Abstract
Leigh syndrome is a fatal neurometabolic disorder caused by defects in mitochondrial function. mTOR inhibition with rapamycin attenuates disease progression in a mouse model of Leigh syndrome (Ndufs4 KO mouse); however, the mechanism of rescue is unknown. Here we identify PKC downregulation as a key event mediating the beneficial effects of rapamycin treatment of Ndufs4 KO mice. Assessing the impact of rapamycin on the brain proteome and phosphoproteome of Ndufs4 KO mice we find that rapamycin restores mitochondrial protein levels, inhibits signaling through both mTOR complexes, and reduces the abundance and activity of multiple protein kinase C (PKC) isoforms. Administration of PKC inhibitors increases survival, delays neurological deficits, prevents hair loss, and decreases inflammation in Ndufs4 KO mice. Thus, PKC may be a viable therapeutic target for treating severe mitochondrial disease.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
INTRODUCTION
Mitochondria are essential organelles, generating 80–90% of cellular ATP via respiratory metabolism1. Genetic impairment of the mitochondrial respiratory chain can lead to several mitochondrial diseases with diverse clinical etiology, but often associated with encephalopathy, neuroinflammation, and neurodegeneration. Leigh syndrome is regarded as the most common childhood mitochondrial disease and is characterized by bilateral lesions in the brainstem and basal ganglia. Mutations in both nuclear and mitochondrially encoded components of the electron transport chain, such as Ndufs4 of Complex I (C-I), have been identified as causal for Leigh Syndrome2. Knockout of the Ndufs4 gene in mice (Ndufs4 KO) leads to rapid onset of a mitochondrial disease that shares clinical features of Leigh Syndrome including neurodegeneration, neuroinflammation, progressive lesions in the brain, and substantially shortened lifespan3.
The mechanistic target of rapamycin (mTOR) is a serine/threonine kinase that controls cell growth and metabolism and is hyperactivated in animal models of mitochondrial disease4–6. mTOR functions in two distinct complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). We previously reported that the mTORC1 inhibitor rapamycin alleviates neuropathic symptoms in Ndufs4 KO mice and more than doubles their life expectancy4. Rapamycin treatment also induces a metabolic shift in this context, reducing accumulation of glycolytic intermediates and lactic acid, and suppressing the extremely low body fat of Ndufs4 KO mice4. To further explore the mechanisms of rapamycin-mediated rescue in Ndufs4 KO mice, we used mass spectrometry-based proteomics to elucidate changes in the brain proteome and phosphoproteome.
RESULTS AND DISCUSSION
Rapamycin similarly remodels the brain proteome in wild-type and Ndufs4 deficient mice
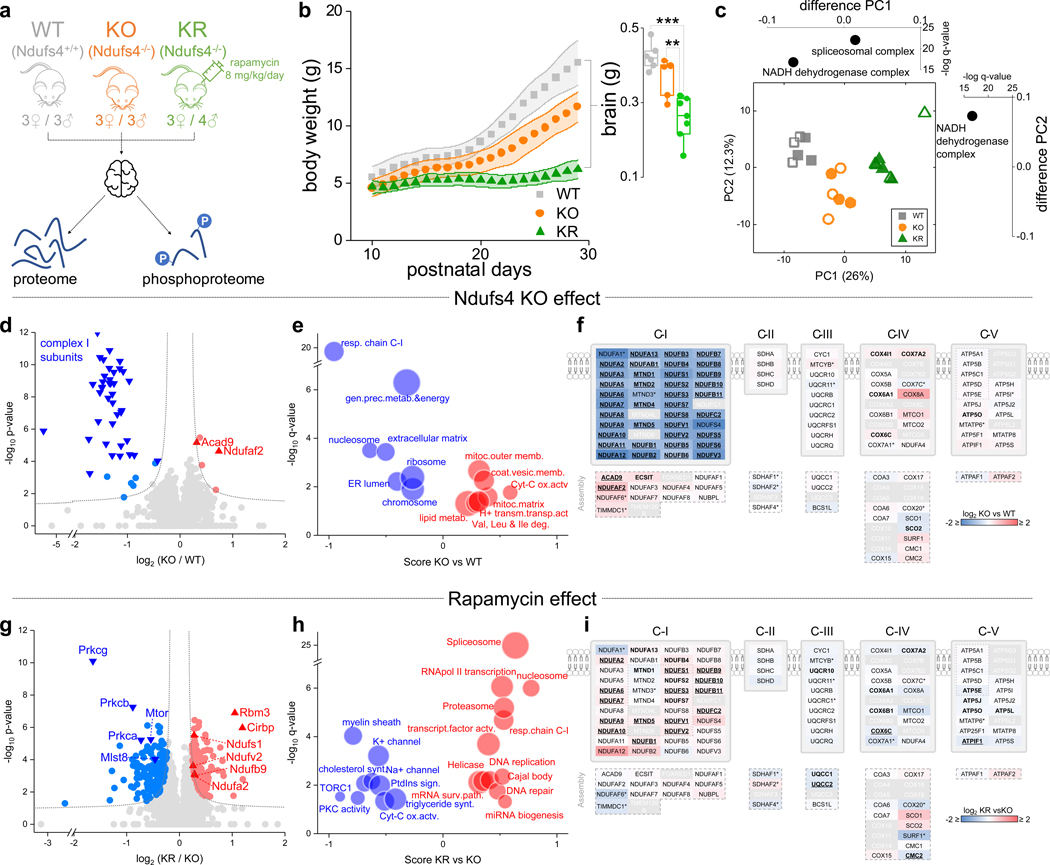
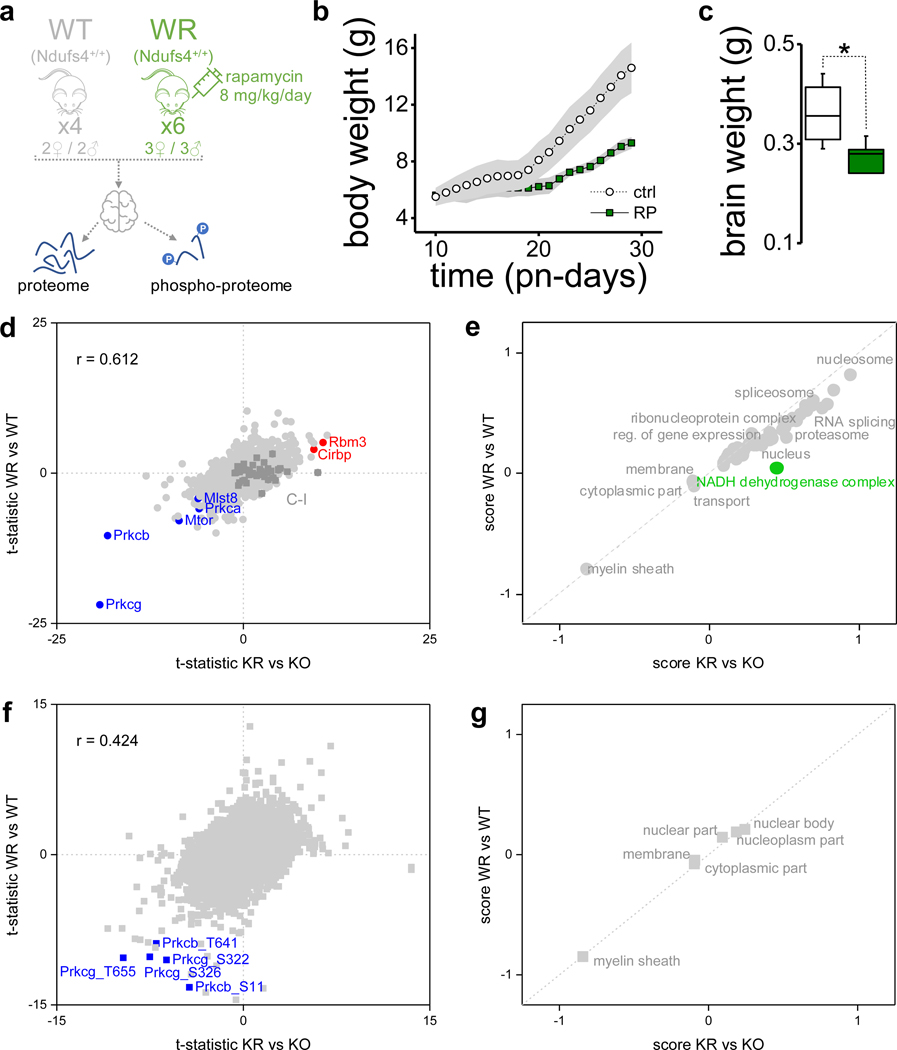
We analyzed brain tissue of 30-day old vehicle-treated wild-type (WT), and Ndufs4 KO mice treated with a daily intraperitoneal injection of 8 mg/kg/day rapamycin (KR) or vehicle (KO) for 20 days (Fig. 1a). Postnatal day 30 is approximately one week before the appearance of neurodegeneration and behavioral abnormalities in Ndufs4 KO mice3, 4. As expected, rapamycin-treated Ndufs4 KO mice had lower body weight and brain weight relative to vehicle-treated mice4 (Fig. 1b).
Fig. 1: Rapamycin remodels the brain proteome in Ndufs4 deficient mice.
a, Experimental design (N = 6 mice for the wild-type (WT) and Ndufs4 KO (KO) groups, N = 7 mice for the rapamycin-treated Ndufs4 KO (KR) group). b, Mice body weight from the three experimental groups (mean ± s.d.). Inner box plot shows total brain weight at the end of the experimental trial (30 days) (t-test: ** p < 0.01; *** p < 0.001; N = 6–7 mice). Box plots include the median line, the box denotes the interquartile range (IQR), whiskers denote ±1.5 × IQR. c, PCA analysis of log2 transformed normalized protein abundance data (hollow/solid symbols indicates female/male samples respectively). Side graphs indicate significantly enriched GO slim terms in the loadings for each component. d,e, Volcano plots comparing protein abundance in KO vs. WT groups (genotype effect; N = 6 mice) at the level of individual proteins (d) or representative biological terms (e). f, Mapping of protein abundance differences in individual subunits of the respiratory chain between the KO and WT groups. Assembly subunits of each mitochondrial respiratory complex are displayed at the bottom of each plot. LFQ normalized abundance data was used, except for subunits with asterisk in which intensity iBAQ values were used. Subunits with significant changes are in bold (p-value < 0.05) and underlined if FDR q-value < 0.05. In grey are subunits with no abundance information. g,h, Volcano plots comparing protein abundance in rapamycin-treated KR vs. KO groups (rapamycin effect; N = 6–7 mice) at the level of individual proteins (g) or representative biological terms (h). In (d) and (g), dotted lines indicate cut-off for significant changes: t-test FDR q-value < 0.05 and artificial within groups variance S0 = 0.1). (e) and (h) show significantly enriched annotation terms from 1D-enrichment analysis of log2 differences between groups (Wilcoxon-Mann-Whitney test, FDR q-value < 0.05); dot size is proportional to the number of proteins within that term. i, Same as in (f) but between KR and KO groups.
