Abstract
The surge of SARS-CoV-2 has created a wave of pandemic around the globe due to its high transmission rate. To contain this virus, researchers are working around the clock for a solution in the form of vaccine. Due to the impact of this pandemic, the economy and healthcare have immensely suffered around the globe. Thus, an efficient vaccine design is the need of the hour. Moreover, to have a generalised vaccine for heterogeneous human population, the virus genomes from different countries should be considered. Thus, in this work, we have performed genome-wide analysis of 10,664 SARS-CoV-2 genomes of 73 countries around the globe in order to identify the potential conserved regions for the development of peptide based synthetic vaccine viz. epitopes with high immunogenic and antigenic scores. In this regard, multiple sequence alignment technique viz. Clustal Omega is used to align the 10,664 SARS-CoV-2 virus genomes. Thereafter, entropy is computed for each genomic coordinate of the aligned genomes. The entropy values are then used to find the conserved regions. These conserved regions are refined based on the criteria that their lengths should be greater than or equal to 60 nt and their corresponding protein sequences are without any stop codons. Furthermore, Nucleotide BLAST is used to verify the specificity of the conserved regions. As a result, we have obtained 17 conserved regions that belong to NSP3, NSP4, NSP6, NSP8, RdRp, Helicase, endoRNAse, 2’-O-RMT, Spike glycoprotein, ORF3a protein, Membrane glycoprotein and Nucleocapsid protein. Finally, these conserved regions are used to identify the T-cell and B-cell epitopes with their corresponding immunogenic and antigenic scores. Based on these scores, the most immunogenic and antigenic epitopes are then selected for each of these 17 conserved regions. Hence, we have obtained 30 MHC-I and 24 MHC-II restricted T-cell epitopes with 14 and 13 unique HLA alleles and 21 B-cell epitopes for the 17 conserved regions. Moreover, for validating the relevance of these epitopes, the binding conformation of the MHC-I and MHC-II restricted T-cell epitopes are shown with respect to HLA alleles. Also, the physico-chemical properties of the epitopes are reported along with Ramchandran plots and Z-Scores and the population coverage is shown as well. Overall, the analysis shows that the identified epitopes can be considered as potential candidates for vaccine design.
Keywords: B-cell epitopes, Conserved regions, Epitopes, SARS-CoV-2, Synthetic vaccine, T-cell epitopes
1. Introduction
In late December 2019, China witnessed the rise of novel coronavirus, SARS-CoV-2, whose exponential rise took everyone by surprise due to its high transmission rate. Later, the origin of the virus was linked to coronaviridae family which also includes SARS-CoV-1 and MERS-CoV (Zhou et al., 2020). By the start of March, number of people from different countries around the globe were found to be infected with the virus. Hence, W.H.O. declared it as a pandemic thereby forcing authorities to adopt counter measures to limit the spread of SARS-CoV-2 among the masses. People infected with SARS-CoV-2can exhibit mild and moderate symptoms, ranging from asymptomatic to high body temperature along with cough, sore throat and may also pose life threatening situations in people suffering from other diseases such as diabetes, cardiovascular diseases etc.. Some of the most widely accepted methods to curb the spread of the virus were nationwide lock down, mandatory face masks and social distancing (Wang et al., 2020). But soon the rising number of cases around the globe nearing to 28.8 million (Worldometer, 2020) made it evident that precautionary measures would not be enough to handle the situation. Therefore, more emphasis is given to the vaccine design for this contagious virus. It is worth mentioning here that the vaccine design for SARS-CoV-2 commenced as soon as the Chinese scientists observed what they called mysterious pneumonia and uploaded the virus sequence on January 10.
The traditional approach of vaccine design involves the use of attenuated microorganism or inactivated components of a pathogen (Mortimer, 1978) for immunization against infectious agents. This method has been able to curb the morbidity and mortality rate to significant amount in the past two decades. But these classical methods also present some major challenges such as long time consumption in fertilization of the microorganism along with an additional risk of auto immunization. Moreover, research has also been conducted on making mRNA and DNA vaccines which have been proven to eliminate any unwanted reactions. However, they also come with set of challenges as they essentially carry very less antigenicity (Rakib et al., 2020). On the other hand, rational vaccine design (Purcell et al., 2007) based on chemical synthetic approaches such as peptide-based (Parvizpour et al., 2020) which involves locating the epitope region inside the viral genes and utilizing them to evoke the immune response inside the host body with minimal microbial component can prove to be more effective for viruses like Influenza which form a resistance against vaccines while going through evolution every year. In this regard, a peptide based vaccine has been designed by Naz et al. (2020b) against the Sarcoptes scabiei virus for highly immunogenic epitopes. Also, extracting the genomic features of a virus genome can be significant while designing a reliable vaccine as shown by Bhattacharya et al. (2020b). In this work, they proposed vaccine for Aeromonas hydrophila virus by targeting Outer membrane proteins (Omps). As we know viruses show constant evolution, it is very difficult to design a vaccine to combat them. Hence, conserved region extraction approach can be adopted while targeting a virus so that the potential epitopes do not get influenced by the virus evolution. A similar work was performed by Tamar and Ruth (2007) for Influenza virus in which they proposed epitopes belonging to the conserved part of the virus protein, hence giving a long lasting cellular and humoral response against the virus. Another study has been performed by Tosta et al. (2020) for 2 strains of yellow fever. In this study, a multi epitope-based vaccine was obtained through a consensus of the common epitopes in both the strains in order to generate a more homogenous response in the different strains of yellow fever.
As is evident from the literature, epitope-based vaccines present several advantages. Considering this, many researches have proposed epitope-based vaccines in order to combat the threat as posed by SARS-CoV-2 virus. Motivated by the fact that spike (S) and nucleocapsid (N) protein region of SARS-CoV-2 is similar to that of SARS-CoV, Ahmed et al. (2020) have proposed an epitope based vaccine derived from the S and N protein part of SARS-CoV and mapped them with the SARS-CoV-2. As a result, they obtained potential T-cell and B-cell epitopes for the 120 SARS-CoV-2 genomes. Recently, many works like (Chen et al., 2020; Rakib et al., 2020; Yadav et al., 2020) have proposed design of epitopes through targeting the S gene of SARS-CoV-2 which resulted in identification of linear, conformational B-cell and T-cell epitopes which are used to stimulate the immunogenic response. Another similar work has been proposed by Noorimotlagh et al. (2020) in which sets of B-cell and T-cell epitopes from the S and N proteins with high antigenicity and without allergenic property or toxic effects are presented. However, Grifoni et al. (2020b) showed that apart from S and N proteins, a majority of T-cells interact with SARS-CoV-2 in membrane (M) and other ORF proteins as well. Thus, apart from S protein, MHC-I restricted T-cell epitopes derived from M, NSP6, ORF3a or N proteins can also be considered for vaccine design. Gupta et al. (2020) have identified T-cell and B-cell epitopes using a web resource “CoronVR” for epitope-based vaccine design. Another approach followed by Poran et al. (2020) involves mass spectrometry-based profiling of individual HLA alleles to predict peptide binding to diverse allele sets resulting in the prediction of MHC-II restricted T-cell epitopes from SARS-CoV-2 proteins. However, the high mutability of SARS-CoV-2 within the Spike protein (Islam et al., 2020; Korber et al., 2020) region has increased the complexity in vaccine design. To mitigate this probem, Crooke et al. (2020) excluded the genomes which were not matching with the reference genome and then aligned the remaining SARS-CoV-2 genomes with the reference genome to predict 41 T-cell epitopes (5 HLA class I, 36 HLA class II) and 6 B-cell epitopes as the possible targets for epitope-based vaccine design. Vaxign and Vaxign-ML reverse vaccinology tools have been used by Ong et al. (2020) to predict potential vaccine candidates for COVID-19. In this regard, they have identified epitopes in NSP3, 3CL-Pro, NSP8, NSP9 and NSP10 coded proteins which can be considered to be potential vaccine design. Other works like (Baruah and Bose, 2020; Bency and Helen, 2020; Bhatnager et al., 2020; Bhattacharya et al., 2020a; Grifoni et al., 2020a; Kar et al., 2020; Kwarteng et al., 2020; Lim et al., 2020; Naz et al., 2020a; Singh et al., 2020; Vashi et al., 2020) have also explored different epitopes in SARS-CoV-2 for vaccine design.
In the literature discussed so far, analysis of virus proteins have been performed for the prediction of epitopes. However, the primary reason for structural change in virus proteins are due to genetical mutations. Motivated by this fact, in this work we have analysed 10,664 available SARS-CoV-2 genomes of 73 countries around the globe to identify the potential conserved regions in virus genomes to predict the immunogenic and antigenic epitopes in order to facilitate epitope based vaccine design. It is to be noted that these identified conserved genetic regions are such places for which the corresponding protein sequences are unchanged. For this purpose, multiple sequence alignment technique Clustal Omega (ClustalO) (Sievers and Higgins, 2014) is used to align the sequences. Following this, entropy is calculated for the aligned sequences to find the conserved regions. Further, these conserved regions are refined based on the criteria that (a) their lengths should be greater than or equal to 60 nt and (b) corresponding protein sequences should not have stop codons. Moreover, Nucleotide BLAST (Johnson et al., 2008) is used to verify the specificity of the conserved regions. As a result, we have obtained 17 conserved regions belonging to NSP3, NSP4, NSP6, NSP8, RdRp, Helicase, endoRNAse, 2’-O-RMT, Spike glycoprotein, ORF3a protein, Membrane glycoprotein and Nucleocapsid protein. Finally, these conserved regions are used to identify the T-cell and B-cell epitopes with their corresponding immunogenic and antigenic scores. This resulted in 30 MHC-I and 24 MHC-II restricted T-cell epitopes with 14 and 13 unique HLA alleles and 21 B-cell epitopes for the 17 conserved regions. Furthermore, the binding conformation of the MHC-I and MHC-II restricted T-cell epitopes are shown with respect to HLA alleles to judge their relevance. Additionally, their physico-chemical properties are also reported along with Ramchandran plots and Z-Scores.
2. Materials and methods
2.1. Data preparation
We have used the reference sequence of SARS-CoV-2 genome (NC_045512.2)2 and 44,583 protein sequences from the National Center for Biotechnology (NCBI) to map the SARS-CoV-2 proteins. The details of these protein sequences are provided in the supplementary Table S1. These protein sequences are only used to identify the SARS-CoV-2 coded proteins and their starting and ending coordinates in the reference sequence in order to have the correct protein sequence for which the corresponding structure of the protein exists. Therefore, to map the SARS-CoV-2 proteins, the reference sequence along with reading frame concept have been considered which works with dividing the sequence of nucleotides in the reference sequence into a set of successive, and non-overlapping triplets. There are three reading frames, Frame 1, Frame 2 and Frame 3. Frame 1 starts from the first nucleotide of a reference sequence, Frame 2 starts from the second nucleotide and Frame 3 starts from the third nucleotide and create the triplets. For each frame, these triplets are then converted into the corresponding proteins following the codon table.3 Finally, we have obtained 25 unique proteins which are matched to the different reading frames for which the structures are available. Moreover, 10,664 complete or near complete SARS-CoV-2 genomes are collected from Global Initiative on Sharing All Influenza Data (GISAID)4 in fasta format. The maximum and average length of the 10,664 virus genomes are 29,903 and 29,821 bp respectively. Please note that the maximum length of the virus genome has been fixed based on the reference genome. These genomes are then aligned to find the conserved regions. Their corresponding coded proteins are extracted as well. For the alignment of sequences, High Performance Computing (HPC) facility of NITTTR, Kolkata has been used. The HPC cluster has a master node with dual Intel Xeon Gold 6130 Processor having 32 Cores, 2.10 GHz, 22 MB L3 Cache and 128 GB DDR4 RAM and 2 GPU and 4 CPU computing nodes with dual Intel Xeon Gold 6152 Processor having 44 Cores, 2.1 GHz, 30 MB L3 Cache and 192 GB DDR4 RAM each, while GPU nodes have NVIDIA Tesla V100 GPU with 16 GB memory each. Multiple sequence alignment was performed using the 2 GPU and 4 CPU computing nodes.
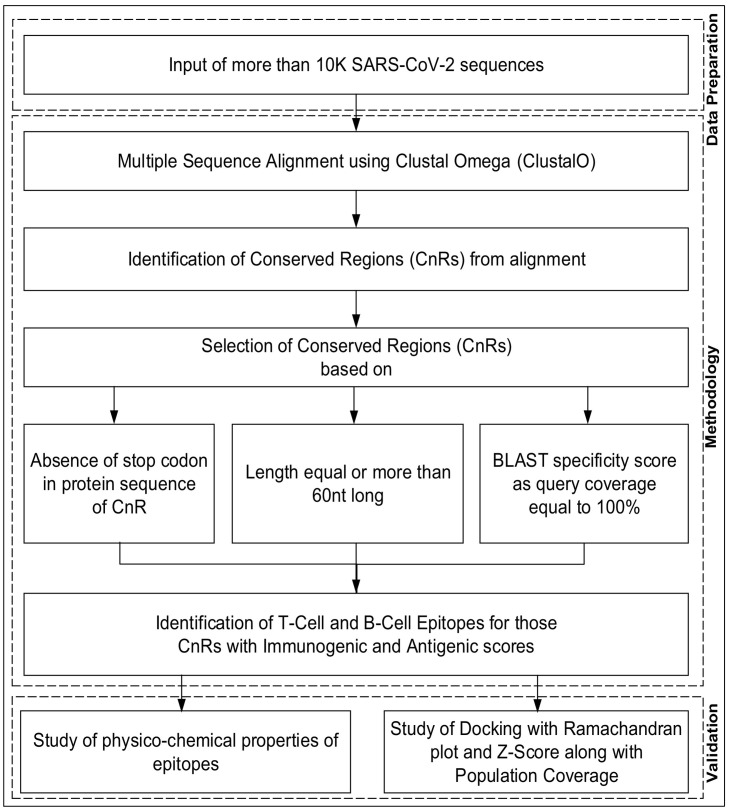
2.2. Pipeline of the workflow
The pipeline of the workflow is shown in Fig. 1 . In order to identify the conserved regions (CnRs) which are not affected by genetic mutations, initially a multiple sequence alignment technique known as ClustalO is performed on 10,664 SARS-CoV-2 genomes. ClustalO is chosen due to its high speed and accuracy. Execution of ClustalO is a multi step process. This involves the pairwise alignment using k-tuple method followed by which each sequence is clustered with the help of pairwise distance using sequence embedding also known as modified mBed method. Next, k-means clustering and construction of guide tree using Unweighted Pair Group Method with Arithmetic Mean (UPGMA) is performed. Finally, multiple alignment is carried out.with the help of HHAlign package provided by HH-Suite (Sievers and Higgins, 2014).
| (1) |
where S x y indicates the frequency of each residue x occurring at position y and 5 represents the four possible residues as nucleotide plus gap. To identify the conserved regions (CnRs) for each alignment technique, a minimum segment length of 15 is considered with maximum average entropy as 0.2. Further, maximum entropy per position is taken as 0.2 with no gaps after finding the consensus sequence for the 10,664 genomic sequences. All these values are taken after following the literature. Thereafter, a filtering criteria is adopted for the conserved regions based on the criteria (a) that their length are > = 60 nt and (b) their corresponding proteins are devoid of any stop codons. In addition to this, Nucleotide BLAST is used to determine the specificity of the conserved regions. Subsequently, the T-cell and B-cell epitopes were identified from these filtered CnRs. In this regard, IEDB5 and ABCPred6 are used to predict the T-cell and B-cell epitopes along with their corresponding immunogenic scores. To predict the MHC-I and MHC-II restricted T-cell epitopes, IEDB recommended NetMHCPan EL 4.17 and Consensus Approach8 (Sidney et al., 2008) are respectively used whereas for the prediction of B-cell epitopes, ABCPred is used. Thereafter, by using these predicted epitopes, antigenic scores are evaluated by VaxiJen2.0.9 In order to validate the identified T-cell epitopes, their conformational 2D non-covalent structures are studied using LigPlot+ (Wallace et al., 1995). On the other hand, BepiPred2.0 server10 (Jespersen et al., 2017) is used for the verification of the predicted B-cell epitopes. Also, the 3D structures of all the predicted epitopes are obtained with the help of Chimera (Pettersen et al., 2004) and their chemical orientations are visualised using ChemSketch (Spessard, 1998). Furthermore, the physico-chemical properties are evaluated with the help of Pfeature server11 (Pande et al., 2019). Also, for docking of the T-cell epitopes with their respective HLA alleles Autodock Vina (Rauf, 2015) is used whereas their structural properties are reported using Ramachandran plot with the help of PyMod 3 (Janson and Paiardini, 2020) (both are plugins of PyMOL (Yuan et al., 2017) software). Finally, ProSA12 (Wiederstein and Sippl, 2007) is used for Z-Score evaluation.
