Significance
Pancreatic cancer utilizes autophagy to survive stress and promote therapeutic resistance. However, the metabolic contribution of autophagy in PDAC has not been fully elucidated. We report that SLC7A11 function requires autophagy machinery for proper membrane localization in PDAC, allowing proper cystine transport. Our findings continue to support the use of autophagy inhibition as a therapeutic strategy in pancreatic cancer by demonstrating that this leads to inactivation of SLC7A11 and disruption of cysteine homeostasis.
Keywords: pancreatic ductal adenocarcinoma, SLC7A11, autophagy, lysosome, pancreatic cancer
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the deadliest forms of cancer and is highly refractory to current therapies. We had previously shown that PDAC can utilize its high levels of basal autophagy to support its metabolism and maintain tumor growth. Consistent with the importance of autophagy in PDAC, autophagy inhibition significantly enhances response of PDAC patients to chemotherapy in two randomized clinical trials. However, the specific metabolite(s) that autophagy provides to support PDAC growth is not yet known. In this study, we demonstrate that under nutrient-replete conditions, loss of autophagy in PDAC leads to a relatively restricted impairment of amino acid pools, with cysteine levels showing a significant drop. Additionally, we made the striking discovery that autophagy is critical for the proper membrane localization of the cystine transporter SLC7A11. Mechanistically, autophagy impairment results in the loss of SLC7A11 on the plasma membrane and increases its localization at the lysosome in an mTORC2-dependent manner. Our results demonstrate a critical link between autophagy and cysteine metabolism and provide mechanistic insights into how targeting autophagy can cause metabolic dysregulation in PDAC.
Despite progress in cancer therapy, the prognosis for pancreatic ductal adenocarcinoma (PDAC) remains extremely poor with a 5-y survival rate of just 9% and it is predicted to become the second leading cause of cancer death in the United States by 2030 (1–3). The PDAC tumor microenvironment is highly desmoplastic and is composed of heterogeneous cell types, as well as an exuberant extracellular matrix. Together, this leads to poor perfusion and extreme hypoxia, creating a nutrient-limited environment with impaired drug penetration (4, 5). Another hallmark of PDAC is elevated basal autophagy which plays multiple protumorigenic roles, including promoting immune evasion and supporting its metabolic demand in this austere microenvironment (6–10). Therefore, clinical strategies have been employed to inhibit autophagy in PDAC patients using lysosomal inhibitors such as chloroquine or hydroxychloroquine (11, 12).
While autophagy can support diverse metabolic processes through the degradation of various cargo, how it supports PDAC metabolism has not been fully elucidated. In the present study, we found that autophagy has a selective role in sustaining cysteine (Cys) pools in PDAC. One of the major mechanisms of Cys homeostasis is through the import of cystine (the oxidized dimer of cysteine) through system xc−, a cystine/glutamate antiporter composed of SLC7A11 (xCT) and SLC3A2 (13). Recently, it was shown that both Cys and SLC7A11 are critical for PDAC growth (14). Here, we report that under low Cys conditions, SLC7A11 utilizes autophagy machinery to allow localization at the plasma membrane. Moreover, we demonstrate that loss of autophagy increases phosphorylation of SLC7A11 by mTORC2, and it remains primarily localized at the lysosome where its cystine import function is impaired. In summary, we identify a mechanism of Cys homeostasis in PDAC where the function of SLC7A11 is coordinately sustained by autophagic machinery and mTORC2 activity based on intracellular Cys levels.
Results
Loss of Autophagy Abrogates Intracellular Cysteine Levels in PDAC.
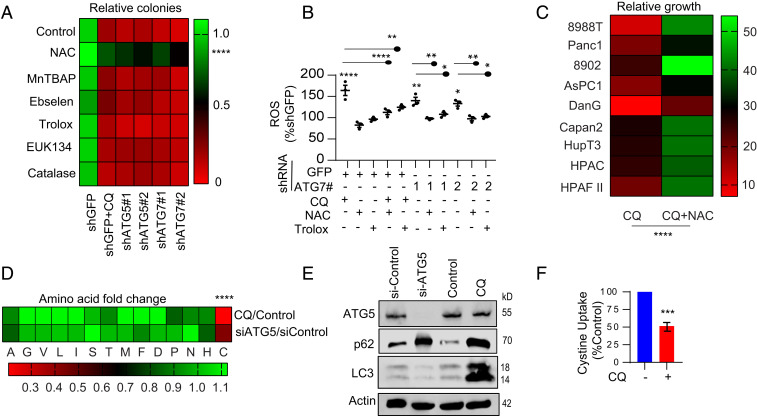
We previously identified that inhibiting autophagy in PDAC results in an accumulation of reactive oxygen species (ROS) and concomitant administration of N-acetyl-l-cysteine (NAC) could rescue the increased ROS (6). To further understand the role of autophagy in redox homeostasis, we inhibited autophagy pharmacologically (chloroquine, CQ) or genetically (RNAi to multiple autophagy-related ATG genes) and assessed the ability of multiple antioxidants to rescue clonogenic growth (Fig. 1A). Consistent with our prior work, NAC was also able to significantly rescue growth in the setting of genetic or pharmacologic autophagy inhibition. In contrast, none of the other antioxidants could rescue colony formation despite restoring ROS levels (Fig. 1B). We observed that NAC was able to rescue proliferation in the setting of autophagy inhibition across a panel of pancreatic cancer cell lines (Fig. 1C). One possibility was that NAC was replenishing intracellular Cys pools that might have been compromised by autophagy inhibition. Strikingly, we found that under nutrient-replete conditions, there was a dramatic drop in intracellular Cys levels when autophagy was inhibited by genetic and pharmacological means while other amino acid pool sizes showed minimal effects (Fig. 1 D and E). We first investigated whether this was due to impaired cystine uptake. Interestingly, we observed that autophagy inhibition markedly diminished cystine uptake (Fig. 1F and SI Appendix, Fig. S1A).
