Abstract
Asymptomatic surveillance testing together with COVID-19-related research can lead to positive SARS-CoV-2 tests resulting not from true infections, but non-infectious, non-hazardous by-products of research (amplicons). Amplicons can be widespread and persistent in lab environments and can be difficult to distinguish for true infections. We discuss prevention and mitigation strategies.
Asymptomatic surveillance testing together with COVID-19-related research can lead to positive SARS-CoV-2 tests resulting not from true infections, but non-infectious, non-hazardous by-products of research (amplicons). Amplicons can be widespread and persistent in lab environments and can be difficult to distinguish for true infections. We discuss prevention and mitigation strategies.
Introduction
On June 17, a positive SARS-CoV-2 test result was reported at a university associated with this study. The individual had no known exposure to any infected person and the incidence rate in Massachusetts at the time was 0.004% (COVID-19 Response Reporting). Further, the individual had no symptoms prior to the test, nor would they develop symptoms in subsequent days. Over the next day, two more employees who worked in the same research laboratory tested positive. They too did not exhibit any symptoms. All three persons worked with an amplicon, i.e., an amplified DNA product, of the N2 epitope of the SARS-CoV-2 nucleocapsid (N) gene. We note that these amplicons are not contained in live viruses and cannot cause clinical disease. As such, the researchers did not believe they were truly SARS-CoV-2 infected, and numerous messages started to come to the offices of the university’s Health Services (UHS) and Environmental Health and Safety (EH&S), voicing a concern that this might be a case of amplicon contamination, a known issue in clinical labs (Aslanzadeh, 2004).
In an attempt to determine whether these were true SARS-CoV-2 infections, two additional tests that targeted different regions of the viral N gene (N1 and N3) were performed on each individual—all were negative. Although this suggested the individuals might not have had COVID-19, uncertainty regarding the source of the originally detected viral genetic material and the delay between the initial and follow-up real-time quantitative polymerase chain reaction (RT-qPCR) tests led local health authorities to instruct the university to handle the cases as standard infections. The individuals were instructed to isolate, and all their traced contacts were put into quarantine.
Even though these positive test results were a significant burden on the individuals, the contact tracing teams, and both UHS and EH&S, they did not initially pursue additional follow-up on these three cases. From June to November 2020, four of the five universities participating in this paper identified approximately 300 positive cases. Each case triggered a chain of immediate reactions from local authorities (CDC, 2020a), including asking the employees to go into isolation, shutting down lab spaces for cleaning, and putting multiple close contacts from those research communities and the general public in quarantine. As part of this study, we were able to follow up with 39 out of the 300 cases (IRB20-0581). Suspiciously, all cases were clustered in individuals that work in or were in close proximity to laboratories that leverage synthetic DNA of the SARS-CoV-2 virus for various research objectives. Indeed, there have been multiple reports of asymptomatic researchers who worked with or near non-infectious SARS-CoV-2 nucleic acids and subsequently tested positive during SARS-CoV-2 surveillance screening (Robinson-McCarthy et al., 2021). Critically though, previous work was circumstantial, not definitively showing that none of these cases were SARS-CoV-2 infection. Given the fact that a significant percentage of cases are asymptomatic (Chau et al., 2020), ruling a case as amplicon contamination because no symptoms developed can easily miss true infections.
Thus, in order to determine whether or not these 39 cases were true COVID-19 infection or cases of amplicon contamination, we performed a series of follow-up tests as described below. We stress that amplicon contamination is not a public health risk or likely to lead to positive tests in the general public. Instead, it is specific to environments that work with viral genetic material. Indeed, all researchers were amplifying DNA that would be detected by their universities’ SARS-CoV-2 screening program. However, given the large number of cases in a relatively small set of schools, the amount of resources being spent on these cases, and the disrupted research activity, we considered it critical to determine, by follow-up testing, whether or not these were true asymptomatic COVID-19 infections or amplicon contamination. Raising the awareness of research universities and health authorities to amplicon contamination and establishing protocols for prevention and mitigation of future cases are urgent.