Fig. 1.
Pipeline of the workflow.
3. Results and discussion
3.1. Selection of CnRs
Results of the experiment which are carried out according to the flowchart in Fig. 1 are discussed in this section. Initially, 10,644 SARS-CoV-2 genomes are aligned using multiple sequence alignment technique, ClustalO. Subsequently, we have obtained 408 conserved regions (CnRs) which is followed by mapping of the CnRs to 11 coding regions, ORF1ab, Spike, ORF3a, Envelope, Membrane, ORF6, ORF7a, ORF7b, ORF8, Nucleocapsid and ORF10. The corresponding protein sequence for each CnR has been taken according to the reading frame it belongs to. For example, the protein sequence for the CnR belonging to Spike region is taken from Reading Frame 2 while that belonging to ORF3a is taken from Reading Frame 1. Next, the 408 CnRs are refined according to (a) their length should be greater than or equal to 60 nt and (b) their corresponding proteins do not have any stop codons. BLAST specificity score as query coverage equal to 100% is also considered for this refinement. This resulted in 17 CnRs which are shown in Table 1 along with their corresponding protein sequences, lengths, blast specificity scores, percentage of BLAST specificity scores as query coverage, coding regions with their starting and ending coordinates, lengths and coded proteins as well. These 17 CnRs belong to the coding regions which code NSP3, NSP4, NSP6, NSP8, RdRp, Helicase, endoRNAse, 2’-O-RMT, Spike glycoprotein, ORF3a protein, Membrane glycoprotein and Nucleocapsid protein. These protein sequences of each conserved region are then used for the prediction of MHC-I and MHC-II restricted T-cell and B-cell epitopes along with their respective immunogenic and antigenic scores. It is to be noted that the immunogenic and antigenic scores are scaled within the range of 0–1 to bring the scores of all the epitopes for different CnRs to a uniform scale. These scores are mentioned throughout the paper and the actual scores are given as Supplementary in an excel file.
Table 1.
Conserved Regions (CnRs) as derived from 10,664 SARS-CoV-2 genomes with associated details.
| DNA sequence of | Protein | Length | BLAST Specificity | % of BLAST Specificity | Coding | Starting | Ending | Length | Coded |
|---|---|---|---|---|---|---|---|---|---|
| Conserved Region (CnR) | Sequence | of CnR | Score of CnR | Score as Query Coverage | Region (CR) | Coordinate | Coordinate | of Protein | Proteins |
| 7622-GTTAATTGTGATACATTCTGTGCTGGTAGTACATTTATTAGTGATGAAGTTGCGAGAGAC-7681 | VNCDTFCAGSTFISDEVARD | 60 | 111 | 100 | ORF1ab | 266 | 21,552 | 21,287 | NSP3 |
| 8918-TTACCTAGAGTTTTTAGTGCAGTTGGTAACATCTGTTACACACCATCAAAACTTATAGAGTACACTG-8984 | LPRVFSAVGNICYTPSKLIEYT | 67 | 124 | 100 | ORF1ab | 266 | 21,552 | 21,287 | NSP4 |
| 11277-ATACTAGTTTGTCTGGTTTTAAGCTAAAAGACTGTGTTATGTATGCATCAGCTGTAGTGTTACTAATCCTTATGACAGCAAGAACTGTGTATGATGATGGTGCTAGGAGAGTGTGGACA-11395 | TSLSGFKLKDCVMYASAVVLLILMTARTVYDDGARRVWT | 119 | 220 | 100 | ORF1ab | 266 | 21,552 | 21,287 | NSP6 |
| 12,438-CCTTGAACATAATACCTCTTACAACAGCAGCCAAACTAATGGTTGTCATACCAGACTATAAC-12499 | LNIIPLTTAAKLMVVIPDYN | 62 | 115 | 100 | ORF1ab | 266 | 21,552 | 21,287 | NSP8 |
| 13,924-TGGTATGATTTTGTAGAAAACCCAGATATATTACGCGTATACGCCAACTTAGGTGAACGTGTACGCCAAGCTTTGTT-14000 | WYDFVENPDILRVYANLGERVRQAL | 77 | 143 | 100 | ORF1ab | 266 | 21,552 | 21,287 | RdRp |
| 15,607-TTACAACACAGACTTTATGAGTGTCTCTATAGAAATAGAGATGTTGACACAGACTTTGTGAATGAGTTTTACGCAT-15682 | LQHRLYECLYRNRDVDTDFVNEFYA | 76 | 141 | 100 | ORF1ab | 266 | 21,552 | 21,287 | RdRp |
| 16,730-TTTCATGGGAAGTTGGTAAACCTAGACCACCACTTAACCGAAATTATGTCTTTACTGGTTATCGTGTAACTAAAAACAGTAAAGTACAAAT-16820 | SWEVGKPRPPLNRNYVFTGYRVTKNSKVQ | 91 | 169 | 100 | ORF1ab | 266 | 21,552 | 21,287 | Helicase |
| 17,215-ATAGATAAATGTAGTAGAATTATACCTGCACGTGCTCGTGTAGAGTGTTTTGATAAATTCAAAGTGAATTCAACATTAGAACAGTATGT-17303 | IDKCSRIIPARARVECFDKFKVNSTLEQY | 89 | 165 | 100 | ORF1ab | 266 | 21,552 | 21,287 | Helicase |
| 17,612-ATAAGCTTAAAGCACATAAAGACAAATCAGCTCAATGCTTTAAAATGTTTTATAAGGGTG-17671 | KLKAHKDKSAQCFKMFYKG | 60 | 111 | 100 | ORF1ab | 266 | 21,552 | 21,287 | Helicase |
| 19,851-GGACATTGCTGCTAATACTGTGATCTGGGACTACAAAAGAGATGCTCCAGCACATATATCTACTATTGGTGTTTGTTCTATGACT-19935 | DIAANTVIWDYKRDAPAHISTIGVCSMT | 85 | 158 | 100 | ORF1ab | 266 | 21,552 | 21,287 | endoRNAse |
| 20,757-TGCAACATTACCTAAAGGCATAATGATGAATGTCGCAAAATATACTCAACTGTGTCAATATTTAAACAC-20825 | ATLPKGIMMNVAKYTQLCQYLN | 69 | 128 | 100 | ORF1ab | 266 | 21,552 | 21,287 | 2′- O- RMT |
| 23,732-ACAGAAATTCTACCAGTGTCTATGACCAAGACATCAGTAGATTGTACAATGTACATTTGTGGTGATT-23798 | TEILPVSMTKTSVDCTMYICGD | 67 | 124 | 100 | Spike | 21,563 | 25,381 | 3819 | Spike glycoprotein |
| 24,406-TCAAGATGTGGTCAACCAAAATGCACAAGCTTTAAACACGCTTGTTAAACAACTTAGCTCCAA-24468 | QDVVNQNAQALNTLVKQLSS | 63 | 117 | 100 | Spike | 21,563 | 25,381 | 3819 | Spike glycoprotein |
| 25,990-TGTGTTGTATTACACAGTTACTTCACTTCAGACTATTACCAGCTGTACTCAACTCAATTGAGTACAGA-26057 | CVVLHSYFTSDYYQLYSTQLST | 68 | 126 | 100 | ORF3a | 25,393 | 26,217 | 825 | ORF3a protein |
| 26,560-TTAAAAAGCTCCTTGAACAATGGAACCTAGTAATAGGTTTCCTATTCCTTACATGGATTTGTCTTCTACAATTTGCCTA-26638 | KKLLEQWNLVIGFLFLTWICLLQFA | 79 | 147 | 100 | Membrane | 26,523 | 27,188 | 666 | Membrane glycoprotein |
| 27,129-AACTATAAATTAAACACAGACCATTCCAGTAGCAGTGACAATATTGCTTTGCTTGTACAGT-27189 | NYKLNTDHSSSSDNIALLVQ | 61 | 113 | 100 | Membrane | 26,523 | 27,188 | 666 | Membrane glycoprotein |
| 28,518-ACCAAATTGGCTACTACCGAAGAGCTACCAGACGAATTCGTGGTGGTGACGGTAAAATGAAA-28579 | QIGYYRRATRRIRGGDGKMK | 62 | 115 | 100 | Nucleocapsid | 28,274 | 29,530 | 1257 | Nucleocapsid protein |
3.2. Identification of T-cell epitopes
To predict the epitopes from the 17 CnRs, the corresponding protein sequences are used as inputs to the prediction tools. IEDB recommended NetMHCPan EL 4.1 (Reynisson et al., 2020) prediction method is used for the prediction of MHC-I restricted T-cell epitopes targeting 27 unique HLA alleles. For each CnR, this resulted in the selection of 5 best HLA epitopes as the representative of good binders in the form of immunogenic scores. Their antigenic scores are evaluated using VaxiJen2.0 (Doytchinova and Flower, 2007). A cut-off of 0.4 is maintained in VaxiJen 2.0. Any epitope beyond this cut-off are considered to be antigenic. Thus, a total of 85 epitopes of length 9–10 mer each are obtained with their corresponding immunogenic and antigenic scores. Subsequently, for each of the 17 CnRs, the most immunogenic and antigenic MHC-I restricted T-cell epitopes are selected. As a result, 30 MHC-I restricted T-cell epitopes are identified and reported in Table 2 . Out of these 30 epitopes, in terms of scores, the most immunogenic MHC-I restricted T-cell epitopes are DTDFVNEFY bounded to HLA-A*01:01 allele and the most antigenic epitope is IPARARVECF bounded to HLA-B*07:02 allele belonging to RdRp and Helicase coded proteins respectively.
Table 2.
List of most Immunogenic and Antigenic Epitopes for MHC-I, MHC-II restricted T-cell and B-cell Epitopes for 17 CnRs.
| Protein |
Coded |
Type | MHC-I restricted T-cell |
MHC-II restricted T-cell |
B-cell |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sequence | Proteins | Epitope | Alleles | Scaled Score of |
Epitope | Alleles | Scaled Score of |
Epitope | Scaled Score of |
||||
| Immunogenicity | Antigenicity | Immunogenicity | Antigenicity | Immunogenicity | Antigenicity | ||||||||
| VNCDTFCAGSTFISDEVARD | NSP3 | Immunogenic | STFISDEVAR | HLA-A*68:01 | 0.91 | 0.23 | FCAGSTFISDEVARD | HLA-DRB3*01:01 | 0.99 | 0.03 | CDTFCAGSTFISDEVA | 0.61 | 0.22 |
| Antigenic | DTFCAGSTF | HLA-A*26:01 | 0.51 | 0.42 | DTFCAGSTFISDEVA | HLA-DQA1*03:01/DQB1*03:02 | 0.88 | 0.12 | |||||
| LPRVFSAVGNICYTPSKLIEYT | NSP4 | Immunogenic | YTPSKLIEY | HLA-A*26:01 | 0.86 | 0.46 | VGNICYTPSKLIEYT | HLA-DRB1*07:01 | 1.00 | 0.88 | AVGNICYTPSKLIEYT | 0.28 | 1.00 |
| Antigenic | FSAVGNICY | HLA-A*01:01 | 0.84 | 0.81 | |||||||||
| TSLSGFKLKDCVMYASAVVLLILMTARTVYDDGARRVWT | NSP6 | Immunogenic | TVYDDGARR | HLA-A*68:01 | 0.99 | 0.02 | ASAVVLLILMTARTV | HLA-DRB1*01:01 | 0.99 | 0.56 | LILMTARTVYDDGARR | 0.69 | 0.23 |
| Antigenic | ILMTARTVY | HLA-B*15:01 | 0.87 | 0.78 | SGFKLKDCVMYASAVV | 0.47 | 0.55 | ||||||
| LNIIPLTTAAKLMVVIPDYN | NSP8 | Immunogenic | TTAAKLMVV | HLA-A*68:02 | 0.72 | 0.52 | LNIIPLTTAAKLMVV | HLA-DRB1*08:02 | 0.99 | 0.71 | LNIIPLTTAAKLMVVI | 0.00 | 0.86 |
| Antigenic | NIIPLTTAAK | HLA-A*68:01 | 0.54 | 0.83 | |||||||||
| WYDFVENPDILRVYANLGERVRQAL | RdRp | Immunogenic | VENPDILRVY | HLA-B*44:03 | 0.95 | 0.30 | ILRVYANLGERVRQA | HLA-DRB3*02:02 | 0.98 | 0.44 | YDFVENPDILRVYANL | 0.50 | 0.13 |
| Antigenic | RVYANLGER | HLA-A*31:01 | 0.76 | 0.63 | DILRVYANLGERVRQA | 0.19 | 0.51 | ||||||
| LQHRLYECLYRNRDVDTDFVNEFYA | RdRp | Immunogenic | DTDFVNEFY | HLA-A*01:01 | 1.00 | 0.48 | RNRDVDTDFVNEFYA | HLA-DQA1*01:01/DQB1*05:01 | 0.83 | 0.53 | HRLYECLYRNRDVDTD | 0.83 | 0.38 |
| Antigenic | YRNRDVDTDFVNEFY | HLA-DQA1*01:01/DQB1*05:01 | 0.79 | 0.58 | YRNRDVDTDFVNEFYA | 0.17 | 0.71 | ||||||
| SWEVGKPRPPLNRNYVFTGYRVTKNSKVQ | Helicase | Immunogenic | YVFTGYRVTK | HLA-A*68:01 | 0.74 | 0.65 | NYVFTGYRVTKNSKV | HLA-DRB1*07:01 | 1.00 | 0.66 | SWEVGKPRPPLNRNYV | 0.97 | 0.00 |
| Antigenic | PPLNRNYVFTGYRVTK | 0.61 | 0.73 | ||||||||||
| IDKCSRIIPARARVECFDKFKVNSTLEQY | Helicase | Immunogenic | KVNSTLEQY | HLA-A*30:02 | 0.94 | 0.52 | ECFDKFKVNSTLEQY | HLA-DRB3*02:02 | 0.97 | 0.18 | CSRIIPARARVECFDK | 0.83 | 0.71 |
| Antigenic | IPARARVECF | HLA-B*07:02 | 0.56 | 1.00 | KCSRIIPARARVECF | HLA-DPA1*02:01/DPB1*14:01 | 0.93 | 0.64 | |||||
| KLKAHKDKSAQCFKMFYKG | Helicase | Immunogenic | KSAQCFKMF | HLA-B*57:01 | 0.85 | 0.47 | AHKDKSAQCFKMFYK | HLA-DPA1*02:01/DPB1*05:01 | 0.70 | 0.60 | HKDKSAQCFK | 0.69 | 0.57 |
| Antigenic | KSAQCFKMFY | HLA-B*57:01 | 0.59 | 0.51 | |||||||||
| DIAANTVIWDYKRDAPAHISTIGVCSMT | endoRNAse | Immunogenic | DAPAHISTI | HLA-B*51:01 | 0.59 | 0.51 | IWDYKRDAPAHISTI | HLA-DRB3*01:01 | 1.00 | 0.47 | ANTVIWDYKRDAPAHI | 1.00 | 0.56 |
| Antigenic | NTVIWDYKR | HLA-A*68:01 | 0.88 | 0.68 | WDYKRDAPAHISTIG | HLA-DRB3*01:01 | 0.99 | 0.65 | |||||
| ATLPKGIMMNVAKYTQLCQYLN | 2′- O- RMT | Immunogenic | KYTQLCQYL | HLA-A*24:02 | 0.79 | 0.74 | PKGIMMNVAKYTQLC | HLA-DRB3*02:02 | 0.98 | 0.42 | PKGIMMNVAKYTQLCQ | 0.64 | 0.41 |
| Antigenic | |||||||||||||
| TEILPVSMTKTSVDCTMYICGD | Spike glycoprotein | Immunogenic | EILPVSMTK | HLA-A*68:01 | 0.95 | 0.98 | PVSMTKTSVDCTMYI | HLA-DRB3*01:01 | 0.80 | 0.85 | LPVSMTKTSVDCTMYI | 0.50 | 1.00 |
| Antigenic | TEILPVSMTKTSVDC | HLA-DRB1*08:02 | 0.63 | 1.00 | |||||||||
| QDVVNQNAQALNTLVKQLSS | Spike glycoprotein | Immunogenic | ALNTLVKQL | HLA-A*02:03 | 0.81 | 0.20 | QDVVNQNAQALNTLV | HLA-DRB1*13:02 | 0.91 | 0.19 | NQNAQALNTLVKQLSS | 0.50 | 0.18 |
| Antigenic | NAQALNTLV | HLA-B*51:01 | 0.18 | 0.41 | DVVNQNAQALNTLVK | HLA-DRB1*13:02 | 0.82 | 0.22 | |||||
| CVVLHSYFTSDYYQLYSTQLST | ORF3a protein | Immunogenic | FTSDYYQLY | HLA-A*01:01 | 0.99 | 0.36 | LHSYFTSDYYQLYST | HLA-DPA1*01:03/DPB1*04:01 | 1.00 | 0.23 | LHSYFTSDYYQLYSTQ | 0.78 | 0.35 |
| Antigenic | YYQLYSTQL | HLA-A*24:02 | 0.84 | 0.56 | HSYFTSDYYQLYSTQ | HLA-DPA1*01:03/DPB1*04:01 | 0.99 | 0.29 | |||||
| KKLLEQWNLVIGFLFLTWICLLQFA | Membrane glycoprotein | Immunogenic | KLLEQWNLV | HLA-A*02:01 | 0.88 | 0.47 | WNLVIGFLFLTWICL | HLA-DPA1*01:03/DPB1*02:01 | 0.99 | 1.00 | LVIGFLFLTWICLLQF | 0.53 | 0.90 |
| Antigenic | LVIGFLFLTW | HLA-B*57:01 | 0.64 | 0.87 | |||||||||
| NYKLNTDHSSSSDNIALLVQ | Membrane glycoprotein | Immunogenic | SSDNIALLV | HLA-A*01:01 | 0.50 | 0.51 | NYKLNTDHSSSSDNI | HLA-DRB3*02:02 | 0.85 | 0.26 | YKLNTDHSSSSDNIAL | 0.22 | 0.35 |
| Antigenic | SSSDNIALL | HLA-A*68:02 | 0.27 | 0.53 | |||||||||
| QIGYYRRATRRIRGGDGKMK | Nucleocapsid protein | Immunogenic | GYYRRATRR | HLA-A*31:01 | 0.57 | 0.24 | IGYYRRATRRIRGGD | HLA-DRB1*11:01 | 0.99 | 0.53 | GYYRRATRRIRGGDGK | 0.61 | 0.31 |
| Antigenic | IGYYRRATR | HLA-A*31:01 | 0.44 | 0.71 | |||||||||
Similarly, MHC-II restricted T-cell epitopes targeting a different set of 27 unique alleles are predicted using consensus approached as approved by IEDB which resulted in 85 epitopes of length 15 mer each along with their corresponding immunogenic and antigenic scores. Eventually, the most immunogenic and antigenic MHC-II restricted T-cell epitopes are selected for each of the 17 CnRs resulting in 24 MHC-II restricted T-cell epitopes which are reported in Table 2. On the basis of immunogenic scores, VGNICYTPSKLIEYT, NYVFTGYRVTKNSKV, IWDYKRDAPAHISTI and LHSYFTSDYYQLYST are found to be most immunogenic where the first two are bounded to HLA-DRB1*07:01 while the rest two are bounded to HLA-DRB3*01:01 and HLA-DPA1*01:03/DPB1*04:01 alleles respectively and they belong to NSP4, Helicase, endoRNAse and ORF3a coded proteins respectively. On the other hand, the most antigenic epitopes are TEILPVSMTKTSVDC and WNLVIGFLFLTWICL from the Spike and Membrane glycoproteins corresponding to HLA-DRB1*08:02 and HLA-DPA1*01:03/DPB1*02:01 alleles respectively. All the 85 MHC-I and MHC-II restricted T-cell epitopes along with their HLA alleles are provided in the supplementary as an excel file.
