Significance
Schlafen-11 (SLFN11) inactivation leads to chemoresistance of a broad range of DNA-damaging agents. We uncover an expanded Ataxia Telangiectasia- and Rad3-related (ATR)-mediated signaling network that overcomes chemoresistance by an unbiased genome-wide RNAi screen in SLFN11-knockout cells and is validated with clinically developing ATR/CHK1 inhibitors. ATR inhibition induces CDT1 phosphorylation, leading to mitotic catastrophe cell death in SLFN11-deficient cells. We identify a key role of SLFN11 for CDT1 degradation by binding to DDB1–CUL4CDT2 ubiquitin ligase during DNA damage, thereby blocking replication reactivation by which SLFN11-deficient cells cause chemoresistance. Our findings provide a rationale for combination therapy with ATR inhibitor and DNA-targeted drugs, and reveal how SLFN11 irreversibly arrests replication during DNA damage acting as a cofactor of CUL4CDT2 ubiquitin ligase.
Keywords: SLFN11, CDT1, CUL4, ATR/CHK1 inhibitor
Abstract
Schlafen-11 (SLFN11) inactivation in ∼50% of cancer cells confers broad chemoresistance. To identify therapeutic targets and underlying molecular mechanisms for overcoming chemoresistance, we performed an unbiased genome-wide RNAi screen in SLFN11-WT and -knockout (KO) cells. We found that inactivation of Ataxia Telangiectasia- and Rad3-related (ATR), CHK1, BRCA2, and RPA1 overcome chemoresistance to camptothecin (CPT) in SLFN11-KO cells. Accordingly, we validate that clinical inhibitors of ATR (M4344 and M6620) and CHK1 (SRA737) resensitize SLFN11-KO cells to topotecan, indotecan, etoposide, cisplatin, and talazoparib. We uncover that ATR inhibition significantly increases mitotic defects along with increased CDT1 phosphorylation, which destabilizes kinetochore-microtubule attachments in SLFN11-KO cells. We also reveal a chemoresistance mechanism by which CDT1 degradation is retarded, eventually inducing replication reactivation under DNA damage in SLFN11-KO cells. In contrast, in SLFN11-expressing cells, SLFN11 promotes the degradation of CDT1 in response to CPT by binding to DDB1 of CUL4CDT2 E3 ubiquitin ligase associated with replication forks. We show that the C terminus and ATPase domain of SLFN11 are required for DDB1 binding and CDT1 degradation. Furthermore, we identify a therapy-relevant ATPase mutant (E669K) of the SLFN11 gene in human TCGA and show that the mutant contributes to chemoresistance and retarded CDT1 degradation. Taken together, our study reveals new chemotherapeutic insights on how targeting the ATR pathway overcomes chemoresistance of SLFN11-deficient cancers. It also demonstrates that SLFN11 irreversibly arrests replication by degrading CDT1 through the DDB1–CUL4CDT2 ubiquitin ligase.
Schlafen-11 (SLFN11) is an emergent restriction factor against genomic instability acting by eliminating cells with replicative damage (1–6) and potentially acting as a tumor suppressor (6, 7). SLFN11-expressing cancer cells are consistently hypersensitive to a broad range of chemotherapeutic drugs targeting DNA replication, including topoisomerase inhibitors, alkylating agents, DNA synthesis, and poly(ADP-ribose) polymerase (PARP) inhibitors compared to SLFN11-deficient cancer cells, which are chemoresistant (1, 2, 4, 8–17). Profiling SLFN11 expression is being explored for patients to predict survival and guide therapeutic choice (8, 13, 18–24).
The Cancer Genome Atlas (TCGA) and cancer cell databases demonstrate that SLFN11 mRNA expression is suppressed in a broad fraction of common cancer tissues and in ∼50% of all established cancer cell lines across multiple histologies (1, 2, 5, 8, 13, 25, 26). Silencing of the SLFN11 gene, like known tumor suppressor genes, is under epigenetic mechanisms through hypermethylation of its promoter region and activation of histone deacetylases (HDACs) (21, 23, 25, 26). A recent study in small-cell lung cancer patient-derived xenograft models also showed that SLFN11 gene silencing is caused by local chromatin condensation related to deposition of H3K27me3 in the gene body of SLFN11 by EZH2, a histone methyltransferase (11). Targeting epigenetic regulators is therefore an attractive combination strategy to overcome chemoresistance of SLFN11-deficient cancers (10, 25, 26). An alternative approach is to attack SLFN11-negative cancer cells by targeting the essential pathways that cells use to overcome replicative damage and replication stress. Along these lines, a prior study showed that inhibition of ATR (Ataxia Telangiectasia- and Rad3-related) kinase reverses the resistance of SLFN11-deficient cancer cells to PARP inhibitors (4). However, targeting the ATR pathway in SLFN11-deficient cells has not yet been fully explored.
SLFN11 consists of two functional domains: A conserved nuclease motif in its N terminus and an ATPase motif (putative helicase) in its C terminus (2, 6). The N terminus nuclease has been implicated in the selective degradation of type II tRNAs (including those coding for ATR) and its nuclease structure can be derived from crystallographic analysis of SLFN13 whose N terminus domain is conserved with SLFN11 (27, 28). The C terminus is only present in the group III Schlafen family (24, 29). Its potential ATPase activity and relationship to chemosensitivity to DNA-damaging agents (3–5) imply that the ATPase/helicase of SLFN11 is involved specifically in DNA damage response (DDR) to replication stress. Indeed, inactivation of the Walker B motif of SLFN11 by the mutation E669Q suppresses SLFN11-mediated replication block (5, 30). In addition, SLFN11 contains a binding site for the single-stranded DNA binding protein RPA1 (replication protein A1) at its C terminus (3, 31) and is recruited to replication damage sites by RPA (3, 5). The putative ATPase activity of SLFN11 is not required for this recruitment (5) but is required for blocking the replication helicase complex (CMG-CDC45) and inducing chromatin accessibility at replication origins and promoter sites (5, 30). Based on these studies, our current model is that SLFN11 is recruited to “stressed” replication forks by RPA filaments formed on single-stranded DNA (ssDNA), and that the ATPase/helicase activity of SLFN11 is required for blocking replication progression and remodeling chromatin (5, 30). However, underlying mechanisms of how SLFN11 irreversibly blocks replication in DNA damage are still unclear.
Increased RPA-coated ssDNA caused by DNA damage and replication fork stalling also triggers ATR kinase activation, promoting subsequent phosphorylation of CHK1, which transiently halts cell cycle progression and enables DNA repair (32). ATR inhibitors are currently in clinical development in combination with DNA replication damaging drugs (33, 34), such as topoisomerase I (TOP1) inhibitors, which are highly synergistic with ATR inhibitors in preclinical models (35). ATR inhibitors not only inhibit DNA repair, but also lead to unscheduled replication origin firing (36), which kills cancer cells (37, 38) by inducing genomic alterations due to faulty replication and mitotic catastrophe (33).
The replication licensing factor CDT1 orchestrates the initiation of replication by assembling prereplication complexes (pre-RC) in G1-phase before cells enter S-phase (39). Once replication is started by loading and activation of the MCM helicase, CDT1 is degraded by the ubiquitin proteasomal pathway to prevent additional replication initiation and ensure precise genome duplication and the firing of each origin only once per cell cycle (39, 40). At the end of G2 and during mitosis, CDT1 levels rise again to control kinetochore-microtubule attachment for accurate chromosome segregation (41). Deregulated overexpression of CDT1 results in rereplication, genome instability, and tumorigenesis (42). The cellular CDT1 levels are tightly regulated by the damage-specific DNA binding protein 1 (DDB1)–CUL4CDT2 E3 ubiquitin ligase complex in G1-phase (43) and in response to DNA damage (44, 45). How CDT1 is recognized by CUL4CDT2 in response to DNA damage remains incompletely known.
In the present study, starting with a human genome-wide RNAi screen, bioinformatics analyses, and mechanistic validations, we explored synthetic lethal interactions that overcome the chemoresistance of SLFN11-deficient cells to the TOP1 inhibitor camptothecin (CPT). The strongest synergistic interaction was between depletion of the ATR/CHK1-mediated DNA damage response pathways and DNA-damaging agents in SLFN11-deficient cells. We validated and expanded our molecular understanding of combinatorial strategies in SLFN11-deficient cells with the ATR (M4344 and M6620) and CHK1 (SRA737) inhibitors in clinical development (33, 46, 47) and found that ATR inhibition leads to CDT1 stabilization and hyperphosphorylation with mitotic catastrophe. Our study also establishes that SLFN11 promotes the degradation of CDT1 by binding to DDB1, an adaptor molecule of the CUL4CDT2 E3 ubiquitin ligase complex, leading to an irreversible replication block in response to replicative DNA damage.
Results
Genome-Wide RNAi Screen Identifies the ATR/CHK1 Pathway as Therapeutic Targets in SLFN11-Deficient Cells.