Fig. 1.
Autophagy inhibition selectively decreases intracellular cysteine. (A) Autophagy was inhibited pharmacologically (CQ) or genetically (ATG5/7 knockdown) in 8988T cells and treated with indicated antioxidants followed by assessing clonogenic growth after 10 d. Only NAC was able to rescue the number of colonies in autophagy-inhibited cells (P < 0.0001). (B) Measurement of ROS in 8988T cells with ATG7 knockdown supplemented with NAC or trolox for 24 h (n = 3 experiments). (C) PDAC cells were treated with CQ, +/− NAC, and relative cell proliferation on day 5 was quantified. Data were normalized to control. (D) Changes in amino acids after CQ treatment or si-ATG5 in 8988T cells. (E) Immunoblot analysis using the indicated antibodies to assess knockdown and autophagy inhibition was performed on 8988T cells used in D. Heatmap represents mean of five independent experiments in A, three independent experiments in C, and D. (F) Cystine starved (16 h) 8988T cells treated with or without CQ were incubated with FITC-labeled cystine (20 min, 37 °C) and the uptake of cystine was analyzed (n = 51 cells). Data: mean ± SEM, P values: two-tailed unpaired t test (A, C, D, and F); P values: one-way ANOVA with Tukey’s post hoc test (B). ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
Autophagy Inhibition Leads to SLC7A11 Inactivation.
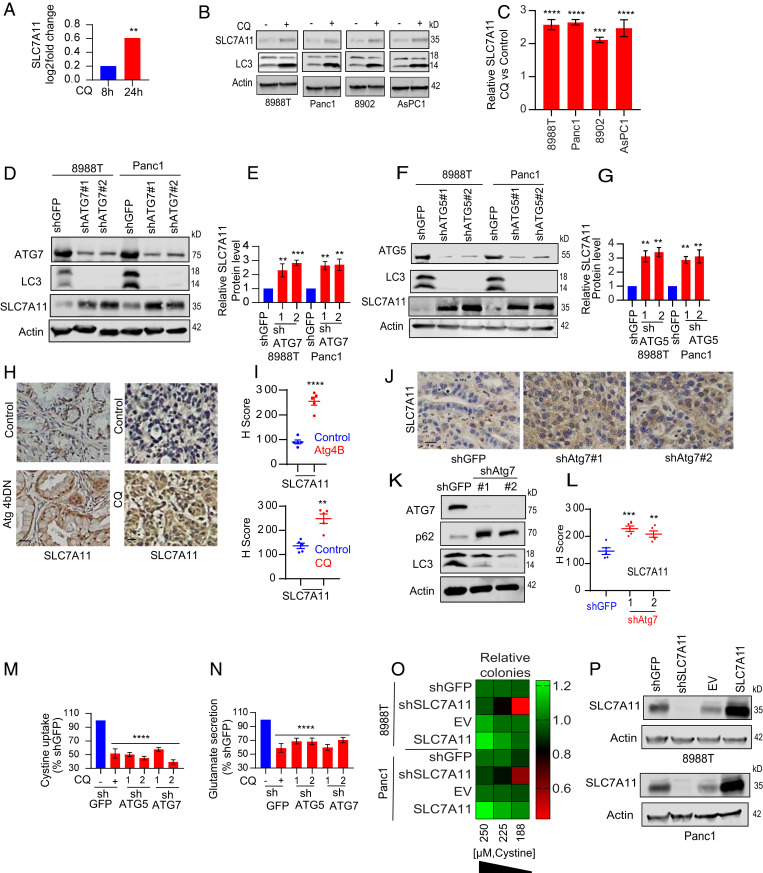
Given the critical role of system xc− in the import of cystine (13–16), we hypothesized that autophagy loss may impair the expression of this glutamate/cystine antiporter. Surprisingly, pharmacological autophagy inhibition in PDAC cells actually increased the levels of the SLC7A11 subunit of system xc− as shown by our quantitative mass spectrometry-based proteomics analysis (Fig. 2A). We confirmed this finding by immunoblot analysis in multiple PDAC lines where autophagy was inhibited pharmacologically using CQ (Fig. 2 B and C) or genetically by knockdown of multiple autophagy genes (ATG7: Fig. 2 D and E; ATG5: Fig. 2 F and G). We next validated our in vitro findings in PDAC-bearing mice expressing a dominant negative (DN) mutant of Atg4B, treated with CQ (Fig. 2 H and I and SI Appendix, Fig. S1B), or with Atg7 suppression in the tumor by RNAi, that also showed elevation in SLC7A11 expression (Fig. 2 J–L).
Fig. 2.