3.3. Identification of B-cell epitopes
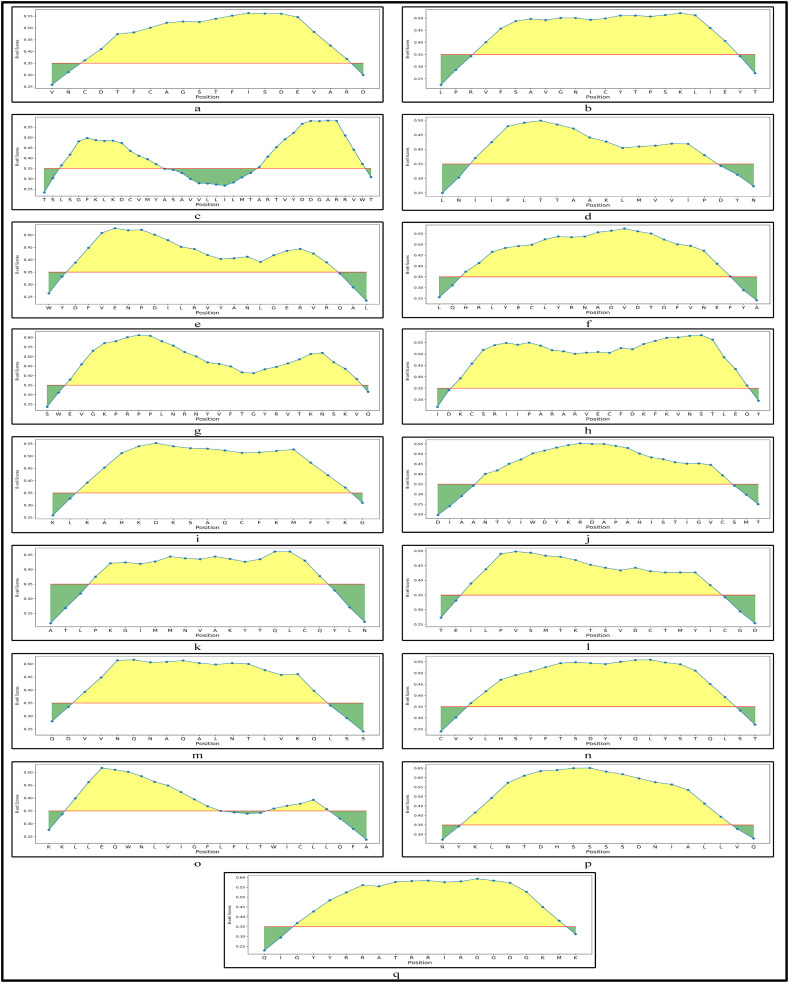
Once, the MHC-I and MHC-II are obtained, the prediction of B-cell epitopes which are responsible for antigen productions are carried out using ABCPred (Saha and Raghava, 2007). An threshold of 0.5 is considered in ABCPred where only the epitopes greater than this threshold are considered to be immunogenic. Their antigenic scores are evaluated using VaxiJen server as well with the same cut-off of 0.4. As a result, we have identified 23 linear B-cell epitopes of length of 16 mer along with their immunogenic and antigenic scores for the 17 CnRs. Among these 23, 21 B-cell epitopes are selected as the most immunogenic and antigenic as shown in Table 2. These 21 B-cell epitopes are also verified using BepiPred 2.0 server and their corresponding graphical representations are shown in Fig. 2 where the red line represents the threshold which is set to 0.35 and the total green and yellow regions indicate a protein sequence. As can be seen from Table 2, in terms of scores, the most immunogenic B-cell epitope is ANTVIWDYKRDAPAHI while the most antigenic epitopes are AVGNICYTPSKLIEYT and LPVSMTKTSVDCTMYI. Their graphical representations are shown respectively in Fig. 2(j), (b) and (l). These three epitopes belong to coded proteins endoRNAse, NSP4 and Spike glycoprotein respectively. All the 23 B-cell epitopes are provided in the supplementary as an excel file.
Fig. 2.
Graphical representation of B-cell epitopes for 17 CnRs belonging to (a) NSP3 (b) NSP4 (c) NSP6 (d) NSP8 (e) RdRp (f) RdRp (g) Helicase (h) Helicase (i) Helicase (j) endoRNAse (k) 2’-O-RMT (l) Spike glycoprotein (m) Spike glycoprotein (n) ORF3a protein (o) Membrane glycoprotein (p) Membrane glycoprotein and (q) Nucleocapsid protein.
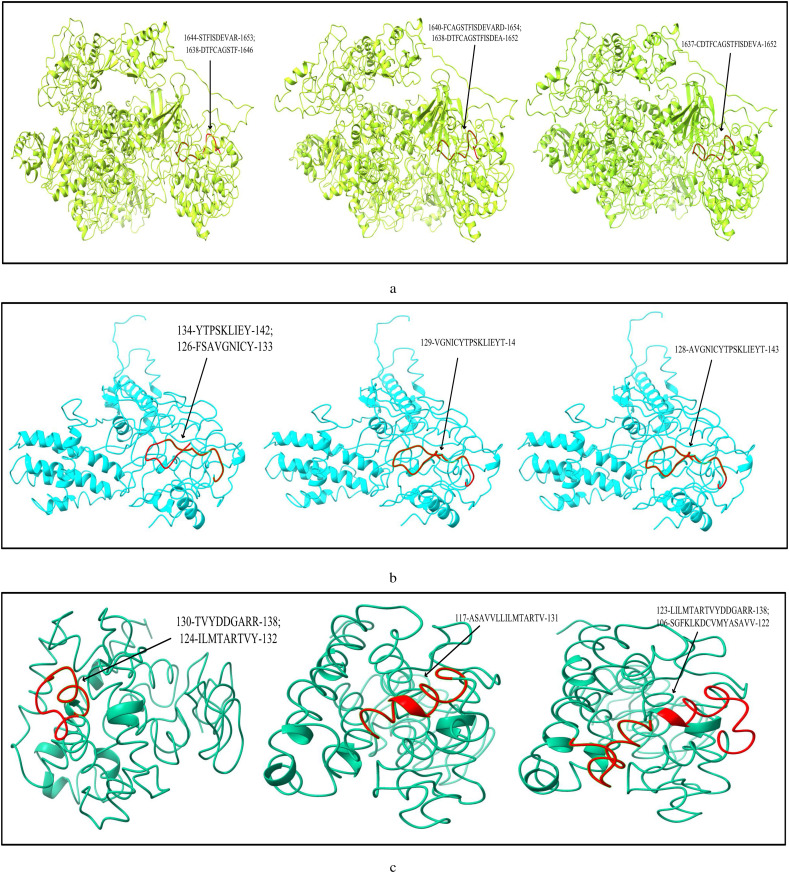
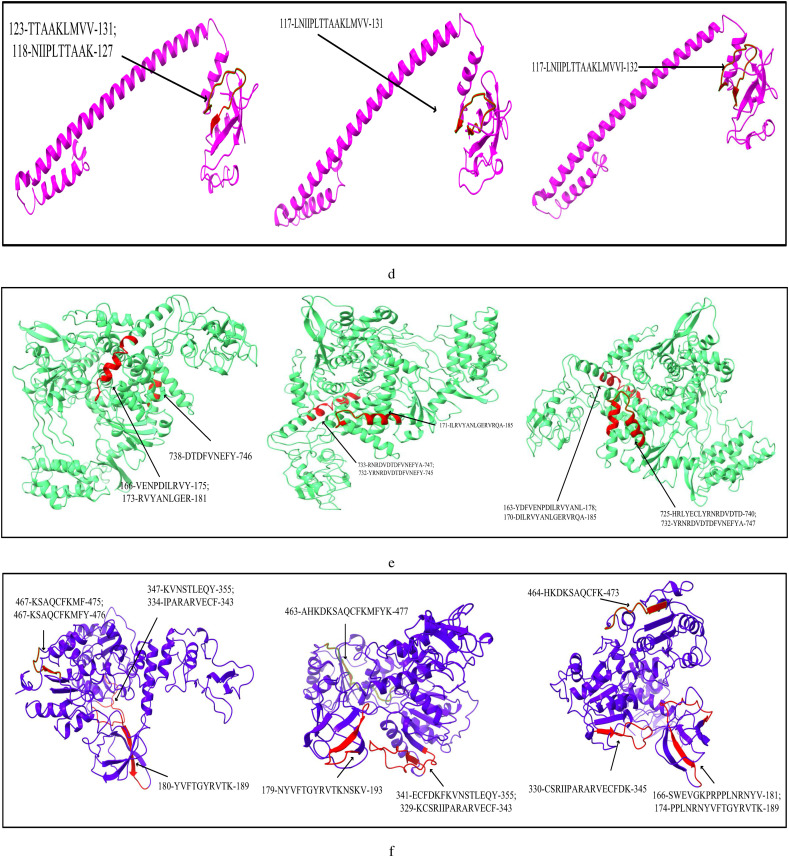
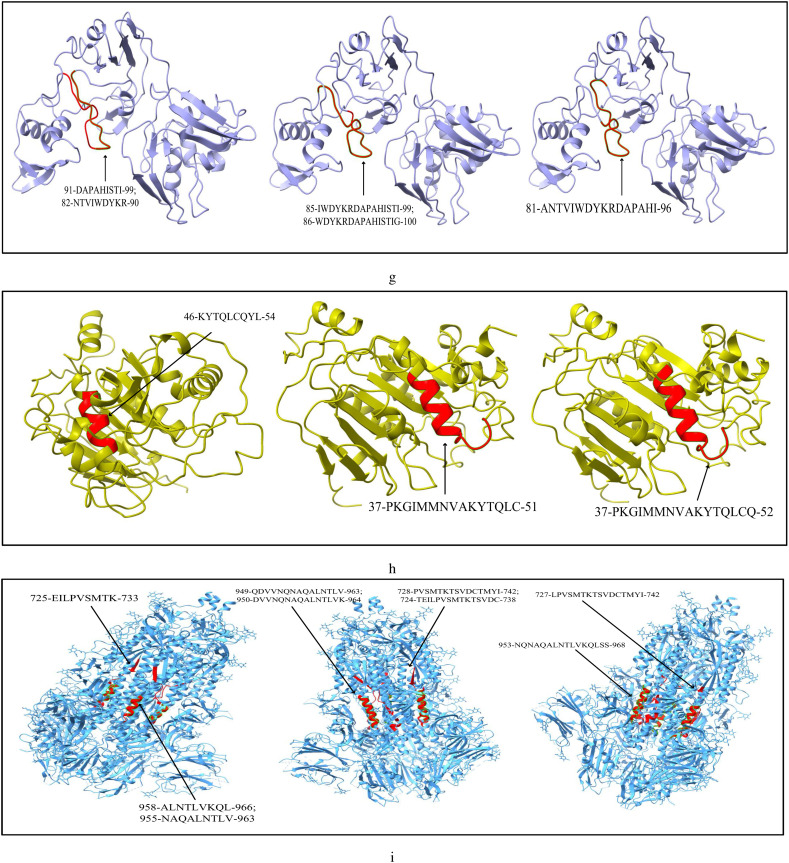
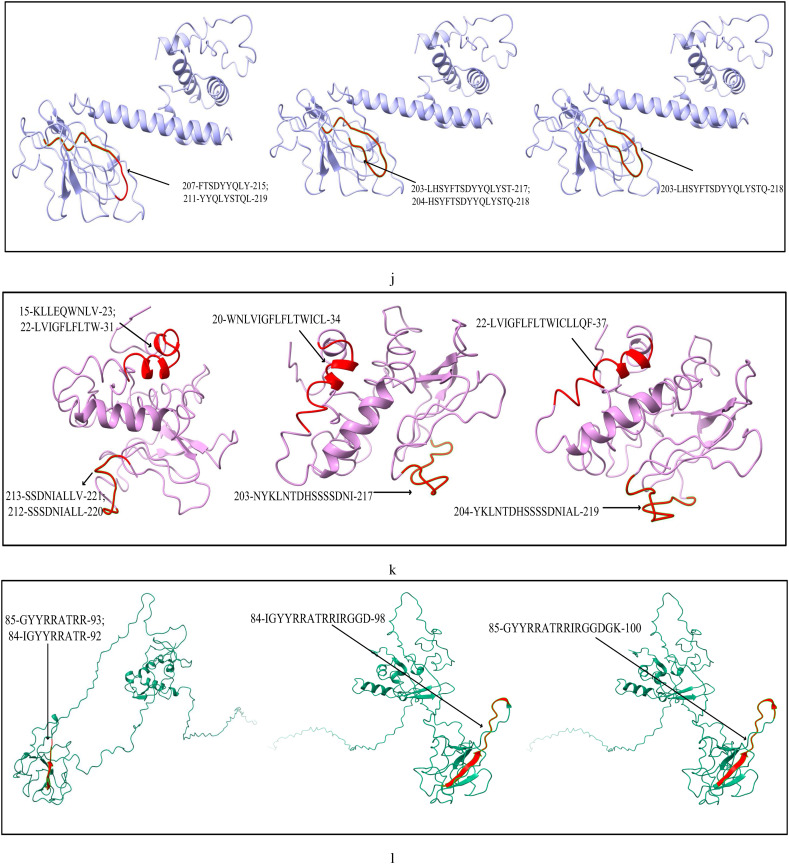
The list of most immunogenic and antigenic epitopes for each of the 17 CnRs are summarised in Table 3 . For better understanding, these epitopes are underlined in Fig. 3 . The red lines, green lines and the blue lines indicate the MHC-I, MHC-II T-cells and B-cell epitopes respectively. The 3D structures of the epitopes summarised in Table 3 are further highlighted in Fig. 4 using ChimeraX. Moreover, for the ease of the readers, all the details related to the 408 CCnRs, 85 MHC-I and MHC-II restricted T-cell epitopes and 23 B-cell epitopes are provided in the supplementary as an excel file, the link of which is given in Table S1. Furthermore, a summary of T-cell and B-cell epitopes identified in the literature (Baruah and Bose, 2020; Bency and Helen, 2020; Bhatnager et al., 2020; Bhattacharya et al., 2020a; Chen et al., 2020; Crooke et al., 2020; Grifoni et al., 2020a; Gupta et al., 2020; Kar et al., 2020; Kwarteng et al., 2020; Lim et al., 2020; Naz et al., 2020a; Ong et al., 2020; Poran et al., 2020; Rakib et al., 2020; Singh et al., 2020; Vashi et al., 2020; Yadav et al., 2020) are presented in Table 4 while the details of all the epitopes are given in the supplementary as an excel file. Table 3, Table 4 thus provide an overview of the epitopes identified so far.