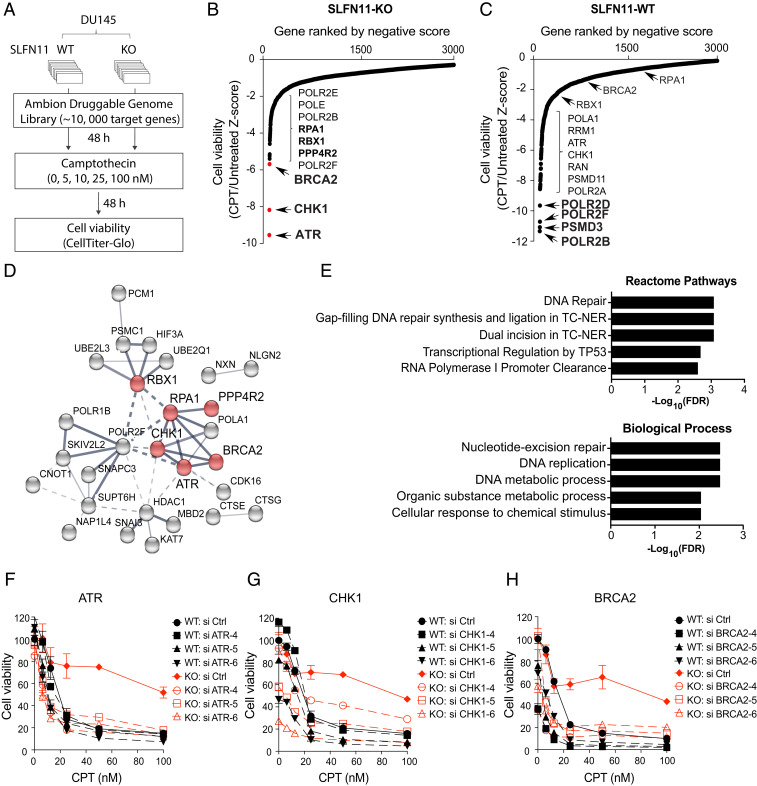
To identify clinically actionable therapeutic targets to overcome the chemoresistance of SLFN11-deficient cells, we carried out an unbiased genome-wide RNAi chemosensitization screen using transient transfection with a short-interfering RNA (siRNA) library and subsequent exposure to the TOP1 inhibitor and replication stress inducer CPT (Fig. 1A) (48). The screen was performed in isogenic DU145 SLFN11 WT and knockout (KO) prostate cancer cells, based on their prior characterization with respect to SLFN11 (4–6, 30) and with a druggable siRNA genome library targeting ∼10,418 human genes (three siRNAs per gene) (SI Appendix, SI Materials and Methods). SLFN11-WT and KO cells were transfected in a 384-well plate format with the siRNA library and subsequently treated for 2 d with a range of CPT concentrations (0, 5, 10, 25, and 100 nM). After evaluating cell viability using a luminescence-based cell number assay, responses were converted to z-scores from three-individual siRNA-transfected wells (Dataset S1). The normalized z-score data in SLFN11-WT and -KO cells were plotted for each gene. Negative z-scores were considered synthetic-lethal interactions with CPT treatment. ATR, CHK1, BRCA2, and RPA1 were identified as the most potent hits in SLFN11-KO cells (Fig. 1B), whereas POLR2B, PSMD3, POLR2F, and POLR2D were found as significant hits in SLFN11-WT cells (Fig. 1C). Gene ontology (GO)-term enrichment analysis with the top 50 hit genes revealed that the hit genes in SLFN11-KO cells form a cluster associated with DNA repair (Fig. 1 D and E), whereas hit genes in SLFN11-WT cells were enriched for RNA processing (SI Appendix, Fig. S1 A and B).
Fig. 1.
Genome-wide RNAi screen identifies ATR pathway as a synergistic target to overcome chemoresistance in SLFN11-deficient cells. (A) Schematic overview of genome-wide RNAi screen workflow in DU145 WT and SLFN11 KO cells. (B and C) Ranked distribution plots of z-scores obtained from cell viability (CPT-treated/untreated) in SLFN11-KO (B) and -WT (C) cells (Dataset S1). Black dots: Individual siRNA targeted genes. Red dots: Hit genes selected for further validation. (D) Protein interaction network with RNAi screen hits in SLFN11-KO cells generated by STRING analysis. Line thickness represents the strength of data confidence. (E) GO analysis of molecular network in SLFN11-KO cells. (F–H) Validation of chemosensitivity in SLFN11-WT and KO cells. Cells were transfected with three siRNAs targeting ATR, CHK1, and BRCA2 and then treated with the indicated concentrations of CPT for 72 h. Cell viability was analyzed by CellTiter-Glo (Promega). Error bars represent SD (n = 3).
To validate the prevalence of the ATR/CHK1 pathway in SLFN11-deficient cells, we tested three independent siRNAs targeting ATR, CHK1, and BRCA2 in SLFN11-WT and -KO cells. Silencing of ATR, CHK1, or BRCA2 reversed CPT resistance of SLFN11-KO cells (Fig. 1 D–F and SI Appendix, Fig. S1 C–E). These findings demonstrate that inhibition of the ATR/CHK1 and BRCA2 pathways could provide a strategy to overcome resistance to TOP1 inhibitors in SLFN11-deficient cancer cells.
Clinical Inhibitors of the ATR/CHK1 Pathway Reverse the Resistance of SLFN11-Deficient Cells to a Broad Range of Clinically Used DNA-Damaging Agents.
Given the prevalence of replicative DNA stress in cancer cells and in response to treatment with chemotherapeutic agents, targeting ATR with small-molecule inhibitors is actively being pursued in clinical trials (33). To validate and extend our siRNA screening, we tested two ATR inhibitors in clinical development: M4344 (VX-803) and M6620 (VX-970; berzosertib) in SLFN11-KO and -WT cells. In the absence of CPT, both ATR inhibitors showed single agent activity regardless of SLFN11 expression and M4344 showed greater potency than M6620 (SI Appendix, Fig. S2A).
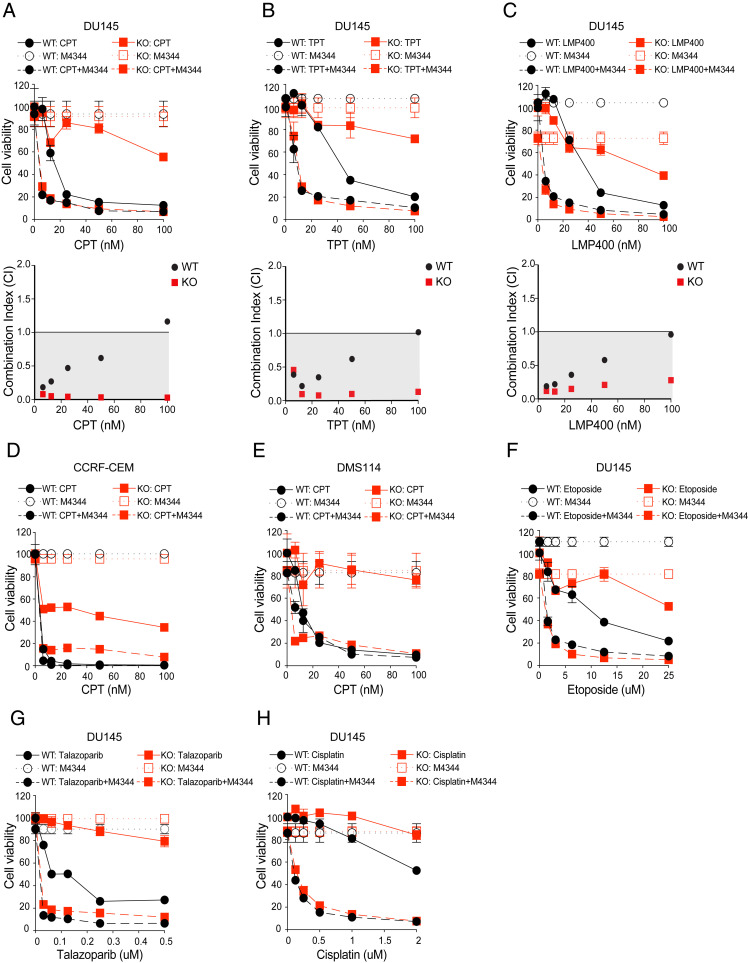
For combination studies, we selected a nontoxic dose (25 nM) of M4344 to avoid secondary effects related to cell death. Consistent with the ATR targeting siRNA results, combining M4344 and CPT markedly sensitized the SLFN11-KO cells and reversed their CPT resistance to a sensitivity level close to the SLFN11-WT cells (Fig. 2A). The synergistic and resistance-reversing effects of the ATR inhibitor M4344 with CPT was observed in additional isogenic human cancer cell lines from different histological background: Leukemia CCRF-CEM (Fig. 2D) (4–6) and small-cell lung cancer DMS114 (Fig. 2E). To substantiate the clinical applicability of our findings, we next tested the combination of M4344 with two clinical TOP1 inhibitors, topotecan (TPT) and indotecan (LMP400) (48) in SLFN11-WT and -KO cells. Consistent with the data with CPT, M4344 significantly resensitized SLFN11-KO cells to both TPT and LMP400, exhibiting strong synergistic cytotoxicity (Fig. 2 B and C).