Loss of autophagy inactivates SLC7A11 function. (A) Quantitative mass spectromentry-based proteomics showing SLC7A11 levels in Panc1 cells treated with CQ at indicated time points. Experiment was done in biological triplicate, P value represents Benjamini–Hochberg corrected. (B) Lysates from different PDAC cells treated with CQ were immunoblotted for indicated proteins and the relative SLC7A11’s expression was quantified (n = 3) in C. Autophagy was genetically inhibited by knocking down ATG7 (D) and ATG5 (F) in 8988T, Panc1 cells, and the level of SLC7A11 (n = 5) was quantified by Western blotting, respectively, in E and G. (H) Tumor (n = 5) sections from mice expressing Atg4b DN or CQ treatment were immunohistochemically analyzed for SLC7A11 followed by their quantification, respectively (I). P values: two-tailed unpaired t test. (J) IHC images of SLC7A11 in murine KPC [KrasLSL-G12D/+, Trp53lox/+, p48-cre+ (p48 is also known as Ptf1a)] PDAC tumors with Atg7 knockdown (n = 5). (K) Immunoblot confirming Atg7 knockdown and quantification of IHC data for SLC7A11 (L). SLC7A11 activity was quantified by measuring cystine uptake (M) and glutamate secretion (N). (O) Overexpression and knockdown of SLC7A11 in 8988T, Panc1 cells were assessed for clonogenic growth under various cystine concentrations along with Western blotting for SLC7A11 in these cells (P). Data: mean ± SEM, P values: one-way ANOVA with Tukey’s post hoc test (except in A). ****P < 0.0001, ***P < 0.001, **P < 0.01. (Scale bar, 15 µm.)
Taken together, our data demonstrate that despite an increase in SLC7A11, there was a significant decrease in cystine import and intracellular Cys pools. This suggested that autophagy inhibition might lead to a physiological situation where the activity of the transporter may be compromised. To assess this, we measured the cellular uptake of cystine (Fig. 2M) and glutamate efflux (Fig. 2N), which revealed that SLC7A11’s activity was reduced in the setting of autophagy inhibition. One possible explanation for the reduced activity of SLC7A11 was that it was not properly localized to the plasma membrane. Indeed, time-lapse video imaging in response to CQ treatment in PDAC cells showed the rapid translocation of SLC7A11 away from the plasma membrane into the cytosol (SI Appendix, Fig. S1C and Movies S1 and S2). Consistent with recent studies (14, 17), cystine and SLC7A11 were observed to be critical for PDAC growth (Fig. 2 O and P).
Loss of Autophagy Leads to SLC7A11 Translocation to Lysosome.
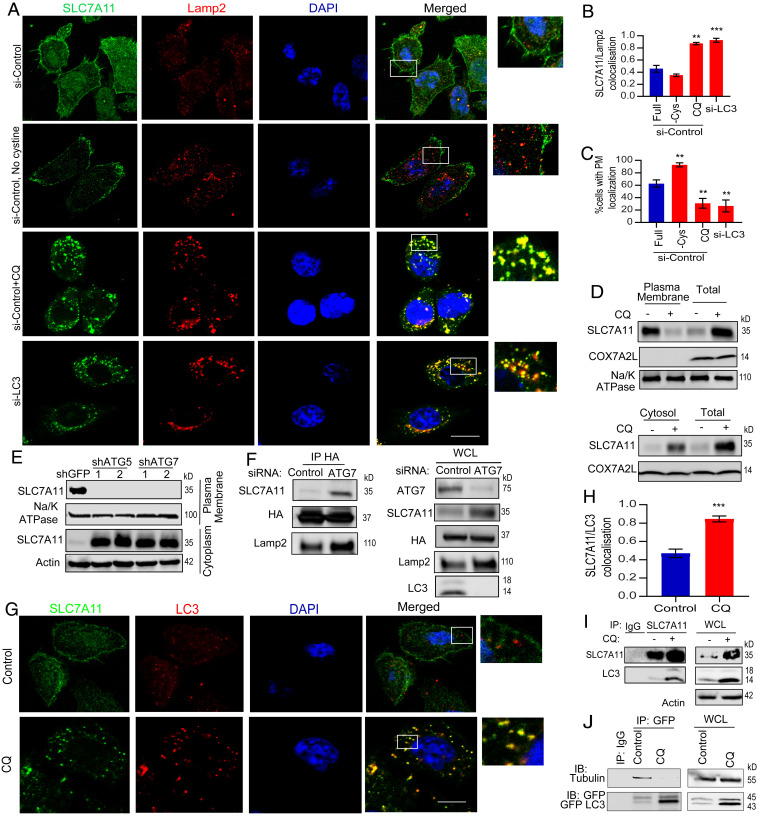
We quantified the plasma membrane distribution of SLC7A11 after both genetic and pharmacologic autophagy inhibition at various phases of the autophagy pathway by immunofluorescence (Fig. 3 A–C and SI Appendix, Fig.S1 D and E) and biochemical fractionation (Fig. 3 D and E). This demonstrated that upon loss of autophagy, there is translocation of SLC7A11 from the plasma membrane to the cytosol. Through a series of colocalization studies, we found that autophagy inhibition results in a significant fraction of SLC7A11 colocalized with the lysosomal membrane marker Lamp2 (Fig. 3 A–C). We further demonstrated the lysosomal enrichment of SLC7A11 after autophagy inhibition by immunopurification of lysosomes (Fig. 3F and SI Appendix, Fig. S2A). Interestingly, cystine deprivation in autophagy intact conditions revealed increased SLC7A11 localization on plasma membrane by immunofluorescence (Fig. 3A) and biochemical fractionation (SI Appendix, Fig. S2B). Taken together, these results indicate that the plasma membrane localization of SLC7A11 is based on Cys demand and is autophagy dependent. To rule out the translocation of a nutrient transporter as a general phenomenon of autophagy inhibition, we assessed the localization of an unrelated solute carrier SLC30A (importer of Zn/Mn) (SI Appendix, Fig. S2C). In contrast to SLC7A11, SLC30A did not show altered subcellular localization in response to CQ. Additionally, there was no significant change in general endocytosis in response to autophagy inhibition as evidenced by the uptake of transferrin (SI Appendix, Fig. S2D). Recently, Roy et al. showed that retromer regulated the translocation of a glucose transporter GLUT1 in an autophagy-dependent manner (18). However, knockdown of key components of the retromer, TBC1D5 and VPS35, did not cause any change in SLC7A11 localization (SI Appendix, Fig. S2E), indicating that the retromer was not involved in SLC7A11 translocation on and off the plasma membrane.