Table 3.
Summary of most Immunogenic and Antigenic Epitopes for MHC-I, MHC-II restricted T-cell and B-cell Epitopes for 17 CnRs.
| Coded | MHC-I restricted T-cell | MHC-II restricted T-cell | B-cell |
|---|---|---|---|
| Proteins | Epitopes | Epitopes | Epitopes |
| NSP3 | STFISDEVAR | FCAGSTFISDEVARD | CDTFCAGSTFISDEVA |
| DTFCAGSTF | DTFCAGSTFISDEVA | ||
| NSP4 | YTPSKLIEY | VGNICYTPSKLIEYT | AVGNICYTPSKLIEYT |
| FSAVGNICY | |||
| NSP6 | TVYDDGARR | ASAVVLLILMTARTV | LILMTARTVYDDGARR |
| ILMTARTVY | SGFKLKDCVMYASAVV | ||
| NSP8 | TTAAKLMVV | LNIIPLTTAAKLMVV | LNIIPLTTAAKLMVVI |
| NIIPLTTAAK | |||
| RdRp | VENPDILRVY | ILRVYANLGERVRQA | YDFVENPDILRVYANL |
| RVYANLGER | DILRVYANLGERVRQA | ||
| RdRp | DTDFVNEFY | RNRDVDTDFVNEFYA | HRLYECLYRNRDVDTD |
| YRNRDVDTDFVNEFY | YRNRDVDTDFVNEFYA | ||
| Helicase | YVFTGYRVTK | NYVFTGYRVTKNSKV | SWEVGKPRPPLNRNYV |
| PPLNRNYVFTGYRVTK | |||
| Helicase | KVNSTLEQY | ECFDKFKVNSTLEQY | CSRIIPARARVECFDK |
| IPARARVECF | KCSRIIPARARVECF | ||
| Helicase | KSAQCFKMF | AHKDKSAQCFKMFYK | HKDKSAQCFK |
| KSAQCFKMFY | |||
| endoRNAse | DAPAHISTI | IWDYKRDAPAHISTI | ANTVIWDYKRDAPAHI |
| NTVIWDYKR | WDYKRDAPAHISTIG | ||
| 2’-O-RMT | KYTQLCQYL | PKGIMMNVAKYTQLC | PKGIMMNVAKYTQLCQ |
| Spike glycoprotein | EILPVSMTK | PVSMTKTSVDCTMYI | LPVSMTKTSVDCTMYI |
| TEILPVSMTKTSVDC | |||
| Spike glycoprotein | ALNTLVKQL | QDVVNQNAQALNTLV | NQNAQALNTLVKQLSS |
| NAQALNTLV | DVVNQNAQALNTLVK | ||
| ORF3a protein | FTSDYYQLY | LHSYFTSDYYQLYST | LHSYFTSDYYQLYSTQ |
| YYQLYSTQL | HSYFTSDYYQLYSTQ | ||
| Membrane glycoprotein | KLLEQWNLV | WNLVIGFLFLTWICL | LVIGFLFLTWICLLQF |
| LVIGFLFLTW | |||
| Membrane glycoprotein | SSDNIALLV | NYKLNTDHSSSSDNI | YKLNTDHSSSSDNIAL |
| SSSDNIALL | |||
| Nucleocapsid protein | GYYRRATRR | IGYYRRATRRIRGGD | GYYRRATRRIRGGDGK |
| IGYYRRATR |
Fig. 3.
MHC-I, MHC-II restricted T-cell and B-cell epitopes underlined in the protein sequences of 17 CnRs for (a) NSP3 (b) NSP4 (c) NSP6 (d) NSP8 (e) RdRp (f) RdRp (g) Helicase (h) Helicase (i) Helicase (j) endoRNAse (k) 2’-O-RMT (l) Spike glycoprotein (m) Spike glycoprotein (n) ORF3a protein (o) Membrane glycoprotein (p) Membrane glycoprotein and (q) Nucleocapsid protein.
Fig. 4.
Modelling of MHC-I, MHC-II restricted T-cell and B-cell epitopes for 17 CnRs belonging to (a) NSP3 (b) NSP4 (c) NSP6 (d) NSP8 (e) RdRp (f) Helicase (g) endoRNAse (h) 2’-O-RMT (i) Spike glycoprotein (j) ORF3a protein (k) Membrane glycoprotein (l) Nucleocapsid protein.
Table 4.
List of proposed epitopes for SARS-CoV-2 as given in the literature.
| Source | Coded Proteins | MHC-I restricted T-cell Epitopes | MHC-II restricted T-cell Epitopes | B-cell Epitopes |
|---|---|---|---|---|
| Baruah and Bose (2020) | Spike glycoprotein | YLQPRTFLL | NA | CVNLTTRTQLPPAYTN |
| GVYFASTEK | NVTWFHAIHVSGTNG | |||
| EPVLKGVKL | SFSTFKCYGVSPTKLND | |||
| Bency and Helen (2020) | Spike glycoprotein | KIADYNYKL | VVFLHVTYV | MDLEGKQGNFKNL |
| CYGVSPTKL | IGINITRFQ | YYVGYLQPR | ||
| VVVLSFELL | FNCYFPLQS | NITNLCPFGE | ||
| Bhatnager et al. (2020) | Spike glycoprotein | LTDEMIAQY | VASQSIIAYTMSLGA | KEEQIGKCSTR |
| LLTDEMIAQY | LTDEMIAQYTSALLA | ELGKYEQYGPGPGKWP | ||
| IPFAMQMAY | VLNDILSRLDKVEAE | IRAGPGPGGNC | ||
| Bhattacharya et al. (2020a) | Spike glycoprotein | SQCVNLTTR | IHVSGTNGT | SQCVNLTTRTQLPPAYTNSFTRGVY |
| YTNSFTRGV | VYYHKNNKS | FSNVTWFHAIHVSGTNGTKRFDN | ||
| GVYYHKNNK | LVRDLPQGF | DPFLGVYYHKNNKSWME | ||
| Chen et al. (2020) | Spike glycoprotein | LSPRWYFYY | IKLDDKDPN | EVRQIAPGQTGKIADY |
| RSRNSSRNS | RSGARSKQR | GCLIGAEHVNNSYECD | ||
| IGYYRRATR | RIGMEVTPS | FAMQMAYRFNGIGVTQ | ||
| Crooke et al. (2020) | Membrane glycoprotein | ATSRTLSYY | TLSYYKLGASQRVAG | EVTPSGTWL |
| RLFARTRSM | RTLSYYKLGASQRVA | KLDDKDPNFK | ||
| YANRNRFLY | ASFRLFARTRSMWSF | KTFPPTEPKKDKKKKADETQALPQ | ||
| Grifoni et al. (2020a) | Spike glycoprotein | NLTTRTQL | TQDLFLPFFSNVTWF | DAVDCALDPLSETKCTLKSFTVEKGIYQTSN |
| LPPAYTNSF | SLLIVNNATNVVIKV | VCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVI | ||
| KVFRSSVLH | LPFFSNVTWFHAIHV | GTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGS | ||
| Gupta et al. (2020) | Spike glycoprotein | VRFPNITNL | NVTWFHAIHV | GDEVRQIAPGQTGKIADYNYKLP |
| YQPYRVVVL | ||||
| PYRVVVLSF | ||||
| Kar et al. (2020) | Spike glycoprotein | QIITTDNTF | INITRFQTLLALHRS | FSYTESLAGKREMAII |
| YQPYRVVVL | GINITRFQTLLALHR | HAGPGPGPY | ||
| FTISVTTEI | GWTFGAGAALQIPFA | KMGPGPGTRFA | ||
| Kwarteng et al. (2020) | 1*Nucleocapsid protein | KTFPPTEPK | AQFAPSASAFFGMSR | AGLPYGANK |
| SSPDDQIGY | IAQFAPSASAFFGMS | SKQLQQSMSSADS | ||
| SSPDDQIGYY | PQIAQFAPSASAFFG | RRIRGGDGKMKDL | ||
| Lim et al. (2020) | Spike glycoprotein | YLQPRTFLL | VVLSFELLHAPATVC | SQCVNLTTRTQLPPAYTNSFTRGVY |
| KIADYNYKL | QQLIRAAEIRASANL | DPFLGVYYHKNNKSWME | ||
| SIIAYTMSL | GNYNYLYRLFRKSNL | NLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN | ||
| Naz et al. (2020a) | Spike glycoprotein | GVYFASTEK | EFVFKNIDGYFKIYS | YNSASFSTFKCYGVSPTKLNDLCFT |
| STQDLFLPF | QPYRVVVLSFELLHA | |||
| KTSVDCTMY | MTKTSVDCTMYICGD | |||
| Ong et al. (2020) | NSP3 | STNVTIATY | ISNSWLMWLIINLVQ | EDEEEGDCEEEEFEPSTQYEYGTEDDYQGKPLEFGATS |
| RMYIFFASF | LAYILFTRFFYVLGL | EEEQEEDWLDDD | ||
| AEWFLAYIL | AAIMQLFFSYFAVHF | VGQQDGSEDNQ | ||
| Poran et al. (2020) | Spike glycoprotein | YQPYRVVVL | TPPIKDFGGFNFSQILPDPSKPSKR | NA |
| FVFLVLLPL | EIDRLNEVAKNLNESLIDLQELGKY | |||
| CVADYSVLY | EKGIYQTSNFRVQPTESIVRFPNIT | |||
| Rakib et al. (2020) | Spike glycoprotein | WTAGAAAYY | LIVNNATNV | RTQLPPAYTNS |
| CNDPFLGVY | IVNNATNVV | SGTNGTKRFDN | ||
| GAAAYYVGY | SKTQSLLIV | LTPGDSSSGWTAG | ||
| Singh et al. (2020) | Nucleocapsid protein | AQFAPSASA | AQFAPSASAFFGMSR | KEDLKFP |
| GDAALALLL | GDAALALLLLDRLNQ | IKLDDKDPNFKDQ | ||
| GMSRIGMEV | ASAFFGMSRIGMEVT | PPTEPKKDKKKKADETQALPQRQKKQQTVT | ||
| Vashi et al. (2020) | Spike glycoprotein | RTQLPPAY | MFVFLVLLPLVSSQC | PPAYTNSFTRGVYY |
| RTQLPPA | MFVFLVLLPLVSSQCVN | HVSGTNGTKRFDN | ||
| LPPAYTNSF | QGNFKNLREFVFKNI | YYHKNNKSWMES | ||
| Yadav et al. (2020) | Spike glycoprotein | GVYFASTEK | NA | HRSYLTPGDSSSGWTA |
| FEYVSQPFL | FPNITNLCPFGEVFNA | |||
| WTAGAAAYY | EVIQIAPGQTGKIADY |
3.4. Study of Physico-chemical properties
The significance of the epitopes reported in this paper are shown through their physico-chemical properties. For each property, the physico-chemical values lie between 0 and 1. The physico-chemical properties for MHC-I, MHC-II restricted T-cells and B-cell epitopes belonging to the 17 CnRs are reported respectively in Table 5, Table 6, Table 7 . As reported in Table 5, MHC-I restricted T-cell epitope STFISDEVAR has a positively charged value of 0.1, a negatively charged value of 0.2, polarity of 0.3, non-polarity of 0.4, aliphaticity of 0.3, aromaticity of 0.1, acidicity of 0.2, basicity of 0.1, hydrophobicity of 0.5, hydrophilicity of 0.1, a neutral value of 0.5, hydroxylic value of 0.3 and sulphur content is 0. For the other epitopes, their physico-chemical properties are reported in tables as well.
Table 5.