Fig. 2.
Clinical inhibitor of the ATR pathway consistently resensitizes SLFN11-deficient cells to clinical DNA damaging agents. (A–C, Upper) DU145 SLFN11-WT and -KO cells were treated with M4344 (25 nM) and the indicated concentrations of the TOP1 inhibitors CPT, TPT, and indotecan (LMP400) for 72 h. Cell viability was analyzed by CellTiter-Glo. Error bars represent SD (n = 3). (Lower) Combination Index (CI) versus Fa (fraction affected) calculated from cell viability data. (D) Combination treatment of M4344 (1 nM) and the indicated concentrations of CPT in the isogenic WT and SLFN11-KO leukemic lymphoblasts CCRF-CEM cells. (E) Combination treatment of M4344 (25 nM) and the indicated concentrations of CPT in small-cell lung cancer DMS114 cells. (F–H) DU145 cells were treated with M4344 (25 nM) and the indicated concentrations of etoposide, talazoparib, and cisplatin for 72 h. Cell viability was analyzed by CellTiter-Glo. Error bars represent SD (n = 3).
Because SLFN11 expression is a dominant determinant of response not just for TOP1 inhibitors, but also for a broad range of DNA-targeted drugs that induce replicative stress as their chemotherapeutic mechanism (6), we tested additional drug combinations in SLFN11-KO and -WT cells. As shown in Fig. 2 F and H, M4344 increased the cytotoxicity and reversed resistance of the SLFN11-KO to the clinically used TOP2 inhibitor etoposide, PARP inhibitor talazoparib and cisplatin in SLFN11-KO cells, while the synergy was less in SLFN11-WT cells.
To further establish that the synergistic effect of M4344 was related to ATR, we tested a structurally different ATR inhibitor (M6620, VX-970, berzosertib), which is undergoing clinical trials in combination with TPT (34). M6620 also strongly reversed the chemoresistance of SLFN11-KO cells to all tested DNA-damaging agents, albeit at slightly higher concentration (100 nM) compared with 25 nM of M4344 (SI Appendix, Fig. S2 B–G).
CHK1, a key downstream protein kinase for the ATR-mediated DNA repair pathways, can also be targeted by selective small molecule inhibitors for anticancer treatment (47). Given that ATR activates CHK1, we hypothesized that the CHK1 inhibitor in clinical trials, SRA737, could also overcome chemoresistance in SLFN11-KO cells. Accordingly, SRA737 reversed the resistance of SLFN11-KO cells (SI Appendix, Fig. S2 H–O). Together, our findings show that combining clinical ATR/CHK1 inhibitors with widely using clinical DNA-damaging agents is highly effective for overcoming the chemoresistance of cancer cells that do not express SLFN11.
ATR Inhibition Leads to Mitotic and Genomic Damage in Cells Lacking SLFN11.
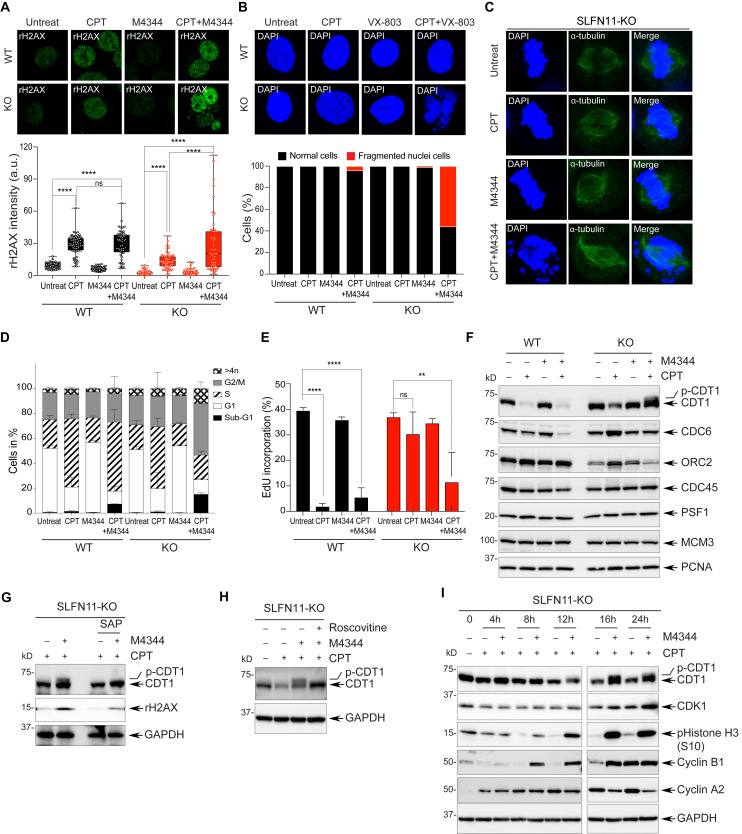
To assess how the combination of ATR inhibitors sensitizes SLFN11-deficient cells, we assessed the phosphorylation status of signaling molecules of the ATR-mediated DDR pathway in SLFN11-WT and -KO cells after treatment with M4344 and CPT. As expected, M4344 preferentially decreased the CPT-induced autophosphorylation of ATR, and phosphorylation of CHK1, CHK2, and RPA32 in both SLFN11-WT and -KO cells, demonstrating that M4344 efficiently blocks the CPT-induced DDR (SI Appendix, Fig. S3A). In addition, combination with M4344 and CPT enhanced γH2AX, a marker of DNA damage and apoptosis (49) in SLFN11-KO cells to levels comparable to SLFN11-WT cells, consistent with increased DNA damage by M4344 under conditions that resensitized the SLFN11-KO cells to CPT (Fig. 3A and SI Appendix, Fig. S3A). In addition, single-cell analyses by immunofluorescence staining of cells treated with M4344 and CPT showed that the strong γH2AX signals originated from multifragmented small nuclei in SLFN11-KO cells (Fig. 3A). SLFN11-KO cells exhibited over 50% multinucleated cells, whereas SLFN11-WT cells showed less than 10% (Fig. 3B). These results indicate that ATR inhibition by M4344 might lead to mitotic catastrophe with nuclear fragmentation in SLFN11-KO cells treated with CPT.
Fig. 3.
Inhibition of ATR leads to increased DNA damage and chromosomal defects with CDT1 hyperphosphorylation in SLFN11-deficient cells treated with CPT. (A) Confocal immunofluorescence staining of γH2AX in DU145 SLFN11-WT and -KO cells treated with CPT (100 nM) and M4344 (25 nM) for 24 h. (Upper) Representative images (γH2AX green). (Magnification, 63×). (Lower) γH2AX signal quantified by ImageJ. Error bars represent SD (n ≥ 50); ****P < 0.0001 Student's t test; ns, not significant. (B) Confocal immunofluorescence staining of nuclei in cells treated with CPT and M4344 for 24 h. (Upper) Representative images (magnification, 63×) of nuclei/single cell, labeled with DAPI (blue). (Lower) Percentages of multinucleated cells after drug treatment (n = 100). (C) Confocal immunofluorescence staining of metaphase alignment in SLFN11-KO cells after treatment with CPT and M4344 for 16 h. Mitotic spindles were stained with α-tubulin (green) and chromosomes with DAPI (blue). (Magnification, 63×). (D) Cell cycle distribution analyzed by flow cytometry after treatment with CPT and M4344 for 24 h. Data are presented as mean values. Error bars represent SD (n = 3). (E) Relative EdU incorporation analyzed by flow cytometry. Cells were treated with CPT and M4344 for 24 h and pulsed with Edu (10 µM) for 30 min prior to harvesting. Error bars represent SD (n = 3). **P < 0.009, ****P < 0.0001 Student’s t test. (F) Protein expression levels of DNA replication initiation factors after treatment with CPT and M4344 for 24 h. (G) Retardation of CDT1 electrophoretic migration in response to combined treatment with CPT and M4344 in SLFN11-KO cells is due to hyperphosphorylation. SLFN11-KO cells were treated with CPT and M4344 for 24 h and then incubated with SAP (1 U/µL). (H) CDT1 hyperphosphorylation in response to combined treatment with CPT and M4344 is mediated by CDK. SLFN11-KO cells were treated with CPT, M4344 and roscovitine (20 µM) for 24 h and then analyzed by Western blotting. (I) CDT1 is hyperphosphorylated in time-dependent manner in response to combined treatment with CPT and M4344.