Fig. 3.
Autophagy inhibition promotes SLC7A11 localization to lysosomes. (A) Confocal microscopy images showing SLC7A11 and Lamp2 colocalization analysis after autophagy inhibition or cystine starvation (overnight) in 8988T followed by quantification of SLC7A11/Lamp2 colocalization (B) and %cells with SLC7A11 present on the plasma membrane (PM) was quantified (n = 90 cells from three experiments, C). (D) SLC7A11 distribution in plasma membrane and cytoplasmic fractions was analyzed in 8988T cells where autophagy was inhibited by CQ treatment (extracted fractions were diluted to half and immunoblotted along with total proteins of whole cell) or ATG5/7 knockdown by RNAi (E). (F) Immunopurified lysosomes from 8988T stably expressing pLJC5-Tmem192-3xHA were analyzed for SLC7A11 with and without autophagy inhibition. (G) Confocal microscopy images of SLC7A11 after CQ treatment (24 h, n = 35 cells) in 8988T cells. (H) Colocalization analysis of SLC7A11/LC3 was performed from cells in G. Data are mean ± SD; P value was obtained by two-tailed unpaired t test. (I) Immunoprecipitation analysis of SLC7A11 to study interaction of SLC7A11 with LC3. (J) Western blots showing interaction analysis between tubulin with GFP-LC3 in 8988T cells lacking endogenous LC3 after CQ treatment. Data: mean ± SEM, P values: one-way ANOVA with Tukey’s post hoc test (except in H). ***P < 0.001, **P < 0.01. (Scale bar, 15 µm.)
SLC7A11 Requires LC3-Microtubule Association for Its Localization to the Plasma Membrane.
Based on our data, we speculated that lipidated LC3 might be essential to transport SLC7A11 along microtubules, as has been shown previously with autophagosomes trafficking toward the lysosome (19–22). Consistent with this, we could detect a complex between SLC7A11 and LC3, as well as SLC7A11 was present in PDAC autophagosomes (Fig. 3 G–I). Paradoxically, inhibition of either autophagosome formation (LC3 knockdown) or autophagosome degradation (CQ) both resulted in decreased membrane-localized SLC7A11 and increased presence at the lysosome (Fig. 3A). It was unclear how CQ, or knockdown of LC3, had a similar effect on SLC7A11 positioning, given that they have opposite impacts on LC3 levels (Fig. 3A and SI Appendix, Fig. S2F). We hypothesized that while CQ treatment leads to an increase in lipidated LC3, this likely remains sequestered in lysosomes, preventing interaction with microtubules. Indeed, CQ treatment markedly inhibited the interaction of LC3 with tubulin even though the amount of total LC3 is increased (Fig. 3J and SI Appendix, Fig. S2G). Thus, any alteration that effectively impairs the ability of LC3 to associate with microtubules will inhibit SLC7A11 localization at the plasma membrane.
Autophagy Inhibition Leads to SLC7A11 Inactivation through an mTOR-Mediated Process.
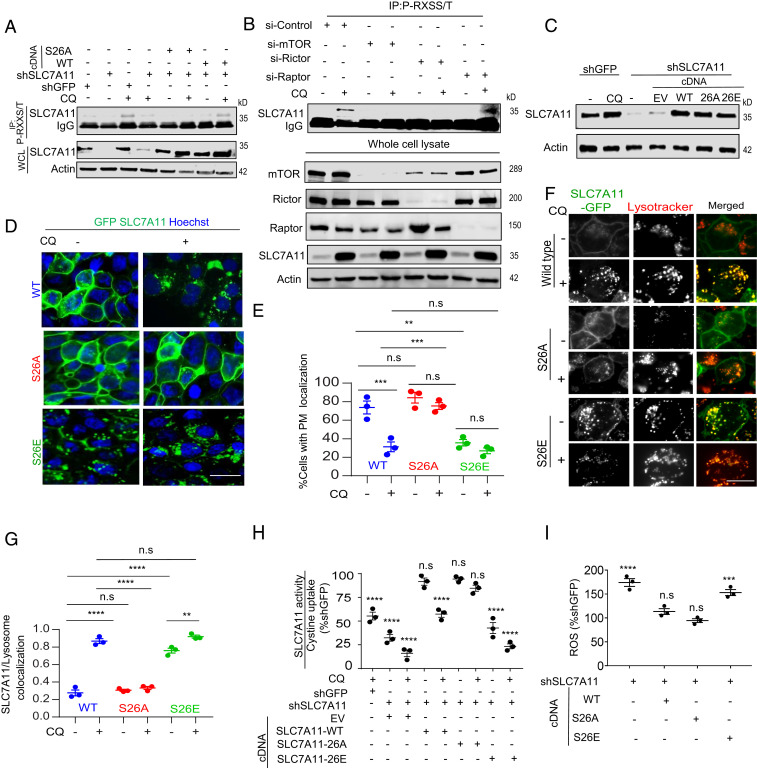
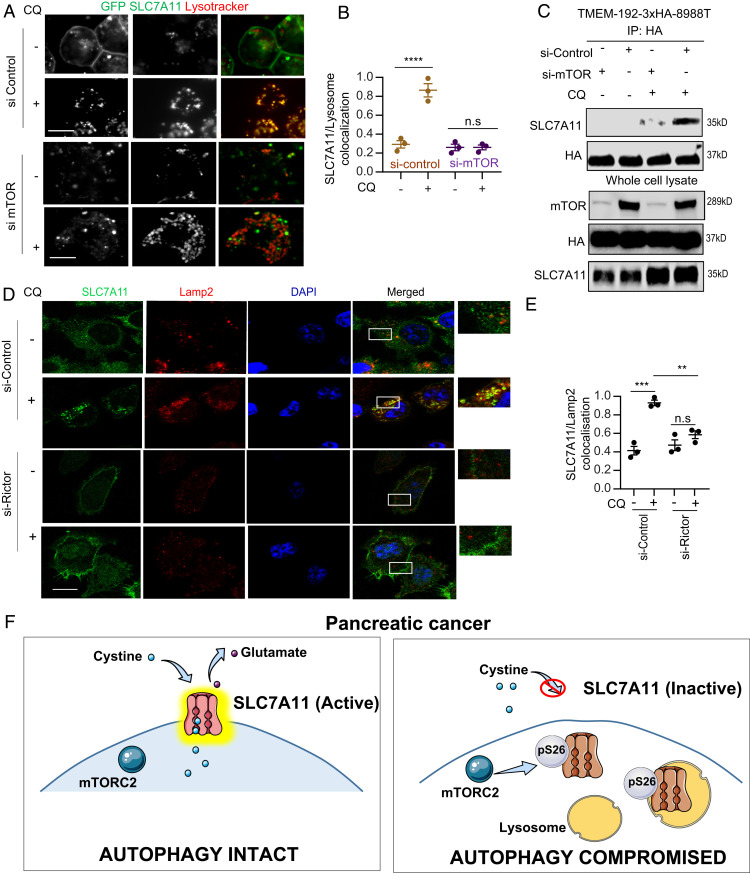