List of physico-chemical properties of MHC-I restricted T-cell epitopes.
| MHC-I restricted T-cell epitopes | Positively charged | Negatively charged | Polarity | Non polarity | Aliphaticity | Aromaticity | Acidicity | Basicity | Hydrophobicity | Hydrophilicity | Neutral | Hydroxylic | Sulphur content |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STFISDEVAR | 0.1 | 0.2 | 0.3 | 0.4 | 0.3 | 0.1 | 0.2 | 0.1 | 0.5 | 0.1 | 0.5 | 0.3 | 0 |
| DTFCAGSTF | 0 | 0.111 | 0.444 | 0.444 | 0.222 | 0.222 | 0.111 | 0 | 0.667 | 0 | 0.556 | 0.333 | 0.111 |
| YTPSKLIEY | 0.111 | 0.111 | 0.444 | 0.333 | 0.333 | 0.222 | 0.111 | 0.111 | 0.444 | 0.222 | 0.333 | 0.222 | 0 |
| FSAVGNICY | 0 | 0 | 0.333 | 0.556 | 0.444 | 0.222 | 0 | 0 | 0.556 | 0.111 | 0.222 | 0.111 | 0.111 |
| TVYDDGARR | 0.222 | 0.222 | 0.222 | 0.333 | 0.333 | 0.111 | 0.222 | 0.222 | 0.333 | 0.222 | 0.444 | 0.111 | 0 |
| ILMTARTVY | 0.111 | 0 | 0.333 | 0.556 | 0.444 | 0.111 | 0 | 0.111 | 0.778 | 0.111 | 0.222 | 0.222 | 0.111 |
| TTAAKLMVV | 0.111 | 0 | 0.222 | 0.667 | 0.556 | 0 | 0 | 0.111 | 0.889 | 0.111 | 0.222 | 0.222 | 0.111 |
| NIIPLTTAAK | 0.1 | 0 | 0.2 | 0.6 | 0.6 | 0 | 0 | 0.1 | 0.8 | 0.3 | 0.2 | 0.2 | 0 |
| VENPDILRVY | 0.1 | 0.2 | 0.1 | 0.5 | 0.5 | 0.1 | 0.2 | 0.1 | 0.5 | 0.3 | 0.2 | 0 | 0 |
| RVYANLGER | 0.222 | 0.111 | 0.111 | 0.444 | 0.444 | 0.111 | 0.111 | 0.222 | 0.333 | 0.333 | 0.222 | 0 | 0 |
| DTDFVNEFY | 0 | 0.333 | 0.222 | 0.333 | 0.111 | 0.333 | 0.333 | 0 | 0.444 | 0.111 | 0.444 | 0.111 | 0 |
| YVFTGYRVTK | 0.2 | 0 | 0.4 | 0.4 | 0.3 | 0.3 | 0 | 0.2 | 0.5 | 0.2 | 0.3 | 0.2 | 0 |
| KVNSTLEQY | 0.111 | 0.111 | 0.444 | 0.222 | 0.222 | 0.111 | 0.111 | 0.111 | 0.333 | 0.222 | 0.444 | 0.222 | 0 |
| IPARARVECF | 0.2 | 0.1 | 0.1 | 0.6 | 0.5 | 0.1 | 0.1 | 0.2 | 0.7 | 0.3 | 0.1 | 0 | 0.1 |
| KSAQCFKMF | 0.222 | 0 | 0.333 | 0.444 | 0.111 | 0.222 | 0 | 0.222 | 0.556 | 0.222 | 0.222 | 0.111 | 0.222 |
| KSAQCFKMFY | 0.2 | 0 | 0.4 | 0.4 | 0.1 | 0.3 | 0 | 0.2 | 0.5 | 0.2 | 0.2 | 0.1 | 0.2 |
| DAPAHISTI | 0.111 | 0.111 | 0.222 | 0.556 | 0.556 | 0 | 0.111 | 0.111 | 0.667 | 0.222 | 0.333 | 0.222 | 0 |
| NTVIWDYKR | 0.222 | 0.111 | 0.222 | 0.333 | 0.222 | 0.222 | 0.111 | 0.222 | 0.444 | 0.333 | 0.222 | 0.111 | 0 |
| KYTQLCQYL | 0.111 | 0 | 0.667 | 0.222 | 0.222 | 0.222 | 0 | 0.111 | 0.444 | 0.111 | 0.333 | 0.111 | 0.111 |
| EILPVSMTK | 0.111 | 0.111 | 0.222 | 0.556 | 0.444 | 0 | 0.111 | 0.111 | 0.667 | 0.222 | 0.333 | 0.222 | 0.111 |
| ALNTLVKQL | 0.111 | 0 | 0.222 | 0.556 | 0.556 | 0 | 0 | 0.111 | 0.667 | 0.222 | 0.222 | 0.111 | 0 |
| NAQALNTLV | 0 | 0 | 0.222 | 0.556 | 0.556 | 0 | 0 | 0 | 0.667 | 0.222 | 0.222 | 0.111 | 0 |
| FTSDYYQLY | 0 | 0.111 | 0.667 | 0.222 | 0.111 | 0.444 | 0.111 | 0 | 0.333 | 0 | 0.444 | 0.222 | 0 |
| YYQLYSTQL | 0 | 0 | 0.778 | 0.222 | 0.222 | 0.333 | 0 | 0 | 0.333 | 0 | 0.444 | 0.222 | 0 |
| KLLEQWNLV | 0.111 | 0.111 | 0.111 | 0.556 | 0.444 | 0.111 | 0.111 | 0.111 | 0.556 | 0.222 | 0.222 | 0 | 0 |
| LVIGFLFLTW | 0 | 0 | 0.1 | 0.9 | 0.6 | 0.3 | 0 | 0 | 0.9 | 0 | 0.2 | 0.1 | 0 |
| SSDNIALLV | 0 | 0.111 | 0.222 | 0.556 | 0.556 | 0 | 0.111 | 0 | 0.556 | 0.111 | 0.333 | 0.222 | 0 |
| SSSDNIALL | 0 | 0.111 | 0.333 | 0.444 | 0.444 | 0 | 0.111 | 0 | 0.444 | 0.111 | 0.444 | 0.333 | 0 |
| GYYRRATRR | 0.444 | 0 | 0.333 | 0.222 | 0.222 | 0.222 | 0 | 0.444 | 0.222 | 0.444 | 0.222 | 0.111 | 0 |
| IGYYRRATR | 0.333 | 0 | 0.333 | 0.333 | 0.333 | 0.222 | 0 | 0.333 | 0.333 | 0.333 | 0.222 | 0.111 | 0 |
Table 6.
List of physico-chemical properties of MHC-II restricted T-cell epitopes.
| MHC-I restricted T-cell epitopes | Positively charged | Negatively charged | Polarity | Non polarity | Aliphaticity | Aromaticity | Acidicity | Basicity | Hydrophobicity | Hydrophilicity | Neutral | Hydroxylic | Sulphur content |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FCAGSTFISDEVARD | 0.067 | 0.2 | 0.267 | 0.467 | 0.333 | 0.133 | 0.2 | 0.067 | 0.533 | 0.067 | 0.467 | 0.2 | 0.067 |
| DTFCAGSTFISDEVA | 0 | 0.2 | 0.333 | 0.467 | 0.333 | 0.133 | 0.2 | 0 | 0.6 | 0 | 0.533 | 0.267 | 0.067 |
| VGNICYTPSKLIEYT | 0.067 | 0.067 | 0.4 | 0.4 | 0.4 | 0.133 | 0.067 | 0.067 | 0.533 | 0.2 | 0.333 | 0.2 | 0.067 |
| ASAVVLLILMTARTV | 0.067 | 0 | 0.2 | 0.733 | 0.667 | 0 | 0 | 0.067 | 0.867 | 0.067 | 0.2 | 0.2 | 0.067 |
| LNIIPLTTAAKLMVV | 0.067 | 0 | 0.133 | 0.733 | 0.667 | 0 | 0 | 0.067 | 0.867 | 0.2 | 0.133 | 0.133 | 0.067 |
| ILRVYANLGERVRQA | 0.2 | 0.067 | 0.133 | 0.533 | 0.533 | 0.067 | 0.067 | 0.2 | 0.467 | 0.267 | 0.2 | 0 | 0 |
| RNRDVDTDFVNEFYA | 0.133 | 0.267 | 0.133 | 0.333 | 0.2 | 0.2 | 0.267 | 0.133 | 0.4 | 0.267 | 0.333 | 0.067 | 0 |
| YRNRDVDTDFVNEFY | 0.333 | 0.067 | 0.267 | 0.333 | 0.133 | 0.2 | 0.067 | 0.333 | 0.4 | 0.333 | 0.2 | 0.067 | 0.133 |
| NYVFTGYRVTKNSKV | 0.2 | 0 | 0.333 | 0.333 | 0.267 | 0.2 | 0 | 0.2 | 0.4 | 0.333 | 0.267 | 0.2 | 0 |
| ECFDKFKVNSTLEQY | 0.133 | 0.2 | 0.333 | 0.267 | 0.133 | 0.2 | 0.2 | 0.133 | 0.4 | 0.2 | 0.4 | 0.133 | 0.067 |
| KCSRIIPARARVECF | 0.267 | 0.067 | 0.2 | 0.467 | 0.4 | 0.067 | 0.067 | 0.267 | 0.6 | 0.333 | 0.133 | 0.067 | 0.133 |
| AHKDKSAQCFKMFYK | 0.333 | 0.067 | 0.267 | 0.333 | 0.133 | 0.2 | 0.067 | 0.333 | 0.4 | 0.333 | 0.2 | 0.067 | 0.133 |
| IWDYKRDAPAHISTI | 0.2 | 0.133 | 0.2 | 0.467 | 0.4 | 0.133 | 0.133 | 0.2 | 0.533 | 0.267 | 0.267 | 0.133 | 0 |
| WDYKRDAPAHISTIG | 0.2 | 0.133 | 0.2 | 0.467 | 0.4 | 0.133 | 0.133 | 0.2 | 0.467 | 0.267 | 0.333 | 0.133 | 0 |
| PKGIMMNVAKYTQLC | 0.133 | 0 | 0.267 | 0.533 | 0.4 | 0.067 | 0 | 0.133 | 0.6 | 0.267 | 0.2 | 0.067 | 0.2 |
| PVSMTKTSVDCTMYI | 0.067 | 0.067 | 0.467 | 0.4 | 0.267 | 0.067 | 0.067 | 0.067 | 0.667 | 0.133 | 0.4 | 0.333 | 0.2 |
| TEILPVSMTKTSVDC | 0.067 | 0.133 | 0.4 | 0.4 | 0.333 | 0 | 0.133 | 0.067 | 0.667 | 0.133 | 0.467 | 0.333 | 0.133 |
| QDVVNQNAQALNTLV | 0 | 0.067 | 0.267 | 0.467 | 0.467 | 0 | 0.067 | 0 | 0.533 | 0.2 | 0.333 | 0.067 | 0 |
| DVVNQNAQALNTLVK | 0.067 | 0.067 | 0.2 | 0.467 | 0.467 | 0 | 0.067 | 0.067 | 0.533 | 0.267 | 0.267 | 0.067 | 0 |
| LHSYFTSDYYQLYST | 0.067 | 0.067 | 0.667 | 0.2 | 0.133 | 0.333 | 0.067 | 0.067 | 0.333 | 0.067 | 0.467 | 0.333 | 0 |
| HSYFTSDYYQLYSTQ | 0.067 | 0.067 | 0.733 | 0.133 | 0.067 | 0.333 | 0.067 | 0.067 | 0.267 | 0.067 | 0.533 | 0.333 | 0 |
| WNLVIGFLFLTWICL | 0 | 0 | 0.133 | 0.8 | 0.533 | 0.267 | 0 | 0 | 0.867 | 0.067 | 0.133 | 0.067 | 0.067 |
| NYKLNTDHSSSSDNI | 0.133 | 0.133 | 0.4 | 0.133 | 0.133 | 0.067 | 0.133 | 0.133 | 0.2 | 0.333 | 0.467 | 0.333 | 0 |
| IGYYRRATRRIRGGD | 0.333 | 0.067 | 0.2 | 0.4 | 0.4 | 0.133 | 0.067 | 0.333 | 0.267 | 0.333 | 0.333 | 0.067 | 0 |
Table 7.
