Significance
The mitotic cell cycle (MCC) sustains plant organ growth by producing new cells in the meristems. Multiple mechanisms of gene expression regulation determine the dynamics of gene expression across the successive phases of the cell cycle. At the transcriptional level, only E2F and MYB3R transcription factors (TFs) have been identified as controlling gene expression dynamics in the MCC. In this work, we analyze the transcriptome of cells in the G2/M phases of the cell cycle and identify SCL28, a previously uncharacterized GRAS TF, as a regulator of the MCC in plants.
Keywords: cell cycle, root meristem, Arabidopsis
Abstract
Gene expression is reconfigured rapidly during the cell cycle to execute the cellular functions specific to each phase. Studies conducted with synchronized plant cell suspension cultures have identified hundreds of genes with periodic expression patterns across the phases of the cell cycle, but these results may differ from expression occurring in the context of intact organs. Here, we describe the use of fluorescence-activated cell sorting to analyze the gene expression profile of G2/M cells in the growing root. To this end, we isolated cells expressing the early mitosis cell cycle marker CYCLINB1;1-GFP from Arabidopsis root tips. Transcriptome analysis of these cells allowed identification of hundreds of genes whose expression is reduced or enriched in G2/M cells, including many not previously reported from cell suspension cultures. From this dataset, we identified SCL28, a transcription factor belonging to the GRAS family, whose messenger RNA accumulates to the highest levels in G2/M and is regulated by MYB3R transcription factors. Functional analysis indicates that SCL28 promotes progression through G2/M and modulates the selection of cell division planes.
The mitotic cell cycle (MCC) is a unidirectional sequence of precisely regulated events. During the S phase (“synthesis”), the chromosomes are replicated, while in the M phase (“mitosis”), the replicated chromosomes are segregated between the two daughter cells. These phases are preceded by the G1 and G2 preparative phases, during which cells grow and check for DNA integrity, among other processes. After nuclear division, cytokinesis divides the cytoplasm to yield two daughter cells.
Plants rely on the precise selection of cell division planes to shape their organs during development (1). The phragmoplast, a structure made of cytoskeletal components, membranes, and cell wall-synthetizing enzymes, assembles during cytokinesis the cell plate that partitions the cytoplasm (2). Before that, the preprophase band (PPB), an array of microtubule, actin filaments, and other proteins, predicts the position of the cortical division zone (2). Mutants with defects in PPB organization lose precision in cell division orientation. It has been proposed that the PPB controls division plane orientation during symmetric cell division by stabilizing mitotic spindle orientation (3). Consistent with this hypothesis, pharmacological disturbances of spindle orientation affect division plane selection (4), highlighting the mitotic spindle role during the selection of the orientation of the division plane. The correct functioning of the PPB, mitotic spindle, and phragmoplast is thus critical to define cell division plane orientation and complete mitosis and cytokinesis successfully. For example, mutants in microtubule-binding proteins (5) or in mitosis-specific cell wall-synthetizing enzymes (6) have defects in cytokinesis, yielding misplaced, curved, and/or incomplete cell walls.
Each step of the MCC has its own particular set of protein activities required for the specific molecular processes that occur in each phase. The S phase is enriched in proteins involved in DNA replication, such as histones and DNA polymerases (7). On the other hand, cells in the M phase are depleted for S phase-specific molecules and enriched in those required for chromosome segregation and cytokinesis, such as motor proteins (8) needed for chromosome segregation and cell wall-synthetizing enzymes specialized in de novo cell wall biosynthesis (6). These gene expression profiles must be reconfigured rapidly, as estimates of cell cycle time in plant root meristematic cells are between 15 and 20 h (9). A variety of mechanisms are deployed to modulate gene expression, including protein phosphorylation, ubiquitination, and degradation (10); protein localization (11); and nuclear sequestration of messenger RNAs (mRNAs) (12). In addition to these posttranscriptional mechanisms, regulation at the transcriptional level plays a predominant role in modulating the activity of cell cycle-associated genes (13, 14). This is evidenced by genome-wide expression analysis in Arabidopsis cell cycle-synchronized cell suspension cultures (CSCs) that identified ∼1,100 genes with peaks in transcription in G1, S, G2, or M (15).
Despite this large number of genes with periodic expression patterns, only two families of transcription factors (TFs), E2F and MYB3R, have been identified as controlling gene expression dynamics in the MCC. In plants and animals, E2F TFs have been mainly classified as regulators of the G1/S transition and S phase progression as they control the onset and progression of DNA replication, DNA repair, and chromatin condensation (16–20). The second family of TFs are the R1R2R3-type MYB (MYB3R) proteins. They regulate the expression of many G2/M-specific genes, such as CYCB1;1, CDKB2;1, and KNOLLE, through binding to a specific cis-acting regulatory sequence known as Mitotic Specific Activator (MSA) element (21–24). The Arabidopsis genome contains five genes that encode MYB3R TFs (25). MYB3R4 acts as a transcriptional activator of genes expressed in G2/M with critical roles in mitosis and cytokinesis (22). By contrast, MYB3R3 and MYB3R5 act as transcriptional repressors of a similar set of genes (14, 26). Finally, MYB3R1 seems to act redundantly as an activator or a repressor depending on the developmental context (26). It has been proposed that activator and repressor MYB3Rs, which recognize the same DNA sequences, act in a coordinated rather than in a competitive manner. In this way, activator MYB3Rs are expressed and function during G2/M in meristematic cells, while repressor MYB3Rs repress G2/M-specific genes during G1/S and in postmitotic cells (14). It has been shown that only a fraction of the genes with expression peaks in G2/M are down-regulated in a myb3r4 myb3r1 double mutant (21), indicating that other transcriptional regulators remain to be identified.
Cell cycle-synchronized plant CSCs have been used extensively in genome-wide studies of gene expression dynamics across the phases of the cell cycle (24, 27, 28). It is a concern that the chemicals used for cell cycle synchronization might provoke transcriptional responses not directly related to the cell cycle. To address this issue, here, we describe a strategy to profile the transcriptome of Arabidopsis root cells in the G2/M phase of the cell cycle using fluorescence-activated cell sorting (FACS) of a GFP reporter line of a mitotic cyclin. From these data, we identified a previously uncharacterized GRAS TF, SCL28, whose expression peaks in G2/M. The phenotypical and molecular characterization of SCL28 indicates that it controls progression through G2/M and the selection of cell division planes.
Results
Transcriptome Analysis of Cells in G2/M Captures Cell Cycle Expression Profiles.
To analyze the transcriptome in the G2/M phases of the MCC, we isolated root meristem cells expressing the CYCB1;1-GFP reporter (29) (hereafter, “G2/M cells”) by FACS (30). This reporter has been extensively used for monitoring cellular proliferation in various organs (31) and is expressed in late G2 and mitosis up to anaphase (11, 32, 33). As control, whole-root meristems were collected by microdissection (SI Appendix, Fig. S1). Subsequently, transcriptome analysis was performed on RNA isolated from three biological replicates of both samples.