Recently, it was shown that mTORC2 inhibits SLC7A11 activity by phosphorylation on the S26 site of SLC7A11 (15). Indeed, we were able to detect the phosphorylation of SLC7A11 which was increased upon inhibition of autophagy by CQ treatment (Fig. 4A). Consistent with the importance of mTORC2, knockdown of Rictor, but not Raptor, significantly inhibited the phosphorylation (Fig. 4B). We confirmed that the S26 site was the mTORC2 target by demonstrating that the phosphorylation was completely abrogated by mutating the S26 site to an alanine (Fig. 4A). To further study the role of phosphorylation in SLC7A11 function, we generated the phosphomimetic S26E mutant and expressed this mutant as well as the corresponding alanine mutant in cells with a GFP tag to study its localization (Fig 4 D–G) or untagged in cells where the endogenous SLC7A11 was suppressed by RNAi to study its transport functions (Fig 4 A, C, H, and I). Interestingly, we found the SLC7A11-S26E mutant showed less plasma membrane localization in comparison to the wild type, or the S26A mutant and exhibited higher lysosomal localization, similar to the situation where autophagy was inhibited (Fig. 4 D–G). Consistent with its predominantly lysosomal localization, the SLC7A11-S26E mutant failed to import cystine (Fig. 4H) and could not reduce the level of reactive oxygen species upon knockdown of endogenous SLC7A11 (Fig. 4I). Moreover, knockdown of mTOR (Fig. 5 A–C) itself or Rictor (Fig. 5 D and E), prevented SLC7A11 colocalization with the lysosome in the setting of autophagy inhibition. Taken together, these data show that SLC7A11 activity is regulated by subcellular localization. Its transport function necessitates plasma membrane localization, requiring intact autophagy, and this is impaired upon autophagy inhibition through phosphorylation by mTORC2, leading to translocation to the lysosome (Fig. 5F).
Fig. 4.
Inhibition of autophagy in PDAC leads to mTOR-mediated inhibition of SLC7A11. (A) 8988T cells stably expressing RNAi-resistant SLC7A11-S26A/wild-type proteins with the endogenous expression suppressed by RNAi, were treated with CQ and the lysates were immunoprecipitated with P-RXXS/T-beads and probed for SLC7A11. (B) 8988T cells with knockdown of mTOR, Rictor, or Raptor were treated with or without CQ and the lysates were immunoprecipitated by P-RXSS/T and analyzed for SLC7A11 phosphorylation. (C) Lysates of 8988T expressing the indicated mutants of SLC7A11 (wild type, S26A, and S26E) in the setting of knockdown of the endogenous SLC7A11 were analyzed for SLC7A11 by Western blotting. (D) GFP-SLC7A11-wild type (WT)/S26A/S26E 8988T cells were treated with CQ followed by analysis of SLC7A11 distribution at plasma membrane distribution (E) (n = 90 cells, three experiments). (F) Autophagy was inhibited by CQ treatment in 8988T cells expressing SLC7A11 mutants and the imaging was performed for GFP-SLC7A11/lysotracker followed by colocalization analysis (G) (n = 41 cells from three experiments). (H) Cystine uptake level was measured to assay for SLC7A11 activity in 8988T cells expressing the indicated mutant with or without autophagy inhibition (n = 3). (I) Indicated mutant bearing 8988T cells were assayed for ROS analysis (n = 3). Data: mean ± SEM, P values: one-way ANOVA with Tukey’s post hoc test. ****P < 0.0001, ***P < 0.001, **P < 0.01, n.s., not significant: P > 0.5. (Scale bar, 15 µm.)