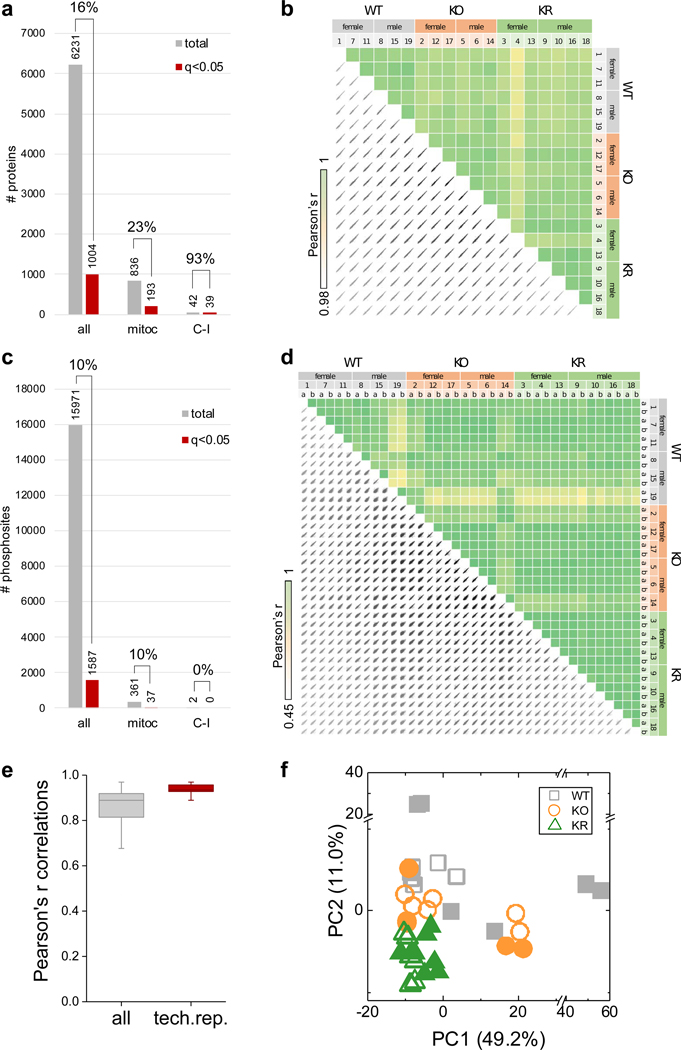
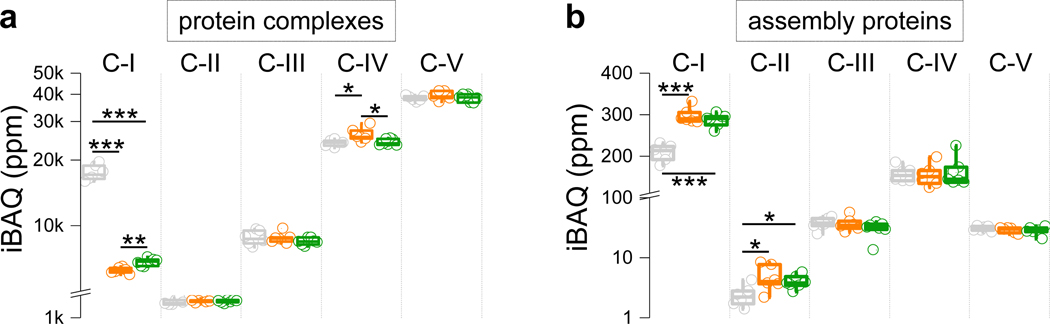
We quantified 6,231 proteins (Supplementary Table 1) with high reproducibility (Extended Data Fig. 1a), of which 16% (1,004 proteins) showed significant changes in abundance between groups (ANOVA test, FDR q-value < 0.05). The mitochondrial proteome showed substantial changes in response to Ndufs4 loss, driven primarily by altered abundance of C-I proteins (Extended Data Fig. 1a). In agreement, principal component analysis (PCA) revealed changes to C-I proteins as the major discriminant among groups (Fig. 1c). No partitioning between sexes was observed either with or without rapamycin treatment (Fig. 1c). Most C-I proteins decreased substantially in Ndufs4 KO mouse brains (Fig. 1d–f and Extended Data Fig. 2), except for two C-I assembly proteins (Acad9 and Ndufaf2) (Fig. 1d,f). Rapamycin treatment caused a modest increase of several C-I subunits (Fig. 1g–i and Extended Data Fig. 2), but is not sufficient to fully restore C-I function4. In addition to the decrease in C-I abundance in Ndufs4 KO brain, we observed changes in other respiratory chain protein complexes (Fig. 1f,i and Extended Data Fig. 2). In particular, Ndufs4 KO mice showed an overall increase in the cytochrome c oxidase complex (C-IV or COX), which was reverted by rapamycin treatment (Extended Data Fig. 2a).
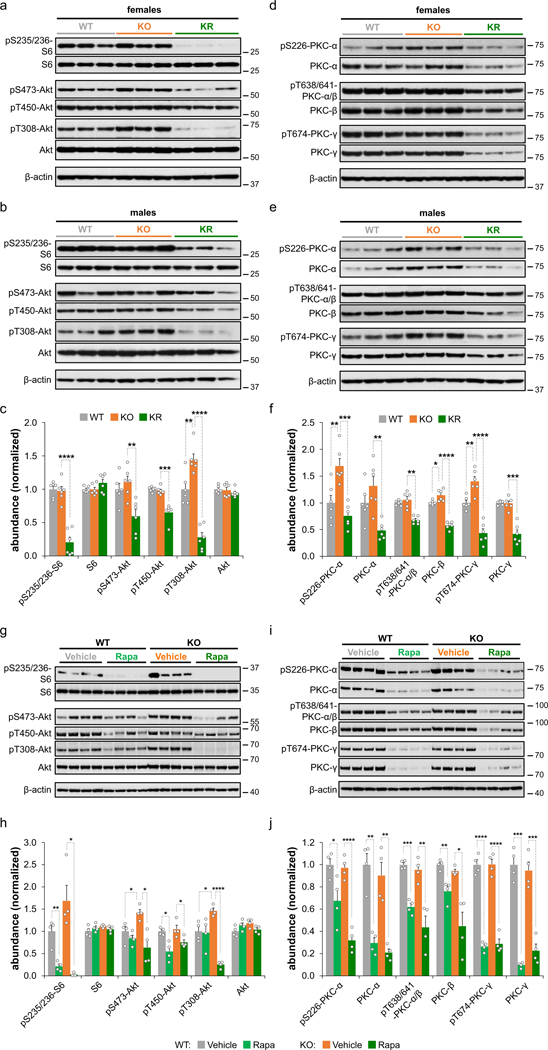
Rapamycin treatment of Ndufs4 KO mice caused additional changes in the brain proteome, including a significant decrease of the mTORC1 and mTORC2 core subunits mTOR and mLST8 (Fig. 1g). Although rapamycin is a specific inhibitor of mTORC17, 8, chronic rapamycin treatment has been reported to inhibit mTORC2 in mice9, 10, which is also supported by our phosphoproteomic data where we observed a reduction in phosphorylation of the mTORC2 targets11, 12 Akt and protein kinase C (PKC) (Extended Data Fig. 3a–f). Notably, all conventional PKC isoforms (PKC-α, PKC-β, and PKC-γ) were significantly decreased (Fig. 1g and Extended Data Fig. 3d–f). Conventional PKC isoforms’ stability is compromised when the activation loop, turn motif, or hydrophobic region are unphosphorylated, leading to its degradation via the ubiquitin-proteasome pathway13–15. Thus, decreased PKC phosphorylation upon rapamycin treatment may promote its instability and degradation. We observed similar results in brain extracts of rapamycin-treated mice at 50 days of age (Extended Data Fig. 3g–j), an age at which untreated Ndufs4 KO mice already display severe neuropathic symptoms and are nearing their median lifespan4. Our data suggest that inhibition of mTORC2 upon chronic rapamycin treatment reduces PKC abundance and activity, something that has been previously demonstrated by ablating mTORC2 function in vitro11 and also in vivo12, 15.
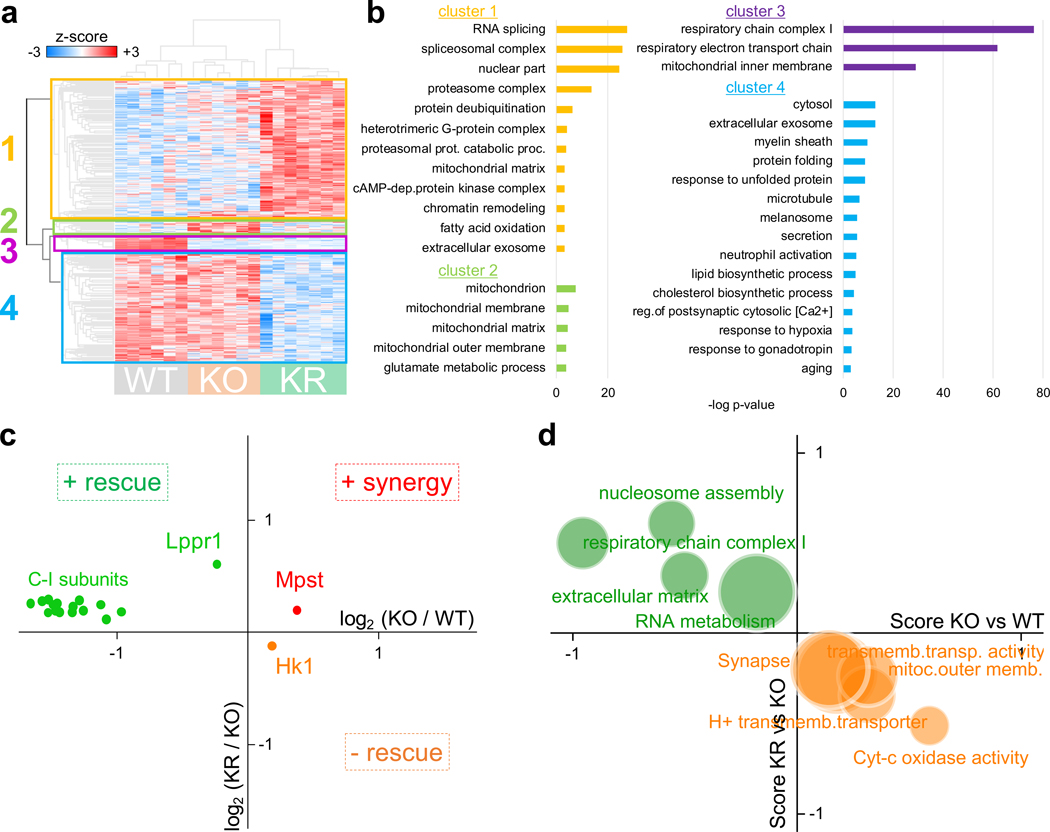
To further explore the potential impact of rapamycin on mitochondrial function in the context of C-I deficiency, we performed hierarchical clustering of protein abundance changes which grouped proteins into 4 clusters (Fig. 2a). Clusters 1 and 4 contain protein changes associated with rapamycin treatment, cluster 3 includes protein changes associated with Ndufs4 ablation, and cluster 2 is composed of proteins that increased in Ndufs4 KO mice and were restored to normal levels with rapamycin (Supplementary Table 2). This latter cluster was significantly enriched in mitochondrial proteins (Fig. 2b). Among these, we found Hk1, the hexokinase responsible for glucose utilization in brain, which mostly localizes to the outer mitochondrial membrane (OMM) to promote the energetically favorable coupling of glycolysis to oxidative phosphorylation16 (Fig. 2c and Supplementary Table 2). This agrees with our prior observations indicating that rapamycin shifts metabolism away from glycolysis in Ndufs4 KO mice4. We found other OMM proteins significantly enriched in cluster 2, especially proteins involved in mitochondrial fission (e.g. Mff), and also proteins involved in other mitochondrial functions such as glutamate metabolism and cytochrome-C oxidase activity (Fig. 2b,d and Supplementary Table 2). Among proteins which decreased in Ndufs4 KO mice and returned to normal levels with rapamycin we found Lppr1 (Fig. 2c), a neuronal growth promoter17. We also found that increased levels of Mpst in Ndufs4 KO mice, a neuroprotectant mitochondrial enzyme involved in hydrogen sulfide production18, were further enhanced by rapamycin (Fig. 2b).
