Significance
Histone H3Q5ser and H3K4me3 can coexist as a dual modification pattern to regulate active gene transcription. However, the molecular impact of H3Q5ser on H3K4me3 recognition and catalysis remains largely unexplored. Here, we profile the influence of H3Q5ser on H3K4 methylation readers, writers, and erasers. Interestingly, all tested H3K4me3 readers bind H3K4me3Q5ser. Furthermore, while the activity of the H3K4 methyltransferase, MLL1, is unaffected by H3Q5ser, corresponding H3K4 methylation erasers are profoundly inhibited by the presence of the mark. Collectively, this work suggests that H3Q5ser plays an augmenting role when coupled with the installation of H3K4 methylation so as to potentiate downstream outputs via chromatin readers and erasers. The present work exemplifies the importance of modification crosstalk in gene regulation.
Keywords: histone modification, H3Q5 serotonylation, H3K4me3, modification crosstalk, designer chromatin
Abstract
Serotonylation of glutamine 5 on histone H3 (H3Q5ser) was recently identified as a permissive posttranslational modification that coexists with adjacent lysine 4 trimethylation (H3K4me3). While the resulting dual modification, H3K4me3Q5ser, is enriched at regions of active gene expression in serotonergic neurons, the molecular outcome underlying H3K4me3–H3Q5ser crosstalk remains largely unexplored. Herein, we examine the impact of H3Q5ser on the readers, writers, and erasers of H3K4me3. All tested H3K4me3 readers retain binding to the H3K4me3Q5ser dual modification. Of note, the PHD finger of TAF3 favors H3K4me3Q5ser, and this binding preference is dependent on the Q5ser modification regardless of H3K4 methylation states. While the activity of the H3K4 methyltransferase, MLL1, is unaffected by H3Q5ser, the corresponding H3K4me3/2 erasers, KDM5B/C and LSD1, are profoundly inhibited by the presence of the mark. Collectively, this work suggests that adjacent H3Q5ser potentiates H3K4me3 function by either stabilizing H3K4me3 from dynamic turnover or enhancing its physical readout by downstream effectors, thereby potentially providing a mechanism for fine-tuning critical gene expression programs.
The eukaryotic genome is packaged in the form of chromatin (1). The basic repeating unit of this nucleoprotein complex is the nucleosome, which is comprised of an octameric core of histone proteins, two copies each of H2A, H2B, H3, and H4, around which 146 base pair (bp) of double-stranded DNA is wrapped. Histones are elaborated by numerous posttranslational modifications (PTMs, or marks), which are installed and removed by chromatin-modifying enzymes, commonly referred to as writers and erasers (2). Histone PTMs, alone or in combination, can alter the local activity of chromatin, either by affecting the intrinsic structure and/or stability of the nucleoprotein complex, or by acting as targeting vectors for nuclear factors that contain so-called reader domains that bind to marks in specific sequence contexts (3). Thus, by acting as dynamic signaling hubs, histone marks play an essential role in epigenetic regulation, and, not surprisingly, dysregulation of these modifications by abnormal inputs or outputs is frequently associated with disease (4–6).
One of the most studied epigenetic marks is trimethylation of lysine 4 on histone H3 (H3K4me3). This evolutionarily conserved PTM is associated with active transcription and is highly enriched at promoter regions and transcription start sites (7–9). The H3K4me3 mark is installed by a group of dedicated writer enzymes that include the mixed-lineage leukemia (MLL) family of S-adenosyl methionine (SAM)-dependent protein lysine methyltransferases (10). H3K4me2/3 is known to be dynamic, and its removal is catalyzed by lysine histone demethylases, of which there are several subfamilies (11), including the Jumonji C (JmjC) domain-containing proteins KDM5A/B/C/D and LSD1/2 (12). It is well established that H3K4me3 regulates gene expression through recruitment of nuclear factors that contain reader domains specific for the mark. These include ATP-dependent chromatin remodeling enzymes, as well as several transcription factors (13). The latter include the general transcription factor complex, TFIID, which engages the mark through its plant homeodomain (PHD) finger domain-containing TAF3 subunit, thereby stimulating preinitiation complex formation (14, 15).
Recently, we uncovered a PTM on the histone H3 tail whereby the monoamine serotonin (5-hydroxytryptamine, 5-HT) is covalently added to the side-chain of glutamine-5 by the writer enzyme, Transglutaminase 2 (TGM2) (16) (Fig. 1A). Notably, the H3Q5ser mark was found to cooccur with neighboring H3K4me3 on the same histone tail (Fig. 1B). This dual mark (i.e., H3K4me3Q5ser) is enriched in euchromatin and correlates strongly with permissive gene expression during neuronal differentiation. Moreover, the dual mark was found to potentiate the interaction with TFIID compared to H3Kme3 alone, offering a potential mechanistic link to active transcription.