Fig. 5.
mTORC2 and autophagy regulate the subcellular localization of SLC7A11 to control its activity. (A) Images of GFP-SLC7A11 expressing 8988T cells were analyzed for colocalization with lysosomes when mTOR expression was suppressed, followed by their quantification (B) (n = 71 cells, three experiments). (C) SLC7A11 was immunoblotted using immunopurified lysosome from mTOR-suppressed pLJC5-TMEM-192-3HA-8988T cells +/− CQ. (D) 8988T cells were immunostained for SLC7A11 and Lamp2 followed by analysis for colocalization (E) in Rictor knockdown cells after +/− CQ treatment. (n = 3 independent experiments). (F) Schematic representation showing the role of autophagy-mediated SLC7A11 regulation to modulate cysteine metabolism. Data: mean ± SEM, P values: one-way ANOVA with Tukey’s post hoc test. ****P < 0.0001, ***P < 0.001, **P < 0.01, n.s, not significant: P > 0.5. (Scale bar, 15 µm.)
Discussion
Autophagy is conventionally regarded as a lysosome-based catabolic pathway. Here, we discover an unconventional role of autophagy machinery that is utilized for promoting uptake of cystine from the extracellular environment via promoting localization of SLC7A11 at the plasma membrane. Our work shows that inhibition of autophagy specifically impairs Cys uptake and metabolism in PDAC. This critical metabolic role further supports our previous studies that established autophagy inhibition as a key therapeutic strategy in PDAC (8–10). While the link to Cys metabolism appears to be prominent in PDAC, we cannot rule out other metabolic functions of autophagy in these tumors, given the complexities of cellular metabolism. Indeed, in other tumor types such as lung cancer, autophagy has been shown to be important for supporting the mitochondrial substrate supply and nucleotide pools (23). This highlights that the physiological importance of autophagy can vary in different cancers even if they have the same oncogenic drivers, such as Kras, and may reflect the unique biology and metabolic needs of a given tumor type.
We used genetic and pharmacological means to impair the function of key proteins involved in the process of initiation, elongation, and lysosomal fusion steps of autophagy and demonstrated the significance of the autophagosome/tubulin interaction as a conduit for SLC7A11 translocation. This is in agreement with a previous report from Kimura et al. where it was shown that microinjection of anti-LC3 antibody prevented the autophagosome movement in the cytoplasm (21). Although it is well established that SLC7A11 is an importer of cystine (13–16) as part of system xc−, its requirement for LC3 machinery to regulate its transport function and proper localization in a Cys-dependent manner had not been previously identified. In PDAC, we found that the regulation of the transport functions of SLC7A11 relies on the localization to the plasma membrane which requires autophagy machinery to lipidate LC3 and mediate its interaction with microtubules. Furthermore, inhibition of autophagy promotes an inhibitory phosphorylation by mTORC2 and results in lysosomal localization of SLC7A11, which prevents extracellular cystine uptake. Future studies will be required to understand the precise mechanisms connecting autophagy inhibition to mTORC2 kinase activity. Additionally, the mechanism of microtubule transport from the plasma membrane to lysosome and the role of SLC7A11 on the lysosome remain to be defined.
One mechanism of how autophagy supports PDAC growth is through maintaining the nutrient transport function of SLC7A11, as it provides the ability of the cell to coordinate Cys availability and proliferation. Cys plays a critical role in sustaining the antioxidant pool that is essential for maintaining tumor growth and proliferation (14, 24). Recent work has shown that shortage of extracellular Cys can be compensated by transulfuration activity to support the growth of neuroblastoma cells (25). However, Badgley et al. showed that loss of Cys was not being compensated by any salvage pathway in PDAC, thereby exposing its metabolic vulnerability (14). Our work also supports targeting Cys metabolism in PDAC and provides an alternative mechanism to do so through inhibition of autophagy, which would impair SLC7A11 activity and deplete Cys pools.
Materials and Methods
Cell Culture.
8988T, Panc1, 8902, AsPC1, DanG, Capan2, HupT3, HPAC, and HPAF II were obtained from ATCC or DSMZ and cultured in DMEM (Dulbecco's Modified Eagle Medium) with 10% FBS (fetal bovine serum) and antibiotic antimycotic. Cystine starvation experiments were carried out in media with dialyzed FBS (Fisher Scientific 26-400-044). All the cell lines were routinely tested for mycoplasma by PCR and periodically authenticated by fingerprinting, visual inspection, and storage in a centralized cell bank.
SLC7A11 Activity.
Cystine uptake was analyzed following the sodium nitroprusside-based protocol previously described (15). In tandem with the cystine uptake, the extracellular glutamate secretion was quantified using Glutamate-Glo (Promega J7021) following the manufacturer’s protocol. Data were normalized by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) based cell number quantification and was represented as %control. For analysis of fluorescent cystine uptake, equal numbers of PDAC cells were starved of cystine by culturing in media without cystine and supplemented with dialyzed serum overnight followed by incubation with BioTracker cystine-FITC (fluorescein isothiocyanate) live cell dye (1 µM, 20 min). Following this, cells were washed with PBS (phosphate-buffered saline) and imaging was done immediately. Data were normalized by cell number quantification by Hoechst-based nuclear staining and represented as %control.
Transferrin Uptake.