Fig. 2: Rapamycin restores the abundance level of several mitochondrial proteins in Ndufs4 KO mice.
a, Heat map and hierarchical clustering of proteins with significant changes in abundance (ANOVA test, FDR q-value < 0.05; N = 6–7 mice). b, Significantly enriched GO terms from main clusters of panel (a) (hypergeometric test, p-value < 0.001). c,d, Scatter plot of individual protein abundances (c) and enriched GO terms (d) showing significant changes (t-test for proteins and Wilcoxon-Mann-Whitney test for GO terms, FDR q-value < 0.05) between KO vs. WT and KR vs. KO groups, in the same direction (i.e. rapamycin enhances Ndufs4 KO changes) or in opposite directions (i.e. rapamycin rescues Ndufs4 KO changes).
In order to determine whether the rapamycin-associated changes observed in the brain proteome were specific to the Ndufs4 KO condition, we assessed the effect of the same rapamycin treatment in wild-type mice (Extended Data Fig. 4 and Supplementary Table 3). Rapamycin evoked a similar response in the brain of WT mice, with mTOR and PKC isoforms significantly reduced (Extended Data Fig. 4d,e). This is consistent with prior observations in the hippocampus of rapamycin-treated Tsc1+/− mice19. Several additional changes in the brain proteome of Ndufs4 KO and WT mice were observed upon rapamycin treatment. These include an increase in the abundance of the cold-inducible RNA-binding proteins Cirbp and Rbm3 (Extended Data Fig. 4d), which have been reported to have neuroprotective effects20, 21. Rapamycin also increased histone abundance (i.e. nucleosome) and up-regulated processes associated with mTOR inhibition like mRNA splicing and proteasomal degradation, while down-regulating others such as myelination (Extended Data Fig. 4e). In contrast, C-I proteins increased slightly in KO but not in WT upon rapamycin treatment (Extended Data Fig. 4d,e). We do not know the cause or mechanisms for this selective control, but hypothesize that other complexes in the respiratory chain may impose an abundance threshold for C-I. These results demonstrate that chronic rapamycin treatment elicits a core response in the brain that is mostly independent of mitochondrial disease.
Rapamycin reduces PKC activity in the mouse brain
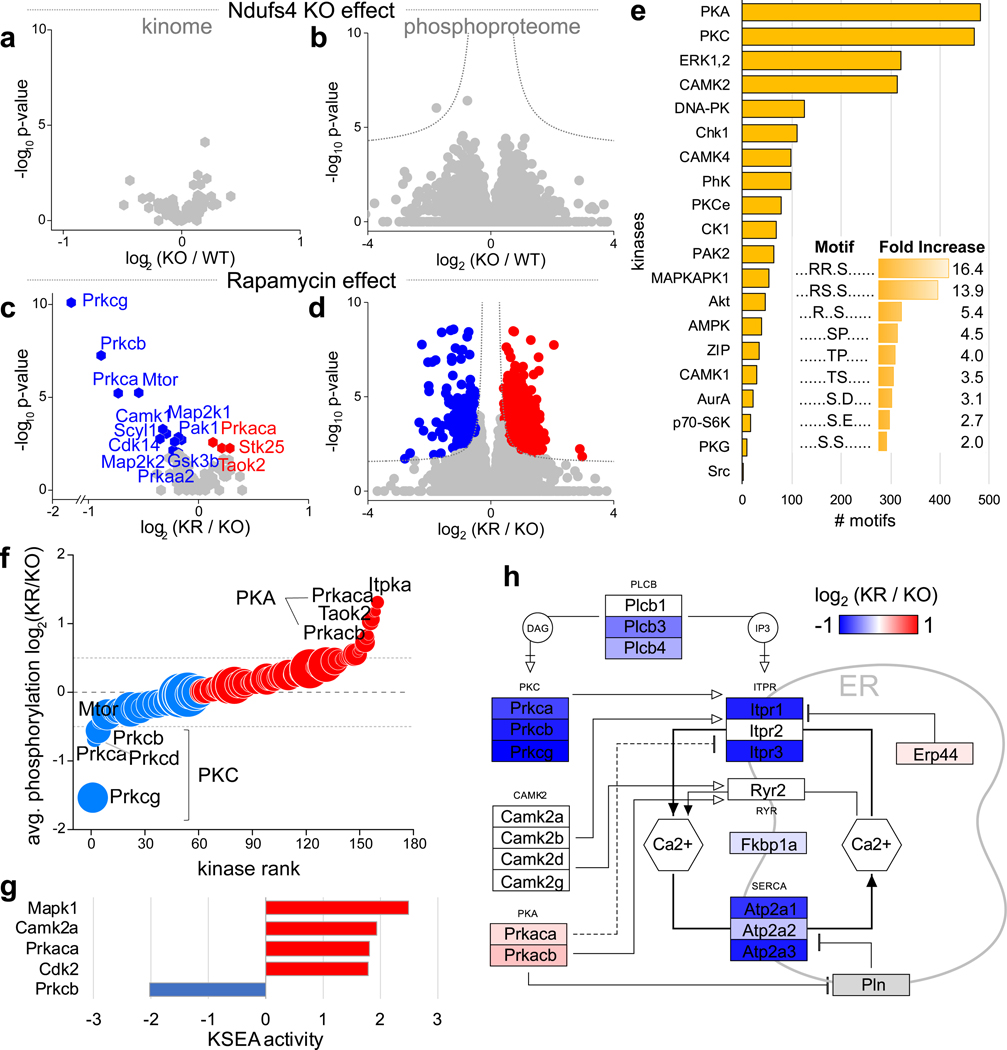
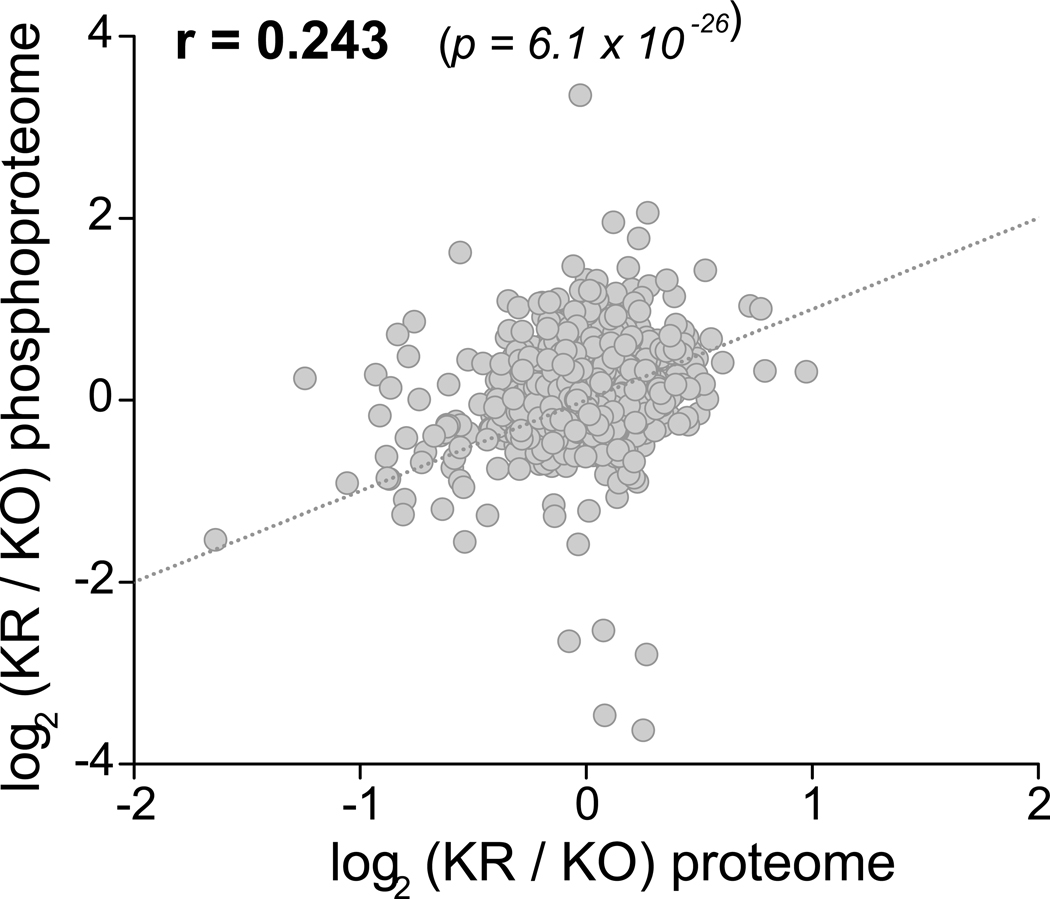
Given that mTOR functions as a kinase, we also sought to assess changes in global protein phosphorylation upon treatment with rapamycin (Fig. 3). We quantified 15,971 phosphorylation sites (Supplementary Table 4) in 3,091 proteins with high reproducibility (Extended Data Fig. 1c–f); 26% of these sites have not been previously reported. Globally, the phosphoproteome was altered to a lesser extent than the proteome (10% phosphosites with significant changes; ANOVA test, FDR q-value < 0.05). Ndufs4 KO induced few changes in brain protein phosphorylation, consistent with the minimal changes in kinase abundance (Fig. 3a,b). Most of the changes were associated with rapamycin treatment (Fig. 3c,d) and correlated with changes in protein abundance (Extended Data Fig. 5), indicating that chronic rapamycin treatment largely rewires the signaling network in the mouse brain by changing protein levels. As in the proteome, these changes were mostly independent of the Ndufs4 KO background (Extended Data Fig. 4f,g and Supplementary Table 5). Of all the phosphorylation sites detected by our analysis, only 2% belong to mitochondrial proteins, and very few changed significantly after rapamycin treatment (Extended Data Fig. 1c). These involve proteins related to mitochondrial transport, metabolism, and fission (Supplementary Table 6). The low occurrence of phosphorylation events on mitochondrial proteins is consistent with other studies22 and suggests that mitochondrial function may be largely regulated by changes in protein and substrate abundance or by other post-translational modifications. However, there is increasing evidence that protein phosphorylation may be important in some mitochondrial functions23, 24.