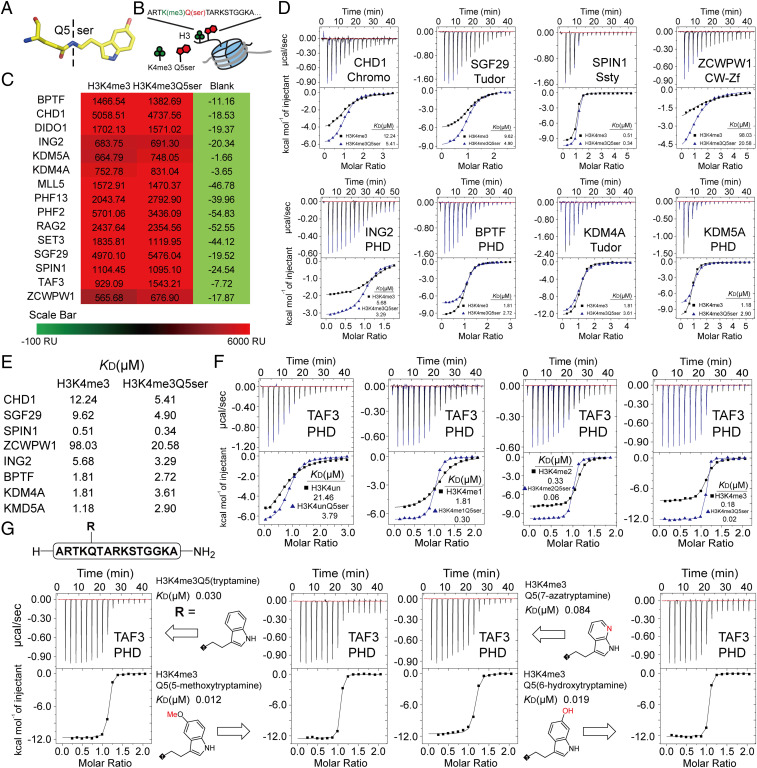
Fig. 1.
Profiling of H3K4me3 readers in response to H3K4me3Q5ser dual modification. (A) Structure of the Q5ser modification. Carbon atoms are shown in yellow; nitrogen atoms are shown in blue; oxygen atoms are shown in red. (B) The coexistence mode of the H3K4me3Q5ser dual modification. (C) SPRi signal of H3(1–15)K4me3 and H3(1–15)K4me3Q5ser peptides toward H3K4me3 readers. The values of SPRi signals are the blanked average signal during 250–300 s of the association curve in SI Appendix, Fig. S1A. (D) Titration curves and fitting curves of H3(1–15)K4me3 and H3(1–15)K4me3Q5ser peptides titrated into H3K4me3 readers. KD values are provided (see SI Appendix, Table. S1 for fitting parameters and error). (E) Summary of binding affinities determined in D. (F) ITC curves of TAF3PHD binding to histone H3 unmodified vs. Q5ser, K4me1 vs. K4me1Q5ser, K4me2 vs. K4me2Q5ser, and K4me3 vs. K4me3Q5ser peptides. KD values are provided (see SI Appendix, Table S1 for fitting parameters and error). (G) ITC curves of TAF3PHD binding to histone H3K4me3Q5(ser-analog) peptides. KD values are provided (see SI Appendix, Table S1 for fitting parameters and error).
While the discovery of H3Q5ser represented an example of monoaminylation of a histone target, the intricacies of H3K4me3–Q5ser biochemical crosstalk (i.e., how each mark influences the others installation and/or removal) and its epigenetic consequences remain largely unexplored. Elucidating the role of local epigenetic environments in regulating the communication between histone readers, writers, and erasers is critical to understanding how spatial and temporal changes in gene expression are achieved (17). In this study, we profiled H3K4me3 readers in the context of adjacent H3Q5ser. All tested H3K4me3 readers tolerate the H3K4me3Q5ser dual modification. Of note, the PHD finger of TAF3 favors H3K4me3Q5ser, and this binding preference relies on the Q5ser modification regardless of H3K4 methylation states. Additionally, and perhaps most strikingly, we provide evidence that while the H3K4me3 writer, MLL1, shows indiscriminate activity toward H3Q5ser-modified substrates, the H3K4me2/3 demethylases LSD1 and KDM5B/C are inhibited by the presence of Q5ser on the same histone tail. Taken together, our data elucidate the consequences of histone H3 serotonylation and suggest a role for H3Q5ser in stabilizing the active H3K4me3 mark.
Results
Histone H3K4me3 Readers Tolerate the H3K4me3Q5ser Dual Modification.
We systematically profiled H3K4me3 reader interactions in the context of H3Q5ser. We expressed and purified 15 representative H3K4me3 reader proteins, including those containing PHD finger domains, tudor domains, chromo domains, Ssty domains, and CW-Zf domains (SI Appendix, Table S2). We evaluated the influence of H3Q5ser on H3K4me3 reader binding through a carbene-based surface plasmon resonance imaging (SPRi) platform. The histone H3(1–15)K4me3 and H3(1–15)K4me3Q5ser peptides were immobilized on the same chip, and purified histone reader domains were detected as the flow phase. Our SPRi results demonstrated that all tested H3K4me3 readers can tolerate the H3Q5ser modification (Fig. 1C). We also found that H3Q5ser had no obvious influence on the binding kinetics of H3K4me3 readers (SI Appendix, Fig. S1A). This indicates that H3Q5ser is a permissive histone mark for H3K4me3 reader interactions.