Results
To determine the cause of the RT-qPCR positive tests, we analyzed all ≈300 positive cases that arose in four out of the five universities associated with this study between June 17, 2020 and November 11, 2020. Several dozen individuals who tested positive during routine active surveillance testified they did not have any interactions with known COVID-19-positive persons and were extremely careful in following COVID-19 CDC guidelines in public. We followed-up with 39 of those individuals (by consent; IRB20-0581) via performing different combinations of tests including RT-qPCR with multiple viral targets, whole-genome sequencing, and serological tests for the presence of SARS-CoV-2 immunoglobulin G (IgG) and/or IgM antibodies (Table S1). Repeat testing of the original samples was usually not possible because specimens were not retained by testing facilities after processing because of their large testing volume. Therefore, all reported repeat tests were performed on freshly collected samples 1–3 days after the original positive test results; antibody tests were performed 33 days on average after the initial test result.
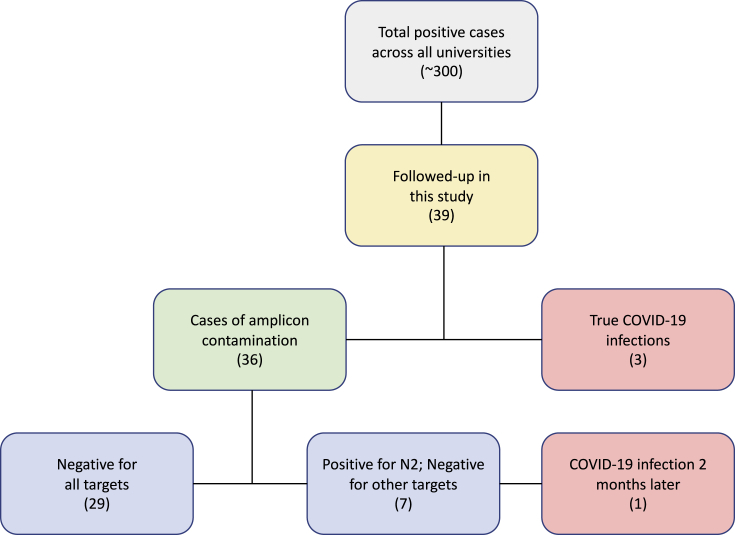
All 39 primary positive tests were obtained by RT-qPCR assays designed to detect the N2 and/or N1 viral genes in anterior nares swabs (Figure 1). Broken down by cases, 29 individuals were negative on all follow-up tests. Another seven individuals had a positive RT-qPCR follow-up test for N2 but were negative for all other tested targets (i.e., N1, N3, E, and/or RdRp) (Figure 1). Notably, the negative qPCR result for 25 of the 36 (29 + 7) cases was obtained as little as 1–3 days after their initial positive test. We also note that those individuals exhibited high Ct values for N2 already in their initial positive test (36.7 ± 1.7; for cases where Ct values were reported), already suggesting that those individuals might not have been true COVID-19 infections. Sealing the case that the vast majority of these cases were amplicon contamination, 18 of the 19 individuals were seronegative when tested for SARS-CoV-2 IgG/M antibodies ∼30 days after the initial positive result (Table S1).
Figure 1.
A flowchart describing the breakdown of cases in this study
A total of 300 cases were identified in 4 out of 5 universities associated with this study between June 17, 2020 and November 11, 2020. Of these cases, 39 were followed up on by RT-qPCR and/or serological tests. Twenty-nine were negative for the N2 target as well as all other tested targets within 1–3 days of the original test; seven cases were positive for N2 in follow-up PCR tests but negative for all other tested targets. One of those seven cases developed symptoms several weeks after the original positive test and was diagnosed as a true COVID-19 infection by PCR of the N1 and N2 targets.
It is extremely important to note that not all individuals were positive due to amplicon contamination. We identified a total of four true positive cases in our cohort. One asymptomatic individual did not receive a follow-up qPCR test but was positive for SARS-CoV-2 antibodies ∼2 months after the initial positive result. Two of the individuals had repeat tests that were positive on secondary tests with different viral targets, and one of the two individuals was symptomatic. The initial Ct value for the asymptomatic COVID-19 infected individual was 31.3, considerably lower than the Ct in each of the amplicon contamination cases (36.3 ± 2.01 on average). The fourth individual was more complicated. This originally asymptomatic individual was tested on September 24 as positive for N2 with a high Ct value (37.4) and negative for N1 in the same test. An antibody test a month later (October 22) came back negative, strongly arguing this was a case of amplicon contamination. However, on November 10 the individual became symptomatic and tested positive for N1 and N2. This individual is therefore one who was initially positive due to amplicon contamination but then contracted COVID-19 afterward (Figure 1).