Fig. 3: Rapamycin reduces PKC activity in Ndufs4 KO mouse brain.
a, Volcano plot comparing the abundance of protein kinases between KO and WT groups (genotype effect). Kinases with significant changes are colored and labeled (t-test FDR q-value < 0.05; N = 6–7 mice). b, Volcano plot comparing phosphosite abundance differences between KO and WT groups (genotype effect; N = 12 samples; 6 mice and duplicated IMAC enrichment and LC-MS/MS analysis). Dotted lines indicate cut-off for significant changes: t-test FDR q-value < 0.05 and artificial within groups variance S0 = 0.1). c,d, Same as in (a) and (b) respectively, but comparing differences between KR and KO groups (rapamycin effect; N = 12–14 samples; 6–7 mice and duplicated IMAC enrichment and LC-MS/MS analysis). e, Enriched kinase motifs in significantly changing phosphosites (Fisher test, FDR q-value < 0.05). Inner graph shows overrepresented linear sequence motifs. f, Average rapamycin-induced changes in phosphorylation of kinases. Dot size is proportional to the number of phosphosites considered for each kinase. Only kinases with n ≥ 3 sites are shown. g, Kinase Substrate Enrichment Analysis (KSEA) showing kinases with significant changes in activity upon rapamycin treatment (Kolmogorov–Smirnov test, p-value < 0.05). h, Mapping of protein abundance changes in proteins involved in intracellular calcium homeostasis between the Ndufs4 KO mice treated (KR) and untreated (KO) with rapamycin. Only significant changes are colored (t-test p-value > 0.05; N = 12–14 samples; 6–7 mice and duplicated IMAC enrichment and LC-MS/MS analysis), non-quantified proteins are shown in grey.
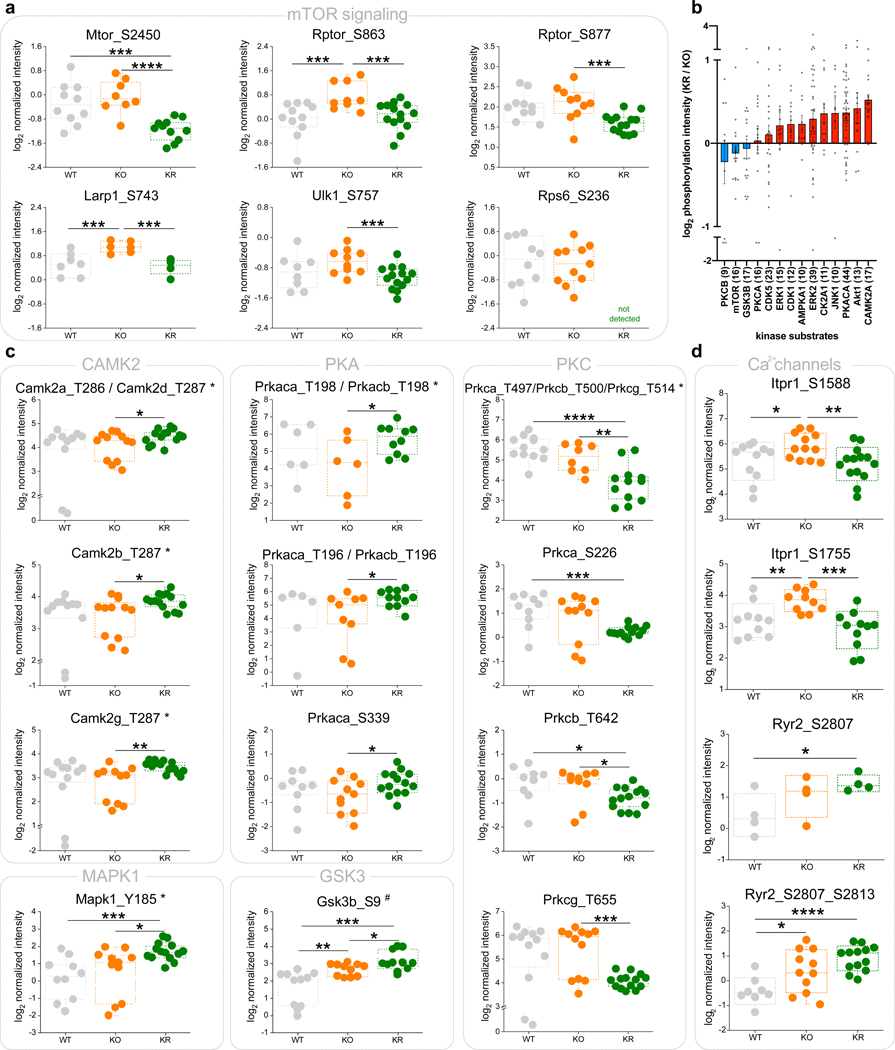
We detected several changes in protein phosphorylation with rapamycin treatment in Ndufs4 KO mice, including a decrease in phosphorylation of several mTOR complex members and known mTOR substrates (Extended Data Fig. 6a). To associate putative kinases targeting these phosphosites, we identified linear sequence motifs that were enriched in the set of rapamycin-regulated phosphosites (Fisher-test, FDR q-value <0.05). We found an overrepresentation of basophilic motifs (e.g. RRxS* and RSxS*; R = arginine, S = serine, S* = phosphoserine, x = any residue), which are typically targeted by kinases in the AGC (protein kinase A (PKA) and PKC) and CamK (CAMK2) families (Fig. 3e). PKC and PKA themselves also exhibited significant changes upon rapamycin treatment, with PKC decreasing and PKA increasing in protein abundance (Fig. 3c) and phosphorylation levels (Fig. 3f). Global phosphorylation state has been recently correlated with kinase activity25, suggesting that the observed changes in PKA and PKC phosphorylation may reflect differences in their activity.
To directly assess changes in kinase activity, we applied kinase-substrate enrichment analysis (KSEA)26, revealing that chronic treatment with rapamycin repressed PKC-β and induced PKA and CAMK2 activity (Fig. 3g). This result was further supported by direct inspection of phosphorylation changes of known substrates of these kinases (Extended Data Fig. 6b) and the kinase activation loop sites (Extended Data Fig. 6c). Given the decrease in phosphorylation of all conventional PKC isoforms (i.e. PKC-α, PKC-β, and PKC-γ), which require calcium for activation, we investigated whether rapamycin treatment also affected proteins involved in calcium homeostasis. We found that rapamycin reverted the hyperphosphorylation on the activation site of Itpr1 (Extended Data Fig. 6d), the main endoplasmic reticulum calcium release channel in the brain, and also decreased its levels and those of several calcium homeostasis related proteins (Fig. 3h). These results suggest a tighter control of intracellular calcium homeostasis in Ndufs4 KO mice brains upon chronic rapamycin treatment which may preclude the activation of conventional PKCs and downstream effectors27, 28.
Rapamycin attenuates inflammation in Ndufs4 KO brain
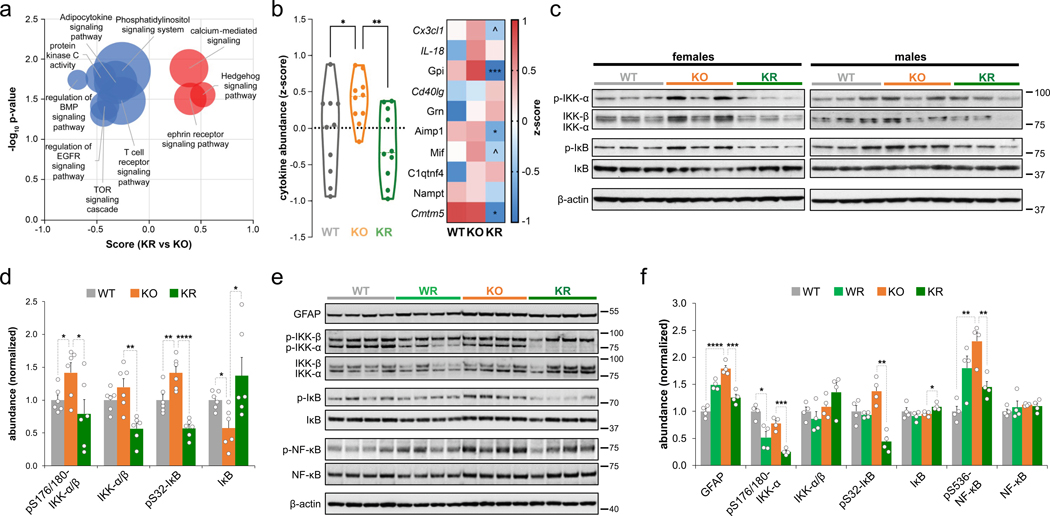
Among the multiple PKC isoforms with reduced abundance upon rapamycin treatment, PKC-β showed the most significant decrease in activity (Fig. 3g). PKC-β mediates activation of the NF-κB pathway via IKK-α and IκB phosphorylation and plays a key role in activation of the immune system and inflammatory response29. In addition to inhibition of PKC-β upon rapamycin treatment, we observed a global decrease in phosphorylation of proteins from pro-inflammatory signaling pathways (e.g. adipocytokine signaling system, T cell receptor signaling pathway, BMP signaling pathway) (Fig. 4a) and a decrease in abundance (Fig. 3c) together with an increase of inhibitory phosphorylation (Extended Data Fig. 6c) of the inflammatory regulator Gsk3b30. Rapamycin treatment also reduced brain cytokine levels (Fig. 4b). Western blot analysis of brain extracts confirmed downregulation of PKC-β phosphorylation (Extended Data Fig. 3d–f) as well as phosphorylation of IKK-ɑ (Fig. 4c,d). Consistent with the role of IKK-ɑ in activating NF-κB by phosphorylating the NF-κB suppressor IκB and promoting its degradation, we observed rapamycin treatment decreased IκB phosphorylation while increasing total IκB levels compared to vehicle-treated mice (Fig. 4c,d).
Fig. 4: Rapamycin attenuates inflammation in Ndufs4 KO mice brain.
a, Volcano plot showing significantly enriched signaling pathways from 1D-enrichment analysis of phosphorylation changes between KR and KO groups (Wilcoxon-Mann-Whitney test, p-value < 0.05). Relative enrichment at the protein level was used to prevent overrepresentation of multiple sites from the same protein. Dot size is proportional to the number of proteins for each annotated term. b, Cytokine abundance changes in the brain. Cytokine classification according to the Uniprot keyword “Cytokine”. Left, violin plot of combined z-score normalized values of median abundance of individual cytokines (each point represents an individual cytokine and the dashed line indicates median z-score value for each group; N = 10). Zero mean score is indicated by a dotted line. Right, individualized information in a heatmap. LFQ normalized abundance data was used, except for cytokines in italics where iBAQ values were used (N = 6–7 mice). Significant changes are indicated for the comparison between KR and KO groups. c, Western blot analysis of proteins in the NF-κB pathway using brain extracts from P30 wild-type (WT) and Ndufs4 KO mice treated daily with vehicle (KO) or rapamycin (KR) from P10 to P30. Each lane corresponds to a brain lysate from a single mouse. d, Densitometry of western blot from (c) (relative to actin) normalized to WT levels (N = 6 mice). e, Western blot analysis of proteins in the NF-κB pathway using brain extracts from P50 male wild-type mice treated daily with vehicle (WT) or rapamycin (WR) and Ndufs4 KO mice treated daily with vehicle (KO) or rapamycin (KR) from P10 to P50. Each lane corresponds to a brain lysate from a single mouse. f, Densitometry of western blot from (e) (relative to actin) normalized to WT levels (N = 4 mice). T-test significance p-values are indicated (^ p < 0.1; * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001).