To further confirm our SPRi results, and to quantitatively analyze binding affinities, we selected eight representative H3K4me3 readers and performed isothermal titration calorimetry (ITC). ITC results revealed that all tested H3K4me3 readers can tolerate the H3Q5ser modification, and the majority of these readers preferred H3Q5ser (Fig. 1 D and E and SI Appendix, Table S1). Royal family proteins (e.g., the chromo domain of CHD1, the tudor domain of SGF29, the Ssty domain of SPIN1), the CW-Zf domain of ZCWPW1, and the PHD finger domain of ING2 showed two- to fivefold enhancements of binding affinities toward the H3K4me3Q5ser dual modification (Fig. 1 D and E).
Conversely, the addition of H3Q5ser resulted in a two- to threefold weakening of KDM4A, BPTF, and KDM5A binding to H3K4me3 (Fig. 1 D and E). These results indicate that the H3Q5ser modification slightly modulates the recognition of numerous H3K4me3 readers in vitro. To complement ITC assessments, we analyzed available H3K4me3 reader complex structures. In all H3K4me3 reader–H3K4me3 histone peptide complex structures, the H3Q5 residue stretches out of the reader surface instead of inserting into the binding pocket (SI Appendix, Fig. S1B). This surface binding mode renders no obvious steric clash upon serotonylation of H3Q5, consistent with our findings that the presence of the mark results in little disturbance of binding affinities.
The PHD Finger of TAF3 Favors the H3K4me3Q5ser Dual Modification.
Among the tested histone H3K4me3 readers, the PHD finger domain of TAF3 displayed obvious preference toward the H3Q5ser modification (Fig. 1C). The H3K4me3 mark alone is known to engage the PHD finger of the TAF3 subunit (TAF3PHD) of TFIID (15). Our previous work showed that the H3K4me3Q5ser dual modification is favored by TAF3 in cells and facilitates the recruitment of the TFIID complex to dual-modified genomic regions (16). In our ITC results, the presence of the H3Q5ser resulted in a five- to ninefold enhancement of binding affinities regardless of H3K4 methylation states (Fig. 1F).
To explore the contribution of the H3Q5ser modification to enhanced TAF3PHD binding, we synthesized a series of H3K4me3Q5(ser-analog) peptides, followed by detection of binding affinities by ITC (SI Appendix, Fig. S1C). We found that 1) TAF3PHD bound to the H3(1–15)K4me3Q5ser peptide with a detected affinity of KD = 0.018 μM (Fig. 1F); 2) H3(1–15)K4me3Q5(tryptamine), which deletes the hydroxyl group in H3Q5ser, resulted in a slight decrease of binding affinity (KD = 0.030 μM) (Fig. 1G); 3) H3(1–15)K4me3Q5(5-methoxytryptamine), which replaces the hydroxyl group in H3Q5ser with a methoxyl group, resulted in a slight increase in binding affinity (KD = 0.012 μM) (Fig. 1G); 4) H3(1–15)K4me3Q5(7-azatryptamine), which disrupts the indole ring, resulted in significant decrease of binding affinity (KD = 0.084 μM) (Fig. 1G); and 5) changing the hydroxyl group position in H3Q5ser did not change the binding affinity [H3(1–15)K4me3Q5(6-hydroxytryptamine)], KD = 0.019 μM) (Fig. 1G). Collectively, these results demonstrate that such increased binding affinity relies, in part, on the electronics imparted by the H3Q5ser modification.
H3Q5ser Does Not Affect the Activity of the H3K4me Writer, MLL1.
Next, we asked whether the presence of H3Q5ser influences the activities of the key H3K4me3 writer MLL1. We prepared a recombinant version of the MLL1 core complex (composed of MLL1, WDR5, RbBP5, ASH2L, and DPY30 subunits) and compared its enzymatic activities on both unmodified histone H3(1–15) and H3(1–15)Q5ser peptides using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry. Our results indicate that the MLL1 core complex can catalyze H3K4 methylation on both unmodified and serotonylated peptides without obvious differences (Fig. 2 A and B).