The identification of four true positive individuals among the group of suspected cases of amplicon contamination shows how critical it is to perform follow-up tests to determine the true nature of the test result. Further, the situation in this last individual highlights the importance of identifying cases of amplicon contamination rapidly. In accordance with CDC guidance, testing programs exclude individuals who have tested positive for 90 days after the positive result due to persistent RT-qPCR after SARS-CoV-2 infection (Cevik et al., 2020; Wang et al., 2020). These individuals, subsequent to amplicon contamination, can contract COVID-19 and be missed because they are not tested. Luckily, this person was asked to return to routine testing once his serological test returned, and shortly thereafter he became symptomatic, presented for clinical evaluation, and was tested positive. As part of our mitigation strategy, we argue that all individuals identified as having positive tests due to amplicon contamination be immediately returned to asymptomatic testing regimes.
Evidence for amplicon
In light of the evidence for amplicon contamination in these cases suggested by Robinson-McCarthy et al. (2021) and demonstrated in this work, we sought to determine how widespread amplicons are in research environments and where in the lab amplicons would be found. Environmental swabs were conducted in 11 locations including lab, office, and kitchen spaces with a total of ≈90 different sites tested. Amplicons were found in high titers (Ct < 30) on a number of sites including centrifuges, pipettes, gel areas, and bench spaces, as well as on other lab equipment including microscopes and incubators (Table S2). Importantly, substantial concentrations were also found on doorknobs, lab notebooks, pens, glasses, and computer keyboards with Ct values ranging from 25.8 to 42.6 with a mean of 32.1 ± 4.9.
The presence of amplicons on doorknobs and bench spaces in neighboring labs, in which SARS-CoV-2 work is not conducted, highlight how individuals in adjoining spaces were amplicon positive. The presence of amplicons on personal belongings could help explain two surprising cases. In one case, a researcher extracted large amounts of DNA encoding for the viral N2 protein from bacteria. The same day that individual tested positive, their roommate, who does not work in a research lab, also tested positive. Both cases were verified as amplicon contamination by the lack of IgG/M antibodies against SARS-CoV-2 and follow-up negative RT-qPCR (against the viral amplicon N1, N2, E, and RdRp) (Table S2). A second case proceeded similarly, but the researcher was performing reverse transcription loop-mediated isothermal amplification (RT-LAMP) and the negative follow-up assays were for N1 and N3 and independently N1 and N2.
Prevention strategies
We believe a large number of the amplicon positive cases are due to research laboratory procedures that lead to amplicon contamination. Amplicon contamination is a well-known issue in laboratories, especially clinical labs (Aslanzadeh, 2004). Clinical labs implement a number of protocols to largely eliminate amplicon contamination (see Dos and don’ts for molecular testing). Indeed, Clinical Laboratory Improvement Amendments (CLIA) labs had trouble with amplicon contaminations many years ago until strict standard operating procedures (SOPs) were put in place to help avoid this type of contamination. These include steps such as performing all pipetting in negative pressure hoods, UV light and bleach cleaning of equipment and work areas, avoiding opening tubes after the amplification reaction, and pipetting below the liquid line or in hoods. In addition, individuals can take a number of steps to lower the chance that amplicon contamination registers during surveillance testing such as testing at the beginning of the day before entering the lab, showering and scrubbing hands with soap and water (not alcohol rub, which does not destroy amplicons) prior to sample collection, and wearing a new pair of disposable gloves when handling test kits, especially if using a self-swab approach (CDC, 2020b). We note that although the protocols being followed by researchers that were exposed to amplicons are safe, the high-cadence testing identified the amplicon contamination as COVID-19 leading to mandatory quarantine, isolation, and cleaning. If high cadency was not occurring, the amplicon contamination would likely either not have an effect or occasionally led to false positives in the assay. But, given the high cadency of SARS-CoV-2 testing in these research labs, stricter SOPs are needed in research labs (more similar to what is done in CLIA labs), to avoid amplicon contamination. As such, a consortium of biosafety committees and EH&S at our universities, in conjunction with local and state public health departments have developed guidance policies that standardize laboratorians training to minimize contamination and recommendations for regular documentation of cleaning within these spaces (see “guidelines for working with amplicons to minimize the risk of amplicon contamination”).