This model was further supported by analysis of 50-day old Ndufs4 KO mice, which exhibit pathological features such as neuroinflammation, weight loss, and moribundity. Western blot analysis of 50-day old Ndufs4 KO brain extracts indicated that rapamycin treatment deactivates mTORC1 and mTORC2 (Extended Data Fig. 3g,h), decreases abundance of phosphorylated and total PKC levels (Extended Data Fig. 3i,j), and downregulates phosphorylation of the inflammation-activating kinase IKK-α, IкB, and NF-кB (Fig. 4e,f). Consistent with the rapamycin-mediated delay of neurodegeneration, levels of the neuroinflammatory marker GFAP decreased upon rapamycin treatment (Fig. 4e,f). These results collectively indicate that rapamycin reduces neuroinflammation and glial activation concomitant with inhibition of PKC.
Inhibition of PKC reduces skin inflammation in Ndufs4 KO mice
To test the hypothesis that chronic rapamycin treatment is improving health in Ndufs4 KO mice through downregulation of PKC signaling, we asked whether pharmacological inhibitors of PKC would be sufficient to reduce inflammation. We treated Ndufs4 KO mice with three different PKC inhibitors: the pan-PKC inhibitors GO6983 and GF109203X, and the PKC-β specific inhibitor ruboxistaurin31, 32.
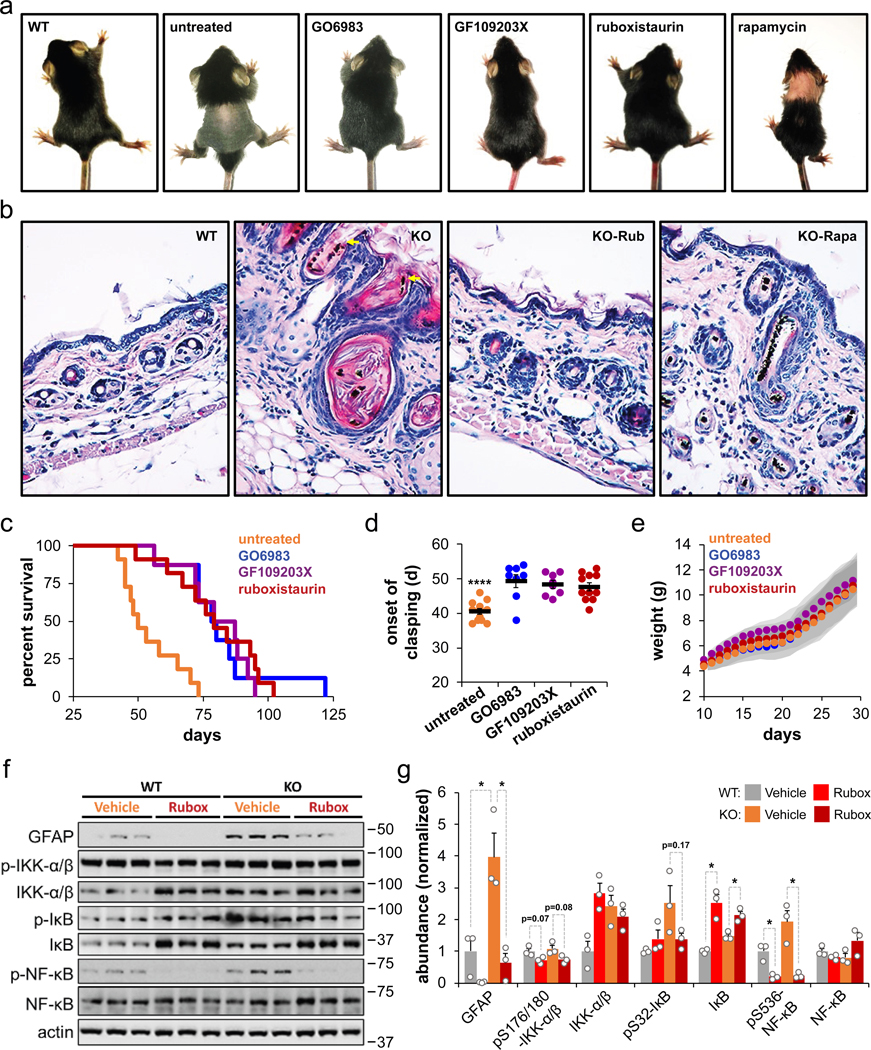
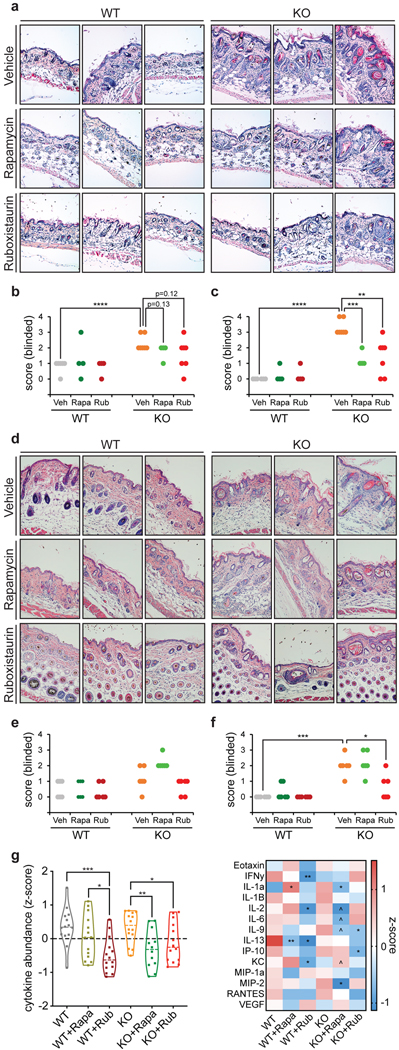
We first examined the effect of PKC inhibition on skin inflammation because prior studies have indicated that Ndufs4 KO mice experience a profound inflammatory burst in skin, which results in hair loss near the time of weaning33. We observed that all three PKC inhibitors largely suppressed this hair loss phenotype, even more potently than rapamycin treatment alone (Fig. 5a and Extended Data Fig. 7). Histological analysis indicated that Ndufs4 KO mice exhibit significant skin abnormalities at weaning (P21), including dilated hair follicles, aberrant keratin features, fractured, bent, or twisted hair shafts, and chronic-active inflammation throughout the dermis characterized by increased infiltration of macrophages, lymphocytes, and neutrophils (Fig. 5b and Extended Data Fig. 8a–c). Ruboxistaurin treatment largely prevented this pathological state and reduced skin inflammation, having a more pronounced effect than rapamycin (Fig. 5b and Extended Data Fig. 8a–c). We made similar observations in skin sections of mice at 30 days of age, and also noticed the presence of atrophied subcutaneous fat in the untreated Ndufs4 KO mice (Extended Data Fig. 8d–f). In addition, we found a general decrease in skin cytokines upon ruboxistaurin and rapamycin treatments (Extended Data Fig. 8g), further supporting an attenuation of the skin inflammatory response by PKC inhibition.
Fig. 5: Inhibition of PKC signaling improves survival of mice with complex I deficiency.
a, Representative images of untreated, PKC inhibitor-treated, and rapamycin-treated Ndufs4 KO mice at postnatal day 21 (P21). WT mice shown for reference. b, Representative images (40X zoom) of H&E staining of skin sections from ~P21 WT and Ndufs4 KO mice treated with vehicle, rapamycin (Rapa), or ruboxistaurin (Rubox) from P10 until ~P21. Yellow arrows represent abnormal features. c, Survival of Ndufs4 KO mice treated with pan-PKC inhibitors GO6983 (N = 8 mice) and GF109203X (N = 8 mice), PKC-β inhibitor ruboxistaurin (N = 11 mice), and untreated control (N = 12 mice). Untreated vs. GO6983 p = 0.0003, untreated vs. GF109203X p = 0.0004, untreated vs. ruboxistaurin p = 0.0003, log-rank test. d, Day of first observed clasping phenotype as a sign of neurodegeneration. **** p < 0.0001, one-way ANOVA with post-hoc Tukey test. e, Weights of Ndufs4 KO mice treated with vehicle or PKC inhibitors. Gray shading represents the standard deviation. f, Western blot analysis of proteins involved in the NF-κB pathway and the neuroinflammatory marker GFAP using brain extracts from P50 WT and Ndufs4 KO male mice treated daily with either vehicle or the PKC-β inhibitor ruboxistaurin from P10 until P50. Each lane corresponds to a brain lysate from a single mouse. g, Densitometry of western blot from (f) (relative to actin) normalized to WT levels (N = 3 mice). T-test significance p-values are indicated (* p < 0.05).
PKC inhibition reduces neuroinflammation and improves survival in Ndufs4 KO mice
We next asked whether PKC inhibition would be sufficient to increase health and survival in Ndufs4 KO mice. All three PKC inhibitors were able to significantly increase survival (Fig. 5c) and delay the onset of neurological symptoms (i.e. clasping; Fig. 5d). Interestingly, inhibition of PKC did not result in the reduced weight and delayed growth characteristics of rapamycin-treatment (Fig. 1b and Fig. 5e).
Neurological symptoms in Ndufs4 KO mice have been found to correlate with neuroinflammation. As expected, vehicle-treated Ndufs4 KO showed a large increase in GFAP in whole brain extracts at 50 days of age (Fig. 5f,g). Treatment with the PKC-β specific inhibitor ruboxistaurin significantly decreased GFAP levels in both WT and Ndufs4 KO mice (Fig. 5f,g), consistent with the delay of clasping (Fig. 5d). Treatment with ruboxistaurin also downregulated the NF-кB inflammatory response in the brain, as indicated by decreased phosphorylation of IKK-α, IкB, and NF-кB, as well as by increases in abundance of total IкB levels (Fig. 5f,g). This was largely independent of genotype as similar effects were observed in ruboxistaurin-treated WT mice (Fig. 5f,g).
Conclusion
Taken together, these results suggest that inhibition of PKC-β and downstream NF-кB mediated inflammation contributes to suppression of mitochondrial disease and increased survival from chronic rapamycin treatment in Ndufs4 KO mice. These effects may be mediated through reduced abundance and activity of mTORC2, a known regulator of PKC. It has been suggested that inhibition of mTORC2 upon chronic rapamycin treatment leads to metabolic dysregulation and detrimental side effects in the context of normative aging in mice34, 35; however, our data raise the possibility that inhibition of mTORC2 may also have beneficial anti-inflammatory and metabolic consequences, at least in the context of severe mitochondrial disease. Recently, it was reported that rapamycin and rapamycin-derivatives can improve outcomes in patients suffering from pediatric Leigh syndrome36 and adult onset MELAS (Mitochondrial Encephalopathy, Lactic acidosis, and Stroke-like episodes) syndrome37. The findings presented here suggest that PKC inhibitors may also have therapeutic value for treating mitochondrial disease and may be particularly useful in patients where side effects associated with mTOR inhibition represent a significant concern.