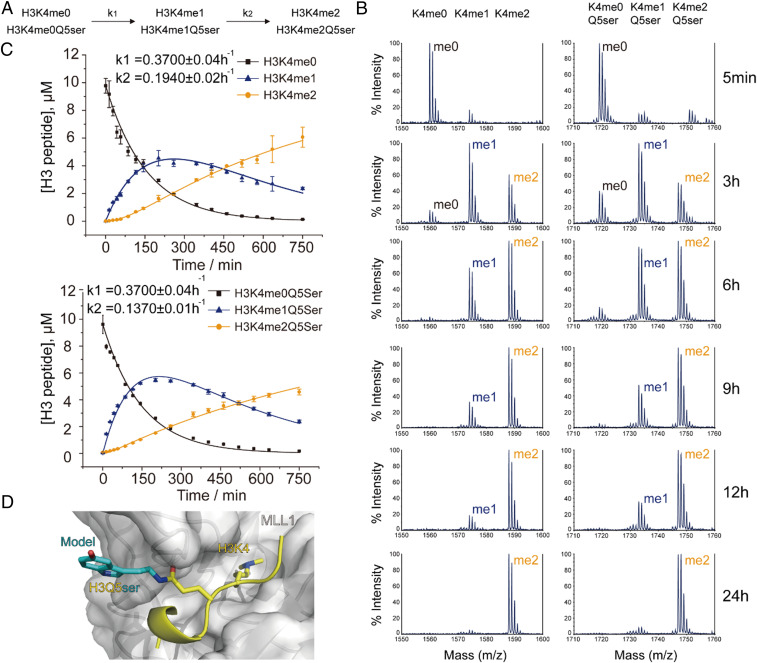
Fig. 2.
Influence of the H3Q5ser modification on the MLL1 complex, an H3K4me3 writer. (A) Kinetic model of two irreversible consecutive reactions catalyzed by the MLL1 complex. k1 and k2 represent the pseudo–first-order rate constants for the conversion of K4me0→K4me1 and K4me1→K4me2, respectively. (B) MALDI-TOF mass spectrometry of MLL1 complex enzymatic assays using H3(1–15)K4me3 and H3(1–15)K4me3Q5ser peptides as substrates. (C) Kinetic fitting of methylation reactions on H3(1–15)un and H3(1–15)Q5ser substrates determined by LC-MS. Summary of rate constants (k1 and k2) derived from globally fitting experimental data are labeled in the panel. (D) The catalytic center of MLL1 in complex with the H3K4 peptide (coordinates were taken from the Protein Data Bank [PDB] entry 2W5Z). H3K4 and Q5 residues are shown as yellow sticks; the binding surface of MLL1 is shown as a white surface. Q5ser modification is modeled as cyan sticks. The H3Q5ser residue stretches out of the MLL1-binding surface.
To further characterize the enzymatic kinetics associated with this methylation reaction, liquid chromatography–mass spectrometry (LC-MS) was employed for quantitative assessments of peptides using a standard curve approach (18) (SI Appendix, Fig. S2). A kinetic model with two irreversible consecutive reactions (Fig. 2A)—SI Appendix, Eqs. 1–3—was applied for global fitting (19). Our analysis indicates that the pseudo–first-order rate constants for the first methylation event (k1) of H3(1–15)un vs. H3(1–15)Q5ser are the same (k1 = 0.3700 h−1), while the k2 value of H3(1–15)Q5ser (k2 = 0.1370 h−1) is slightly lower than that of H3(1–15)un (k2 = 0.1940 h−1) (Fig. 2C). These results suggest that H3Q5ser is a permissive mark for the methyltransferase MLL1 in terms of H3K4me deposition. Subsequent structural modeling suggests the molecular basis for H3Q5 tolerance. When bound to the MLL1 catalytic domain, the H3Q5 residue projects out beyond the binding pocket, with the additional serotonyl group causing no obvious steric clash (20) (Fig. 2D). This detailed kinetic study compliments and confirms data in which MLL1 is able to successfully methylate mononucleosome substrates in vitro regardless of their H3Q5ser modification status (16).
H3Q5ser Inhibits H3K4me2/3 Demethylation.