Hypothesizing that M4344 triggered abnormal replication and the premature entry of SLFN11-KO cells in S-phase, leading to improper chromosome segregation, we measured ssDNA, which is known to arise in response to replication stress (36) in cells treated with M4344 and CPT. To do so, we stained the cells with bromodeoxyuridine (BrdU) antibodies without denaturation following incorporation of BrdU (SI Appendix, Fig. S3B). Combination treatment produced high levels of ssDNA in SLFN11-KO cells compared to SLFN11-WT cells (SI Appendix, Fig. S3B), which is consistent with the generation of abnormal replication intermediates by inhibition of ATR in the SLFN11-KO cells. M4344 also disrupted chromosome assembly to microtubules in metaphase in the SLFN11-KO cells (Fig. 3C). These results suggest that mitotic defects induced by M4344 promote cell death by replication catastrophe in SLFN11-deficient cells following replication stress.
We next analyzed cell cycle distribution and apoptotic cell death. M4344 triggered apoptotic cell death in the SLFN11-KO cells treated with CPT (SI Appendix, Fig. S3 C and E), with levels of cleaved PARP and caspase-3 comparable to SLFN11-WT cells. M4344 also produced an accumulation of cells with G2/M arrest and sub-G1 content (apoptotic fragmented cells) in the SLFN11-KO cells, while it had minimal impact on the S-phase arrest produced by CPT in the SLFN11-WT cells (Fig. 3D and SI Appendix, Fig. S3 D and E). To examine DNA replication dynamic in response to prolonged (24 h) ATR inhibition, we used pulse-EdU incorporation. SLFN11-KO cells treated with M4344 and CPT significantly reduced their EdU incorporation while in the absence of M4344, EdU incorporation was not significantly inhibited by CPT (Fig. 3E). In contrast, SLFN11-WT cells showed a profound and similar suppression of EdU incorporation in response to CPT both in the absence and presence of M4344 (Fig. 3D and SI Appendix, Fig. S3D). Under those conditions, M4344 reduced cyclins E, A2, and B1 levels and increased histone H3 phosphorylation, a marker of chromatin condensation (SI Appendix, Fig. S3E). These results imply that ATR inhibition results in replication catastrophe and abnormal mitosis in SLFN11-KO cells, while ATR inhibition is ineffective in the SLFN11-proficient cells, which irreversibly arrest replication independently of ATR.
ATR Inhibition Leads to CDT1 Hyperphosphorylation by Cyclin-Dependent Kinases and Stabilization of Hyperphosphorylated CDT1 in Cells Lacking SLFN11.
Because ATR can regulate replication origin firing in response to replication stress to allow cells to compensate for stalled replication forks and finish genome duplication (36, 37, 50), we investigated whether ATR inhibition was associated with molecular alterations that could explain the catastrophic effects on both DNA replication and mitosis in SLFN11-KO cells. As shown in Fig. 3F, the levels of CDT1 (Cdc10-dependent transcript 1 protein), a licensing factor for the pre-RC (39, 51) were differentially affected by M4344 in SLFN11-KO and SLFN11-WT cells in response to CPT. While CDT1 levels were profoundly reduced in the SLFN11-WT cells treated with CPT alone or in combination with 4344, in the SLFN11-KO cells treated with CPT, CDT1 levels remained high and were even higher in the presence of M4344 (Fig. 3F). Additionally, CDT1 immunoblotting showed a slower migrating band in the SLFN11-KO cells treated with M4344 in the presence of CPT. We confirmed these results by fluorescence-conjugated quantification analysis (SI Appendix, Fig. S3F) and in a different isogenic CCRF-CEM SLFN11-WT and -KO cells (SI Appendix, Fig. S3G). Together, these experiments show that CDT1 degradation by CPT-induced damage is reduced in the absence of SLFN11 and that ATR inhibition further enhances CDT1 persistence in cells lacking SLFN11.
Next, we examined the molecular process leading to CDT1 electrophoretic retardation induced by ATR inhibition with M4344 in SLFN11-KO cells. To test whether CDT1 hyperphosphorylation was responsible for the electrophoretic shift, we treated the samples with shrimp alkaline phosphatase (SAP). As shown in Fig. 3G, SAP suppressed the upper smeared signal, indicating that CDT1 hyperphosphorylation is responsible for the electrophoretic shift. Consistent with this conclusion, we also found that the cyclin-dependent kinase (CDK) inhibitor roscovitine prevented the CDT1 upper shifted band (Fig. 3H). These results indicate that inhibiting ATR with M4344 releases the CDKs, which in turn induce the hyperphosphorylation of CDT1 in SLFN11-KO cells.
CDT1 is not only a well-established replication licensing factor, but also a mitosis regulator for chromosome segregation by promoting the attachment of microtubules to kinetochores depending on its phosphorylation (41, 52, 53). We therefore hypothesized that CDT1 hyperphosphorylation and unscheduled stabilization could be associated with the mitotic catastrophe produced by M4344 in CPT-treated SLFN11-KO cells. To explore this possibility, we monitored the time-course of CDT1 hyperphosphorylation and cell cycle regulatory molecules. Histone H3 phosphorylation, which is associated with chromosome condensation and mitosis was coincidentally induced by M4344 in the CPT-treated SLFN11-KO cells (Fig. 3I), implying that hyperphosphorylation of CDT1 in G2/M cells might be involved in the irregular chromosomal distribution during metaphase (Fig. 3C). Collectively, these results suggest a relationship between CDT1 hyperphosphorylation and the mitotic defects caused by ATR inhibition. Hence, we propose that the combinations of ATR inhibitors and DNA-damaging agents targeting replication, such as CPTs, induce mitotic catastrophe in SLFN11-deficient cells through abnormal partitioning of chromatin and transfer of underreplicated DNA into daughter cells, and that hyperphosphorylation of CDT1 by CDKs due to ATR inhibition may contribute to this process.
Delayed CDT1 Degradation Leads to Chemoresistance through Replication Recovery in SLFN11-Deficient Cells.
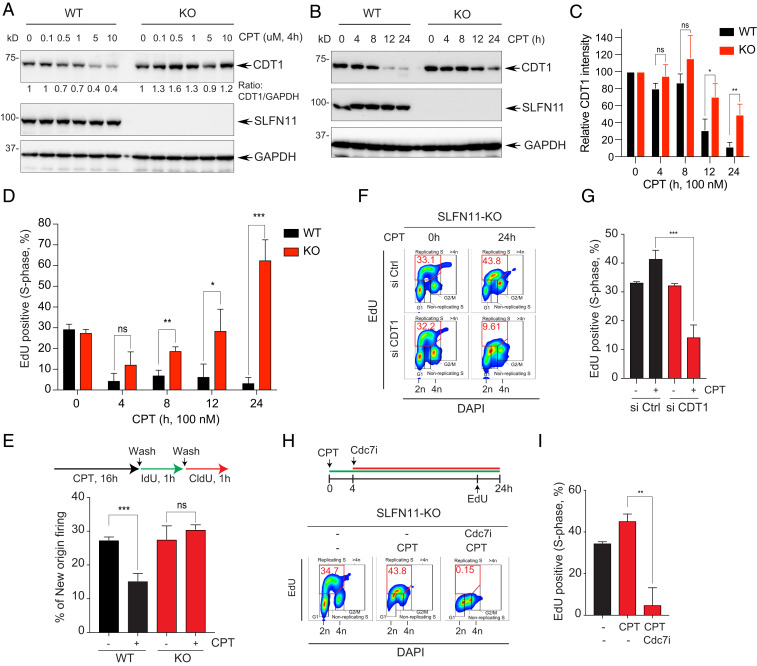
Given that CDT1 is rapidly degraded upon completion of its licensing role during normal cell cycle and in response to radiation-mediated DNA damage to keep genomic integrity (40, 45, 54), we examined whether alterations of CDT1 stability could be involved in the response to CPT and be modulated by SLFN11 and ATR. As shown in Fig. 3F, we observed the disappearance of CDT1 in response to CPT, particularly in the SLFN11-expressing cells. We confirmed this differential response after short treatment with CPT (Fig. 4A). While CDT1 protein levels remained stable in the SLFN11-KO cells, CDT1 disappeared in a CPT concentration-dependent manner in the SLFN11-WT cells. Time-course experiments also confirmed the limited CDT1 degradation in SLFN11-KO cells (Fig. 4 B and C) and the disappearance of CDT1 at 2 to 4 h at the higher concentrations of CPT in SLFN11-WT DU145 cells (SI Appendix, Fig. S4 A and B). SLFN11-dependent CDT1 suppression was also observed in human small-cell lung cancer DMS114 SLFN11-WT and -KO cells and in human embryonic kidney HEK293 SLFN11-proficient and the HEK293T cells, which are SLFN11-deficient and chemoresistant (SI Appendix, Fig. S4 C–E). These results show that SLFN11 promotes CDT1 degradation. Yet, no difference in half-life of the CDT1 protein was observed in SLFN11-WT and -KO cells treated only with the protein synthesis inhibitor cycloheximide (CHX) (SI Appendix, Fig. S4F), indicating that SLFN11-dependent degradation of CDT1 is a specific response to DNA damage.