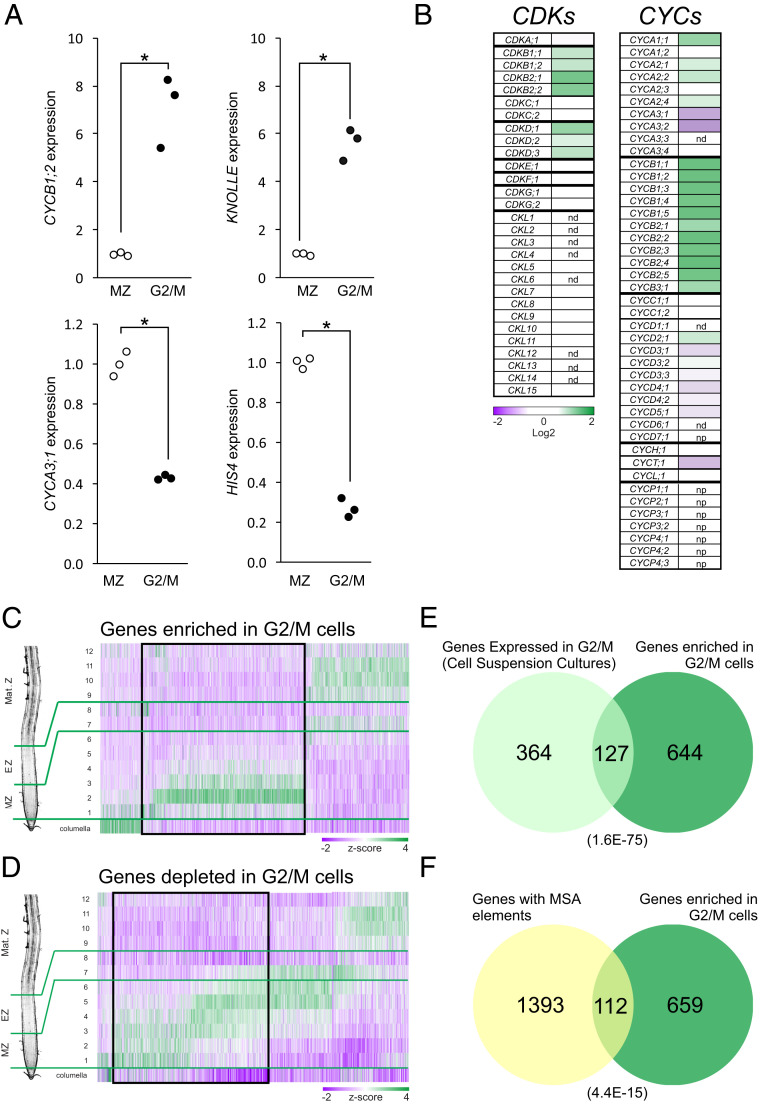
The mitotic and cytokinesis markers CYCB1;2 and KNOLLE were both at least sixfold-enriched in the G2/M cell samples (Fig. 1A). Moreover, expression of the S phase markers HISTONE H4 (HIS4) and CYCA3;1 was depleted threefold in the G2/M cells with respect to the whole-root meristem (Fig. 1A). These results confirm that our approach can accurately capture cell cycle-associated gene expression profiles.
Fig. 1.
Analysis of the transcriptome of cells in the G2/M phase of the MCC. (A) Expression levels of G2/M (CYCB1;2 and KNOLLE) and G1/S (CYCA3;1 and HISTONE H4) marker genes in cells expressing the CYCB1;1:GFP reporter (“G2/M cells”) and in complete microdissected root meristems (“MZ”). Values were normalized to the mean value obtained in complete microdissected root meristems. *P < 0.05 (significant differences; logit-T). (B) Expression levels of CDKs and cyclins in G2/M cells. Values were normalized to the value obtained in complete microdissected root meristems, log2-transformed, and expressed as a heat map. nd, not detected; np, not present in the array. See Dataset S1 for detailed numerical data. (C and D) Expression along the root’s longitudinal axis of genes enriched (C) or depleted (D) in G2/M cells. Data were obtained from the literature (37), and z scores were calculated across samples and expressed as a heat map. Numerals indicate the root segments analyzed, with slices 1 to 6 comprising the root meristem (MZ); 7 and 8, the elongation zone (EZ); and 9 to 12, the maturation zone (Mat. Z). The black boxes highlight the genes expressed preferentially in the root meristematic zone (MZ), which were selected for further work. (E) Intersection of the list of genes expressed in G2/M, as determined in synchronized cell suspension cultures (15) and the list of genes enriched in G2/M cells identified in this work. The number in parentheses indicates the P value for the overlap between the two lists (Fisher’s exact test). (F) Intersection of a list of Arabidopsis genes with MSA elements in their upstream regulatory regions and the list of genes enriched in G2/M cells. The number in parentheses indicates the P value of the overlap between the 2 lists (Fisher´s exact test).
Next, we analyzed the expression levels of cyclins and cyclin-dependent kinases (CDKs) (34) (Fig. 1B and Dataset S1). We found CDKB1;2, CDKB2;1 and CDKB2;2 are enriched in G2/M cells, as previously reported (35). Experiments in CSCs determined that among the D-class of CDKs, only CDKD;1 expression peaked at the G2/M transition. In contrast, all three D-class CDK mRNAs were enriched in G2/M cells.
As observed in CSCs, the expression of all detected CYCB genes was enriched in G2/M cells (Dataset S1). CYCAs of the subgroups 1 and 2 were also enriched in G2/M cells, including CYCA2;4, which was not detected previously in CSCs. By contrast, two CYCAs of subgroup 3, CYCA3;1 and CYCA3;2, were depleted in G2/M cells, whereas CYCA3;4 showed no significant enrichment in G2/M cells, confirming recent data (36). CYCA3;3 was not detected, as expected for a meiosis-specific cyclin gene (33).
Cyclins of the D-type (CYCDs) showed the greatest difference when compared with CSCs. While expression of CYCD5;1, CYCD4;1, CYCD4;2, CYCD7;1, and CYCD6;1 peaked at G1/S in CSCs, we did not detect any statistical enrichment in a particular cell cycle phase. Also, while CYCD3;1 has a clear expression peak in G2/M in synchronized CSC, we found its mRNA depleted in G2/M cells. Finally, although constantly expressed in CSCs, we found CYCD2;1 mRNA enriched in G2/M cells. Interestingly, these differences might reflect disparities between a growing organ versus an undifferentiated cell culture, consistent with a role for the D-type cyclins in integrating mitogenic and differentiating signals with cell cycle progression.
Genome-Wide Analysis of Cell Cycle-Specific Gene Expression Identifies Previously Uncharacterized G2/M-Regulated Genes.
We then mined our dataset for genes whose expression is either enriched or depleted in G2/M cells. Using an enrichment threshold of 1.5-fold, we identified 1,520 probesets (representing 1,578 genes) enriched in G2/M cells (P < 0.05) and 1,567 probesets (representing 1,632 genes) depleted in G2/M cells (P < 0.05) (Dataset S3). Of note was that all members of a previously defined list of Mitosis Specific Genes (35) were at least 1.5-fold enriched in our G2/M cells dataset (Dataset S2).
The expression patterns of genes enriched in G2/M cells were analyzed in developmental stage-specific gene expression database (37), finding that ∼50% of the genes were enriched in the root meristem (Fig. 1C). Likewise, ∼50% of the genes depleted in G2/M cells were expressed in the meristem (Fig. 1D). Genes with a function in cell proliferation are expected to be expressed preferentially in the root meristem, thereby these genes were selected for further analysis (Dataset S4).
Gene ontology (GO)-term enrichment analysis revealed terms related to cell cycle and cell division in both up- and down-regulated genes (SI Appendix, Fig. S2). GO terms related to mitosis and cytokinesis (“microtubule-related processes,” “M-phase of the mitotic cycle,” “cytokinesis,” etc.) were enriched only in the list of G2/M enriched genes. By contrast, the terms “DNA replication,” “response to DNA damage,” and “cell wall modifications” were enriched only in the list of G2/M-depleted genes, indicating that this list includes genes with functions in the G1 and S phases of the cell cycle. This analysis confirms that our approach effectively captured cell cycle expression profiles, both those associated with the G2/M and G1/S transitions. We found a significant overlap between the list of genes expressed in G2/M in synchronized cell suspension cultures (15) and our list of genes enriched in G2/M cells (Fig. 1E). In addition, 644 genes were found to be enriched in G2/M only in our dataset, suggesting that our approach identified a complementary set of genes expressed in this phase of the cell cycle. Next, the occurrence of MSA elements was analyzed in the promoter of genes enriched in G2/M cells, finding that 112 out of 771 genes contained this cis element (Fig. 1F). These data highlight the relevance of this cis-regulatory element in transcriptional regulation during G2/M. Taken together, these results indicate that our approach captures gene expression patterns associated with the phases of the cell cycle and confirm and extend previous data obtained with synchronized cell suspension cultures (15, 35).
