapping complementary oligos 5ʹ-ATT GGC GAC ACC CAG CTT TTC CC-3ʹ and 5ʹ-AAA CGG GAA AAG CTG GGT GTC GC-3ʹ, and mutants were identified in T1 after sequencing the corresponding part of the UVR8 locus.
Arabidopsis plants were transformed using the floral dip method (65) and transgenic lines with a single insertion locus (75% resistance in T2) were selected for homozygosity in T3.
Growth Conditions.
For hypocotyl length measurements, quantification of anthocyanins, analysis of flavonol glycosides, and acclimation assays, seedlings were grown under aseptic conditions on half-strength Murashige and Skoog (MS) medium (Duchefa) supplemented with 1% (wt/vol) sucrose (Applichem). For immunoblot analysis, seedlings were grown on sterile half-strength MS medium (Duchefa) without sucrose. For induction of estradiol-inducible expression under the XVE system, the growth medium was supplemented with 5 μM of β-estradiol (Sigma).
After sowing, seeds were stratified for 2 d at 4 °C in the dark before the start of the light treatments, which were performed at 22 °C as described before (14, 22). White light (3.6 μmol m−2 s−1) was supplied by Osram L18W/30 tubes, which were supplemented with narrowband Philips TL20W/01RS tubes (1.5 μmol m−2 s−1) for UV-B treatment. To analyze etiolated growth in darkness, seeds were exposed to 6-h white light (approximately 60 μmol m−2 s−1) to induce germination before being transferred back to darkness. For monochromatic light treatments, a light-emitting diode (LED) chamber (floraLEDs, CLF Plant Climatics) was used with 5 μmol m−2 s−1 of blue light or 30 μmol m−2 s−1 of red light.
For analysis of vegetative growth, plants were grown in short days (8-h/16-h light/dark cycles) in UV-B lamp-containing GroBanks (CLF Plant Climatics), under white light (120 μmol m−2 s−1) or white light supplemented with UV-B (1.5 μmol m−2 s−1) (66).
For broadband UV-B treatments (acclimation assays, analyses of dimer/monomer status), Philips TL40W/12RS UV-B tubes were used (4, 14).
Supplementary Material
Acknowledgments
We thank Richard Chappuis for excellent technical assistance, Sarah Raffelberg for providing UVR8D96N,D107N in a yeast two-hybrid expression vector, and José Manuel Nunes (BioSC, Geneva) for helpful discussions regarding statistical analyses. This work was supported by the University of Geneva, a Howard Hughes Medical Institute International Research Scholar Award to M.H., the Swiss National Science Foundation (grant 31003A_175774 to R.U.), and the European Research Council under the European Union’s Seventh Framework Programme (grant 310539 to R.U.). R.P. was supported by an iGE3 PhD salary award and K.L. was supported by a European Molecular Biology Organization long-term fellowship (ALTF 493-2015).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017284118/-/DCSupplemental.
Data Availability.
The atomic coordinates of complexes have been deposited with the following Protein Data Bank (PDB) accession codes: UVR8D96N,D107N: 6XZL (https://www.rcsb.org/structure/6XZL), UVR8D96N,D107N,W285A: 6XZM (https://www.rcsb.org/structure/6XZM), and UVR8G101S,W285A: 6XZN (https://www.rcsb.org/structure/6XZN).
References
- 1.Galvão V. C., Fankhauser C., Sensing the light environment in plants: Photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 34, 46–53 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Tilbrook K., et al., The UVR8 UV-B photoreceptor: Perception, signaling and response. Arabidopsis Book 11, e0164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenkins G. I., The UV-B photoreceptor UVR8: From structure to physiology. Plant Cell 26, 21–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzini L., et al., Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Jenkins G. I., Photomorphogenic responses to ultraviolet-B light. Plant Cell Environ. 40, 2544–2557 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Yin R., Ulm R., How plants cope with UV-B: From perception to response. Curr. Opin. Plant Biol. 37, 42–48 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Demarsy E., Goldschmidt-Clermont M., Ulm R., Coping with ‘dark sides of the sun’ through photoreceptor signaling. Trends Plant Sci. 23, 260–271 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Podolec R., Ulm R., Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 45, 18–25 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Hoecker U., The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 37, 63–69 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Rai N., et al., How do cryptochromes and UVR8 interact in natural and simulated sunlight? J. Exp. Bot. 70, 4975–4990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie J. M., et al., Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335, 1492–1496 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu D., et al., Structural basis of ultraviolet-B perception by UVR8. Nature 484, 214–219 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Zeng X., et al., Dynamic crystallography reveals early signalling events in ultraviolet photoreceptor UVR8. Nat. Plants 1, 14006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favory J. J., et al., Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28, 591–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cloix C., et al., C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc. Natl. Acad. Sci. U.S.A. 109, 16366–16370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin R., Arongaus A. B., Binkert M., Ulm R., Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell 27, 202–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau K., Podolec R., Chappuis R., Ulm R., Hothorn M., Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J. 38, e102140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X., et al., Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc. Natl. Acad. Sci. U.S.A. 110, 16669–16674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osterlund M. T., Hardtke C. S., Wei N., Deng X. W., Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Ulm R., et al., Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 1397–1402 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown B. A., et al., A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. U.S.A. 102, 18225–18230 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oravecz A., et al., CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18, 1975–1990 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X., et al., Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24, 4590–4606 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binkert M., et al., UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26, 4200–4213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang T., et al., UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev. Cell 44, 512–523.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Yang Y., et al., UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat. Plants 4, 98–107 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Sharma A., et al., UVR8 disrupts stabilisation of PIF5 by COP1 to inhibit plant stem elongation in sunlight. Nat. Commun. 