Fig. 4.
Defective CDT1 degradation causes replication recovery in SLFN11-deficient cells. (A) DU145 SLFN11-WT and -KO cells were treated with the indicated concentrations of CPT for 4 h. Protein levels were analyzed by Western blotting. (B) DU145 SLFN11-WT and -KO cells were treated with CPT (100 nM) for the indicated times. Levels of protein expression were analyzed by Western blotting. (C) Quantitation of CDT1 as shown in B. Bars show the mean band intensity from triplicate experiments, normalized to GAPDH. *P < 0.0274, **P < 0.0077. (D) Percentage of EdU+ S-phase cells in the time course treatment of CPT was determined by flow cytometry. Error bars represent SD (n = 3). *P < 0.0335, **P < 0.0025, ***P < 0.0005 Student’s t test. (E, Upper) Labeling protocols for the DNA combing assay. Cells were treated with CPT (100 nM) for 16 h and then pulse-labeled sequentially with IdU (100 µM) and CldU (100 µM) for 1 h. (Lower) Frequencies of new origins only labeled by the CldU pulse. Error bars represent SD (n = 3). ***P < 0.0009 Student’s t test. (F) CDT1-dependent replication recovery. SLFN11-KO cells were transfected with siControl (Ctrl) or siCDT1 for 48 h, and then treated with CPT for 24 h. Cells were incubated with EdU (10 µM) for 30 min prior to harvest. The percentage of EdU+ and DAPI+ cells was determined by flow cytometry. (G) Quantification of the EdU+ cells in S-phase. Error bars represent SD (n = 3). ***P < 0.0009 Student’s t test. (H, Upper) Treatment protocol. Cells were treated with CPT for 24 h and with the Cdc7 inhibitor (PHA-767491, 5 µM) after 4 h of CPT treatment. Cells were incubated with EdU (10 µM) for 30 min prior to harvest. Percentage of EdU+ and DAPI+ cells was determined by flow cytometry. (I) Quantification of the EdU+ cells in S-phase. Error bars represent SD (n = 3). **P < 0.008 Student’s t test.
Although both SLFN11-WT and KO cells were arrested in S-phase by CPT (Fig. 3D), SLFN11-WT cells were mostly arrested in early S-phase with accumulation of cyclins E and A, whereas SLFN11-KO cells were arrested in late S-phase with accumulation of cyclins A and B (SI Appendix, Fig. S3 D and E), implying that SLFN11-KO cells treated with CPT maintain some replication. To assess whether the differential CDT1 stability between the SLFN11-WT and -KO cells was coupled with DNA replication, we performed time-course experiments with EdU pulse-labeling. As shown in Fig. 4D, at 4 h DNA synthesis was suppressed by CPT regardless of SLFN11 expression. However, at later time points, DNA synthesis (EdU+ cells) recovered and even rebounded at 24 h in the SLFN11-KO cells, while in the SLFN11-WT cells DNA replication was continuously suppressed (Fig. 4D and SI Appendix, Fig. S4G).
We also examined whether the replication recovery in SLFN11-KO cells was accompanied by the firing of new replication origin using the DNA fiber-combing assays at the time point of the replication recovery phase (i.e., at 16 h of CPT treatment). As shown in Fig. 4E, CPT treatment significantly blocked origin firing in SLFN11-WT cells, whereas new origins in the SLFN11-KO cells were comparable to untreated cells, indicating that the new origin firing activity contributes to the replication recovery of SLFN11-KO cells.
To determine whether the replication recovery of SLFN11-KO cells was dependent on CDT1, we depleted endogenous CDT1 by siRNA before CPT treatment (SI Appendix, Fig. S4H). Silencing of CDT1 significantly reduced the replication recovery of SLFN11-KO cells (Fig. 4 F and G). Because the origins licensed by CDT1 are subsequentially fired by phosphorylation and activation of the Mcm2 helicase by Cdc7 kinase (39, 55), we tested whether replication recovery in SLFN11-KO cells was impaired by inhibition of Cdc7 kinase. We first treated cells with CPT and subsequently added a Cdc7 inhibitor before the replication recovery phase. Replication recovery in SLFN11-KO cells was blocked by the Cdc7 inhibitor (Fig. 4 H and I and SI Appendix, Fig. S4I). We also confirmed that Cdc7 inhibitor could overcome chemoresistance in SLFN11-KO cells (SI Appendix, Fig. S4 J and K). Similarly, chemoresistance to CPT was reversed by the WEE1 inhibitor AZD1775 (SI Appendix, Fig. S4L). Collectively, these data show that SLFN11-mediated replication arrest in response to DNA damage is linked with the degradation of CDT1, and that the absence of CDT1 degradation in SLFN11-deficient cells allows those cells to recover replication and gain resistance against CPT.
SLFN11 Promotes CDT1 Degradation by Binding to DDB1, a Component of the CUL4 Ubiquitin Ligase Complex in Response to Replicative DNA Damage.
To elucidate how SLFN11 induces the down-regulation of CDT1 in cells treated with CPT, we cotreated SLFN11-WT and -KO cells with proteasome inhibitor. MG132 not only blocked the disappearance of CDT1 in the SLFN11-expressing cells but also induced a marked accumulation of CDT1 in both the SLFN11-WT and -KO cells (SI Appendix, Fig. S5A). These results are consistent with the rapid turnover of CDT1 (SI Appendix, Fig. S4F) and demonstrate that SLFN11 induces the proteasomal degradation of CDT1. They are also consistent with the conclusion that CPT induces ubiquitin-mediated CDT1 degradation as previously observed for radiation DNA damage, where CDT1 is proteolyzed following its poly-ubiquitination by the DDB1–CUL4CDT2 E3 ubiquitin ligase complex (44, 45).
DDB1–CUL4CDT2 ubiquitin ligase has been reported to degrade not only CDT1 but other critical regulators involved in DNA replication and cell cycle progression in response to DNA damage (56–58). The CDK inhibitor CDKN1A (p21WAF1/CIP1) and thymine DNA glycosylase, which are known as substrates of the DDB1–CUL4CDT2 ubiquitin ligase, were also degraded more effectively in response to CPT in the SLFN11-WT compared to the -KO cells (SI Appendix, Fig. S4M), indicating that SLFN11 promotes the degradation of replication-related substrates of DDB1–CUL4CDT2.
To determine whether DDB1–CUL4CDT2 E3 ubiquitin ligase mediates CDT1 degradation in response to CPT in SLFN11-expressing cells, we tested the impact of CDT2 depletion. siRNA-mediated CDT2 depletion blocked the degradation of CDT1 in the CPT-treated SLFN11-expressing cells (SI Appendix, Fig. S5B). In addition, MLN4924, a potent inhibitor of the NEDD8-activating enzyme, which is required for the activation of the CUL4 ubiquitin ligase, increased the levels of CDT1 in the CPT-treated SLFN11-expressing cells (SI Appendix, Fig. S5C). MLN4924 also increased the basal level of CDT1, as much as in the cells treated with MG132 (SI Appendix, Fig. S5C). In contrast, a SKP2 inhibitor had no effect (SI Appendix, Fig. S5C), indicating that the SCFSkp2 complex mediating the CDT1 degradation in unperturbed S-phase (39) is not involved in SLFN11-induced degradation of CDT1. Together, these findings demonstrate that DDB1–CUL4CDT2 mediates CDT1 degradation in response to CPT in SLFN11-expressing cells and suggest that SLFN11 may act as a cofactor of the DDB1–CUL4CDT2 E3 ubiquitin ligase complex in response to DNA damage.