List of physico-chemical properties of B-cell epitopes.
| B-cell T-cell epitopes | Positively charged | Negatively charged | Polarity | Non polarity | Aliphaticity | Aromaticity | Acidicity | Basicity | Hydrophobicity | Hydrophilicity | Neutral | Hydroxylic | Sulphur content |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDTFCAGSTFISDEVA | 0 | 0.188 | 0.375 | 0.438 | 0.312 | 0.125 | 0.188 | 0 | 0.625 | 0 | 0.5 | 0.25 | 0.125 |
| AVGNICYTPSKLIEYT | 0.062 | 0.062 | 0.375 | 0.438 | 0.438 | 0.125 | 0.062 | 0.062 | 0.562 | 0.188 | 0.312 | 0.188 | 0.062 |
| LILMTARTVYDDGARR | 0.188 | 0.125 | 0.188 | 0.5 | 0.438 | 0.062 | 0.125 | 0.188 | 0.562 | 0.188 | 0.312 | 0.125 | 0.062 |
| SGFKLKDCVMYASAVV | 0.125 | 0.062 | 0.25 | 0.562 | 0.438 | 0.125 | 0.062 | 0.125 | 0.562 | 0.125 | 0.25 | 0.125 | 0.125 |
| LNIIPLTTAAKLMVVI | 0.062 | 0 | 0.125 | 0.75 | 0.688 | 0 | 0 | 0.062 | 0.875 | 0.188 | 0.125 | 0.125 | 0.062 |
| YDFVENPDILRVYANL | 0.062 | 0.188 | 0.125 | 0.5 | 0.438 | 0.188 | 0.188 | 0.062 | 0.5 | 0.25 | 0.188 | 0 | 0 |
| DILRVYANLGERVRQA | 0.188 | 0.125 | 0.125 | 0.5 | 0.5 | 0.062 | 0.125 | 0.188 | 0.438 | 0.25 | 0.25 | 0 | 0 |
| HRLYECLYRNRDVDTD | 0.25 | 0.25 | 0.25 | 0.188 | 0.188 | 0.125 | 0.25 | 0.25 | 0.312 | 0.312 | 0.312 | 0.062 | 0.062 |
| YRNRDVDTDFVNEFYA | 0.125 | 0.25 | 0.188 | 0.312 | 0.188 | 0.25 | 0.25 | 0.125 | 0.375 | 0.25 | 0.312 | 0.062 | 0 |
| SWEVGKPRPPLNRNYV | 0.188 | 0.062 | 0.125 | 0.5 | 0.438 | 0.125 | 0.062 | 0.188 | 0.438 | 0.5 | 0.188 | 0.062 | 0 |
| PPLNRNYVFTGYRVTK | 0.188 | 0 | 0.25 | 0.438 | 0.375 | 0.188 | 0 | 0.188 | 0.5 | 0.438 | 0.188 | 0.125 | 0 |
| CSRIIPARARVECFDK | 0.25 | 0.125 | 0.188 | 0.438 | 0.375 | 0.062 | 0.125 | 0.25 | 0.562 | 0.312 | 0.188 | 0.062 | 0.125 |
| HKDKSAQCFK | 0.4 | 0.1 | 0.3 | 0.2 | 0.1 | 0.1 | 0.1 | 0.4 | 0.3 | 0.4 | 0.3 | 0.1 | 0.1 |
| ANTVIWDYKRDAPAHI | 0.188 | 0.125 | 0.125 | 0.5 | 0.438 | 0.125 | 0.125 | 0.188 | 0.562 | 0.312 | 0.188 | 0.062 | 0 |
| PKGIMMNVAKYTQLCQ | 0.125 | 0 | 0.312 | 0.5 | 0.375 | 0.062 | 0 | 0.125 | 0.562 | 0.25 | 0.25 | 0.062 | 0.188 |
| LPVSMTKTSVDCTMYI | 0.062 | 0.062 | 0.438 | 0.438 | 0.312 | 0.062 | 0.062 | 0.062 | 0.688 | 0.125 | 0.375 | 0.312 | 0.188 |
| NQNAQALNTLVKQLSS | 0.062 | 0 | 0.375 | 0.375 | 0.375 | 0 | 0 | 0.062 | 0.438 | 0.25 | 0.375 | 0.188 | 0 |
| LHSYFTSDYYQLYSTQ | 0.062 | 0.062 | 0.688 | 0.188 | 0.125 | 0.312 | 0.062 | 0.062 | 0.312 | 0.062 | 0.5 | 0.312 | 0 |
| LVIGFLFLTWICLLQF | 0 | 0 | 0.188 | 0.812 | 0.562 | 0.25 | 0 | 0 | 0.875 | 0 | 0.188 | 0.062 | 0.062 |
| YKLNTDHSSSSDNIAL | 0.125 | 0.125 | 0.375 | 0.25 | 0.25 | 0.062 | 0.125 | 0.125 | 0.312 | 0.25 | 0.438 | 0.312 | 0 |
| GYYRRATRRIRGGDGK | 0.375 | 0.062 | 0.188 | 0.375 | 0.375 | 0.125 | 0.062 | 0.375 | 0.188 | 0.375 | 0.375 | “0.062 “ | 0 |
3.5. Study of docking with Ramachandran plot and Z-score along with population coverage
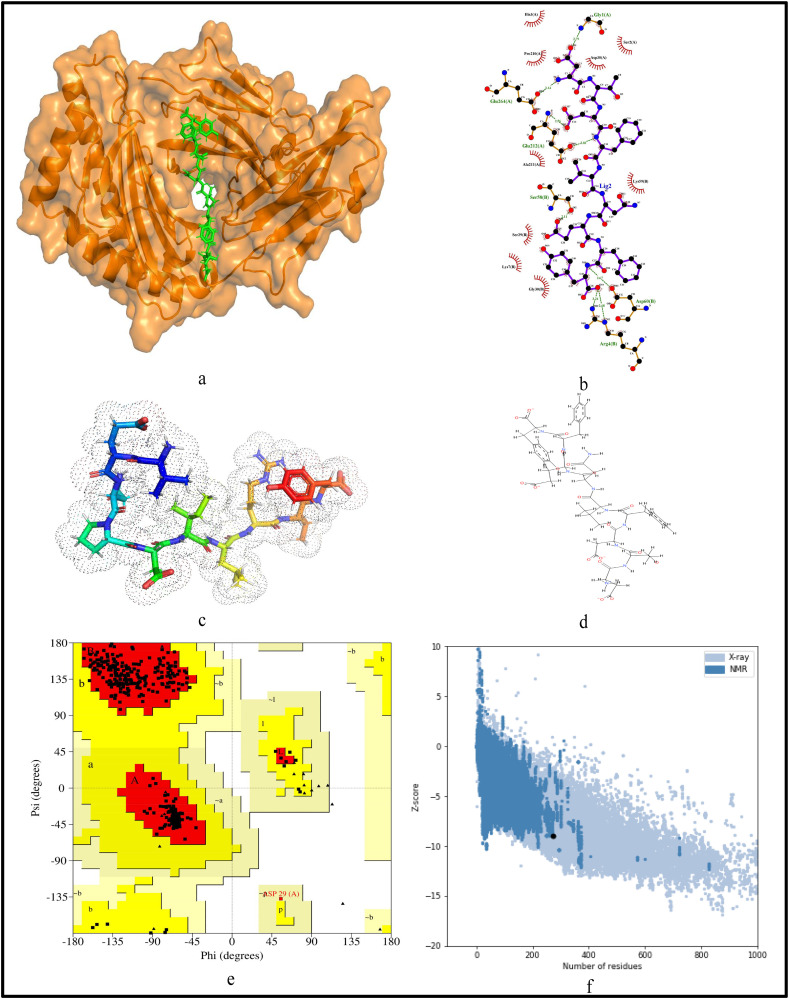
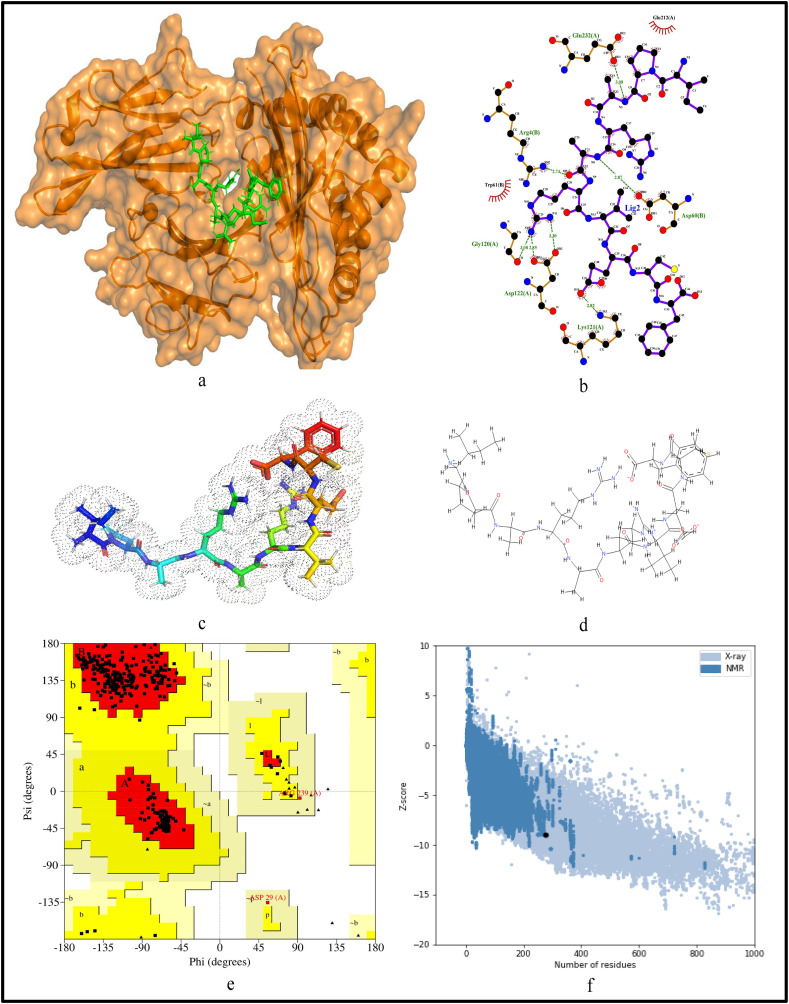
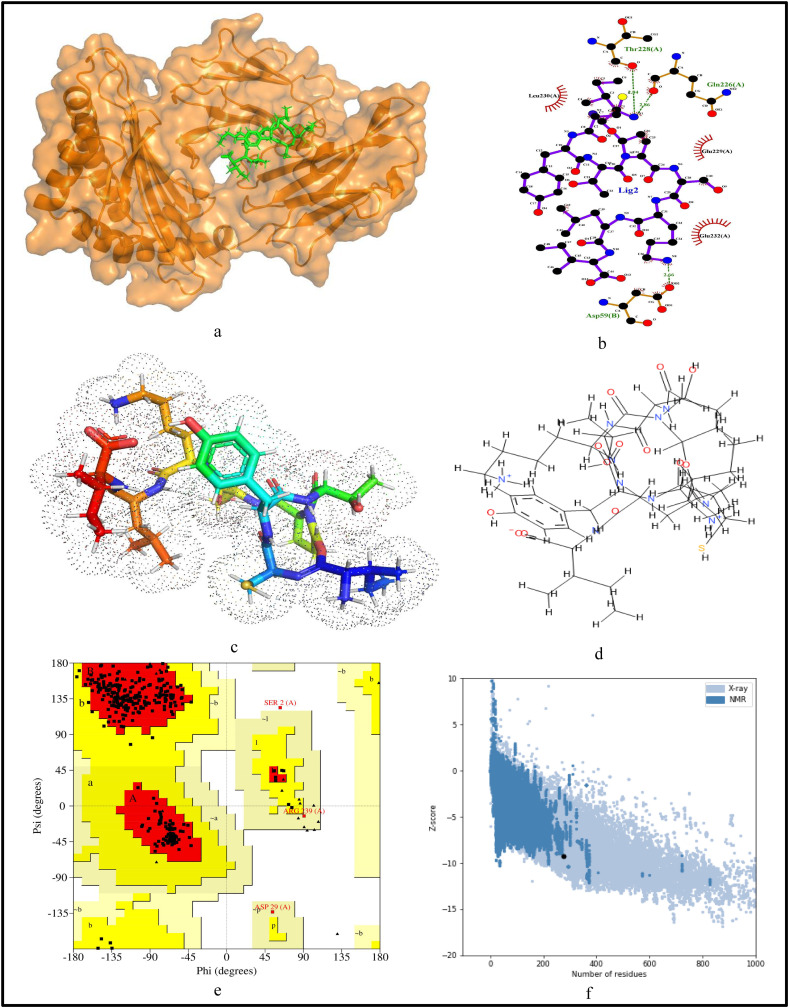
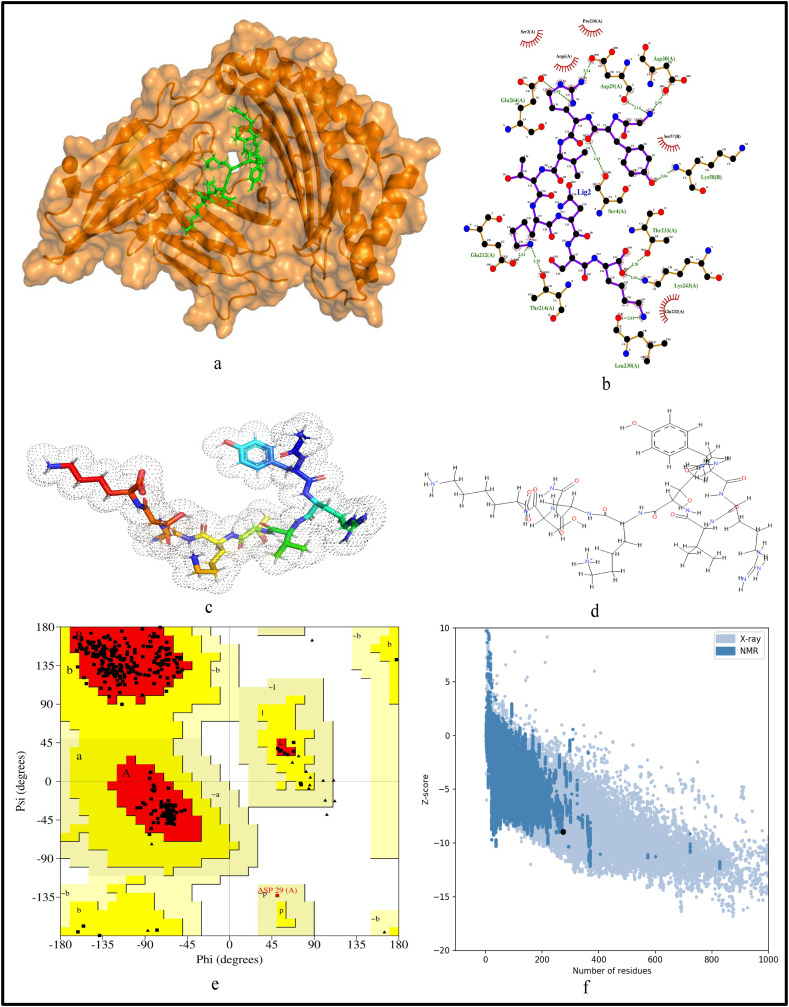
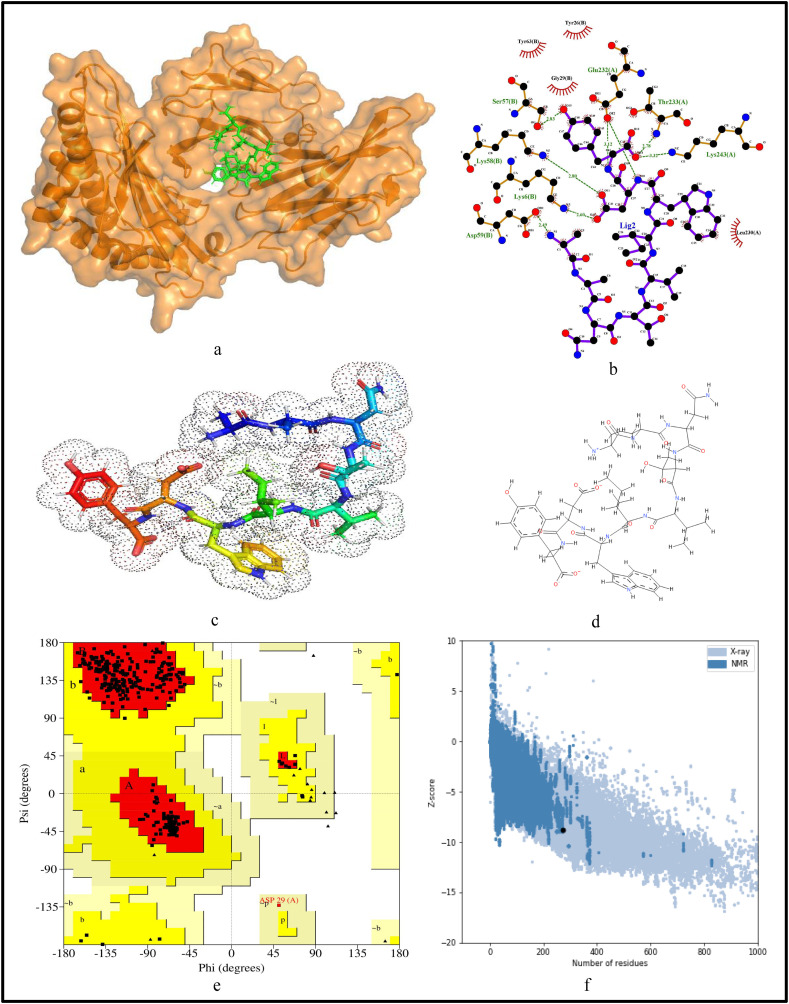
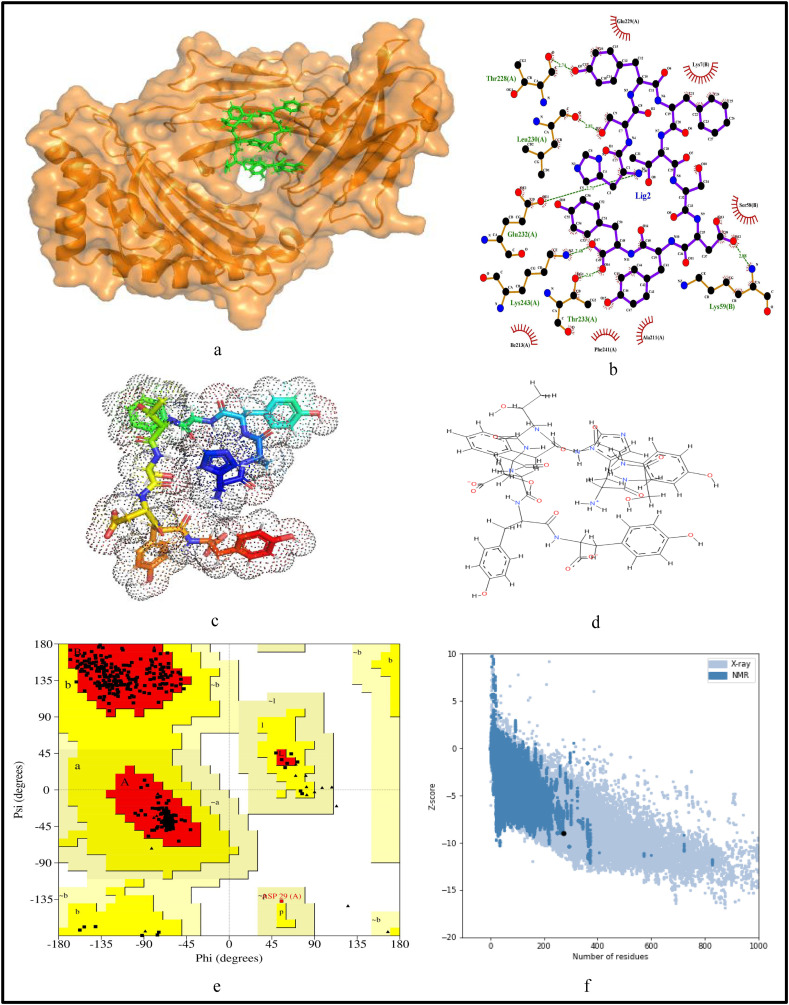
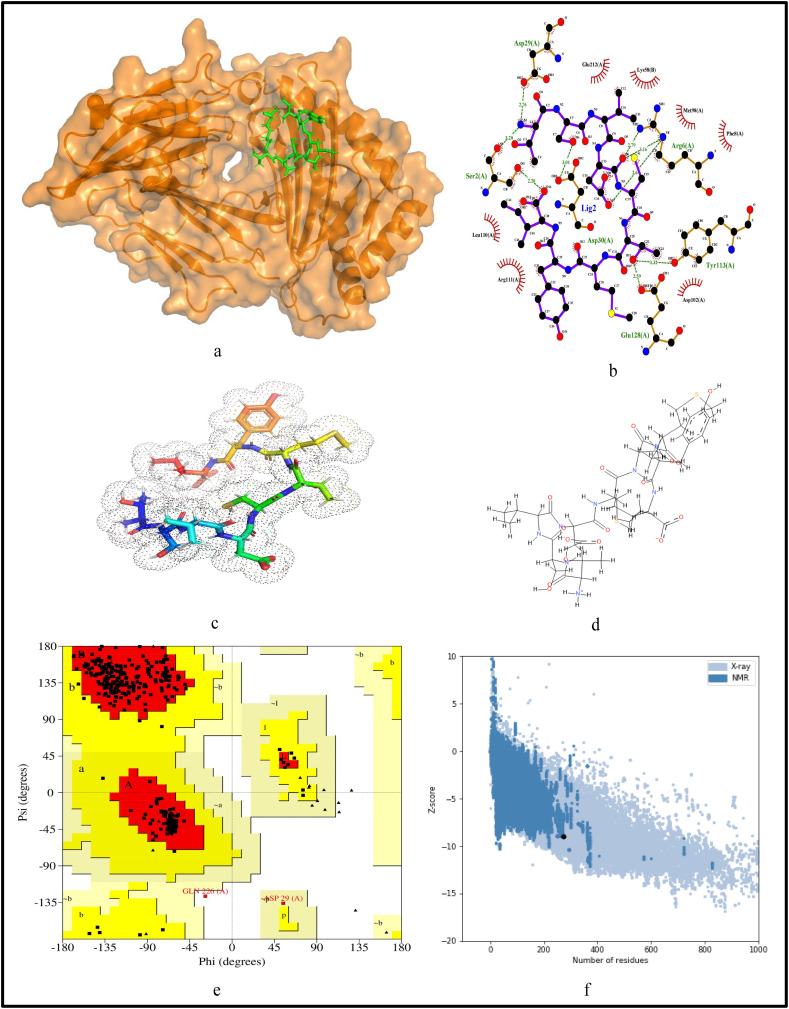
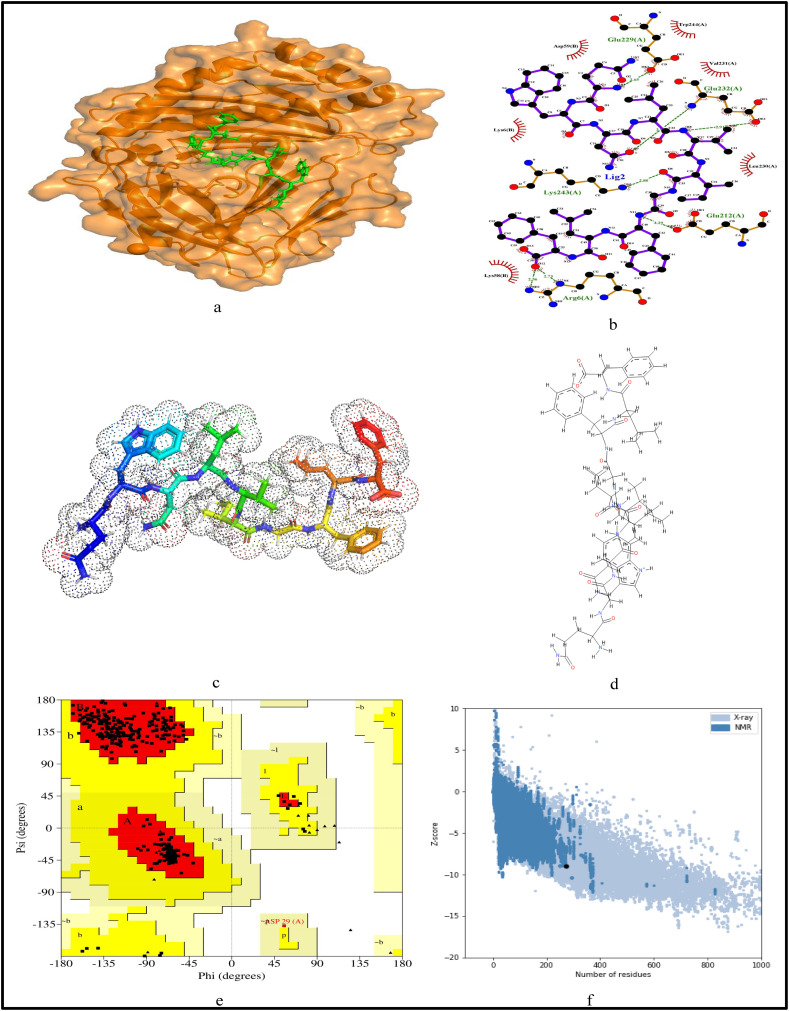
For further validation of the identified MHC-I and MHC-II restricted T-cell epitopes, their conformational 2D non-covalent structures are studied using LigPlot+. To identify the stable binding interaction of each epitope allele pair of the most immunogenic and antigenic epitopes of the 17 CnRs, molecular docking is evaluated using Autodock Vina. For this purpose, the crystal structures of the HLA protein molecule are retrieved from the RCSB Protein Data Bank in PDB format. The docked PDB structures of MHC-I and MHC-II restricted T-cell epitopes (as complex) are respectively reported in supplementary Tables S2 and S3. For the identification of binding energy at the binding groove of alleles with an epitope, the space box centre is set at (0, 0, and 0) for X, Y and Z axes respectively. The size is set at 40 for each of the X, Y and Z dimensions and these analysis are performed at a 0.964 spacing parameter. The finest model was selected by higher binding affinity i.e. lowest docking score generated through Autodock Vina. Moreover, PyMod 3 and ProSA server are used to generate the Ramachandran plot and Z-score respectively. The results of docking and Z-score along with the respective PDB ID13 are given in Table 8 . The results for DTDFVNEFY and IPARARVECF which are the most immunogenic and antigenic MHC-I restricted T-cell epitopes while VGNICYTPSKLIEYT, NYVFTGYRVTKNSKV, IWDYKRDAPAHISTI, LHSYFTSDYYQLYST, TEILPVSMTKTSVDC and WNLVIGFLFLTWICL which are the most immunogenic and antigenic MHC-II restricted T-cell epitopes are shown respectively in Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11, Fig. 12 . In these figures, (a) represents the docking structure of the epitopes as obtained from Autodock Vina where for MHC-I the docking scores are −7.891 and − 8.185 and that for MHC-II the scores are −7.002, −7.170, −7.100, −7.960, −7.269 and − 9.020. It is to be noted that a low docking score shows the efficacy of the identified epitopes as probable vaccine candidates, (b) shows the 2D binding representation of the epitopes with their respective HLA alleles, (c) shows the 3D structures of the identified epitopes, (d) represents the chemical structures of the identified epitopes obtained from ChemSketch, (e) shows the stereochemical quality of the structure through Ramachandran plot which has been evaluated using PyMod 3 where the residues are shown for most favoured regions, additional allowed region, the generously allowed region and 1.3% in the disallowed regions and (f) shows the Z-Score of the identified epitopes where negative values of −8.02 and − 9.72 for MHC-I and − 8.77, −9.01, −8.92, −9.13, −9.42 and − 9.14 for MHC-II verify the stability of the structures of the identified epitopes. Similar structural based evaluation is done for the most immunogenic and antigenic MHC-I and MHC-II restricted T-cell epitopes for the rest of the CnRs and shown in Figs. S1-S46. For the identified B-cell epitopes, the visualization is realised through 3D and chemical structures as shown in Figs. S47-S67. Furthermore, we have also reported the population coverage of the identified MHC-I and MHC-II restricted T-cell epitopes using the IEDB population coverage analysis tool14 in Table 9 . In the table, coverage refers to the projected population coverage, average hit is average number of epitope hits/HLA combinations as recognised by the population and pc90 is the minimum number of epitope hits/HLA combinations as recognised by 90% of the population. For example, coverage, average hit and pc90 of MHC-I restricted T-cell epitopes for World are 86.51%, 3.2 and 0.74 respectively while for MHC-II restricted T-cell epitopes, these values are 44.51%, 0.77 and 0.18 respectively. It is to be noted that MHC-II is present in only three types of cells viz. dendritic cells, macrophages and B cells along with MHC-I. Moreover for MHC-II, HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:01/DQB1*05:01, HLA-DPA1*01:03/DPB1*04:01, HLA-DRB3*01:01, HLA-DPA1*02:01/DPB1*14:01, HLA-DRB3*02:02, HLA-DPA1*02:01/DPB1*05:01 and HLA- DQA1*03:01/DQB1*03:02 alleles are not available and thus not included in the calculation of population coverage.