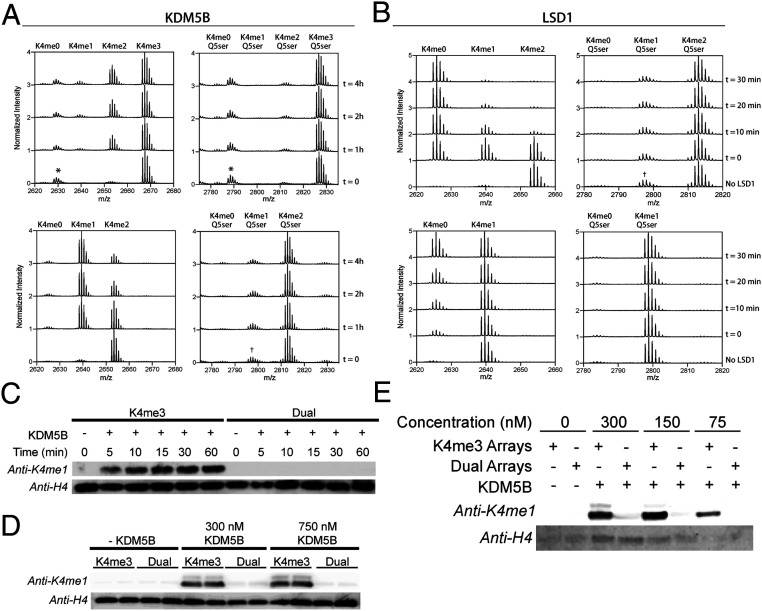
Next, we analyzed the impact of the dual serotonyl mark on demethylation of H3K4me3. To this end, full-length KDM5B, a known eraser for H3K4me3 (21), was expressed in insect cells and purified (SI Appendix, Fig. S3A). Demethylation of H3(1–25)K4me2/3 vs. H3(1–25)K4me2/3Q5ser peptide substrates was then measured by MALDI-TOF (SI Appendix, Fig. S3B). While the H3(1–25)K4me3 peptide was found to be a robust substrate of the enzyme, no demethylation was observed for the dual-modified peptide, even after prolonged reaction times (Fig. 3 A, Top). A similar result was observed for dimethylated peptide substrates, that is, the presence of the serotonyl mark completely blocked demethylation (Fig. 3 A, Bottom). The same trend was also found for the related lysine demethylase, KDM5C (SI Appendix, Fig. S3C). To explore whether this inhibitory effect was more general, we tested lysine-specific demethylase 1 (LSD1), a member of the flavin adenine dinucleotide (FAD)-dependent class of K4-demethylases. Again, the presence of the serotonyl mark on H3Q5 completely blocked demethylation of either H3K4me2 or H3K4me1 peptide substrates (Fig. 3B and SI Appendix, Fig. S3D). In an effort to confirm this observation on a more physiological substrate, designer chromatin arrays consisting of 12 homogeneous mononucleosome substrates were assembled, containing semisynthetic histone H3 bearing either K4me3 or K4me3Q5ser (SI Appendix, Fig. S3E). Consistent with our peptide data, immunoblotting against H3K4me1 revealed robust demethylation of the H3K4me3 arrays, while no demethylation was detected for those containing H3K4me3Q5ser (Fig. 3C). Increasing the amount of enzyme or substrate in the reaction did not alter this result (Fig. 3 D and E). Importantly, control studies confirmed equal recognition of the H3K4me1 and H3K4me1Q5ser epitopes by the antibody (SI Appendix, Fig. S3 F and H). In addition, peptide controls were performed to ensure reaction conditions did not alter each epitope during the course of the reaction (SI Appendix, Fig. S3 G and H).
Fig. 3.
Impact of H3Q5ser modification on LSD1 and KDM5B mediated demethylation of H3K4. (A) MALDI-TOF demethylation assay using H3(1–25)K4me3 vs. H3(1–25)K4me3Q5ser peptide substrates (Top) and H3(1–25)K4me2 vs. H3(1–25)K4me2Q5ser peptide substrates (Bottom). Demethylation of H3(1–25)K4me3 resulting in the H3(1–25)K4me1/2 product is visible after 4 h, while no demethylation is observed for H3(1–25)K4me3Q5ser. *Impurity from peptide synthesis. †Residual K4me1Q5ser species from synthesis of K4me2Q5ser substrates, unable to purify. (B) MALDI-TOF demethylation assay as described in A using recombinant LSD1. LSD1 robustly demethylates native substrates K4me2 (Top) and K4me1 (Bottom), while no demethylation activity is seen for substrates containing Q5ser. (C) Western blot of 60-min time course of KDM5B demethylation of 12-mer chromatin array substrates. In the presence of KDM5B, H3K4me3 arrays are readily demethylated, while no demethylation occurs for H3K4me3Q5ser-containing arrays. (D) Western blot of KDM5B concentration course. Increased amounts of KDM5B do not promote demethylation of H3K4me3Q5ser arrays. (E) Increasing 12-mer array substrates in the KDM5B demethylation assay does not promote demethylation of H3K4me3Q5ser arrays.