10, 4417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y., et al., UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J. 39, e101928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian C., et al., Coordinated transcriptional regulation by the UV-B photoreceptor and multiple transcription factors for plant UV-B responses. Mol. Plant 13, 777–792 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Tavridou E., Pireyre M., Ulm R., Degradation of the transcription factors PIF4 and PIF5 under UV-B promotes UVR8-mediated inhibition of hypocotyl growth in Arabidopsis. Plant J. 101, 507–517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayes S., Velanis C. N., Jenkins G. I., Franklin K. A., UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl. Acad. Sci. U.S.A. 111, 11894–11899 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavridou E., Schmid-Siegert E., Fankhauser C., Ulm R., UVR8-mediated inhibition of shade avoidance involves HFR1 stabilization in Arabidopsis. PLoS Genet. 16, e1008797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruber H., et al., Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 20132–20137 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heijde M., Ulm R., Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc. Natl. Acad. Sci. U.S.A. 110, 1113–1118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren H., et al., Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 116, 4722–4731 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Hara A., Jenkins G. I., In vivo function of tryptophans in the Arabidopsis UV-B photoreceptor UVR8. Plant Cell 24, 3755–3766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heijde M., et al., Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 20326–20331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X., Yang P., Ouyang X., Chen L., Deng X. W., Photoactivated UVR8-COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. PLoS Genet. 10, e1004218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heilmann M., et al., Dimer/monomer status and in vivo function of salt-bridge mutants of the plant UV-B photoreceptor UVR8. Plant J. 88, 71–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin R., Skvortsova M. Y., Loubéry S., Ulm R., COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 113, E4415–E4422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNellis T. W., et al., Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6, 487–500 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Findlay K. M., Jenkins G. I., Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions. Plant Cell Environ. 39, 1706–1714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao X., Liu W., Yang H. Q., Jenkins G. I., A dynamic model of UVR8 photoreceptor signalling in UV-B-acclimated Arabidopsis. New Phytol. 227, 857–866 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Tissot N., Ulm R., Cryptochrome-mediated blue-light signalling modulates UVR8 photoreceptor activity and contributes to UV-B tolerance in Arabidopsis. Nat. Commun. 11, 1323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camacho I. S., et al., Native mass spectrometry reveals the conformational diversity of the UVR8 photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 116, 1116–1125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heilmann M., Christie J. M., Kennis J. T., Jenkins G. I., Mathes T., Photoinduced transformation of UVR8 monitored by vibrational and fluorescence spectroscopy. Photochem. Photobiol. Sci. 14, 252–257 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Liang T., Yang Y., Liu H., Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol. 221, 1247–1252 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Allorent G., et al., UV-B photoreceptor-mediated protection of the photosynthetic machinery in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 113, 14864–14869 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tilbrook K., et al., UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell 28, 966–983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clayton W. A., et al., UVR8-mediated induction of flavonoid biosynthesis for UVB tolerance is conserved between the liverwort Marchantia polymorpha and flowering plants. Plant J. 96, 503–517 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Soriano G., et al., Evolutionary conservation of structure and function of the UVR8 photoreceptor from the liverwort Marchantia polymorpha and the moss Physcomitrella patens. New Phytol. 217, 151–162 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Han X., et al., Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol. Plant 12, 847–862 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Kondou Y., et al., Physiological function of photoreceptor UVR8 in UV-B tolerance in the liverwort Marchantia polymorpha. Planta 249, 1349–1364 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Tokutsu R., Fujimura-Kamada K., Yamasaki T., Matsuo T., Minagawa J., Isolation of photoprotective signal transduction mutants by systematic bioluminescence screening in Chlamydomonas reinhardtii. Sci. Rep. 9, 2820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X., et al., Pivotal roles of Tomato photoreceptor SlUVR8 in seedling development and UV-B stress tolerance. Biochem. Biophys. Res. Commun. 522, 177–183 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Li H., et al., Tomato UV-B receptor SlUVR8 mediates plant acclimation to UV-B radiation and enhances fruit chloroplast development via regulating SlGLK2. Sci. Rep. 8, 6097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernández M. B., Tossi V., Lamattina L., Cassia R., A comprehensive phylogeny reveals functional conservation of the UV-B photoreceptor UVR8 from green algae to higher plants. Front. Plant Sci. 7, 1698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng X.-W., et al., COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G β homologous domain. Cell 71, 791–801 (1992). [DOI] [PubMed] [Google Scholar]
- 59.Oyama T., Shimura Y., Okada K., The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karimi M., Inzé D., Depicker A., GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Curtis M. D., Grossniklaus U., A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu Z., et al., Gene modification by fast-track recombineering for cellular localization and isolation of components of plant protein complexes. Plant J. 100, 411–429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakagawa T., et al., Improved gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Wang Z.-P., et al., Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 66.Arongaus A. B., et al., Arabidopsis RUP2 represses UVR8-mediated flowering in noninductive photoperiods. Genes Dev. 32, 1332–1343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of complexes have been deposited with the following Protein Data Bank (PDB) accession codes: UVR8D96N,D107N: 6XZL (https://www.rcsb.org/structure/6XZL), UVR8D96N,D107N,W285A: 6XZM (https://www.rcsb.org/structure/6XZM), and UVR8G101S,W285A: 6XZN (https://www.rcsb.org/structure/6XZN).