Table 8.
Docking and Z-scores of most Immunogenic and Antigenic MHC-I and MHC-II restricted T-cell epitopes for 17 CnRs.
| MHC-I epitopes | PDB ID | Score from AutoDock Vina | Z-Score | MHC-II epitopes | PDB ID | Score from Autodock Vina | Z-Score |
|---|---|---|---|---|---|---|---|
| STFISDEVAR | 4HWZ:A | −7.905 | −9.09 | FCAGSTFISDEVARD | 2Q6W:B | −8.081 | −8.97 |
| DTFCAGSTF | 2HN7:A | −7.164 | −8.95 | DTFCAGSTFISDEVA | 4Z7U:A; 4Z7U:B | −7.712 | −9.27 |
| YTPSKLIEY | 2HN7:A | −9.143 | −8.95 | VGNICYTPSKLIEYT | 4I5B:B | −7.002 | −8.77 |
| FSAVGNICY | 3BO8:A | −7.356 | −8.98 | ||||
| TVYDDGARR | 4HWZ:A | −8.450 | −9.09 | ASAVVLLILMTARTV | 2G9H:B | −7.899 | −8.97 |
| ILMTARTVY | 1XR9:A | −7.888 | −8.93 | ||||
| TTAAKLMVV | 4HX1:A | −7.704 | −9.42 | LNIIPLTTAAKLMVV | 6CPN:B | −7.764 | −8.95 |
| NIIPLTTAAK | 4HWZ:A | −8.332 | −9.09 | ||||
| VENPDILRVY | 1N2R:A | −7.603 | −8.95 | ILRVYANLGERVRQA | 4H25:B | −7.030 | −8.71 |
| RVYANLGER | 3RL1:A | −7.224 | −8.95 | ||||
| DTDFVNEFY | 3BO8:A | −7.891 | −8.02 | RNRDVDTDFVNEFYA | 1UVQ:A; 1UVQ:B | −8.920 | −8.12 |
| YRNRDVDTDFVNEFY | 1UVQ:A; 1UVQ:B | −7.920 | −8.43 | ||||
| YVFTGYRVTK | 4HWZ:A | −7.902 | −9.12 | NYVFTGYRVTKNSKV | 4I5B:B | −7.170 | −9.01 |
| KVNSTLEQY | 1X7Q:A | −7.741 | −9.01 | ECFDKFKVNSTLEQY | 4H25:B | −7.742 | −8.72 |
| IPARARVECF | 4U1H:A | −8.185 | −9.72 | KCSRIIPARARVECF | 3WEX:A; 3WEX:B | −7.940 | −8.27 |
| KSAQCFKMF | 3VRI:A | −7.390 | −8.91 | AHKDKSAQCFKMFYK | 3WEX:A; 3WEX:B | −7.170 | −9.23 |
| KSAQCFKMFY | 3VRI:A | −7.041 | −8.87 | ||||
| DAPAHISTI | 1E27:A | −7.912 | −8.44 | IWDYKRDAPAHISTI | 2G9H:B | −7.100 | −8.92 |
| NTVIWDYKR | 4HWZ:A | −7.471 | −8.18 | WDYKRDAPAHISTIG | 2G9H:B | −7.899 | −8.43 |
| KYTQLCQYL | 3WL9:A | −7.971 | −8.74 | PKGIMMNVAKYTQLC | 4H25:B | −7.167 | −8.18 |
| EILPVSMTK | 4HWZ:A | −7.771 | −9.09 | PVSMTKTSVDCTMYI | 2G9H:B | −7.186 | −8.98 |
| TEILPVSMTKTSVDC | 6CPN:B | −7.269 | −9.42 | ||||
| ALNTLVKQL | 3OX8:A | −8.424 | −9.30 | QDVVNQNAQALNTLV | 4MDJ:B | −7.932 | −9.21 |
| NAQALNTLV | 1E27:A | −7.933 | −9.21 | DVVNQNAQALNTLVK | 4MDJ:B | −7.751 | −9.42 |
| FTSDYYQLY | 3BO8:A | −8.812 | −8.12 | LHSYFTSDYYQLYST | 3WEX:A; 3WEX:B | −7.960 | −9.13 |
| YYQLYSTQL | 3WL9:A | −7.388 | −8.89 | HSYFTSDYYQLYSTQ | 3WEX:A; 3WEX:B | −7.240 | −9.25 |
| KLLEQWNLV | 3UTQ:A | −7.541 | −8.43 | WNLVIGFLFLTWICL | 3WEX:A; 3WEX:B | −9.020 | −9.14 |
| LVIGFLFLTW | 3VRI:A | −7.442 | −9.01 | ||||
| SSDNIALLV | 3BO8:A | −8.917 | −9.71 | NYKLNTDHSSSSDNI | 6ATF:B | −8.660 | −8.72 |
| SSSDNIALL | 4HX1:A | −7.922 | −8.70 | ||||
| GYYRRATRR | 3RL1:A | −8.977 | −8.60 | IGYYRRATRRIRGGD | 1A6A:B | −9.033 | −8.97 |
| IGYYRRATR | 3RL1:A | −7.618 | −8.60 |
Fig. 5.
Structural analysis for the most immunogenic MHC-I restricted T-cell epitope “DTDFVNEFY” in 17 CnRs (a) Docking structure of MHC-I restricted T-cell epitope (b) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (c) 3D structure of the epitope (d) Chemical structure of the epitope (e) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frames and (f) Z-Score plot.
Fig. 6.
Structural analysis for the most antigenic MHC-I restricted T-cell epitope “IPARARVECFF” in 17 CnRs (a) Docking structure of MHC-I restricted T-cell epitope (b) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (c) 3D structure of the epitope (d) Chemical structure of the epitope (e) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frames and (f) Z-Score plot.
Fig. 7.
Structural analysis for the most immunogenic MHC-II restricted T-cell epitope “VGNICYTPSKLIEYT” in 17 CnRs (a) Docking structure of MHC-II restricted T-cell epitope (b) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (c) 3D structure of the epitope (d) Chemical structure of the epitope (e) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frames and (f) Z-Score plot.
Fig. 8.
Structural analysis for the most immunogenic MHC-II restricted T-cell epitope “NYVFTGYRVTKNSKV” in 17 CnRs (a) Docking structure of MHC-II restricted T-cell epitope (b) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (c) 3D structure of the epitope (d) Chemical structure of the epitope (e) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frames and (f) Z-Score plot.
Fig. 9.
Structural analysis for the most immunogenic MHC-II restricted T-cell epitope “IWDYKRDAPAHISTI” in 17 CnRs (a) Docking structure of MHC-II restricted T-cell epitope (b) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (c) 3D structure of the epitope (d) Chemical structure of the epitope (e) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frames and (f) Z-Score plot.
Fig. 10.
Structural analysis for the most immunogenic MHC-II restricted T-cell epitope “LHSYFTSDYYQLYST” in 17 CnRs (a) Docking structure of MHC-II restricted T-cell epitope (b) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (c) 3D structure of the epitope (d) Chemical structure of the epitope (e) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frames and (f) Z-Score plot.
Fig. 11.
Structural analysis for the most antigenic MHC-II restricted T-cell epitope “TEILPVSMTKTSVDC” in 17 CnRs (a) Docking structure of MHC-II restricted T-cell epitope (b) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (c) 3D structure of the epitope (d) Chemical structure of the epitope (e) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frames and (f) Z-Score plot.
Fig. 12.
Structural analysis for the most antigenic MHC-II restricted T-cell epitope “WNLVIGFLFLTWICL” in 17 CnRs (a) Docking structure of MHC-II restricted T-cell epitope (b) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (c) 3D structure of the epitope (d) Chemical structure of the epitope (e) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frames and (f) Z-Score plot.
Table 9.