SCL28 Is a TF Enriched in G2/M.
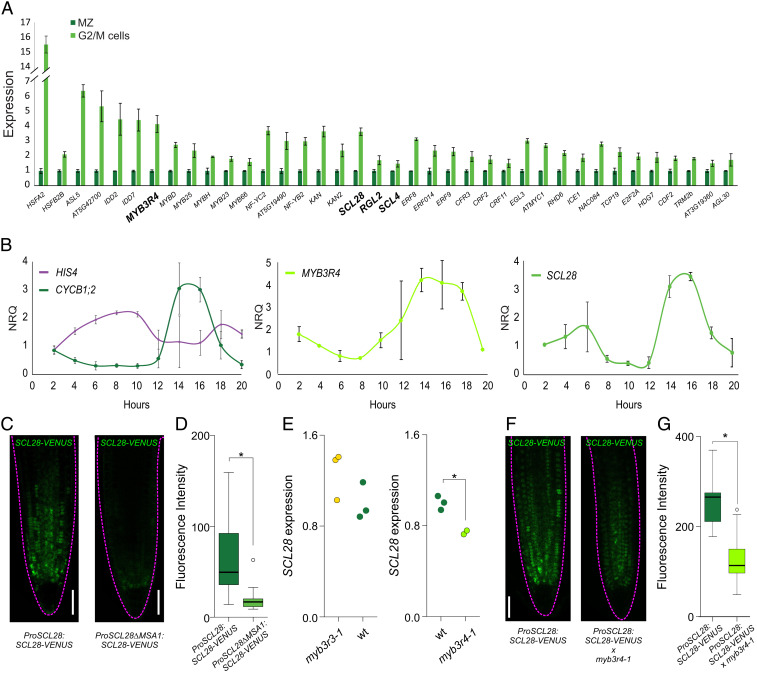
We found 38 TFs enriched in G2/M cells (Fig. 2A and Dataset S5), including the expected MYB3R4. Interestingly, several families of TFs were represented by more than one member, which might reflect the typical functional redundancy found for TFs in plants. We identified three TFs from the GRAS family of TFs enriched in G2/M cells. Among them, SCARECROW-LIKE 28 (SCL28) had the highest enrichment (Fig. 2A and Dataset S5), so we focused on this gene for further analysis.
Fig. 2.
SCL28 expression in the MCC. (A) Enrichment of 38 TFs in G2/M cells when compared to complete meristems. Data shown are means ± SE from the three biological replicates obtained in the transcriptome analysis of G2/M cells and complete microdissected root meristems (MZ). All values were normalized to the mean value obtained in complete microdissected root meristems. (B) Time-course expression analysis in cell cycle-synchronized root tips for S and G2/M marker genes (HIS4 and CYCB1;2, respectively), MYB3R4 and SCL28. Expression was estimated by RT-qPCR, and NRQs are reported. Data shown are means ± SE of three biological replicates. (C, Left) Expression pattern of SCL28 determined by LSCM with a ProSCL28:SCL28-VENUS reporter in 6-d-old roots. (C, Right) A reporter with a deletion in one of the MSA elements (ProSCL28ΔMSA1:SCL28-VENUS) was used to analyze the regulation of SCL28 by MYB3R proteins. (Scale bar, 50 μm.) (D) Quantification of fluorescence intensity in the ProSCL28:SCL28-VENUS and ProSCL28ΔMSA1:SCL28-VENUS reporters. Fluorescence intensity in the root meristem was measured in 20 primary transgenics plants transformed with each construct. *P < 0.05 (significant differences; Student’s t test). (E) Expression of SCL28 in wild-type (wt), myb3r3-1, and myb3r4-1 plants. Expression was estimated by RT-qPCR in three biological replicates and normalized to the mean value obtained in wild-type plants. *P < 0.05 (significant differences; Student’s t test). (F) SCL28 expression analyzed by LSCM with a ProSCL28:SCL28-VENUS reporter in 6-d-old wild-type and myb3r4-1 roots. (Scale bar, 50 μm.) (G) Quantification of fluorescence intensity in wild-type and myb3r4-1 roots expressing the ProSCL28:SCL28-VENUS reporter. Fluorescence intensity in the root meristem was measured in 20 roots. *P < 0.05 (significant differences; Student’s t test).
To confirm high levels of SCL28 transcript in G2/M, we used an experimental assay based on whole-root cell cycle synchronization to analyze the dynamics of expression of the TF during the MCC (38). In this experiment, G1/S and G2/M were recognized by the expression of HIS4 and CYCB1;2, respectively (Fig. 2B). When the expression of MYB3R4 and SCL28 was estimated, a clear peak in G2/M was detected for both TFs (Fig. 2B), confirming that SCL28 expression peaks at this phase of the cell cycle.
To further analyze the spatial and temporal expression pattern of SCL28, we made a translational reporter line by fusing the SCL28 coding sequence to that of the VENUS fluorescent protein under the control of SCL28 upstream regulatory regions (ProSCL28:SCL28-VENUS). In at least 20 independent transgenic lines, the ProSCL28:SCL28-VENUS reporter was expressed in patches in the root meristem, with expression declining in the transition zone to become almost undetectable in the elongation zone (Fig. 2C). Reporters used to monitor the expression of proteins with oscillating activities in the cell cycle usually have patchy expression patterns as cells in meristems are in different phases of the MCC, as we observed for ProSCL28:SCL28-VENUS. Furthermore, the SCL28 was detected within the nucleus, as expected for a transcriptional regulator, and across all cell types in proliferating cells of the root meristem.
To characterize the upstream genes regulating SCL28 expression, we first inspected the promoter and found a cluster of four conserved MSA elements (SI Appendix, Fig. S3 A–C). Genome-wide studies of the MYB3R4-binding sites through DNA-affinity purification and sequencing (39) showed a peak associated to these cis elements in the promoter of SCL28 (SI Appendix, Fig. S3D). To confirm the regulation of SCL28 by MYB3Rs in vivo, we prepared a version of the ProSCL28:SCL28-VENUS reporter in which the first MSA element was deleted (ProSCL28ΔMSA1:SCL28-VENUS). When these constructs were introduced into plants and 20 primary transgenic seedlings for each construct were evaluated, we observed a lower fluorescent signal (Fig. 2 C and D). We then estimated the levels of SCL28 mRNA in different mutant backgrounds (Fig. 2E). We found a minor increase in transcript abundance in the myb3r3-1 background, which has a mutation in the repressor MYB3R3, whereas a significant decrease was observed in myb3r4-1, which carries a mutation in the activator MYB3R4 (Fig. 2E). Finally, we crossed the ProSCL28:SCL28-VENUS reporter with myb3r4-1 and, as expected, found a significant decrease in fluorescent intensity in the mutant (Fig. 2 F and G).
To summarize, our data show that the GRAS TF SCL28 has a peak of expression during G2/M, which is at least partially caused via regulation by MYB3R4. As other conserved regions are found in the regulatory regions of the SCL28 gene (SI Appendix, Fig. S3B), our results cannot exclude the possibility that other regulators contribute to establish its expression pattern.
SCL28 Promotes Plant Organ Growth and Root Meristem Activity.