MATERIALS AND METHODS
Animals and animal care
All studies with mice were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols used were approved by the University of Washington Institutional Animal Care and Use Committee. Mice were provided food, water, and/or gel ad libitum and were maintained on a 12:12-hr light/dark cycle.
Ndufs4+/− mice were generously provided by the Palmiter Laboratory at the University of Washington and backcrossed with C57Bl/6 CR mice. Breeders of heterozygous Ndufs4+/− mice were used and offspring were genotyped by PCR to identify Ndufs4+/+, Ndufs4+/−, and Ndufs4−/− pups. Mice were weaned at 21–28 days of age. Ndufs4−/− mice used in lifespan experiments were housed with one control littermate (Ndufs4+/+ or Ndufs4+/−) for warmth and companionship. Ndufs4−/− mice used in proteomic experiments were housed with one Ndufs4+/+ control littermate for warmth and companionship. Both male and female mice were used for all (phospho)proteomic experiments, with no discernible differences observed in these experiments. Mice were weighed daily, and food and gel were provided on the bottom of cage. Mice used in lifespan experiments were checked daily for signs of clasping and were euthanized if they showed a 30% loss in maximum body weight or loss of righting reflex, were found prostrate, or were generally unresponsive. The minimum sample size for mouse lifespan experiments was determined to be 7 using continuous endpoint, two independent sample study calculations to determine a 30% change in lifespan considering an average lifespan of 50 +/− 10 days for untreated mice with a confidence interval of 0.05 and 80% power.
Drug treatment
Rapamycin (LC Laboratories) and GO6983 (LC Laboratories) were dissolved in DMSO and diluted 100-fold in 5% or 30% PEG-400/5% Tween-80 (vehicle), sterile filtered, aliquoted into 1 mL portions, and stored at −80 °C until needed for use. GF109203X (Cayman Chemical) and ruboxistaurin hydrochloride (Synnovator) were directly dissolved in vehicle containing 1% DMSO. Starting at postnatal day 10 (P10), mice were treated daily between 2:00–6:00 p.m. (via intraperitoneal injection with 29 ½ gauge, 300 μL syringes) using 6.66 μL/g body weight of these solutions for final doses of 10 mg/kg/day (ruboxistaurin), 8 mg/kg/day (rapamycin and GF109203X), or 4 mg/kg/day (GO6983). Control mice were generally treated with vehicle using 6.66 μL/g body weight. For the lifespan experiments control mice were untreated, as it has been reported that untreated Ndufs4−/− mice exhibit identical phenotypes as vehicle-treated Ndufs4−/− mice4, and we also show this here for lifespan, onset of clasping and weight gain (Extended Data Fig. 9).
Sample preparation for proteomic analysis
WT and Ndufs4−/− mice were treated daily with rapamycin or vehicle as described above from P10 until P29. At that age, mice were fasted overnight (for 12 h), treated with rapamycin, and re-fed in the morning for ~4 hours. Unanesthetized mice were then euthanized by cervical dislocation. Brains were immediately isolated, flash frozen in liquid N2 and stored at −80°C. Frozen brains were first weighed and then grinded in liquid nitrogen. About 15 mg of ground tissue were resuspended in 600 μL of lysis buffer composed of 8M urea, 75mM NaCl, 50 mM Tris pH 8.2, and a mix of protease inhibitors (Roche Complete EDTA-free) and phosphatase inhibitors (50 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 1 mM sodium orthovanadate and 50 mM sodium fluoride). Samples were then subjected to 2 cycles of bead beating (1 min beating, 1.5 min rest) with 0.5mm diameter zirconia beads and sonicated for 5 min in ice. Samples were centrifuged at 4°C to remove debris and lysate protein concentration was measured by BCA assay (Pierce). Protein was reduced with 5 mM dithiothreitol (DTT) for 30 min at 55°C and alkylated with 15 mM iodoacetamide in the dark for 30 min at room temperature. The alkylation reaction was quenched by incubating with additional 5 mM DTT for 15 min at room temperature. Samples were diluted five-fold with 50 mM Tris pH 8.2. Proteolytic digestion was performed by adding trypsin at 1:200 enzyme to protein ratio and incubating at 37°C overnight. The digestion was quenched by addition of trifluoroacetic acid to pH 2. Samples were centrifuged to remove insoluble material and peptides were desalted over a 50 mg tC18 SepPak cartridge (Waters). Briefly, cartridges were conditioned with 1 mL of methanol, 3 mL of 100% acetonitrile, 1 mL of 70% acetonitrile, 0.25% acetic acid and 1 mL of 40% acetonitrile, 0.5% acetic acid; and equilibrated with 3 mL of 0.1% trifluoroacetic acid. Then peptide samples were loaded into the cartridges, washed with 3 mL of 0.1% trifluoroacetic acid and 1 mL of 0.5% acetic acid, and then sequentially eluted first with 0.5mL of 40% acetonitrile, 0.5% acetic acid and then with 0.5 mL of 70% acetonitrile, 0.25% acetic acid. 20 μg and 200 μg aliquots of eluted peptides were dried by vacuum centrifugation and stored at −80°C for proteomic and phosphoproteomic analysis, respectively.
Phosphopeptide enrichment
Phosphopeptide enrichment was done by immobilized metal affinity chromatography (IMAC). 200 μg of peptides were resuspended in 150 μl 80% acetonitrile, 0.1% trifluoroacetic acid. To prepare IMAC slurry, Ni-NTA magnetic agarose (Qiagen) was stripped with 40 mM EDTA for 30 min, reloaded with 10 mM FeCl3 for 30 min, washed 3 times and resuspended in 80% acetonitrile, 0.1% trifluoroacetic acid. Phosphopeptide enrichment was performed using a KingFisher Flex robot (Thermo Scientific) programmed to incubate peptides with 150 μl 5% bead slurry for 30 min, wash 3 times with 150 μl 80% acetonitrile, 0.1% trifluoroacetic acid, and elute with 60 μl 1:1 acetonitrile:1% ammonium hydroxide. The eluates were acidified with 30 μl 10% formic acid, 75% acetonitrile, dried by vacuum centrifugation, and stored at −80ᵒC until mass spectrometry analysis. Both sample processing and phosphopeptide enrichment were performed in technical duplicate.
LC-MS/MS analysis
Peptide and phosphopeptide samples were dissolved in 4% formic acid, 3% acetonitrile, loaded onto a 100 μm ID × 3 cm precolumn packed with Reprosil C18 3 μm beads (Dr. Maisch GmbH), and separated by reverse phase liquid chromatography (LC) on a 100 μm ID × 30 cm analytical column packed with 1.9 μm beads of the same material and housed into a column heater set at 50 °C. As peptides eluted off the column they were online analyzed by mass spectrometry (MS).
Peptides for proteome analysis were eluted into a Q-Exactive (Thermo Fisher) mass spectrometer by gradient elution delivered by an EasyII nanoLC system (Thermo Fisher). The gradient was 9–30% acetonitrile in 0.125% formic acid over the course of 90 min. The total duration of the method, including column wash and equilibration was 120 min. All MS spectra were acquired on the orbitrap mass analyzer and stored in centroid mode. Full MS scans were acquired from 300 to 1500 m/z at 70,000 FWHM resolution with a fill target of 3E6 ions and maximum injection time of 100 ms. The 20 most abundant ions on the full MS scan were selected for fragmentation using 2 m/z precursor isolation window and beam-type collisional-activation dissociation (HCD) with 26% normalized collision energy. MS/MS spectra were collected at 17,500 FWHM resolution with a fill target of 5E4 ions and maximum injection time of 50 ms. Fragmented precursors were dynamically excluded from selection for 30 s. Two technical replicate LC-MS/MS runs were performed for proteome analysis.
Phosphopeptides for phosphoproteome analysis were eluted into a Velos Orbitrap (Thermo Fisher) mass spectrometer by gradient elution delivered by an Easy1000 nanoLC system (Thermo Fisher). The gradient was 9–23% acetonitrile in 0.125% formic acid over the course of 90 min. The total duration of the method, including column wash and equilibration was 120 min. Full MS scans were acquired in the orbitrap mass analyzer and recorded in centroid mode. Mass range was 300 to 1500, resolution 60,000 FWHM, fill target 3E6 ions, and maximum injection time 100 ms. Each MS scan was followed by up to 20 data-dependent MS/MS scans on the top 20 most intense precursor ions with 2 m/z isolation window, collision-induced dissociation (CID) with 35% normalized collision energy, and acquired on the ion trap. MS/MS spectra were collected at 17,500 FWHM resolution with a fill target of 5E4 ions and maximum injection time of 50 ms. Fragmented precursors were dynamically excluded from selection for 30 s.
MS data analysis
Acquired mass spectra raw files were searched with MaxQuant v1.6.0.138 against the Uniprot mouse canonical plus isoform protein sequence database (downloaded August 2017, 51434 entries) with common contaminants added. The precursor mass tolerance was set to 6 ppm, and the fragment ion tolerance was set to 20 ppm. A static modification on cysteine residues and variable modifications of methionine oxidation and protein N-terminal acetylation were used for all searches except for phosphopeptide-enriched samples where phosphorylation on serine, threonine and tyrosine residues was also included as variable modification. Trypsin/P was the specified enzyme allowing for up to two missed cleavages. The minimum required peptide length was seven residues. The target-decoy database search strategy was used to guide filtering and estimate false discovery rates (FDR). All data were filtered to 1% FDR at both peptide and protein levels. The “match between runs” option was enabled with a time window of 0.7 min to match identifications between replicates. Proteins with at least two peptides (one of them unique to the protein) were considered identified. Label free quantification (LFQ) and intensity-based absolute quantification (iBAQ) algorithms for protein quantification were selected. LFQ intensity data was used for individual protein comparisons between experimental groups, except for specific analysis including low abundance proteins which had no LFQ intensity values and iBAQ normalized intensities were used instead.
For protein and phosphorylation site analysis we used the generated ‘proteinGroups.txt’ and ‘Phospho(STY)Sites.txt’ tables, respectively, after filtering off contaminants and reverse hits. Protein intensity measurements from technical replicates were aggregated, whereas phosphosite intensities were treated individually due to the stochasticity inherent to data dependent mass-spectrometry sampling of individual phosphopeptides. Perseus v1.6.0.7 software39 was used for statistical (e.g. PCA) and bioinformatic analysis (e.g. 1D and 2D-enrichment annotation analysis40) using log2 transformed data from LFQ normalized protein intensities and median normalized phosphopeptide intensities. Phosphosites were considered localized with a localization probability > 0.75.
Annotations were extracted from Gene Ontology (GO), the Kyoto Encyclopedia of Genes and Genomes (KEGG), and mouse MitoCarta 2.041 databases. GOrilla application was used for GO terms enrichment analysis42, using the whole list of identified proteins as background. Kinase-substrate relationships and kinase-motif sequence information were obtained from the PhosphositePlus database43. Motif extractor tool (motif-x) was used to extract overrepresented motif patterns in sequences surrounding phosphosites (± 7 amino acids)44. Kinase activity was determined using kinase set enrichment analysis (KSEA) which uses Kolmogorov–Smirnov test to assess whether a predefined set of kinase substrates is statistically enriched in phosphosites that are at the two of extremes of a ranked list defined by their differential regulation26. KSEA activity denotes the log10-transformed p‐values from the enrichment analysis of kinase substrates and is signed based on the average sign of all substrates (i.e. if the majority of substrates present increased or reduced phosphorylation, the kinase is predicted as activated (+) or inactivated (−), respectively).