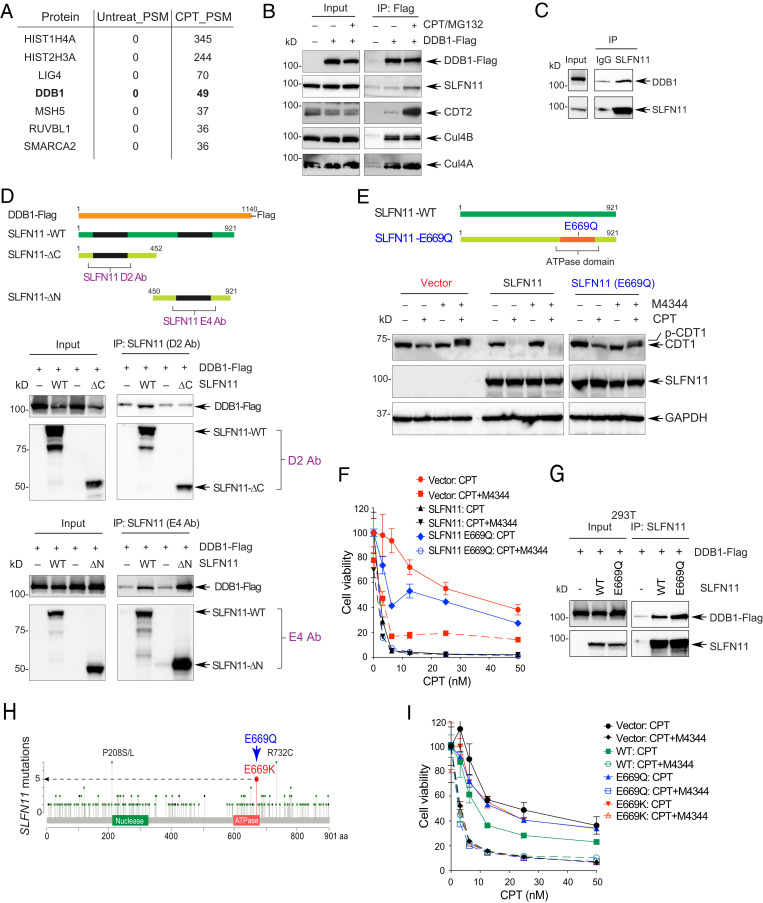
To broadly identify the protein–protein interactions of SLFN11 in CPT-treated cells, we performed proteomic analysis at the time point (16 h) where replication is irreversibly blocked and CDT1 degraded in response to CPT treatment. Mass spectrometry (MS) analysis in DU145 SLFN11-WT cells revealed SLFN11 interaction with the adaptor subunit of the CUL4CDT2 E3 ubiquitin ligase complex DDB1 (Fig. 5A and Dataset S2). The association of SLFN11 with the DDB1–CUL4CDT2 complex was validated by immunoprecipitation (IP) and Western blotting analyses in HEK293 cells transiently transfected with DDB1-Flag. Endogenous SLFN11 was detected in the DDB1-Flag immunocomplexes after combination treatment of CPT and MG132 (Fig. 5B). The endogenous interaction and colocalization of SLFN11 with DDB1 was confirmed by IP and immunofluorescence microcopy with antibodies targeting endogenous SLFN11 and DDB1 in DU145 cells (Fig. 5C and SI Appendix, Fig. S5D). We also found that the interaction between SLFN11 and DDB1 contributed to the recruitment of CDT2 to the DDB1–CUL4 premature complex to form a complete E3 ubiquitin ligase complex after treatment with CPT and MG132 (SI Appendix, Fig. S5E). These findings suggest that SLFN11 might contribute to the activity of the DDB1–CUL4CDT2 E3 ubiquitin ligase complex by binding to DDB1 and acting as a cofactor of CUL4CDT2 upon DNA damage.
Fig. 5.
SLFN11 promotes CDT1 degradation by binding to the CUL4–DDB1 complex in response to replicative DNA damage. (A) Representative interactors of SLFN11. DU145 cells were treated with CPT for 16 h. Nuclear cell lysates were immunoprecipitated with anti-SLFN11 antibody and analyzed by mass spectrometry. (B) HEK293 cells transfected with a DDB1-Flag construct for 2 d were treated with CPT (10 µM) and MG132 (20 µM) for 4 h were lysed and immunoprecipitated with anti-Flag antibody. Interacting proteins were profiled by Western blotting. (C) Interaction between endogenous SLFN11 and DDB1 was assessed by immunoprecipitation with anti-SLFN11 (D2) antibody after treatment with CPT (10 µM) and MG132 (20 µM) for 4 h in DU145 cells. (D, Top) Diagram of DDB1-Flag and SLFN11 deletion mutants. (Middle) 293T cells transfected with DDB1-Flag and indicated SLFN11 constructs were treated with CPT (10 µM) and MG132 (20 µM) for 4 h. Cells were immunoprecipitated with anti-SLFN11 antibody (D2) that detects the N terminus region of SLFN11. (Bottom) Same as above but using anti-SLFN11 antibody (E4) that detects the C terminus region of SLFN11. Interacting proteins were identified by Western blotting. (E) K562 cells stably transfected with vector, SLFN11 and SLFN11-E669Q constructs were treated with CPT (100 nM) and M4344 for 24 h. Proteins were identified by Western blotting. (F) K562 cells stably transfected with vector, SLFN11-WT or SLFN11-E669Q constructs were treated with CPT and M4344 for 72 h. Cell viability was analyzed by CellTiter-Glo. (G) SLFN11-E669Q interacts with DDB1. 293T cells were transiently transfected with DDB1 Flag, SLFN11-WT, and -E669Q for 48 h. Following treatment with CPT (10 µM) and MG132 (20 µM) for 4 h, protein interactions were assessed by IP with anti-SLFN11 (D2) antibody. (H) Diagram of SLFN11 mutations in TCGA. (I) 293T cells transfected with SLFN11-WT, SLFN11-E669Q, or SLFN11-E669K constructs for 48 h, were treated with CPT and M4344 for 72 h. Cell viability was analyzed by CellTiter-Glo.
The C Terminus ATPase/Helicase Domain of SLFN11 Binds and Activates DDB1–CUL4 in Response to Replicative DNA Damage.
To understand how SLFN11 interacts with DDB1, we performed IP with antibodies specifically targeting the N- and C-terminus regions of SLFN11 in 293T cells, which are SLFN11-deficient (2). We transiently cotransfected them with DDB1-Flag and two different SLFN11 expression constructs corresponding to deletions of the N terminus (1 to 452 aa) or the C terminus (450 to 901 aa) (Fig. 5D). The SLFN11 C-terminal deletion-mutant failed to interact with DDB1 whereas the N-terminal deletion-mutant retained the ability to interact with DDB1 (Fig. 5D). Furthermore, transfection of the C-terminal deletion-mutant failed to suppress EdU incorporation in CPT-treated cells (SI Appendix, Fig. S5F). These data indicate that the C-terminal region of SLFN11 is required for the interaction of SLFN11 with DDB1 and suppression of DNA replication.
Given that the C-terminal region of SLFN11 contains a Walker B/ATPase motif critical for the nuclear DDR functions of SLFN11 (3, 5, 30), we tested whether mutation/inactivation of this ATPase motif (E669Q) (5) would affect SLFN11 binding to DDB1. Similar binding was observed after transfection of SLFN11-WT and SLFN11-E669Q (Fig. 5G). To test whether the Walker B/ATPase motif was required for CDT1 degradation, we used human leukemia K562 cells, which are deficient for SLFN11 (25), and which were previously engineered to stably express SLFN11-WT or SLFN11-E669Q (5). SLFN11-WT complementation resulted in CDT1 degradation, whereas complementation with the SLFN11-E669Q mutant failed to induce CDT1 degradation after CPT treatment (Fig. 5E). As previously shown for PARP inhibitors (4), the K562 cells expressing the SLFN11-E669Q mutant failed to reverse the CPT resistance of K562 cells compared with those expressing SLFN11-WT (Fig. 5F). Furthermore, treatment with the ATR inhibitor M4344 significantly abolished the chemoresistance of the SLFN11-E669Q-complemented cells like SLFN11-deficient parental cells (Fig. 5F). Taken together, these results show that the Walker B/ATPase motif of SLFN11 in the C-terminal domain of SLFN11 is critical for the SLFN11-induced degradation of CDT1 and growth arrest in response to CPT.
Although SLFN11 has not yet been the focus of extensive and systematic high-resolution genomic sequencing studies, analysis of the TCGA database revealed that the same residue (E669) used for inactivating the Walker B/ATPase motif of SLFN11 is mutated in five patient samples as E669K mutation (Fig. 5H). To determine whether this patient mutation exhibits the same phenotype as the E669Q mutation, a SLFN11-E669K construct was generated by site-directed mutagenesis and transfected HEK293T cells (SI Appendix, Fig. S5G) to analyze the effects of the E669K mutation on CPT-induced CDT1 degradation and cytotoxicity. The SLFN11-E669K mutant construct was defective in inducing CDT1 degradation compared with SLFN11-WT (SI Appendix, Fig. S5H). It also failed to sensitize cells to CPT (Fig. 5I). From these findings, we conclude that the Walker B/ATPase motif of SLFN11 is required both for chemosensitivity and CDT1 degradation.