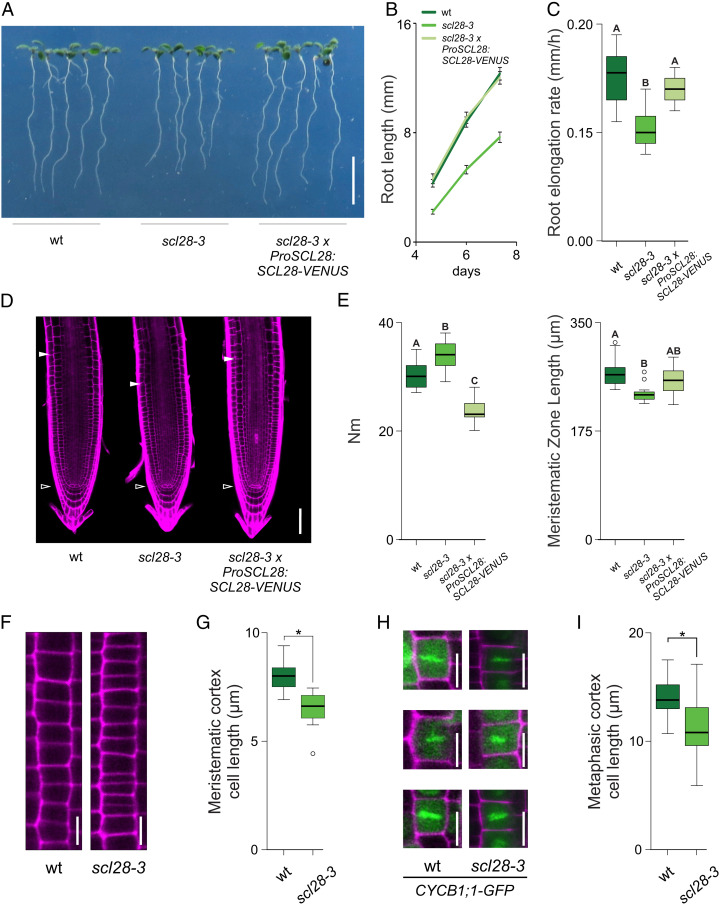
To analyze the function of SCL28 in plant development, we obtained an insertional mutant allele (scl28-3) (SI Appendix, Fig. S4A) in which the SCL28 mRNA was almost undetectable (SI Appendix, Fig. S4B). We found that roots were shorter in scl28-3 than in wild type (Fig. 3 A and B) due to a significant reduction in root growth rate (Fig. 3C), indicating that SCL28 promotes root growth.
Fig. 3.
SCL28 promotes plant organ growth and affects the activity of the meristem. (A) Root phenotype in 10 d-old wild-type (wt), scl28-3, and scl28-3 × ProSCL28:SCL28-VENUS plants. (Scale bar, 1 cm.) (B) Root growth of wild-type, scl28-3, and scl28-3 × ProSCL28:SCL28-VENUS plants. (C) Root-elongation rate (millimeters per hour) of wild-type, scl28-3, and scl28-3 × ProSCL28:SCL28-VENUS plants. Box plots with the measurements from 15 roots are shown. Different letters indicate significant differences (P < 0.05; ANOVA followed by Tukey’s multiple comparison test). (D) Root tip architecture of 6-d-old wild-type, scl28-3, and scl28-3 × ProSCL28:SCL28-VENUS plants examined by LSCM in PI (magenta)-stained plants. The white arrowheads mark the position of the QC and the end of the meristem where cells start to elongate. (Scale bar, 50 μm.) (E) Number of meristematic cortex cells (Nm) and meristematic zone length in wild-type, scl28-3, and scl28-3 × ProSCL28:SCL28-VENUS plants. Box plots with the measurements of 15 roots are shown. Different letters indicate significant differences (P < 0.05; ANOVA followed by Tukey’s multiple comparison test). (F) Detailed view of meristematic cortex cell files in wild-type and scl28-3 plants. (Scale bar, 10 μm.) (G) Average length of cortex cells in the root meristem in wild-type and scl28-3 plants. Box plots with the measurements of 10 cells from 20 roots of each genotype are shown. *P < 0.05 (significant differences from the wild type; Student’s t test). (H) Metaphasic cortex cells in the root meristem in wild-type and scl28-3 plants stained with PI (magenta) identified using the CYCB1;1-GFP (green) reporter. (I) Length of metaphasic cortex cells in the root meristem in wild-type and scl28-3 plants. Box plots with the measurements of at least 20 metaphasic cells found in at least 10 plants of each genotype are shown. *P < 0.05 (significant differences from the wild type; Student’s t test).
Using laser-scanning confocal microscopy (LSCM), we found that compared to wild-type plants, scl28-3 roots have a shorter meristem with a higher number of meristematic cortex cells (Fig. 3 D and E). This was surprising, as the length of the meristem usually correlates with the number of meristematic cells, suggesting that cell expansion in proliferating cells is compromised in scl28-3. Indeed, we found that the average length of meristematic cortex cells in the mutant was reduced by 20% when compared to wild-type plants (Fig. 3 F and G).
In root meristems, cortex cells steadily elongate between mitotic events (40), dividing symmetrically (3) when they reach a length between 12 and 15 μm. As proliferating cells are distributed across all stages of the cell cycle, we measured cell length just before cytokinesis, when cells reach their maximum length. To do this, we crossed the scl28-3 mutant with the CYCB1;1-GFP reporter that colocalizes with chromatin during metaphase (32) (Fig. 3H). Again, we found a significant reduction in cell length at division in the mutant, suggesting the occurrence of premature cell divisions and/or defects in the expansion of mitotic cells (Fig. 3I).
To confirm that the phenotype showed by scl28-3 is due to the inactivation of the SCL28 locus, the mutant was transformed with the ProSCL28:SCL28-VENUS construct, resulting in the recovery of root growth rate and proper meristem structure (Fig. 3 A–E). In addition, we generated a dominant repressor version of SCL28 by fusing the EAR-repression motif (SRDX) (41) to the carboxyl terminus of the protein. Transgenic plants transformed with the ProSCL28:SCL28-SRDX construct, in which SCL28-SRDX is expressed under the control of SCL28 regulatory regions, displayed similar phenotypes to scl28-3, including a reduction in root growth rate, shorter meristems, and shorter meristematic cortex cells (SI Appendix, Fig. S5). Taken together, these results confirm that SCL28 promotes plant organ growth and modulates root meristem activity.
SCL28 Promotes Progression through G2/M.
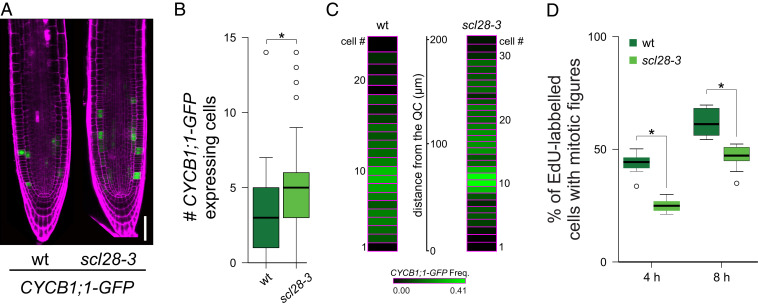
Interestingly, we found a significantly higher number of cells expressing the mitotic reporter CYCB1;1-GFP per file of cortex cells in scl28-3 mutants than in wild-type plants (Fig. 4 A and B). This can be a consequence of a higher number of proliferating cells, a delay in the progression through G2/M, or a combination of both. To distinguish between these possibilities, we performed a more detailed analysis of the cell cycle in scl28-3. The distribution of mitosis and cell cycle properties are not homogeneous along the whole meristem (9, 42, 43). Therefore, we scored the frequency of mitotic cells as a function of the distance from the quiescent center (QC) (Fig. 4C). As previously reported (44, 45), wild-type plants have a very low frequency value of cells expressing CYCB1;1-GFP close to the QC that rises to peak around 80 μm shootward from the QC (Fig. 4C) and declined to 0 at 200 μm. In scl28-3, the frequency of CYCB1;1-GFP–expressing cells displayed a similar profile but with higher values along the complete length of the meristem, suggesting a delay in the G2/M transition in the mutant.