Sample preparation for Western blot
Vehicle-treated Ndufs4+/+ and Ndufs4−/− mice, or ruboxistaurin- or rapamycin-treated Ndufs4+/+ and Ndufs4−/− mice were treated daily from P10 until P27–31 or P47–51. Mice were then fasted overnight (for 12 h), treated with vehicle or small molecule, and re-fed. Unanesthetized mice were then euthanized by cervical dislocation 3–4.5 hours later. Brains were immediately isolated and flash frozen in liquid N2 for Western blot. Brains were ground using a cryogenic homogenizer. Approximately 50 mg of homogenized brain tissue was lysed with 1 mL RIPA buffer containing Pierce™ protease and phosphatase inhibitor tablets (Thermo Fisher) for 30 minutes. Samples were then centrifuged to remove insoluble cell debris, quantified by BCA, diluted to 2 mg/mL, and used for Western blot.
Western blotting
Relative protein levels were determined through Western blotting of ~20 μg protein lysate diluted in 4X Laemmli Sample Buffer, 10X Reducing Agent, and RIPA buffer. The samples were heated at 95 °C for 5 minutes, 70 °C for 10 minutes, or 37 °C for 1 hour, then were loaded onto a NuPage™ 4–12%, 8%, or 10% Bis-Tris MIDI or MINI gel and run at 120V in MOPS running buffer. Protein was transferred to a PVDF membrane using a Trans-Blot® Turbo Transfer System according to manufacturer instructions (an additional 0.2% SDS was added to the transfer buffer). Unless otherwise noted the blots were blocked with 5% BSA or 5% milk in TBST at room temperature for ~1 hour and incubated with primary antibody in 5% BSA or 5% milk in TBST overnight at 4°C followed by ~2 hours at room temperature. Primary antibodies used were: pS235/236-S6 ribosomal protein (1:1000, Cell Signaling Technology 4858), S6 ribosomal protein (1:1000, Cell Signaling Technology 2217), pS473-Akt (1:1000, Cell Signaling Technology 4060), pT308-Akt (1:500, Cell Signaling Technology 9275), pT450-Akt (1:1000, Cell Signaling Technology 9267), Akt (1:1000, Cell Signaling Technology 4691), pS222-PKC-α (1:2000, ABClonal AP0559), PKC-α (1:2000, Cell Signaling Technology 2056), pT638/641-PKC-α/β (1:5000, Cell Signaling Technology 9375), PKC-β (1:500, Cell Signaling Technology 46809), pT674-PKC-γ (1:1000, Abcam ab5797), PKC-γ (1:2000, Santa Cruz Biotechnology sc-166385), pS176/180-IKKα/β Antibody II (1:1000, Cell Signaling Technology 2694), IKK-α (1:200 in 1% milk, Novus Biologicals NB-100–56704), pS32-IκB (1:200 in 1% BSA, Santa Cruz Biotechnology sc-8404), IκB (1:50,000, Abcam ab-32518), pS536-NF-κB p65 (1:100 in 1% BSA, Santa Cruz Biotechnology sc-136548), NF-κB p65 (1:2000 in 1% BSA, Cell Signaling Technology cs-8242), GFAP (1:5000 in 5% milk, Cell Signaling Technology 12389), or β-actin HRP conjugate (1:25000 at room temperature for 2 hours, Cell Signaling Technology 5125). The blots were then rinsed three times at room temperature with TBST for 10 minutes each. The blots were incubated with the secondary antibody in 5% BSA or 5% milk for 1–2 hours. Secondary antibodies used were donkey anti-rabbit IgG HRP conjugate (1:10000 or 1:25,000, Thermo Scientific 31458) or mouse-IgGκ BP HRP conjugate (1:2000, Santa Cruz Biotechnology sc-516102). Blots were thoroughly rinsed with TBST and developed after incubation with Amersham™ ECL™ or Thermo-Fisher SuperSignal™ West Pico PLUS, Femto, or Atto Western Blotting Detection Reagent for 5 minutes. Western blots were imaged using an iBright CL1500 system (Thermo Fisher) and analyzed using Thermo Fisher Connect iBright Analysis Software.
Skin cytokine analysis
Skin protein was extracted from skin tissue of treated mice at ~30 days of age in a RIPA buffer containing protease and phosphatase inhibitors, as described above. Samples were then submitted to Eve Technologies (Calgary, AB, Canada) for multiplex cytokine immunoassay analysis using a mouse cytokine array. Only cytokines that were reliably above the limit of detection were analyzed.
H&E staining
Mice were treated as described above until ~21 or ~30 days of age. Mice were then terminally anesthetized in the morning with ketamine/xylazine. Mice were fixed by perfusion via the left ventricle near the apex of the heart with cold filtered 10% NBF (50 mL) at a flow rate of ≤5 mL/min followed by cold PBS (50 mL). The carcass was then fixed for an additional 24 hours in 10% NBF, the NBF was removed followed by a second fixation with 10% NBF for 24 hours. The NBF was removed, the brain was excised and stored in PBS, and the carcass was stored in 70% ethanol. The carcass was submitted to the Histology and Imaging Core at the University of Washington or Zyagen (San Diego, CA) for H&E of skin using established protocols. Three sections of skin from the top, middle, and bottom sections of the back torso were used. The hair follicle and skin pathology was scored blinded.
Extended Data
Extended Data Fig. 1. Proteome and phosphoproteome analysis statistics.
a, Number of total (all), mitochondrial (mitoc) and Complex I (C-I) proteins quantified (gray) and those with significant changes among WT, KO, and KR experimental groups (ANOVA test, FDR q-value < 0.05) (red). Mitochondrial proteins annotations were extracted from mouse MitoCarta 2.0 database. b, Correlations of log2 transformed LFQ-normalized protein abundance measurements between samples (N = 6–7 mice). c, Same as in (a) but for phosphorylation sites. d, Correlations of log2 transformed median-normalized phosphorylation site intensity values between samples and replicates. Two technical replicates of IMAC phosphopeptide enrichment were performed for each brain sample to increase phosphoproteome coverage (N = 12–14 samples; 6–7 mice and duplicated IMAC enrichment and LC-MS/MS analysis). e, Distribution of Pearson’s r correlation values among all samples and only for technical replicates. Box plots include the median line, the box denotes the interquartile range (IQR), whiskers denote ±1.5 × IQR. f, PCA analysis of log2 transformed median-normalized phosphorylation site intensities data (hollow/solid symbols indicates female/male samples respectively).
Extended Data Fig. 2. Global changes in respiratory chain related proteins.
a,b, Aggregated protein abundance changes in respiratory chain complexes (a) and respiratory chain assembly proteins (b). Box plots include the median line, the box denotes the interquartile range (IQR), whiskers denote ±1.5 × IQR. Sum of relative iBAQ intensities for all members of each complex or protein group were used (N = 6–7 mice). T-test significance p-values are indicated (* p < 0.05; ** p < 0.01; *** p < 0.001).
Extended Data Fig. 3. Western blot analysis of brain extracts from P30 and P50 mice treated with vehicle or rapamycin from P10 to P30 or P50.
a,b, Western blot analysis of mTORC1 and mTORC2 markers of brain lysates from P30 wild-type (WT) and Ndufs4 KO mice treated daily with vehicle (KO) or rapamycin (KR) from P10 to P30. c, Densitometry (relative to actin) of western blot data from (a) and (b) normalized to wild-type levels (N = 6 mice). d,e, Western blot analysis of PKC isoforms of brain lysates from P30 wild-type (WT) and Ndufs4 KO mice treated daily with vehicle (KO) or rapamycin (KR) from P10 to P30. f, Densitometry (relative to actin) of western blot data from (d) and (e) normalized to wild-type levels (N = 6 mice). g,h, Representative WB images and densitometry (relative to actin) normalized to wild-type (WT) levels showing relative phosphorylated and total levels of proteins involved in mTORC1 and mTORC2 (N = 4 mice). i,j, Representative WB images and densitometry (relative to actin) normalized to WT levels showing relative phosphorylated and total levels of PKC proteins (N = 4 mice). Each lane corresponds to a brain lysate from a single mouse. T-test significance p-values are indicated (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001).
Extended Data Fig. 4. Rapamycin exerts similar effects in the brains of wild-type mice to Ndufs4 KO mice.
a, Experimental design to evaluate rapamycin-mediated effects in the brain of wild-type mice. b, Body weight gain in mice from the two experimental groups (mean ± s.d.; N = 4–6 mice). c, Total brain weight at the end of the experimental trial (N = 4–6 mice). T-test significance p-values are indicated (* p < 0.05). Box plots include the median line, the box denotes the interquartile range (IQR), whiskers denote ±1.5 × IQR. d, Comparison of rapamycin mediated changes between wild-type and knock-out mice in individual protein levels. Pearson’s correlation values are indicated. e, 2D-enrichment analysis of GO and KEGG terms comparing the effect of rapamycin between wild-type and knock-out mice in the proteome (Wilcoxon-Mann-Whitney test, FDR q-value < 0.05). f, Comparison of rapamycin mediated changes between wild-type and knock-out mice in phosphorylation sites. Pearson’s correlation values are indicated. g, 2D-enrichment analysis of GO and KEGG terms comparing the effect of rapamycin between wild-type and knock-out mice in the phosphoproteome (Wilcoxon-Mann-Whitney test, p-value < 0.01). In 2D enrichment analysis (panels (e) and (f)) most data points are close to the diagonal dashed line (i.e. identity function), indicating no differences in the effect of rapamycin on wild-type and Ndufs4 KO mice.
Extended Data Fig. 5. Correlation between rapamycin effects in Ndufs4 KO mice at the proteomic (x axis) and phosphoproteomic (y axis) levels.
Pearson’s r coefficient and goodness-of-fit test p-value of linear curve fitted line (dashed line) are indicated.
Extended Data Fig. 6. Phosphorylation changes in brain proteins of Nfus4 KO mice upon rapamycin treatment.
a, Phosphorylation sites on proteins of the mTOR complexes and associated substrates that show significant changes among experimental groups (WT: wild-type; KO: Ndufs4 KO; KR: rapamycin-treated Ndufs4 KO). b, Average (mean ± s.e.m.) changes in phosphorylation of kinase substrates upon rapamycin treatment in Ndufs4 KO mice brain. Each dot represents an individual phosphorylation site substrate. Only kinases with more than 9 substrates found are shown. c, Significant changes in phosphorylation on activity regulatory sites of specific kinases (*activation loop sites, #inhibitory sites). d, Significant changes in activating phosphorylation sites of the main two calcium-release channels from the endoplasmic reticulum. All box plots include the median line, the box denotes the interquartile range (IQR), whiskers denote ±1.5 × IQR. T-test significance p-values are indicated (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; N = 12–14 samples; 6–7 mice and duplicated IMAC enrichment and LC-MS/MS analysis, if no missing values are found).