Discussion
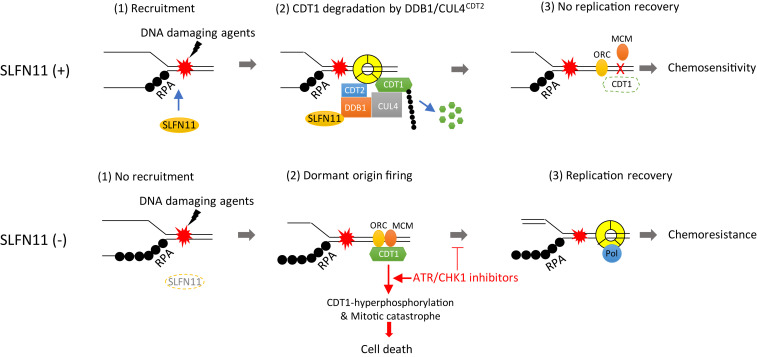
The present study elucidates the coordination of SLFN11 and ATR in the cellular responses to replication stress. Although both are recruited to stressed replication forks by binding to RPA filaments (3, 5, 31, 32, 36), their impact on replication is strikingly different (Fig. 6). While ATR produces a transient replication arrest allowing the cell to repair and resume replication (32), SLFN11 irreversibly blocks replication and cellular proliferation (6). In this study, we relate this divergence to the differential molecular effects of SLFN11 and ATR on the pre-RC and mitotic licensing factor CDT1 and show a previously unsuspected activating role of SLFN11 on the replication-associated DDB1–CUL4CDT2 E3 ligase complex.
Fig. 6.
Proposed model for SLFN11-mediated replication reactivation block and synthetic lethality of ATR/CHK1 inhibitors in SLFN11-negative cancer cells. In SLFN11-proficient cells, SLFN11 is recruited to chromatin by DNA damage. It subsequently binds to DDB1 and promotes the degradation of CDT1 and other substrates as a cofactor of activated DDB1–CUL4CDT2. Consequently, CDT1-mediated replication recovery is irreversibly blocked, leading to sensitivity to DNA-damaging agents. In SLFN11-deficient cells, conversely, DDB1–CUL4CDT2-mediated degradation of CDT1 is reduced. CDT1 then promotes replication recovery by activating dormant origins, resulting in chemoresistance. Treatment of ATR/CHK1 inhibitor reverses chemoresistance of SLFN11-deficient cells by inducing premature replication and mitosis through CDT1 phosphorylation, which induces mitotic catastrophe and cell death.
Overcoming the Chemoresistance of SLFN11-Deficient Cancer Cells by Targeting the ATR/CHK1 Pathway.
SLFN11 is genetically inactivated in ∼50% of all cancer lines and cancer tissues (6, 25) and studies are ongoing in various research institutions to establish clinically whether SLFN11 inactivation determines resistance to anticancer treatments as it does in preclinical models (20). In this study, we demonstrate that inhibition of the ATR pathway in SLFN11-deficient cancer cells overcomes chemoresistance to a broad range of clinical DNA-damaging agents targeting TOP1 (TPT, indotecan [LMP400]), TOP2 (etoposide), PARP (talazoparib), DNA (cisplatin), WEE1 (AZD1775), and Cdc7 (PHA-767491). We first used a genome-wide RNAi screening approach to identify pathways that are synthetic lethal in cells lacking SLFN11 and treated with the TOP1 inhibitor CPT, and found that the top genetics hits were inactivation of ATR as well as depletion of other key components in the ATR pathway. We next validated the clinical applicability of our finding with clinical inhibitors of the ATR/CHK1 pathway combined with various classes of chemotherapeutic DNA-targeted drugs. These results provide a rationale for the clinical testing and potential development of these agents with ATR and CHK1 inhibitors in chemoresistant SLFN11-negative tumors.
Indeed, based on the fact that epigenetic gene silencing is a common cause of SLFN11 inactivation in tumorigenesis, reactivation of SLFN11 by the DNA demethylating drug 5-aza-2’-deoxycytidine, the EZH2 inhibitor EPZ011989 and the HDAC inhibitors entinostat or romidepsin resensitizes SLFN11-deficient cells to clinical DNA-damaging agents (11, 21, 25, 26). Although epigenetic therapies can efficiently reexpress SLFN11, they also reactivate normally silenced genes related to genome instability, carcinogenesis and antiapoptotic pathways (59, 60).
Targeting selectively SLFN11-negative cancer cells with ATR or CHK1 inhibitors is an alternative approach and potential synthetic-lethal strategy to overcome resistance in SLFN11-deficient cancers. We recently reported that the preclinical ATR inhibitor VE-821 can overcome resistance of SLFN11-deficient cancer cell lines to the clinical PARP inhibitors olaparib and talazoparib (4). Here, we demonstrate the importance of the ATR-SLFN11 connection by showing that genetic inactivation of ATR, CHK1, or RPA1 selectively sensitize SLFN11-KO cells to CPT and that clinical inhibitors of ATR and CHK1 selectively sensitize SLFN11-KO cells not only to CPT but also to other Food and Drug Administration-approved clinical anticancer agents (TPT, etoposide, cisplatin, and talazoparib).
Potent and selective ATR inhibitors are in clinical development (33, 61). Given that ATR responds and protects cells against replication stress as a primary sensor, combinations of ATR inhibitors are actively studied with chemotherapeutic drugs that induce replication stress. Here, we tested two clinical ATR inhibitors, M6620 (VE-822, VX-970, berzosertib) and M4344 (also known as VX-803). M4344 is a potent and tight-binding inhibitor of ATR, orally bioavailable and currently undergoing phase I trial (NCT02278250). Both M6620 and M4344 showed high synergy at nontoxic nanomolar concentrations with clinical DNA-damaging agents in SLFN11-deficient cancer cells. Consistent with the ATR inhibitor and genetic screen results, we found that the CHK1 inhibitor SRA737, a new orally bioavailable small molecule in clinical trials (NCT02797964, NCT02797977) (46), also overcame the chemoresistance of SLFN11-KO cells. Considering that several ATR/CHK1 inhibitors are currently in clinical trials (33), our results provide a rationale for their combinations with clinical DNA-damaging agents in multidrug-resistant SLFN11-deficient tumors.
At the molecular level, inhibition of the ATR/CHK1 pathway enhances replication stress induced by DNA-targeted anticancer agents by preventing the transient inactivation of CDKs (SI Appendix, Fig. S6), thereby abrogating the replication and G2 checkpoints, eventually leading to unscheduled replication origin firing (62), defective DNA repair, mitotic catastrophe, and cell death (33, 61). Our study provides a previously unnoted insight regarding the molecular mechanism by which inhibiting ATR impinges on the replication licensing factor CDT1 in SLFN11-KO cells. Indeed, we found that, instead of being degraded in response to DNA damage (40, 45, 54, 63), CDT1 was stabilized and was hyperphosphorylated by inhibiting ATR in cells treated with CPT. We also found that this effect is associated with increased mitotic defects, abnormal chromosome segregation, and subsequent cell death. The accumulation of hyperphosphorylated CDT1 in mitosis could lead to destabilization of CDT1-mediated kinetochore-microtubule attachments and inhibition of rereplication (52, 53). In addition, overexpression of CDT1 in S-phase is known to be cytotoxic due to unscheduled replication origin firing by the inappropriate licensing of dormant origins (42, 64). Hence, we propose that one of the mechanisms of ATR/CHK1 in response to DNA damage is by reducing CDT1 levels transiently during replication stress (Fig. 6).
SLFN11 Overrides ATR and Blocks Replication Irreversibly by Binding DDB1 and Activating the CUL4CDT2-Mediated Degradation of CDT1.
Unexpectedly, by comparing SLFN11-expressing and SLFN11-KO cells, we discovered that SLFN11 promotes the degradation of CDT1 in response to CPT and that this effect is independent of ATR activity. Using molecular and MS analyses, we demonstrate that CDT1 degradation is due to the binding of SLFN11 to DDB1, a critical structural component of the CUL4CDT2 ubiquitin ligase complex. The interaction of SLFN11 with DDB1–CUL4CDT2 is plausible as both are part of replisome complexes (5, 43). We also found that the C-terminal Walker B/ATPase motif of SLFN11 is critical for this functional interaction. This is notable because this same motif is essential for the other activities of SLFN11 in suppressing replication by direct binding to the CMG replication helicase, increasing chromatin accessibility and induction of the immediate early response genes (FOS, JUN, EGR1) (5, 30). Hence, the present study demonstrates that SLFN11 irreversibly arrests replication in cells under replicative stress by at least two different molecular mechanisms: degradation of CDT1 (present study) and arrest of replisomes (5). Degradation of CDT1 may act to prevent the firing of late origins and irreversibly arrest cell cycle progression (Fig. 6). In addition, SLFN11 has been reported to suppress homologous recombination (3), and a recent report showed that SLFN11 can reduce ATR by limiting its translation, implying that SLFN11 deficiency may contribute to alteration of the ATR pathway (27).
In conclusion, the differential activity of ATR inhibitors depending on SLFN11 expression offers opportunities to gain new molecular insights on the molecular cross-talks between the cell survival (ATR) and death (SLFN11) pathways (SI Appendix, Fig. S6) and provides a rationale for the clinical development of ATR inhibitors in combination with chemotherapeutic agents targeting DNA replication.