Fig. 4.
SCL28 promotes progression through G2/M. (A) LSCM images of the CYCB1;1-GFP reporter (green) in 6-d-old wild-type (wt) and scl28-3 roots stained with PI (magenta). (Scale bar, 50 μm.) (B) Cell number per meristematic cortex cell file expressing the CYCB1;1-GFP marker. Box plots with the measurements from 15 roots of each genotype are shown. *P < 0.05 (significant differences from the wild type; Student’s t test). (C) Heat map showing the frequency of CYCB1;1-GFP–positive cells at a given position in the root meristem. The frequency of CYCB1;1-GFP–expressing cells was scored from the cell adjacent to the QC shootward in 30 cortex cell files from 15 plants for each genotype. Each box represents a cell position, and the height of the box corresponds to the average length of cortex cells at that particular position. The distance from the QC in microns is indicated. (D) Cell cycle progression through the G2/M phase. Five-day-old plants were pulse-labeled with EdU for 15 min. Seedlings were washed, transferred to MS, and collected at the indicated time points. Root meristematic cells were double-stained with EdU and DAPI, and cells with mitotic figures were counted. Box plots with the percentage of EdU-labeled cells among those with mitotic figures are shown. *P < 0.05 (significant differences from wild type; Student’s t test).
To further test this hypothesis, we estimated cell cycle length by following 5-ethynyl-2 deoxyuridine (EdU) incorporation as a function of time. As shown in Table 1, total cell cycle length was increased from 19.5 h in wild type to 24.1 h in scl28-3. Overexpressing SCL28 in a β-estradiol inducible line (XVE-SCL28) (46) resulted in root phenotypes opposite to those found in scl28-3 (SI Appendix, Fig. S6), including an acceleration of the cell cycle by 2 h (Table 1).
Table 1.
Cell cycle length is regulated by SCL28
| Genotype or condition | Cell cycle length, h | CI |
| Wild type | 19.5 | 18.0 to 21.4 |
| scl28-3 | 24.1** | 21.6 to 27.3 |
| XVE-SCL28 | 19.4 | 17.6 to 21.6 |
| XVE-SCL28 + β-estradiol | 17.4* | 15.9 to 19.0 |
*P < 0.01; **P < 0.001 (statistically significant differences from the corresponding control genotype or condition).
Next, we made direct measurements of progression through G2/M. To do this, roots were pulse-labeled with EdU, and the labeled S-phase cells were tracked for progression into M phase by scoring Edu-positive chromosomes displaying mitotic condensed chromosomes. In wild type, the percentage of cells with EdU-labeled mitotic figures increased after 4 and 8 h (Fig. 4D). In contrast, scl28-3 plants showed a 30 to 50% reduction in the number of EdU-positive cells with mitotic figures, indicating a delay in G2/M progression. Taken together, our results indicate that SCL28 modulates meristem activity, both by controlling the transition from cell proliferation to elongation and by regulating cell cycle length, promoting the progression through G2/M.
SCL28 Modulates Cell Division Plane Orientation and Phragmoplast Dynamics.
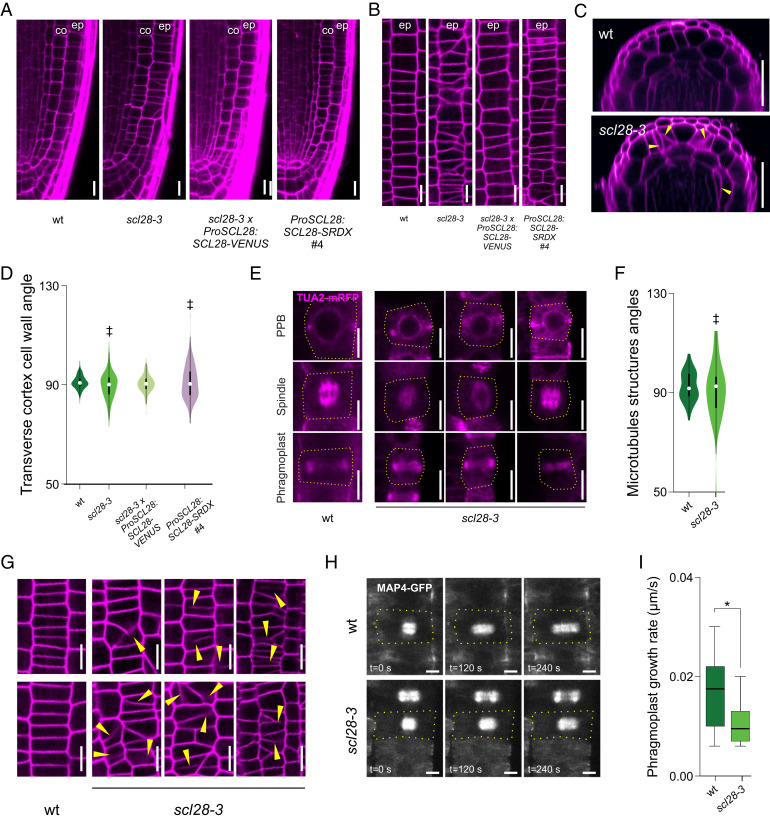
In scl28-3 roots, tissue patterning appeared normal (SI Appendix, Fig. S7), indicating that there are no detectable defects in the precise orientation of cell divisions executed by stem cells. By contrast, cell wall orientation was less regular, and some cell files were distorted in the region of rapid cell proliferation (Fig. 5A). Noteworthy, tangential LSCM optical sections revealed striking distortions in the epidermis (Fig. 5B). As expected, these phenotypes were complemented by expressing ProSCL28:SCL28-VENUS in scl28-3 and phenocopied by ProSCL28:SCL28-SRDX (Fig. 5A and SI Appendix, Fig. S5). Finally, optical cross-sections 100 μm shootward from the QC highlighted the consequences of the misoriented cell walls in scl28-3, as extra cell layers were generated randomly in both the cortex and epidermis (Fig. 5C).
Fig. 5.