Extended Data Fig. 7. Treatment of Ndufs4 KO mice with PKC inhibitors largely prevents the alopecia phenotype at weaning (~P21).
a, Wild-type mice at weaning show no hair loss. b, Untreated Ndufs4 KO mice normally exhibit alopecia (i.e. hair loss) at 21-days old due to a TLR2/4 innate immune response. In contrast, minimal hair loss was observed in 21-day old Ndufs4 KO mice treated with c,d, GF109203X and ruboxistaurin from P10 to P21. e, Some hair loss was observed in 21-day old Ndufs4 KO mice treated with rapamycin from P10 to P21.
Extended Data Fig. 8. Histological analysis of skin pathology at P21 or P30.
a, Representative images (20X zoom) of H&E staining of skin sections of P21 WT and Ndufs4 KO mice treated with vehicle, rapamycin, or ruboxistaurin from P10 to P21. Each picture corresponds to an image from an individual mouse. b, Blinded skin inflammation pathology scores from H&E staining of skin sections of P21 WT and Ndufs4 KO mice treated with vehicle, rapamycin, or ruboxistaurin from P10 to P21. Increasing scores represent increasing severity of pathology. c, Blinded hair follicle pathology scores from H&E staining of skin sections of P21 WT and Ndufs4 KO mice treated with vehicle, rapamycin, or ruboxistaurin from P10 to P21. d, Representative images (20X zoom) of H&E staining of skin sections of P30 WT and Ndufs4 KO mice treated with vehicle, rapamycin, or ruboxistaurin from P10 to P30. Each picture corresponds to an image from an individual mouse. e, Blinded skin inflammation pathology scores from H&E staining of skin sections of P30 WT and Ndufs4 KO mice treated with vehicle, rapamycin, or ruboxistaurin from P10 to P30. Increasing scores represent increasing severity of pathology. f, Blinded hair follicle pathology scores from H&E staining of skin sections of P30 WT and Ndufs4 KO mice treated with vehicle, rapamycin, or ruboxistaurin from P10 to P30. Increasing scores represent increasing severity of pathology. Each point represents the score for an individual mouse. T-test significance p-values are indicated (* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; N = 6 mice). g, Cytokine levels in mouse skin (N = 3–6 mice) of P30 WT and Ndufs4 KO mice treated with vehicle, rapamycin, or ruboxistaurin from P10 to P30. Cytokine levels were measured using a cytokine array and z-score normalized. Left, violin plot of combined z-score normalized values of median abundance of individual cytokines (each point represents an individual cytokine and the dashed line indicates median z-score value for each group; N = 14). Zero mean score is indicated by a dotted line. Right, individualized information in a heatmap. T-test significance p-values are indicated for the different treatments compared to their respective wild-type or knock-out mice values (^ p < 0.1; * p < 0.05; ** p < 0.01; N = 3–6 mice).
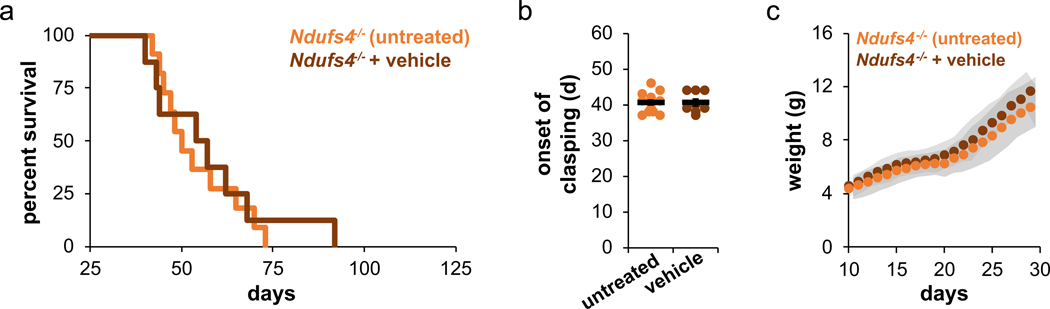
Extended Data Fig. 9. Untreated and vehicle-treated Ndufs4 KO mice exhibit similar symptoms of disease.
a, Vehicle treatment does not alter the lifespan of Ndufs4 KO mice compared to untreated controls. Untreated vs. vehicle p-value = 0.7898, log-rank. b, Vehicle treatment does not alter the onset of clasping of Ndufs4 KO mice compared to untreated controls. c, Vehicle treatment does not alter weight gain of Ndufs4 KO mice compared to untreated controls (mean ± s.d). N = 8 mice for vehicle and N = 11 mice for untreated controls in all plots.
Supplementary Material
ACKNOWLEDGMENTS
We thank Victor V. Pineda, Nicolas J. LeTexier, James Phillips, Jo Tan, Yenna Lee, Tuyet Nguyen, Samy Khessib, Natalie Lim, Chenhao Lu, Surapat Mekvanich, Camille Bodart, Vivian T. Ha, Sydney A. Huff, Danny Kim, Sanchita Narayan, and Azaad O. Zimmermann for assisting with animal experiments. We thank Jessica Snyder and the Histology Imaging Core at the University of Washington for assistance with histology and helpful discussions. We thank Jonathan An for assistance with anesthesia. We thank S. C. Johnson at the Seattle Children's Research Institute for helpful discussions during revisions. This work was supported by NIH grants R01 NS098329 and P30 AG013280 (to M.K. and J.V.), and R35 GM119536 (to J.V.). T.K.I. was supported by a JSPS Postdoctoral Fellowship and a Uehara Memorial Foundation Postdoctoral Research Fellowship. A.S.G. was supported by NIH Ruth L. Kirschstein NRSA fellowship F32 NS110109. S.W.E. was supported by NIH T32 HG00035 Interdisciplinary training grant in Genome Sciences and Samuel and Althea Stroum Endowed Graduate Fellowships. A.S.V. was supported by NIH T32 LM012419 Big Data in Genomics and Neurosciences training grant.
Footnotes
DATA AVAILABILITY
All mass spectrometry raw files and searches have been deposited in the MassIVE repository with dataset identifier PXD012158. Protein and phosphorylation quantification results are provided as Supplementary Tables 1–6. Source data for western blots are provided with this paper.
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Rich P, Chemiosmotic coupling: The cost of living. Nature 421, 583 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Darin N, Oldfors A, Moslemi AR, Holme E, Tulinius M, The incidence of mitochondrial encephalomyopathies in childhood: clinical features and morphological, biochemical, and DNA abnormalities. Ann. Neurol. 49, 377–383 (2001). [PubMed] [Google Scholar]
- 3.Quintana A, Kruse SE, Kapur RP, Sanz E, Palmiter RD, Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc. Natl. Acad. Sci. U.S.A. 107, 10996–11001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson SC et al. , mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ising C et al. , Inhibition of insulin/IGF-1 receptor signaling protects from mitochondria-mediated kidney failure. EMBO Mol Med. 7, 275–287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan NA et al. , mTORC1 Regulates Mitochondrial Integrated Stress Response and Mitochondrial Myopathy Progression. Cell Metab. 26, 419–428.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Loewith R et al. , Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 10, 457–468 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Jacinto E et al. , Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD et al. , Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 22, 159–168 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Lamming DW et al. , Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarbassov DD et al. , Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Guertin DA et al. , Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 11, 859–871 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Gould CM, Newton AC, The life and death of protein kinase C. Curr. Drug Targets 9, 614–625 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y et al. , Sequential posttranslational modifications regulate PKC degradation. Mol. Biol. Cell 27, 410–420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikenoue T et al. , Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signaling. EMBO J. 27, 1919–1931 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moon J-S et al. , mTORC1-induced HK1-dependent glycolysis regulates NLRP3 inflammasome activation. Cell Rep. 12, 102–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Savaskan NE, Bräuer AU, Nitsch R, Molecular cloning and expression regulation of PRG-3, a new member of the plasticity-related gene family. Eur. J. Neurosci. 19, 212–220 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Shibuya N et al. , 3-Mercaptopyruvate sulfurtransferase produces hydrogen sulfide and bound sulfane sulfur in the brain. Antioxid. Redox Signal. 11, 703–714 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Wesseling H, Elgersma Y, Bahn S, A brain proteomic investigation of rapamycin effects in the Tsc1+/− mouse model. Mol. Autism 8, 41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peretti D et al. , RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature 518, 236–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu X, Bührer C, Wellmann S, Cold-inducible proteins CIRP and RBM3, a unique couple with activities far beyond the cold. Cell. Mol. Life Sci. 73, 3839–3859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu R et al. , A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 8, 677–683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebert-Chatelain E et al. , A cannabinoid link between mitochondria and memory. Nature 539, 555–559 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Niemi NM et al. , Pptc7 is an essential phosphatase for promoting mammalian mitochondrial metabolism and biogenesis. Nat. Commun. 10, 3197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hijazi M et al. , Reconstructing kinase network topologies from phosphoproteomics data reveals cancer-associated rewiring. Nat. Biotechnol. 38, 493–502 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Ochoa D et al. , An atlas of human kinase regulation. Mol. Syst. Biol. 12, 888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, Yang W, Li J, Ca2+- and protein kinase c-dependent signaling pathway for nuclear factor-kappaB activation, inducible nitric-oxide synthase expression, and tumor necrosis factor-alpha production in lipopolysaccharide-stimulated rat peritoneal macrophages. J. Biol. Chem. 281, 31337–31347 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Lieu Q et al. , Cathepsin C promotes microglia M1 polarization and aggravates neuroinflammation via activation of Ca2+-dependent PKC/p38MAPK/NF-κB pathway. J. Neuroinflammation 16, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leitges M et al. , Immunodeficiency in protein kinase c beta-deficient mice. Science 273, 788–791 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Jope RS, Yuskaitis CJ, Beurel E, Glycogen synthase kinase-3 (GSK3): inflammation, diseases, and therapeutics. Neurochem. Res. 32, 577–595 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mochly-Rose D, Das K, Grimes KV, Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug Discov. 11, 937–957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aiello LP et al. , Oral protein kinase C β inhibition using ruboxistaurin: efficacy, safety, and causes of vision loss among 813 patients (1,392 eyes) with diabetic retinopathy in the protein kinase C β inhibitor-diabetic retinopathy study and the protein kinase C β inhibitor-diabetic retinopathy study 2. Retina 31, 2084–2094 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Jin Z, Wei W, Yang M, Du Y, Wan Y, Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. 20, 483–498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamming DW et al. , Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell 13, 911–917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang Y et al. , Effects of rapamycin on growth hormone receptor knockout mice. Proc. Natl. Acad. Sci. U.S.A. 115, E1495–E1503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sage-Schwaede A et al. , Exploring mTOR inhibition as treatment for mitochondrial disease. Ann. Clin. Transl. Neurol. 6, 1877–1881 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson SC et al. , mTOR inhibitors may benefit kidney transplant recipients with mitochondrial diseases. Kidney Int. 95, 455–466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox J, Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Tyanova S et al. , The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Cox J, Mann M, 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calvo SE, Clauser KR, Mootha VK, MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eden E et al. , GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hornbeck PV et al. , PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz D, Gygi SP, An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.