Materials and Methods
All materials including cell lines, reagents such as inhibitors, antibodies, DNA oligos for CRISPR/CAS9 editing, RNA interference, and gene cloning can be found in SI Appendix. A genome-wide RNAi screen analysis was performed using the Silencer Select Human Druggable siRNA library (Thermo Fisher Scientific, Grand Island, NY). All detailed methods including RNAi screen, cell viability assay, siRNA transfection, apoptosis assay, immunofluorescence microscopy, flow cytometry, Western blot analysis, DNA combing assay, and mass spectrometry are described in the SI Appendix.
Supplementary Material
Acknowledgments
This research was funded by the National Cancer Institute (Z01-BCC006150). The ATR inhibitor M4344 was provided by Merck KGaA (MCRADA 03199).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015654118/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Barretina J., et al., The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoppoli G., et al., Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl. Acad. Sci. U.S.A. 109, 15030–15035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mu Y., et al., SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 17, 94–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murai J., et al., Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 7, 76534–76550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murai J., et al., SLFN11 blocks stressed replication forks independently of ATR. Mol. Cell 69, 371–384.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murai J., Thomas A., Miettinen M., Pommier Y., Schlafen 11 (SLFN11), a restriction factor for replicative stress induced by DNA-targeting anti-cancer therapies. Pharmacol. Ther. 201, 94–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou C., et al., SLFN11 inhibits hepatocellular carcinoma tumorigenesis and metastasis by targeting RPS4X via mTOR pathway. Theranostics 10, 4627–4643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coussy F., et al., BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci. Transl. Med. 12, eaax2625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marzi L., et al., The indenoisoquinoline TOP1 inhibitors selectively target homologous recombination-deficient and schlafen 11-positive cancer cells and synergize with olaparib. Clin. Cancer Res. 25, 6206–6216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner E. E., et al., Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 Axis. Cancer Cell 31, 286–299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lok B. H., et al., PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin. Cancer Res. 23, 523–535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajapakse V. N., et al., CellMinerCDB for integrative cross-database genomics and pharmacogenomics analyses of cancer cell lines. iScience 10, 247–264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shee K., Wells J. D., Jiang A., Miller T. W., Integrated pan-cancer gene expression and drug sensitivity analysis reveals SLFN11 mRNA as a solid tumor biomarker predictive of sensitivity to DNA-damaging chemotherapy. PLoS One 14, e0224267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allison Stewart C., et al., Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 8, 28575–28587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathkey D., et al., Sensitivity of mesothelioma cells to PARP inhibitors is not dependent on BAP1 but is enhanced by temozolomide in cells with high-Schlafen 11 and low-O6-methylguanine-DNA methyltransferase expression. J. Thorac. Oncol. 15, 843–859 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur S., et al., Identification of schlafen-11 as a target of CD47 signaling that regulates sensitivity to ionizing radiation and topoisomerase inhibitors. Front. Oncol. 9, 994 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwasaki J., et al., Schlafen11 expression is associated with the antitumor activity of trabectedin in human sarcoma cell lines. Anticancer Res. 39, 3553–3563 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Conteduca V., et al., SLFN11 expression in advanced prostate cancer and response to platinum-based chemotherapy. Mol. Cancer Ther. 19, 1157–1164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietanza M. C., et al., Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J. Clin. Oncol. 36, 2386–2394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takashima T., et al., Immunohistochemical analysis of SLFN11 expression uncovers potential non-responders to DNA-damaging agents overlooked by tissue RNA-seq. Virchows Arch., 10.1007/s00428-020-02840-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinhold W. C., Thomas A., Pommier Y., DNA-targeted precision medicine; have we been caught sleeping? Trends Cancer 3, 2–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isnaldi E., et al., Schlafen-11 expression is associated with immune signatures and basal-like phenotype in breast cancer. Breast Cancer Res. Treat. 177, 335–343 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Peng Y., et al., Methylation of SLFN11 promotes gastric cancer growth and increases gastric cancer cell resistance to cisplatin. J. Cancer 10, 6124–6134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luan J., Gao X., Hu F., Zhang Y., Gou X., SLFN11 is a general target for enhancing the sensitivity of cancer to chemotherapy (DNA-damaging agents). J. Drug Target. 28, 33–40 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Tang S. W., et al., Overcoming resistance to DNA-targeted agents by epigenetic activation of Schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors. Clin. Cancer Res. 24, 1944–1953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nogales V., et al., Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget 7, 3084–3097 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M., et al., DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 25, 1047–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J. Y., et al., Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 9, 1165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mavrommatis E., Fish E. N., Platanias L. C., The schlafen family of proteins and their regulation by interferons. J. Interferon Cytokine Res. 33, 206–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murai J., et al., Chromatin remodeling and immediate early gene activation by SLFN11 in response to replication stress. Cell Rep. 30, 4137–4151.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maréchal A., et al., PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell 53, 235–246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saldivar J. C., Cortez D., Cimprich K. A., The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18, 622–636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradbury A., Hall S., Curtin N., Drew Y., Targeting ATR as cancer therapy: A new era for synthetic lethality and synergistic combinations? Pharmacol. Ther. 207, 107450 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Thomas A., et al., Phase I study of ATR inhibitor M6620 in combination with topotecan in patients with advanced solid tumors. J. Clin. Oncol. 36, 1594–1602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jossé R., et al., ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase i inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 74, 6968–6979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toledo L. I., et al., ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Chen Y. H., et al., ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol. Cell 58, 323–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buisson R., Boisvert J. L., Benes C. H., Zou L., Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol. Cell 59, 1011–1024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aladjem M. I., Redon C. E., Order from clutter: Selective interactions at mammalian replication origins. Nat. Rev. Genet. 18, 101–116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pozo P. N., Cook J. G., Regulation and function of Cdt1; a key factor in cell proliferation and genome stability. Genes (Basel) 8, E2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varma D., et al., Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat. Cell Biol. 14, 593–603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liontos M., et al., Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 67, 10899–10909 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Panagopoulos A., Taraviras S., Nishitani H., Lygerou Z., CRL4Cdt2: Coupling genome stability to ubiquitination. Trends Cell Biol. 30, 290–302 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C., A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Hu J., McCall C. M., Ohta T., Xiong Y., Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6, 1003–1009 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Rogers R. F., et al., CHK1 inhibition is synthetically lethal with loss of B-family DNA polymerase function in human lung and colorectal cancer cells. Cancer Res. 80, 1735–1747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dent P., Investigational CHK1 inhibitors in early phase clinical trials for the treatment of cancer. Expert Opin. Investig. Drugs 28, 1095–1100 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Thomas A., Pommier Y., Targeting topoisomerase I in the era of precision medicine. Clin. Cancer Res. 25, 6581–6589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogakou E. P., Nieves-Neira W., Boon C., Pommier Y., Bonner W. M., Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J. Biol. Chem. 275, 9390–9395 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Moiseeva T. N., et al., An ATR and CHK1 kinase signaling mechanism that limits origin firing during unperturbed DNA replication. Proc. Natl. Acad. Sci. U.S.A. 116, 13374–13383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wohlschlegel J. A., et al., Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290, 2309–2312 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Agarwal S., et al., Cdt1 stabilizes kinetochore-microtubule attachments via an Aurora B kinase-dependent mechanism. J. Cell Biol. 217, 3446–3463 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y., Pozo P. N., Oh S., Stone H. M., Cook J. G., Distinct and sequential re-replication barriers ensure precise genome duplication. PLoS Genet. 16, e1008988 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higa L. A., Mihaylov I. S., Banks D. P., Zheng J., Zhang H., Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5, 1008–1015 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Swords R., et al., Cdc7 kinase—A new target for drug development. Eur. J. Cancer 46, 33–40 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Abbas T., et al., PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22, 2496–2506 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shibata E., Dar A., Dutta A., CRL4Cdt2 E3 ubiquitin ligase and proliferating cell nuclear antigen (PCNA) cooperate to degrade thymine DNA glycosylase in S phase. J. Biol. Chem. 289, 23056–23064 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jørgensen S., et al., SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J. Cell Biol. 192, 43–54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taby R., Issa J. P., Cancer epigenetics. CA Cancer J. Clin. 60, 376–392 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Eden A., Gaudet F., Waghmare A., Jaenisch R., Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300, 455 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Lecona E., Fernandez-Capetillo O., Targeting ATR in cancer. Nat. Rev. Cancer 18, 586–595 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Toledo L., Neelsen K. J., Lukas J., Replication catastrophe: When a checkpoint fails because of exhaustion. Mol. Cell 66, 735–749 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Coleman K. E., et al., Sequential replication-coupled destruction at G1/S ensures genome stability. Genes Dev. 29, 1734–1746 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jang S. M., et al., The replication initiation determinant protein (RepID) modulates replication by recruiting CUL4 to chromatin. Nat. Commun. 9, 2782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.