SCL28 modulates cell division plane orientation and phragmoplast expansion rate. (A) Cellular organization of the root meristem in wild-type (wt), scl28-3, scl28-3 × ProSCL28:SCL28-VENUS, and ProSCL28:SCL28-SRDX plants examined by LSCM in PI (magenta)-stained plants. “co” and “ep” indicate the cortex and epidermis cell layers, respectively. (Scale bar, 10 μm.) (B) Detailed view of cell wall orientations in meristematic epidermal cells in wild-type, scl28-3, scl28-3 × ProSCL28:SCL28-VENUS, and ProSCL28:SCL28-SRDX #4 plants. To image meristematic epidermal cells, tangential longitudinal optical sections were obtained by LSCM from PI (magenta)-stained plants. “ep” indicate the epidermis cell layer. (Scale bar, 10 μm.) (C) Optical cross-sections of wild-type and scl28-3 roots 100 μm above the QC obtained by LSCM in PI (magenta)-stained plants. Yellow arrowheads mark misplaced, misoriented, curved, and/or incomplete cell walls found only in scl28-3. (Scale bar, 20 μm.) (D) Combined box and violin plot diagrams of the angles between the transverse cortex cell wall and the main longitudinal axis of the root. The data shown are the measurements of 20 cells in 15 different roots of each genotype. ‡P < 0.05 (significant differences in the variance when compared to wild type; F test). (E) Microtubule arrays (PPB, mitotic spindle, and phragmoplast) in meristematic cortex cells of wild-type and scl28-3 plants. Microtubules are detected thanks to an alpha-tubulin 2 RFP reporter (TUA2-RFP). TUA2-mRFP fluorescence is shown in magenta. The dashed yellow line traces cell outlines. (Scale bar, 10 μm.) (F) Combined box and violin plot diagram of the angles between the mitotic microtubule arrays and the main longitudinal axis of the root. The data shown are the measurements of at least 50 cells of each genotype. ‡P < 0.05 (significant differences in the variances; F test). (G) Incomplete and asymmetric cell walls found in meristematic epidermal cells from scl28-3 roots are indicated with yellow arrowheads. These abnormal cell division planes were not found in wild-type plants. To image meristematic epidermal cells, tangential longitudinal optical sections were obtained by LSCM in PI (magenta)-stained plants. (Scale bar, 10 μm.) (H) Phragmoplast expansion in meristematic cortex cells from wild-type and scl28-3 plants. The images show the moment when the disk-phragmoplast has just formed (time, 0 s) and 120 or 240 s later. Microtubules are visualized with the MAP4-GFP reporter (gray scale). The dashed yellow lines mark cell outlines. (Scale bar, 5 μm.) (I) Velocities of phragmoplast expansion (μm/s) in wild-type and scl28-3 meristematic cortex cells. Box plots with the measurements from 27 cells from each genotype are shown. *P < 0.05 (significant differences from the wild type; Student’s t test).
The symmetric divisions that occur in transit-amplifying cells (TACs) are normally oriented perpendicular to the longitudinal axis of root growth, as shown in measurements of the angle between the transverse cortex cell walls and the longitudinal axis of the root (Fig. 5D). In scl28-3, the mean value was identical to that of wild type, but the variance of the angles showed a significant increase, indicating that SCL28 is required for precise division plane orientation in TACs. While SCL28 overexpression caused no change in the distribution of cell division angles (SI Appendix, Fig. S6G), the scl28-3 phenotypes were complemented with ProSCL28:SCL28-VENUS and phenocopied by ProSCL28:SCL28-SRDX (Fig. 5D and SI Appendix, Fig. S5D), further confirming the role of SCL28 in the determination of cell division angles.
The PPB, the mitotic spindle, and the phragmoplast are structures that contain special arrays of microtubules that contribute to define the cell division planes and execute cytokinesis (2, 47). Mutants with defects in these structures have similar alterations in cell division orientation (3) as those in scl28-3. Using two different reporters to image these structures, TUA2-RFP (Fig. 5E) and MAP4-GFP (SI Appendix, Fig. S8A), we found a strong increase in the variance of the angles they form with the longitudinal axis of the root in the mutant (Fig. 5F and SI Appendix, Fig. S8B).
Among scl28-3 plants, 27 out of 100 roots showed at least 1 incomplete and asymmetric cell wall in their meristematic epidermal cells, while none of these types of wall configurations were found in wild-type plants (Fig. 5 B and G). These defects were transient, as we found no signs of permanent cytokinesis defects in mature root epidermal cells of scl28-3 plants. As these results might indicate that SCL28 modulates cytokinesis dynamics, we investigated phragmoplast expansion rate by time-lapse microscopy following microtubule arrays in plants expressing the MAP4-GFP reporter (Fig. 5 H and I). Phragmoplasts of scl28-3 expand at an initial mean velocity of 0.010 ± 0.001 μm/s, while wild-type phragmoplasts expand about 70% faster (0.017 ± 0.001 μm/s).
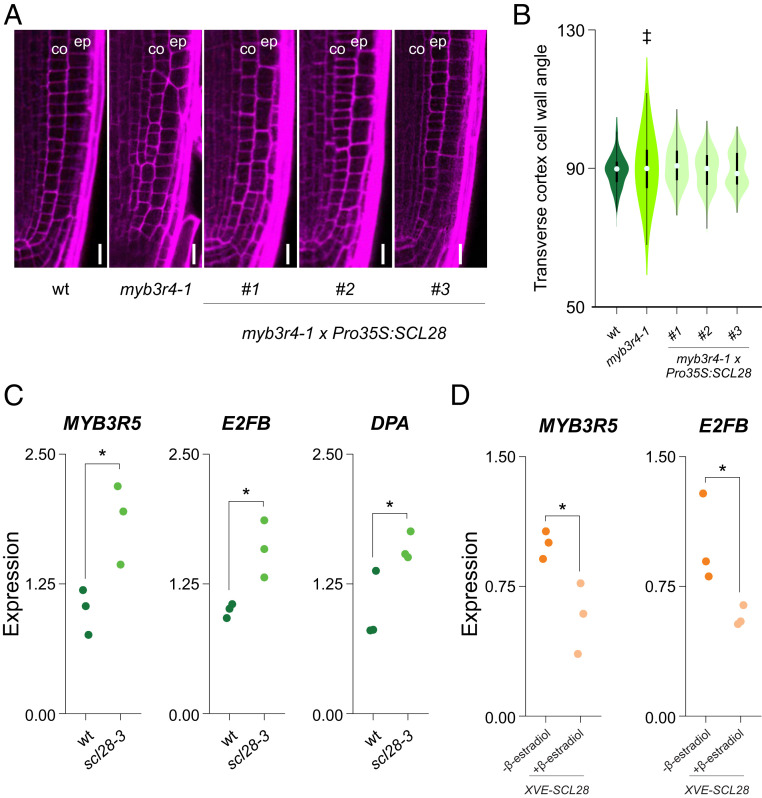
The previous results showed that MYB3R4 controls SCL28 expression in the root meristem (Fig. 2). Further confirming this observation, we found that myb3r4-1 plants have similar defects in the selection of cell division planes as those found in scl28-3 (Fig. 6 A and B). Noteworthy, this phenotype was fully complemented by SCL28 overexpression from the CaMV 35S promoter in plants transformed with a Pro35S:SCL28 construct (Fig. 6 A and B).
Fig. 6.
Regulatory interactions between MYB3R, E2F, and SCL28. (A) Cellular organization of the root meristem in wild-type (wt), myb3r4-1, and myb3r4-1 × Pro35S:SCL28 plants examined by LSCM in PI (magenta)-stained plants. “co” and “ep” indicate the cortex and epidermis cell layers, respectively. (Scale bar, 10 μm.) (B) Combined box and violin plot diagrams of the angles between the transverse cortex cell wall and the main longitudinal axis of the root in wild-type, myb3r4-1, and independent transgenic lines of myb3r4-1 transformed with Pro35S:SCL28. The data are the measurements of 20 cells in 15 different roots of each genotype. ‡P < 0.05 (significant differences in the variances when compared to wild type; F test). (C) Expression of cell cycle regulators in wild-type and scl28-3 plants. Expression was estimated by RT-qPCR in three biological replicates and normalized to the mean value obtained in wild-type plants. *P < 0.05 (significant differences; Student’s t test). (D) Expression of cell cycle regulators in XVE-SCL28 plants grown in MS media supplemented with 0.25 μM β-estradiol. Expression was estimated by RT-qPCR in three biological replicates and normalized to the mean value obtained in mock-treated control plants. *P < 0.05 (significant differences; Student’s t test).
Finally, to provide insight into the SCL28 gene-regulatory network responsible for the phenotypes described here, we first estimated the expression of other cell cycle regulators in scl28-3 plants, including both activator and repressor MYB3Rs and E2Fs and their interacting partners as well as RETINOBLASTOMA-RELATED (RBR) (SI Appendix, Fig. S9). We found that MYB3R5, E2FB, and DPA were up-regulated in scl28-3 (Fig. 6). As expected, MYB3R5 and E2FB were down-regulated by SCL28 overexpression in XVE-SCL28 plants treated with β-estradiol (Fig. 6), suggesting that the GRAS TF might act as a repressor of these two TFs.