PDAC cells under the indicated conditions were analyzed for transferrin uptake as previously reported (18).
Plasma Membrane and Cytoplasm Purification.
Plasma membrane and cytoplasm were purified using the Minute Plasma Membrane Protein Isolation and Cell Fractionation Kit (Invent Biotechnologies Inc. SM-005) according to the manufacturer’s instructions.
Cell Proliferation and Clonogenic Assay.
Cell proliferation and clonogenic assay were performed as previously reported (6, 26). For these assays, CQ was used at 10 μM.
Immunofluorescence Analysis.
Unless otherwise mentioned, antibodies (see SI Appendix) were used against endogenous proteins for analyzing colocalization using a Leica-DM6 and analyzed using LAS-X software (version 2.0.0.14332.2) as previously described (27). Percentage of cells with predominantly plasma membrane expression of GFP-SLC7A11 was quantified by measuring the integrated optical density (threshold filter was set at a value of 40) and area fraction with the ImageJ software (National Institute of Mental Health); nuclear-staining by DAPI/Hoechst was used to normalize cell number. Random visual fields were observed for each group to quantify the immunofluorescence signal for colocalization using JACoP plugin in ImageJ (27). SLC30A10-GFP was gifted by Somshuvra Mukhopadhyay (The University of Texas at Austin, Austin, TX), Addgene 104381.
Immunohistochemistry Analysis.
The mentioned antibodies were used at the manufacturer’s recommended concentration for immunohistochemical (IHC) analysis which was performed according to the previously reported methods (9). We performed color deconvolution of IHC images using ImageJ Fiji software with the help of the “H DAB” vector option and selected a threshold of the antibody staining intensity to quantify the H-score.
Live Cell Imaging.
8988T cells stably expressing GFP-SLC7A11 were treated with CQ and live time-lapse imaging acquisition of spatiotemporal location of GFP-SLC7A11 was performed using the Zeiss LSM-880 Airyscan Fast Live Cell using Zen software at 20-s intervals, 7 slice z-stack, 60 cycles. Images were acquired for 20 min in CQ-treated and control groups. The real-time tracking of GFP-SLC7A11 was analyzed by Imaris software.
Reactive Oxygen Species Detection.
Briefly, 10,000 cells underwent indicated experimental treatments followed by quantification of ROS by DCFDA (2’,7’–dichlorofluorescin diacetate) Cellular ROS Detection Assay Kit (Abcam: ab113851) using manufacturer instructions followed by reading at Ex/Em (excitation/emission wavelength): 485/535 with a BMG Labtech Spectrophotometer. Alteration in ROS level is represented as %control or %shGFP.
Animal Experiments.
Histology sections from murine PDAC tumors with autophagy inhibition expressing the dominant negative Atg4b (9), shAtg7, or CQ (10) treated were obtained from previous studies. Briefly, 25,000 mouse PDAC cells were suspended in a 40-µL solution comprising 50% HBSS (Hank's balanced salt solution) and 50% Matrigel (Corning: 356231) and injected subcutaneously into the lower flank of female NCr nude mice (Taconic) at 6 wk of age. Daily intraperitoneal injection was performed with CQ at 60 mg/kg in 100 µL of PBS or 100 µL of PBS only daily for 3 wk. All mouse experiments were conducted in compliance with ethical regulations approved by the New York University Institutional Animal Care and Use Committee (IACUC-ID:IA16-00507, IA16-01331).
Metabolomics.
Metabolites were extracted by first washing cells with ice-cold 0.9% saline followed by addition of 5:2:5 ratio of methanol:water:chloroform (−20 °C). Cells were scraped into an Eppendorf and vortexed at 4 °C for 10 to 15 min followed by separation of aqueous and inorganic metabolites by centrifugation. Polar metabolites containing amino acids were transferred to an Eppendorf and evaporated by SpeedVac (Savant, Thermo). Metabolite levels were quantified by gas chromatography-mass spectrometry (GC-MS) as previously described (28). Briefly, metabolites were derivatized to form their methoxime-tBDMS (tert-butyldimethylsilyl) derivatives and analyzed on an Agilent 5977B gas chromatograph (GC) interfaced with an Agilent 5977B mass spectrometer (MS). Metabolite levels were normalized for extraction and derivatization efficiency by normalizing the total ion counts to an added norvaline internal standard and cell number. As intracellular Cys levels were difficult to measure by GC-MS, they were measured separately using the cysteine assay kit (MyBioSource: MBS8309602) following the manufacturer’s instructions. In this case the normalization was done to cell count.
Western Blotting.
Freshly pelleted cells were lysed in chilled buffer comprising Cell Lysis Buffer (1×, Cell Signaling Technology: 9803) supplemented with protease inhibitor (1×, Thermo A32953) and phosphatase inhibitor (1×, PhosSTOP, Sigma: 04906837001) and followed as previously described (10).
Immunoprecipitation.
Briefly 1 × 106 cells were lysed in a buffer comprising 50 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 1% Nonidet P-40 supplemented with protease inhibitor (Thermo A32953) and phosphatase inhibitor (PhosSTOP, Sigma 04906837001). For experiments with mTORC1, C2 components, CHAPS (3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate) buffer was used instead of Nonidet P-40. Cell lysates were immunoprecipitated using the recommended concentration of antibody which was already tagged with Dynabead A protein matrix following the instructions from the Dynabeads Protein A Immunoprecipitation Kit (Thermo 10006D). Lysosomal immunopurification was performed in PDAC cells stably expressing pLJC5-Tmem192-3xHA (gifted by David Sabatini, Whitehead Institute for Biomedical Research and Massachusetts Institute of Technology, Cambridge, MA, Addgene 102930) following a previously reported method (10).