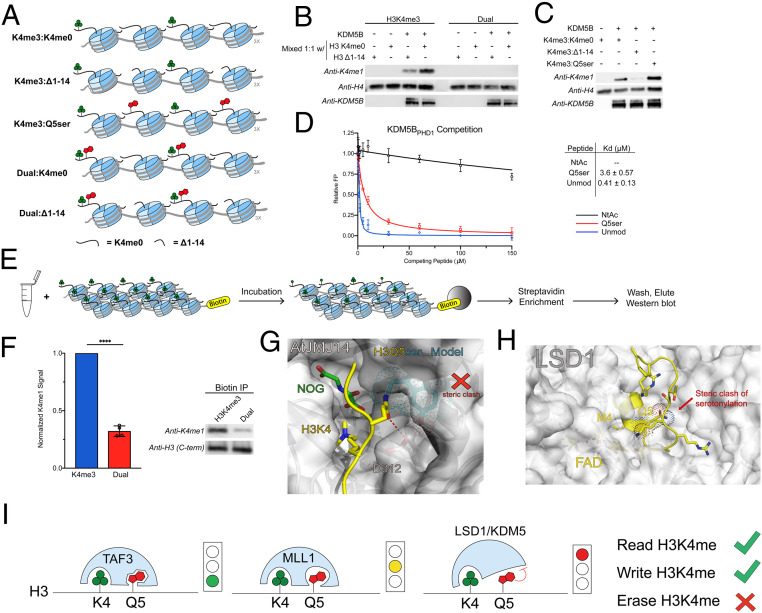
The demethylase activity of KDM5B is known to be stimulated upon binding of unmodified H3K4 peptides to the PHD1 finger contained within the protein, which is embedded in the catalytic JmjC domain (22, 23). To test whether this stimulation might overcome the inhibitory effect of H3Q5ser, designer chromatin arrays were assembled containing a 1:1 mixture of either H3K4me3 or H3K4me3Q5ser mixed with H3K4me0 or truncated H3Δ1–14 (missing the activating H3 tail) (Fig. 4A and SI Appendix, Fig. S4 A and B). Robust stimulation of KDM5B demethylation was observed for the H3K4me3:H3K4me0 substrates, in line with previously proposed mechanisms (Fig. 4B). By contrast, no demethylation was observed for arrays containing H3K4me3Q5ser:H3K4me0. This indicates that additional stimulation of KDM5B provided by nearby H3K4me0 is not sufficient to overcome the inhibitory effect of the serotonyl mark. To investigate whether the presence of H3Q5ser on a neighboring mononucleosome would inhibit stimulation by KDM5BPHD1, designer chromatin arrays were generated containing a 1:1 mixture of H3K4me3 and either H3K4me0, H3Δ1–14, or H3Q5ser (Fig. 4A and SI Appendix, Fig. S4 A and B). Stimulation of KDM5B demethylase activity was found for both H3K4me0 and H3Q5ser (Fig. 4C). This indicates that KDM5BPHD1 is still able to functionally recognize H3Q5ser on a neighboring mononucleosome. To explore this further, we performed fluorescence polarization competition studies using isolated KDM5BPHD1 and histone H3(1–15) peptide substrates (Fig. 4D and SI Appendix, Fig. S4 C–E). Addition of the Q5ser mark to the H3 peptide was found to lower the affinity for KDM5BPHD1 (KD = 0.41 vs. 3.6 μM); however, the effect was modest in comparison to the effect of peptide N-acetylation, which largely abrogated binding to the reader. Analysis of the structure of KDM5BPHD1 in complex with H3K4me0 also suggested the binding event would be tolerant to the H3Q5ser modification (SI Appendix, Fig. S4F). Taken together with our functional data on peptides and array substrates, these results argue that the inhibitory effect of the serotonyl mark on KDM5B activity largely operates at the level of the enzyme active site, i.e., by disrupting substrate binding.
Fig. 4.
KDM5B inhibition of demethylation of semisynthetic 12-mer chromatin arrays. (A) Schematic of mixed 12-mer array substrates. Both H3 tails are modified. (B) H3K4me0 on a neighboring mononucleosome stimulates KDM5B demethylation activity of H3K4me3 but not H3K4me3Q5ser. (C) H3Q5ser on a neighboring mononucleosome does not inhibit stimulation of KDM5B by KDM5BPHD1. (D) Normalized fluorescence polarization of GST-KDM5BPHD1 binding to H3 unmodified (blue) and H3Q5ser (red) peptides. N-terminally acetylated H3 was included as a negative control (black). Errors represent ± SD of n = 3. (E) Experimental flow for chromatin dipping in active, isolated HeLa lysate. (F) Chromatin array dipping in isolated HeLa nuclear lysate followed by streptavidin enrichment shows dramatically less demethylation of H3K4me3Q5ser arrays compared to H3K4me3. Representative Western blot shown from four replicate experiments. t test (two-tailed), 95% confidence; ****, < 0.0001. Errors represent ± SD (G) The catalytic center of JMJ14 (the H3K4me3 eraser in Arabidopsis thaliana) in complex with an H3K4me3 peptide (coordinates were taken from the PDB entry 5YKO). H3K4 and Q5 residues are shown as yellow sticks; the binding surface of JMJ14 is shown as a white surface. The hydrogen bond between H3Q5 and D312 is highlighted with a red dash. Q5ser modification is modeled as cyan sticks. H3Q5ser causes steric clash at the catalytic center of JMJ14. (H) The catalytic center of LSD1 in complex with the histone H3 peptide (coordinates were taken from the PDB entry 2V1D). H3M4 (K4me3 mimic) and Q5 residues are shown as yellow sticks; the binding surface of LSD1 is shown as a white surface. H3Q5ser causes steric clash at the catalytic center of LSD1. (I) Model representing the different degrees of crosstalk between H3K4me3 and H3Q5ser. KDM5/LSD1 are inhibited by Q5ser, MLL1 remains active against Q5ser-modified substrates, albeit with a slower k2, and the reader TAF3 displays increased binding to H3K4me3Q5ser.