Discussion
Cell cycle-synchronized plant cell suspension cultures have been used extensively in genome-wide studies of gene expression dynamics across cell cycle phases (24, 27, 28). However, the manner in which the suspension culture is synchronized needs to be considered. Furthermore, cell suspension cultures are maintained with hormones in rather undifferentiated states, which could hide gene expression patterns relevant to specific organs or tissues. Moreover, the treatments to induce synchrony might provoke transcriptional responses that are not cell cycle-related.
We present here an approach to study gene expression dynamics during the cell cycle based on the isolation of cells from whole roots. We used a G2/M-specific cell cycle reporter, but the method can be implemented using reporters specific to other stages of the cell cycle. Moreover, it should be possible to combine this approach with cell type-specific reporters and multicolor FACS to analyze gene expression during the cell cycle in particular cell types. It should also be feasible to isolate cells from different mutant backgrounds to accurately pinpoint defects in the cell cycle. Finally, the sorted cells might be used for proteomics (48), epigenetic (49), and metabolic studies (50).
To date, members of the E2F and MYB3R family of TFs have been shown to regulate gene expression in plants at the G1/S and G2/M phases, respectively. The roles of these TFs seem to be deeply conserved in evolution. In plants, activator-MYB3R TFs promote the expression of G2/M genes, including those encoding mitotic CYC and CDKs, proteins involved in chromosome segregation and cytokinesis, and as demonstrated here, the TF SCL28. MYB3R has a similar protein domain organization as the animal B-Myb oncoprotein (51) that activates the transcription of genes whose expression peaks at G2/M through binding to Myb sites or CHR elements found in the promoters of the target genes (52). Based on this, it has been proposed that the MYB3R TFs might be the plant counterpart of animal B-Myb, indicating that the regulatory module that promotes the expression of G2/M genes might be conserved in plants and animals (14).
In our transcriptome analysis, we found 38 TFs enriched in G2/M cells. Among them were TFs that have been described to participate in plant development, including 6 members of the Ethylene Response Factor (ERF) family (53). Interestingly, ERF8 and ERF9 have been implicated in controlling cell proliferation in leaves mediating the response to osmotic stress (54).
Three members of the GRAS family of TFs had peak expression during G2/M. Among all GRAS family members SCL28 has a novel function in the control of the cell cycle in the root meristem, with a specific role in promoting progression through G2/M and in modulating the selection of cell division planes and the dynamics of the phragmoplast. Whether this function is conserved in other plant organs and species remains to be established, but it has been reported that a mutant in a putative rice ortholog of Arabidopsis SCL28 has small leaves, short roots, and stems with short cells and distorted cells files (55, 56).
Interestingly, while SCL28 promotes cell proliferation, other members the GRAS family of TFs act in the opposite way. For example, DELLA proteins restrain cell proliferation in the root meristem by enhancing the levels of CDK inhibitors (57) or by modulating the cytokinin–auxin antagonism that sets root meristem size (58).
Cell cycle control in eukaryotes results from a complex regulatory network that operates at both the transcriptional and posttranscriptional levels (59, 60). We describe here how MYB3R4 activates SCL28 expression. In turn, we show that SCL28 represses the expression of MYB3R5 and E2FB, two well-known regulators of the cell cycle. MYB3R5 belongs to the repressor type of MY3BR TFs and restricts cell proliferation by repressing G2/M-specific genes both in proliferating and in nonproliferating cells (14, 61). Functional studies of Arabidopsis leaf development have shown that E2FB, in a complex with RBR, can restrain mitosis in actively proliferating cells (62). It is possible that unchecked expression of MYB3R5 and E2FB in scl28-3 might be responsible for the observed delays in the progression through the MCC phases.
Our results indicate that SCL28 not only promotes progression through G2/M but also modulates the selection of cell division planes, phragmoplast activity, and mitotic cell expansion. Alternative scenarios can explain this varied range of functions. First, SCL28 might regulate specific sets of genes involved in each of these processes, or, alternatively, the cell cycle delays observed in scl28-3 might have an impact on these cellular mechanisms. This is a tenable hypothesis, as several reports link the selection of cell wall orientation with cell cycle progression. For example, the maize tangled1 mutant, defective in the expression of a microtubule binding protein, exhibits delayed metaphases and telophases together with major defects in division plane orientation (63). Also, Arabidopsis mutants in TONNEAU1a fail to form the PPB and have changes in epidermal cell division angles (64) that are complemented by pharmacological or genetic restrictions in cell cycle progression (65). What processes are regulated directly by SCL28 and how they interact with each other will come from further functional, genetic, and genome-wide analyses of the gene-regulatory network.
Considering that the roles of E2F and Myb TFs in cell cycle are conserved between plant and animals, and that GRAS proteins are unique to plants, it is tempting to speculate that SCL28 might constitute a plant-specific gene-regulatory module that was recruited during evolution to cope with the complexity of the plant cell cycle and cytokinesis. Consistent with this hypothesis, SCL28 appears to regulate aspects of the MCC that are critical during plant morphogenesis, such as the dynamics of the phragmoplast and the precise control of the orientation of cell division planes.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana accession Col-0 was used throughout this study. See SI Appendix, Table S1 for a list and description of the mutants and reporter lines used in this study. Plants were grown in long photoperiods (16 h light/8 h dark) using fluorescent bulbs (triphosphor code no. 840; 100 µmol quanta m−2 s−1) at 21 °C. For root analysis, surface-sterilized seeds were sown on Murashige and Skoog (MS) solid medium containing 1× MS salt mixture, 1% sucrose, and 2.3 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.8) in 1% agar. For the induction of the XVE-SCL28 lines, media were supplemented with the indicated concentrations of β-estradiol. Seeds were placed in the dark at 4 °C for 2 d. After stratification, Petri dishes were placed in a vertical orientation inside growth chambers in a continuous light condition at 21 °C. See SI Appendix, Table S2 for a detailed description of the constructs prepared and used in this study. Arabidopsis plants were transformed using the floral dip method.
Whole-root cell cycle synchronization was performed as described (38). Briefly, plants were grown for 6 d on MS medium. In detail, seeds were placed on a piece of sterile Nylon mesh (Nitex catalog no. 03-100/44; Sefar) placed previously on top of the medium. The mesh with the seedlings were transferred to fresh media supplemented with 2 mM hydroxyurea (Sigma-Aldrich) to induce cell cycle synchrony. Then, samples were taken every 2 h, and gene expression was analyzed by RT-qPCR, as described in the Gene Expression Analysis via RT-qPCR section below.
G2/M Cell Transcriptome Analysis.
Arabidopsis root meristems were microdissected from 6-d-old plants, and protoplasts expressing the CYCB1;1-GFP reporter were isolated from root tips as described (66). Total RNA was extracted from both samples using the mirVana miRNA Isolation Kit (Ambion). Microarray analyses using the Affymetrix ATH1 platform were performed on three biological replicates as described (67). Normalized expression estimates were obtained using GC-RMA (Guanine-Cytosine Robust Multi-Array Average) (68), and significant changes were determined using logit-T (69).
Gene Expression Analysis via RT-qPCR.