Post-positive mitigation strategies
Although all amplicon contamination would ideally be eliminated by improved laboratory protocols, it is unlikely that all cases will be eliminated. Therefore, it is necessary to be able to empirically differentiate between COVID-19 viral infection and amplicon contamination. As we show, it is critical to perform follow-up tests on individuals with suspected cases of amplicon contamination. Therefore, we advise that two follow-up tests be conducted; ideally, follow-up tests should be performed with tests that probe different amplicons. Because during the initial phase of the infection viral titer can be low (Cevik et al., 2020) and different amplicons and platforms can be more sensitive than others, we do not recommend retesting the initial sample unless the relative efficiency of each test is known. Instead, we advise waiting 24 h before collecting the follow-up test to help distinguish between low viral titer early in infection and amplicon contamination. This is because viral titer rises significantly after it is initially detectable by qPCR (Cevik et al., 2020; Kissler et al., 2020). Waiting 24 h should also help in cases where the source of amplicon contamination is suspected to be whole-genome amplification instead of a smaller part of the viral genome. Although several other approaches can be useful to determine whether there is amplicon contamination, e.g., qPCR amplification with no RT, we do not advise these as a follow-up strategy because someone can simultaneously have SARS-CoV-2 infection and amplicon contamination.
Methods
RT-qPCR analysis
All RT-qPCR assays were performed by CLIA-approved labs. Several different primer and probe pairs were used to detect the targets listed in Table S1. Reported Ct values are listed in Table S1; no Ct value threshold was applied for positive tests.
Antibody testing
Convalescent serum IgM/G antibody studies were completed on a total of 19 cases. Assays (VITROS Immunodiagnostic Products; Anti-SARS-CoV-2 Reagent Pack assay) were performed on samples collected on days 7–75 after the initial positive test results.
Swab sampling procedure
All test sites are listed in Table S1. At each site, an area of 12 in2 was sampled with a sterile 3/16-inch-thick medical-grade polyurethane swab (Puritan, Guilford; Cat No. 2 5-1607 1PF SC). Swabs were stroked in vertical and diagonal ‘S’ shapes. New pairs of gloves were used for each sample to avoid cross-contamination and were placed in a screw cap 1.5 mL tube pre-aliquoted with transport medium (VMA from AHDC Molecular Diagnostics). Samples were labeled and submitted to AHDC Molecular Diagnostics for analysis by qPCR (no RT) using primers for N1, N2, S, and Orf1 viral genes.
Concluding remarks
Amplicon contamination can lead to spurious results and incorrect clinical outcomes in clinical laboratories—results that are often quickly followed up on because of their clinical significance. In research labs, however, it is typically ignored or seen as a nuisance that is mitigated by repeated experiments or different approaches. High-cadency asymptomatic testing changes the risk in research labs. Now amplicon contamination can lead to positive RT-qPCR tests with clinically relevant follow-up (e.g., isolation and quarantine). As indicated by our study, the large number of cases in a relatively small set of schools highlights how important it is to rapidly distinguish amplicon contamination cases from true infections and how important it is for research labs to follow proper handling guidelines. The amount of resources being spent on these cases and the disrupted research activity burdens universities and the healthcare system and slows down timely COVID-19-related research.
Finally, one key point is that in communities where there is a large amount of biomedical and life science research underway, it is important during the case investigation process to elicit the specific nature of the work when an individual works in a biomedical laboratory. Investigators should ask about an individual’s occupation, whether it involves any research related to SARS-CoV-2 or whether their worksite is in close proximity to a lab where such research is conducted, and if so, inquire about laboratory practices. In addition, all cases should be asked whether a member of the household is involved in SARS-CoV-2 research in any way, or whether they work in a space adjacent to a lab conducting this research. An attempt should be made to understand whether there is potential for amplicon contamination that could yield a positive result with the original PCR assay used. Patients who work in these environments or who have household contacts working in these environments should be offered confirmatory testing with an alternate PCR assay that uses different probes, as discussed above. It should be noted that, although most of our amplicon contamination occurred with N2, there is no reason to believe that amplicon contamination isn’t occurring with all amplified portions of SARS-CoV-2. In summary, we hope these prevention and mitigation strategies will help to alleviate the burden being caused to research universities and hospitals and allow us to focus resources on true positive cases without impacting research and researcher safety. We are hopeful that the lessons learned from this study would apply beyond this pandemic.