Finally, to validate our in vitro observations, we generated active nuclear extracts from HeLa cells and incubated these extracts with biotinylated designer chromatin arrays containing H3K4me3 or H3K4me3Q5ser semisynthetic histones (Fig. 4E and SI Appendix, Fig. S4G). Following incubation, the arrays were captured through streptavidin enrichment and eluted for analysis—an experiment we term “chromatin dipping” (Fig. 4E). Analysis by Western blot showed significantly less demethylation of the H3K4me3Q5ser-containing substrate in comparison to H3K4me3 (Fig. 4F). Interestingly, we did observe a small amount of demethylation of the H3K4me3Q5ser arrays in lysates, which contrasts with the complete lack of activity in the reconstituted biochemical assays with KDM5B. Conceivably, this could result from either the action of other demethylases present in the nuclear lysate and/or from a small amount of histone deserotonylation, affording a viable substrate for the erasers.
Discussion
The functional interplay between histone PTMs has emerged as a key component of epigenetic regulation (24). Indeed, there are now many examples of preexisting marks influencing the installation, readout, or removal of other histone modifications (2). Here, we explore the biochemical crosstalk between the recently identified H3Q5ser mark and the neighboring, and heavily studied, H3K4me3 modification. These two PTMs have been shown to cooccur on the same histone tail, with the dual mark correlating with active transcription (16). Given the known importance of H3K4me3 in transcriptional activation, the presence of the bulky serotonyl mark on the adjacent glutamine might be expected to impact processes involved in gene regulation. The biochemical data presented herein broadly support this assertion in that we observed changes in both binding properties and the stability of H3K4me3 as a function of the neighboring Q5ser mark.
Classically, histone H3K4me3 readers have been found to be specific and sensitive to surrounding modifications, such as H3T3ph and H3T6ph. Interestingly, however, all of the H3K4me3 readers that we tested can tolerate the Q5ser modification. Structurally, the H3Q5 residue stretches beyond the contact surfaces between the histone H3 peptide and H3K4me3 readers. As such, this surface-binding mode renders the Q5ser modification as a permissive mark for H3K4me3 readers. Previously, we found evidence that the H3Q5ser mark potentiates TFIID interactions with histone H3K4me3 (16). In the current study, we show that this effect is, at least in part, due to increased binding between the PHD finger of the TAF3 subunit of TFIID and the H3 tail. Also, this increased binding affinity relies on the Q5ser modification regardless of K4 methylation states.
In addition to studying the effects of H3Q5ser on reader domain interactions, we also examined the impact of the PTM on writers and erasers of adjacent H3K4me3. Consistent with our previous work (16), we did not observe a significant effect on the installation of the methyl mark—our kinetic studies indicate that the presence of the serotonyl group has no impact on the installation of the first methyl group (i.e., H3K4me0→H3K4me1) and only a very minor impact on the subsequent methylation step (i.e., H3K4me1→H3K4me2). Inspection of the available crystal structure of the catalytic SET [Su(var)3-9, Enhancer-of-zeste and Trithorax] domain of MLL1 bound to a substrate peptide suggests that there is no obvious steric clash between the H3Q5ser mark and the active site residues of the enzyme (Fig. 2D).
While this H3K4me3 writer is quite tolerant to the presence of the H3Q5ser mark, the corresponding demethylases, KDM5B and KDM5C, were found to be highly sensitive to the modification. Indeed, our biochemical studies employing purified recombinant enzymes and either peptide or designer chromatin substrates indicate that the serotonyl mark completely blocks the demethylation reaction. These results are further supported by our chromatin-dipping experiments in which we exposed the designer chromatins to nuclear extracts. Conceivably, the inhibitory effect of the serotonyl mark may be due to an inability of the dual-modified substrate to bind properly to the active site of the JmjC catalytic domain and/or a loss of enzyme stimulation associated with binding of the H3 tail to the PHD1 finger within the enzyme (23). As noted above, the H3Q5ser mark does lower the affinity of the nonmethylated H3 peptide for the reader domain. However, we do not believe this binding effect contributes to the inability of the enzyme to demethylate the dual-modified substrate. Indeed, we find in the context of designer chromatin substrates that the isolated H3Q5ser mark is able to stimulate KDM5B activity to the same level as unmodified H3. Additionally, full-length KDM5 contains a PHD3 module that is known to bind K4me2/3 at the C terminus (25). The ITC result of KDM5APHD3 shows little difference in binding affinity in the presence of Q5ser (Fig. 1 D and E). This result indicates that PHD3 binding and recognition of H3K4me2/3 or H3K4me2/3Q5ser is not a driving factor for difference in catalytic activity. Based on this, we favor a mechanism in which the serotonyl mark directly disrupts substrate binding to the KDM5B active site. Unfortunately, there are no structures available for the catalytic domain of any KDM5 family member in complex with a substrate peptide. As an alternative, we turned to the H3K4me3 eraser from Arabidopsis thaliana, JMJ14, whose structure has been solved when bound to a substrate peptide (26). The H3Q5 residue inserts within the binding pocket of KDM5 and forms a hydrogen bond with a conserved aspartic acid residue (Fig. 4G and SI Appendix, Fig. S4H). We thus assume that serotonylation of H3Q5 will result in steric clash between the KDM5 catalytic domain and the N-terminal H3 tail, thereby diminishing enzymatic activities of KDM5 on K4me3. Likewise, examination of the LSD1 structure shows a similar steric clash within the catalytic center (Fig. 4H).