Quantitative Proteomics.
Analysis for quantitative mass spectrometry-based proteomics was performed as previously described (26). All analysis involved an MS3-based TMT (tandem mass tag) method as previously mentioned and mass spectra were processed as described earlier (26, 29).
Statistical Analysis.
All data are presented as the mean ± SEM. The experimental results were analyzed by comparing one-way ANOVA followed by Tukey’s post hoc test until otherwise mentioned. P < 0.05 was considered the level of significance when comparing the values from the experimental treatments to those of the control using GraphPad 7. Each experiment was conducted with biological replicates and repeated multiple times. All attempts at replication were successful and no data were excluded. Mice were randomly allocated to experimental groups and the investigators were not blinded during the experiments.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute Grants R01CA157490, R01CA188048, P01CA117969, and R35CA232124; NIH Grant R01GM095567; and the Lustgarten Foundation, and SU2C (Stand-Up-To-Cancer) (to A.C.K.); and NIH/National Institute of General Medical Sciences Grant R01 GM132129 (to J.A.P.). R.S.B. is a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2348-18). We thank Paul Mischel (University of California, San Diego) for sharing pLVX, pLVX-SLC7A11, and pLVX-SLC7A11-S26A constructs. Fig. 5F was created with the help of Servier Medical Art, which is licensed under a Creative Commons Attribution 3.0 Unported License.
Footnotes
Competing interest statement: A.C.K. is a founder and has financial interests in Vescor Therapeutics, LLC. A.C.K. is an inventor on patents pertaining to Kras-regulated metabolic pathways, redox control pathways in pancreatic cancer, targeting GOT1 as a therapeutic approach and the autophagic control of iron metabolism. A.C.K. is on the science advisory board of Rafael/Cornerstone Pharmaceuticals. A.C.K. is a consultant for Deciphera and Abbvie. J.D.M. is an inventor on a patent pertaining to the autophagic control of iron metabolism.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021475118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Rahib L., et al., Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74, 2913–2921 (2014). [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society . Cancer Facts & Figures 2020. Atlanta: American Cancer Society; 2020. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2020/cancer-facts-and-figures-2020.pdf. Accessed 21 January 2020.
- 4.Feig C., et al., The pancreas cancer microenvironment. Clin. Cancer Res. 18, 4266–4276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponz-Sarvise M., et al., Identification of resistance pathways specific to malignancy using organoid models of pancreatic cancer. Clin. Cancer Res. 25, 6742–6755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang S., et al., Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sousa C. M., et al., Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang A., et al., Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 4, 905–913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang A., et al., Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 8, 276–287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto K., et al., Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581, 100–105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karasic T. B., et al., Effect of gemcitabine and nab-paclitaxel with or without hydroxychloroquine on patients with advanced pancreatic cancer: A phase 2 randomized clinical trial. JAMA Oncol. 5, 993–998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeh H. J., et al., A randomized phase II preoperative study of autophagy inhibition with high-dose hydroxychloroquine and gemcitabine/nab-paclitaxel in pancreatic cancer patients. Clin. Cancer Res. 26, 3126–3134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassi M. T., et al., Identification and characterisation of human xCT that co-expresses, with 4F2 heavy chain, the amino acid transport activity system xc-. Pflugers Arch. 442, 286–296 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Badgley M. A., et al., Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 368, 85–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Y., et al., mTORC2 regulates amino acid metabolism in cancer by phosphorylation of the cystine-glutamate antiporter xCT. Mol. Cell 67, 128–138.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato H., Tamba M., Ishii T., Bannai S., Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 274, 11455–11458 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Daher B., et al., Genetic ablation of the cystine transporter xCT in PDAC cells inhibits mTORC1, growth, survival, and tumor formation via nutrient and oxidative stresses. Cancer Res. 79, 3877–3890 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Roy S., Leidal A. M., Ye J., Ronen S. M., Debnath J., Autophagy-dependent shuttling of TBC1D5 controls plasma membrane translocation of GLUT1 and glucose uptake. Mol. Cell 67, 84–95.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geeraert C., et al., Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J. Biol. Chem. 285, 24184–24194 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korolchuk V. I., et al., Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura S., Noda T., Yoshimori T., Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 33, 109–122 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Pedrotti B., Ulloa L., Avila J., Islam K., Characterization of microtubule-associated protein MAP1B: Phosphorylation state, light chains, and binding to microtubules. Biochemistry 35, 3016–3023 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Guo J. Y., et al., Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 30, 1704–1717 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kshattry S., et al., Enzyme-mediated depletion of l-cyst(e)ine synergizes with thioredoxin reductase inhibition for suppression of pancreatic tumor growth. NPJ Precis. Oncol. 3, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J., et al., Transsulfuration activity can support cell growth upon extracellular cysteine limitation. Cell Metab. 30, 865–876.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biancur D. E., et al., Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat. Commun. 8, 15965 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukhopadhyay S., et al., Autophagy protein Ulk1 promotes mitochondrial apoptosis through reactive oxygen species. Free Radic. Biol. Med. 89, 311–321 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Parker S. J., et al., Selective alanine transporter utilization creates a targetable metabolic Niche in pancreatic cancer. Cancer Discov. 10, 1018–1037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mancias J. D., Wang X., Gygi S. P., Harper J. W., Kimmelman A. C., Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.