Population coverage of identified MHC-I and MHC-II restricted T-cell epitopes.
| Population/Area | MHC-I |
MHC-II |
MHC-I,II combined |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Coverage | Average Hit | PC90 | Coverage | Average Hit | PC90 | Coverage | Average Hit | PC90 | |
| Central Africa | 55.58% | 1.2 | 0.23 | 43.39% | 0.79 | 0.18 | 74.85% | 1.95 | 0.4 |
| Central America | 1.4% | 0.01 | 0.1 | 40.68% | 0.83 | 0.17 | 41.51% | 0.85 | 0.17 |
| China | 67.08% | 2.05 | 0.3 | 22.09% | 0.37 | 0.13 | 74.3% | 1.94 | 0.39 |
| East Africa | 66.54% | 1.71 | 0.3 | 47.58% | 0.74 | 0.19 | 82.41% | 2.41 | 0.57 |
| East Asia | 90.57% | 3.78 | 1.05 | 39.35% | 0.69 | 0.16 | 94.28% | 3.48 | 1.51 |
| Europe | 92.32% | 3.69 | 1.12 | 50.99% | 0.87 | 0.2 | 96.19% | 4.2 | 1.55 |
| India | 65% | 2.66 | 0.29 | 48.81% | 0.87 | 0.2 | 81.96% | 3.18 | 0.55 |
| North Africa | 73.32% | 2.36 | 0.37 | 45.93% | 0.88 | 0.18 | 85.36% | 3.06 | 0.68 |
| North America | 88.88% | 3.28 | 0.9 | 48.42% | 0.84 | 0.19 | 94.21% | 3.66 | 1.31 |
| Northeast Asia | 66.82% | 2.02 | 0.3 | 22.09% | 0.37 | 0.13 | 74.11% | 1.9 | 0.39 |
| Oceania | 75.87% | 3.1 | 0.41 | 24.34% | 0.29 | 0.13 | 81.74% | 2.35 | 0.55 |
| South America | 77.81% | 3.11 | 0.45 | 39.08% | 0.76 | 0.16 | 86.26% | 3.38 | 0.73 |
| Southeast Asia | 70.97% | 2.4 | 0.34 | 19.56% | 0.29 | 0.12 | 76.64% | 1.89 | 0.43 |
| Southwest Asia | 74.72% | 2.53 | 0.4 | 27.29% | 0.46 | 0.14 | 80.72% | 2.75 | 0.52 |
| United States | 88.87% | 3.26 | 0.9 | 48.66% | 0.85 | 0.19 | 94.23% | 3.65 | 1.31 |
| West Africa | 65.7% | 1.91 | 0.29 | 37.69% | 0.64 | 0.16 | 78.63% | 2.44 | 0.47 |
| West Indies | 85.79% | 3.07 | 0.7 | 48.8% | 0.83 | 0.2 | 92.55% | 3.54 | 1.18 |
| World | 86.51% | 3.2 | 0.74 | 44.51% | 0.77 | 0.18 | 92.45% | 3.53 | 1.17 |
In this study, we have identified MHC-I and MHC-II restricted T-cell and B-cell epitopes using computational methods and tools for potential vaccine design. To summarise, the main advantages of this work can be listed as (i) genome-wide analysis of 10,664 SARS-CoV-2 genomes from 73 countries around the globe to find the conserved regions and (ii) use of latest tools like PyMod 3, NetMHCpan EL 4.1 and BepiPred 2.0 for computational purposes to identify potential epitopes.
4. Conclusion
Current impact of SARS-CoV-2 is giving rise to more evolutionary approaches towards epitope-based vaccine design. In this paper, we have identified highly immunogenic as well as antigenic T-cell and B-cell epitopes from conserved regions by analysing 10,644 SARS-CoV-2 genomes from 73 countries around the globe. In this regard, we have identified 408 CnRs from the aligned SARS-CoV-2 sequence. These conserved regions are filtered based on the criteria that their lengths should be greater than or equal to 60 nt, their corresponding protein sequences are devoid of any stop codons and BLAST specificity score as query coverage is 100%. As a result, 17 CnRs are obtained belonging to NSP3, NSP4, NSP6, NSP8, RdRp, Helicase, endoRNAse, 2’-O-RMT, Spike glycoprotein, ORF3a protein, Membrane glycoprotein and Nucleocapsid protein. These CnRs are then used to identify the T-cell and B-cell epitopes. Based on their scores, the most immunogenic and antigenic epitopes are then selected for each of these 17 CnRs resulting in 30 MHC-I and 24 MHC-II restricted T-cell epitopes with 14 and 13 unique HLA alleles and 21 B-cell epitopes. Moreover, to judge the relevance of these epitopes, their binding conformation are shown with respect to HLA alleles. Furthermore, their physico-chemical properties are reported along with Ramchandran plots and Z-Scores and the population coverage is shown as well. Thus, the reported epitopes can be considered as potential candidates for the design of epitope-based synthetic vaccine.
Ethics approval and consent to participate
The ethical approval or individual consent was not applicable.
Availability of data and materials
The aligned 10,664 Indian SARS-CoV-2 genomes with the reference sequence and the final results of this work are available at ‘http://www.nitttrkol.ac.in/indrajit/projects/COVID-EpitopeVaccine-Global/’. Moreover, the SARS-CoV-2 genomes used in this work are publicly available at GISAID database.
Consent for publication
Not applicable.
Funding
This work has been partially supported by CRG short term research grant on COVID-19 (CVD/2020/000991) from Science and Engineering Research Board (SERB), Department of Science and Technology, Govt. of India.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
Acknowledgement
We thank all those who have contributed sequences to GISAID database.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104823.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed S., Quadeer A., McKay M. Preliminary identification of potential vaccine targets for the covid-19 coronavirus (sars-cov-2) based on sars-cov immunological studies. Viruses. 2020;12 doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah V., Bose S. Immunoinformatics-aided identification of t cell and b cell epitopes in the surface glycoprotein of 2019-ncov. J. Med. Virol. 2020;92 doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bency J., Helen M. Novel epitope based peptides for vaccine against sars-cov-2 virus: immunoinformatics with docking approach. Int. J. Res. Med. Sci. 2020;8:2385. doi: 10.18203/2320-6012.ijrms20202875. [DOI] [Google Scholar]
- Bhatnager R., Bhasin M., Arora J., Dang A. Epitope based peptide vaccine against sars-cov2: an immune-informatics approach. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1787227. [DOI] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A., Patra P., Ghosh P., Sharma G., Patra B., Lee S.S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (sars-cov-2): Immunoinformatics approach. J. Med. Virol. 2020;92 doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sharma A., Sharma G., Patra P., Mondal N., Patra B., Lee S.S., Chakraborty C. Computer aided novel antigenic epitopes selection from the outer membrane protein sequences of Aeromonas hydrophila and its analyses. Infect. Genet. Evol. 2020;82 doi: 10.1016/j.meegid.2020.104320. [DOI] [PubMed] [Google Scholar]
- Chen H.Z., Tang L.L., Yu X.L., Zhou J., Chang Y.F., Wu X. Bioinformatics analysis of epitope-based vaccine design against the novel sars-cov-2. Infect. Dis. Poverty. 2020;9 doi: 10.1186/s40249-020-00713-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke S.N., Ovsyannikova I.G., Kennedy R.B., Poland G.A. Immunoinformatic identification of b cell and t cell epitopes in the sars-cov-2 proteome. Sci. Rep. 2020;10:14179. doi: 10.1038/s41598-020-70864-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I., Flower D. Vaxijen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8(4) doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to sars-cov-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S., Mateus J., Dan J., Moderbacher C., Rawlings S., Sutherland A., Premkumar L., Jadi R., Marrama D., Silva A., Frazier A., Carlin A., Greenbaum J., Peters B., Krammer F., Smith D., Crotty S., Sette A. Targets of t cell responses to sars-cov-2 coronavirus in humans with covid-19 disease and unexposed individuals. Cell. 2020;181 doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A.K., Khan M.S., Choudhury S., Mukhopadhyay A., Sakshi Rastogi A., Thakur A., Kumari P., Kaur M., Shalu Saini C., Sapehia V., Barkha Patel P.K., Mare K.T., Kumar M. Coronavr: A computational resource and analysis of epitopes and therapeutics for severe acute respiratory syndrome coronavirus-2. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Hoque M., Rahman M., Alam A.S.M., Akther M., Puspo J., Akter S., Sultana M., Crandall K., Hossain M. Genome-wide analysis of sars-cov-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson G., Paiardini A. PyMod 3: a complete suite for structural bioinformatics in PyMOL. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa849. [DOI] [PubMed] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. Bepipred-2.0: improving sequence-based b-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T. Ncb blast: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar T., Narsaria U., Basak S., Deb D., Castiglione F., Mueller D., Srivastava A. A candidate multi-epitope vaccine against sars-cov-2. Sci. Rep. 2020;10:10895. doi: 10.1038/s41598-020-67749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E., Bhattacharya T., Foley B., Hastie K., Parker D., Partridge C., Evans T., Freeman T., Silva C., Mcdanal L., Perez H., Tang A., Wyles M. Tracking changes in sars-cov-2 spike: evidence that d614g increases infectivity of the covid-19 virus. Cell. 2020;182 doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwarteng A., Asiedu E., Sakyi S.A., Asiedu S.O. Targeting the sars-cov2 nucleocapsid protein for potential therapeutics using immuno-informatics and structure-based drug discovery techniques. Biomed. Pharmacother. 2020:132. doi: 10.1016/j.biopha.2020.110914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.X., Lim J., Jazayeri S.D., Poppema S., Poh C.L. Development of multi-epitope peptide-based vaccines against sars-cov-2. Biom. J. 2020 doi: 10.1016/j.bj.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer E. Immunization against infectious disease. Science (New York, N.Y.) 1978;200:902–907. doi: 10.1126/science.347579. [DOI] [PubMed] [Google Scholar]
- Naz A., Shahid F., Butt T., Awan F., Ali A., Malik D. Designing multi-epitope vaccines to combat emerging coronavirus disease 2019 (covid-19) by employing immuno-informatics approach. Front. Immunol. 2020;11:1663. doi: 10.3389/fimmu.2020.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz S., Ahmad S., Walton S., Abbasi S. Multi-epitope based vaccine design against sarcoptes scabiei paramyosin using immunoinformatics approach. J. Mol. Liq. 2020:84–91. doi: 10.1016/j.molliq.2020.114105. [DOI] [Google Scholar]
- Noorimotlagh Z., Karami C., Mirzaee S.A., Kaffashian M., Mami S., Azizi M. Immune and bioinformatics identification of t cell and b cell epitopes in the protein structure of sars-cov-2: a systematic review. Int. Immunopharmacol. 2020;86 doi: 10.1016/j.intimp.2020.106738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E., Wong M.U., Huffman A., He Y. Covid-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande A., Patiyal S., Lathwal A., Arora C., Kaur D., Dhall A., Mishra G., Kaur H., Sharma N., Jain S., Usmani S., Agrawal P., Kumar R., Kumar V., Raghava G. Computing wide range of protein/peptide features from their sequence and structure. bioRxiv. 2019 doi: 10.1101/599126. [DOI] [PubMed] [Google Scholar]
- Parvizpour S., Pourseif M., Razmara J., Rafi M., Omidi Y. Epitope-based vaccine design: a comprehensive overview of bioinformatics approaches. Drug Discov. Today. 2020:25. doi: 10.1016/j.drudis.2020.03.006. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. Ucsf chimera — A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Poran A., Harjanto D., Malloy M., Arieta C., Rothenberg D., Lenkala D., Buuren M., Addona T., Rooney M., Srinivasan L., Gaynor R. Sequencebased prediction of sars-cov-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic t cell epitopes. Genome Med. 2020;12 doi: 10.1186/s13073-020-00767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell A., McCluskey J., Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 2007;6:404–414. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- Rakib A., Saad A., Sami S., Mimi N.J., Chowdhury M., Eva T., Nainu F., Paul A., Shahriar A., Tareq A., Laam N.U., Chakraborty S., Shil S., Mily D.T., Hadda T.B., Almalki F., Emran T. Immunoinformatics-guided design of an epitope-based vaccine against severe acute respiratory syndrome coronavirus 2 spike glycoprotein. Comput. Biol. Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf M. Ligand docking and binding site analysis with pymol and autodock/vina. Int. J. Basic Appl. Sci. 2015;4:168–177. doi: 10.14419/ijbas.v4i2.4123. [DOI] [Google Scholar]
- Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. Netmhcpan-4.1 and netmhciipan-4.0: improved predictions of mhc antigen presentation by concurrent motif deconvolution and integration of ms mhc eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Raghava G. Prediction methods for b-cell epitopes. Methods Mol. Biol. (Clifton, N.J.) 2007;409:387–394. doi: 10.1007/978-1-60327-118-9_29. [DOI] [PubMed] [Google Scholar]
- Sidney J., Dow C., Mothé B., Sette A., Peters B. A systematic assessment of mhc class ii peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Higgins D. Clustal omega. Curr. Protoc. Bioinformatics. 2014;48:3.13.1–3.13.16. doi: 10.1002/0471250953.bi0313s48. [DOI] [PubMed] [Google Scholar]
- Singh A., Thakur M., Sharma L., Chandra K. Designing a multi-epitope peptide based vaccine against sars-cov-2. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-73371-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spessard G.O. Acd labs/logp db 3.5 and chemsketch 3.5. J. Chem. Inf. Comput. Sci. 1998;38:1250–1253. doi: 10.1021/ci980264t. [DOI] [Google Scholar]
- Tamar Y.B., Ruth A. Epitope-based vaccine against influenza. Expert Rev. Vaccines. 2007;6:939–948. doi: 10.1586/14760584.6.6.939. [DOI] [PubMed] [Google Scholar]
- Tosta S.F.O., Passos M.S., Kato R., Salgado A., Jaiswal A.K., Jaiswal X., Soares S.C., Azevedo V., Giovanetti M., Tiwari S., Alcantara L.C.J. Multi-epitope based vaccine against yellow fever virus applying immunoinformatics approaches. J. Biomol. Struct. Dyn. 2020:1–17. doi: 10.1080/07391102.2019.1707120. [DOI] [PubMed] [Google Scholar]
- Vashi Y., Jagrit V., Kumar S. Understanding the b and t cells epitopes of spike protein of severe respiratory syndrome coronavirus-2: A computational way to predict the immunogens. Infect. Genet. Evol. 2020;84:104382. doi: 10.1016/j.meegid.2020.104382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski A.R., Thornton J.M. Ligplot: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tian H., Zhang L., Zhang M., Guo D., Wu W., Zhang X., Kan G.L., Jia L., Huo D., Liu B., Wang X., Sun Y., Wang Q., Yang P., MacIntyre C.R. Reduction of secondary transmission of sars-cov-2 in households by face mask use, disinfection and social distancing: a cohort study in Beijing, China. BMJ Glob. Health. 2020;5 doi: 10.1136/bmjgh-2020-002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M., Sippl M.J. Prosa-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worldometer Coronavirus Disease 2019 (covid-19) Cases in India. 2020. https://www.worldometers.info/coronavirus/country/india/ Accessed: 2020-07-18.
- Yadav P.D., Potdar V., Choudhary M.L., Nyayanit D.A., Agrawal M., Jadhav S.M., Majumdar T.D., Aich A.S., Basu A., Abraham P., Cherian S.S. Full-genome sequences of the first two sars-cov-2 viruses from India. Indian J. Med. Res. 2020;151 doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Chan H.S., Hu Z. Using pymol as a platform for computational drug design. WIREs Comput. Mol. Sci. 2017;7 [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C., Chen H., Chen J., Luo Y., Guo H., Jiang R., Liu M., Chen Y., Shen X., Wang X., Zheng X., Zhao K., Chen Q., Deng F., Liu L.L., Yan B., Zhan F., Wang Y., Xiao G., Shi Z. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The aligned 10,664 Indian SARS-CoV-2 genomes with the reference sequence and the final results of this work are available at ‘http://www.nitttrkol.ac.in/indrajit/projects/COVID-EpitopeVaccine-Global/’. Moreover, the SARS-CoV-2 genomes used in this work are publicly available at GISAID database.