In summary, we show here that the H3Q5ser modification is a permissive mark for known H3K4me3 readers, with either clearly enhanced (e.g., TAF3PHD and ZCWPW1CW-Zf) or slightly reduced (e.g., BPTFPHD and KDM4ATudor) affinities. While the H3Q5ser mark has a minimal impact on the activity of the H3K4me3/2 writer, MLL1, the same was not true for the reverse reaction, demethylation of H3K4me3. A clear implication of our biochemical studies is that the H3Q5ser modification is expected to simultaneously protect the active H3K4me3 mark from being erased, while enhancing binding to key components of the basal transcription apparatus, e.g., TFIID (Fig. 4I). Thus, we suggest that this two-pronged effect would provide a powerful mechanism for sustaining and augmenting critical gene expression programs during cellular differentiation. The present work thus serves as a foundation for future studies geared toward testing this hypothesis in vivo. Also, other monoaminyl marks have recently been identified [e.g., the H3Q5 dopaminyl modification (27)] and may elicit similar or distinct effects on binding. The dual modifications of other H3K4me3Q5(monoaminyl) states will be explored in future studies.
Materials and Methods
Materials and general laboratory methods, peptide synthesis of serotonylated peptides, protein expression and purification, Western blotting, isothermal titration calorimetry, surface plasma resonance imaging, MALDI-TOF analysis of enzymatic activities, LC-MS analysis of enzymatic activities, pre-steady state kinetics, peptide synthesis of fluorescent polarization anisotropy peptides, generation of peptide thioesters, preparation of modified synthetic histones, octamer refolding, DNA preparations, 12-mer array assembly, KDM5B assays on 12-mer chromatin arrays, fluorescence anisotropy peptide-binding assays, HeLa nuclear lysate isolation, chromatin dipping, antibody epitope controls, and additional necessary information are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Jinsong Zhu and Dr. Mo Yang at the National Center for Nanoscience and Technology for the assistance in SPR imaging. This work was supported by grants from the National Science Foundation of China (31725014, 91753203) and the State Key Research Development Program of China (2020YFA0803300, 2016YFA0500700) (to H.L.). Additionally, this work was supported by grants from the NIH [R37-GM086868 and PO1-CA196539 (to T.W.M.); DP1-DA042078, R01-MH116900, and R21-DA044767 (to I.M.); F32 GM129935-03 (to K.N.C.); R01-CA204639 and R01-CA129325 (to R.G.R.)].
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. P.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016742118/-/DCSupplemental.
References
- 1.McGinty R. K., Tan S., Nucleosome structure and function. Chem. Rev. 115, 2255–2273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruthenburg A. J., Li H., Patel D. J., Allis C. D., Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenuwein T., Allis C. D., Translating the histone code. Science 293, 1074–1080 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Allis C. D., Jenuwein T., The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Chi P., Allis C. D., Wang G. G., Covalent histone modifications—Miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venkatesh S., Workman J. L., Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Shilatifard A., The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos-Rosa H., et al., Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Bernstein B. E., et al., Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. U.S.A. 99, 8695–8700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hess J. L., MLL: A histone methyltransferase disrupted in leukemia. Trends Mol. Med. 10, 500–507 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Greer E. L., Shi Y., Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klose R. J., Kallin E. M., Zhang Y., JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Sims R. J. 3rd, et al., Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 280, 41789–41792 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vermeulen M., et al., Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Lauberth S. M., et al., H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021–1036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrelly L. A., et al., Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567, 535–539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feil R., Fraga M. F., Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 13, 97–109 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Xu L., Wang X., Jiao Y., Liu X., Assessment of potential false positives via orbitrap-based untargeted lipidomics from rat tissues. Talanta 178, 287–293 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Patel A., Dharmarajan V., Vought V. E., Cosgrove M. S., On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 284, 24242–24256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Southall S. M., Wong P. S., Odho Z., Roe S. M., Wilson J. R., Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol. Cell 33, 181–191 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Klein B. J., et al., The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 6, 325–335 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakravarty S., Essel F., Lin T., Zeigler S., Histone peptide recognition by KDM5B-PHD1: A case study. Biochemistry 54, 5766–5780 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., et al., The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B. Protein Cell 5, 837–850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides T., Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Wang G. G., et al., Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459, 847–851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z., et al., Structure of the Arabidopsis JMJ14-H3K4me3 complex provides insight into the substrate specificity of KDM5 subfamily histone demethylases. Plant Cell 30, 167–177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lepack A. E., et al., Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 368, 197–201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.