Acknowledgments
We thank the COVID Testing Laboratory at the Animal Health Diagnostic Center at Cornell University, Ithaca, NY. We also thank the many staff of the affiliated university health services, environmental health and safety units, and research laboratories who responded to these cases and helped to accumulate the data necessary for this manuscript. M.S. is supported by R01 GM120122-01. D.D. is an European Molecular Biology Organization (EMBO) long-term fellow (ALTF 1146-2018), Human Frontier Science Foundation (HFSP) fellow (LT000232/2019-L), and Rothschild fellow.
Author contributions
D.D., D.H.H., and M.S. analyzed the data and wrote the manuscript; S.F., H.L.G., S.J., C.M.K., L.L., S.M., S.E.M., V.M.B., G.T.N., J.T.P., K.K., H.L., C.W.S., J.E.T., and D.H.H. collected data, performed experiments, and/or supervised the experiments. M.S. supervised the work.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100005.
Supplemental information
Follow-up RT-qPCR and serology test results for 39 cases (IRB20-0581). RT-qPCR test results are indicated with respect to the order of tested targets; for example, case ID 6 was negative for both N1 and N3 targets in a follow-up RT-qPCR test; case ID 10 was negative for N1 and N3 but positive for N2 in follow-up RT-qPCR tests. RT-qPCR follow-up tests were performed 1–3 days after the initial positive test unless stated otherwise in the notes (the only such case is case ID 26). For specific dates of each RT-qPCR test and Ct values, refer to the “test details” tab. Specific information about the assays that were used for each test are not provided to avoid case identification; ND means “not detected;” missing values indicate no test was done on these cases.
References
- Aslanzadeh J. Preventing PCR amplification carryover contamination in a clinical laboratory. Ann. Clin. Lab. Sci. 2004;34:389–396. [PubMed] [Google Scholar]
- CDC Investigating a COVID-19 Case. 2020. https://www.cdc.gov/coronavirus/2019-ncov/php/contact-tracing/contact-tracing-plan/investigating-covid-19-case.html
- CDC Coronavirus Disease 2019 (COVID-19) 2020. https://www.cdc.gov/coronavirus/2019-ncov/index.html
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. The Lancet Microbe. 2020;0:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau N.V.V., Thanh Lam V., Thanh Dung N., Yen L.M., Minh N.N.Q., Hung L.M., Ngoc N.M., Dung N.T., Man D.N.H., Nguyet L.A., et al. The natural history and transmission potential of asymptomatic SARS-CoV-2 infection. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Response Reporting https://www.mass.gov/info-details/covid-19-response-reporting
- Dos and don’ts for molecular testing. https://www.who.int/teams/global-malaria-programme/case-management/diagnosis/nucleic-acid-amplification-based-diagnostics/dos-and-don-ts-for-molecular-testing
- Kissler S.M., Fauver J.R., Mack C., Tai C., Shiue K.Y., Kalinich C.C., Jednak S., Ott I.M., Vogels C.B.F., Wohlgemuth J., et al. Viral dynamics of SARS-CoV-2 infection and the predictive value of repeat testing. medRxiv. 2020 doi: 10.1101/2020.10.21.20217042. [DOI] [Google Scholar]
- Robinson-McCarthy L.R., Mijalis A.J., Filsinger G.T., de Puig H., Donghia N.M., Schaus T.E., Rasmussen R.A., Ferreira R., Lunshof J.E., Chao G., et al. Anomalous COVID-19 tests hinder researchers. Science. 2021;371:244–245. doi: 10.1126/science.abf8873. [DOI] [PubMed] [Google Scholar]
- Wang J., Hang X., Wei B., Li D., Chen F., Liu W., Yang C., Miao X., Han L. Persistent SARS-COV-2 RNA positivity in a patient for 92 days after disease onset: A case report. Medicine (Baltimore) 2020;99:e21865. doi: 10.1097/MD.0000000000021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Follow-up RT-qPCR and serology test results for 39 cases (IRB20-0581). RT-qPCR test results are indicated with respect to the order of tested targets; for example, case ID 6 was negative for both N1 and N3 targets in a follow-up RT-qPCR test; case ID 10 was negative for N1 and N3 but positive for N2 in follow-up RT-qPCR tests. RT-qPCR follow-up tests were performed 1–3 days after the initial positive test unless stated otherwise in the notes (the only such case is case ID 26). For specific dates of each RT-qPCR test and Ct values, refer to the “test details” tab. Specific information about the assays that were used for each test are not provided to avoid case identification; ND means “not detected;” missing values indicate no test was done on these cases.