Total RNA was isolated from root tissue using Tripure isolation reagent (Roche). Approximately 150 root tips (∼2 mm) from 6-d-old plants were collected for each sample. The numbers of biological replicates are specified in the legends to figures 1, 2, 6, S4, S6, and S9. Total RNA (0.5 µg) was treated with RQ1 RNase-free DNase (Promega). First-strand complementary DNA synthesis was performed using “Moloney Murine Leukemia Virus Reverse Transcriptase” (M MLV) (Invitrogen). PCR was performed in an “AriaMx” thermal cycler (Agilent) using SYBR Green I (Roche) to monitor double-stranded DNA synthesis. Normalized relative quantities (NRQs) were obtained using the qBase method (70), with RPS26C and PP2A as reference genes for normalization across samples. When indicated, NRQ values were normalized to the mean value obtained in wild-type plants. Melting curve analyses at the end of the process and “no template controls” were performed to ensure product-specific amplification without primer–dimer artifacts. Primer sequences are given in SI Appendix, Table S3.
Confocal Microscopy.
LSCM was performed throughout the study using a Plan Apochromat 20×, 0.8-NA lens on a Zeiss LSM880 microscope. Roots were stained with 15 μg/mL propidium iodide (PI) (Sigma), rinsed, mounted in water, and visualized after excitation by a 488-nm laser line for RFP, GFP, and PI or by a 514-nm laser line for VENUS. The fluorescence emission was collected between 590 and 700 nm for PI and RFP, 496 and 542 nm for GFP, and 524 and 570 nm for VENUS. Cellular parameters and fluorescence-signal intensity were analyzed with using Fiji Is Just ImageJ (71). Fluorescence-intensity measurements were performed as described (72). Briefly, mean fluorescence was measured using FIJI in a region of interest of 120 × 300 μm including the meristem starting at the QC.
Phragmoplast expansion was evaluated from time-lapse movies obtained by LSCM from wild-type and scl28-3 plants transformed with the MAP-GFP reporter. Plants were grown for 5 d in MS medium using chambered coverglasses (Nunc-Lab-Tek) (40). The experimental setup allows long-term imaging with minimum perturbation as the roots grow between the coverglass and the block of solid MS medium and the chambers are placed directly on the microscope stage. Anaphases were located in meristematic cortex cells and when disk-phragmoplasts were assembled, images were taken every 20 s for 4 min to follow phragmoplast expansion. Images were taken in medial longitudinal sections of cortex cells that capture the full diameter of the phragmoplast, and only cells with no z-drift in the time course were taken into account for subsequent measurements. The diameter of the phragmoplast was measured in these time-lapse microscopies and the velocity of phragmoplast expansion (microns per second) was obtained from phragmoplast diameter (microns) vs. time (seconds) plots.
Analysis of Root Phenotypes.
For root-elongation measurements, seedlings were grown vertically for the indicated number of days. Starting from day 5 after germination until the end of the experiment, a dot was drawn at the position of the root tip. Finally, plates were photographed, and the root length was measured over time. Root growth rate, expressed in millimeters per hour, was estimated from root length (millimeters) vs. plant age (days after sowing) plots.
Meristematic zone length and meristematic cell number were determined from LSCM images using the file of cortex cells. The meristematic zone was defined as the region from the QC up to the cell that was twice the length of the immediately preceding cell (73).
To quantify the orientation of symmetric cell divisions, the angle between the transverse walls and the root main longitudinal axis was measured in meristematic cortex cells from 6 d-old-seedlings stained with PI and imaged by LSCM. To measure the orientation of microtubule arrays (PPB, spindle, and phragmoplast), the TUA2-RFP (74) or MAP4-GFP (75) reporters were used.
The InfoStat software (https://www.infostat.com.ar/) was used to perform statistical analyses. The tests applied in each experiment are indicated in the legend of figures 2, 3, 4, 5, 6, S4, S5, S6, S8, and S9.
EdU-Incorporation Assays.
Estimations of cell cycle length were performed as described using EdU (76). Briefly, seeds were germinated on half-strength MS. Five days after germination, plantlets were transferred to EdU-supplemented (10 μM; Sigma-Aldrich) medium and harvested after 3, 6, 9, and 12 h of incubation. The percentage of EdU-positive nuclei was estimated, and total cell cycle length was estimated from plots of the percentage of EdU-positive nuclei vs. time. Progression through G2/M of wild-type and scl28-3 mutants were analyzed using pulse labeling with EdU. Briefly, 5-d-old seedlings were grown on MS plates and then transferred to liquid MS medium with 10 µM EdU, followed by a 15 min incubation. After washing with MS medium, seedlings were transferred again to MS medium and collected after 4 and 8 h. Later, nuclei were stained with DAPI, and cells with mitotic figures were counted. Data are presented as the percentages of EdU-labeled cells among those with mitotic figures.
Sequence Analysis.
VISTA plots and promoter sequences were obtained from Phytozome Version 12.1 (https://phytozome.jgi.doe.gov/pz/portal.htm). Multiple sequence alignments of the conserved promoter regions were performed using T-Coffee (tcoffee.crg.cat) (77).
Supplementary Material
Acknowledgments
We thank Arantxa Rojas and other members of the P.N.B, L.D.V., and J.P. laboratories for comments and discussions. We thank Peter Doerner for CYCB1;1-GFP, Dirk Inzé for TUA2-RFP, and Moritz Nowack for SMB-GFP. We thank the technical support from Diego Aguirre for plant work and Enrique Morales for microscopy. C.G. was supported by fellowships from National Scientific and Technical Research Council (CONICET) and Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación (ANPCyT). R.E.R. and J.P. are members of CONICET. L.D.V. was supported by Research Foundation Flanders Grant G010820N. The majority of the studies were supported by grants to R.E.R. (ANPCyT Grants PICT2015-3758 and PICT2017-2762).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005256118/-/DCSupplemental.
Data Availability.
Accession numbers (Arabidopsis Genome Initiative locus identifiers) for the genes described here are provided in SI Appendix, Table S3. Microarray data have been deposited in the Gene Expression Omnibus database (78).
References
- 1.Rasmussen C. G., Bellinger M., An overview of plant division-plane orientation. New Phytol. 219, 505–512 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Smertenko A., et al., Plant cytokinesis: Terminology for structures and processes. Trends Cell Biol. 27, 885–894 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Schaefer E., et al., The preprophase band of microtubules controls the robustness of division orientation in plants. Science 356, 186–189 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Kosetsu K., et al., Cytoplasmic MTOCs control spindle orientation for asymmetric cell division in plants. Proc. Natl. Acad. Sci. U.S.A. 114, E8847–E8854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mir R., Morris V. H., Buschmann H., Rasmussen C. G., Division plane orientation defects revealed by a synthetic double mutant phenotype. Plant Physiol. 176, 418–431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu F., et al., Arabidopsis CSLD5 functions in cell plate formation in a cell cycle-dependent manner. Plant Cell 28, 1722–1737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaubet N., Flénet M., Clément B., Brignon P., Gigot C., Identification of cis-elements regulating the expression of an Arabidopsis histone H4 gene. Plant J. 10, 425–435 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Vanstraelen M., Inzé D., Geelen D., Mitosis-specific kinesins in Arabidopsis. Trends Plant Sci. 11, 167–175 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Rahni R., Birnbaum K. D., Week-long imaging of cell divisions in the Arabidopsis root meristem. Plant Methods 15, 30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inzé D., De Veylder L., Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77–105 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Boruc J., et al., Systematic localization of the Arabidopsis core cell cycle proteins reveals novel cell division complexes. Plant Physiol. 152, 553–565 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W., Wightman R., Meyerowitz E. M., Cell cycle control by nuclear sequestration of CDC20 and CDH1 mRNA in plant stem cells. Mol. Cell 68, 1108–1119.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berckmans B., De Veylder L., Transcriptional control of the cell cycle. Curr. Opin. Plant Biol. 12, 599–605 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi K., et al., MYB3Rs, plant homologs of Myb oncoproteins, control cell cycle-regulated transcription and form DREAM-like complexes. Transcription 6, 106–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menges M., Hennig L., Gruissem W., Murray J. A., Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53, 423–442 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Ramirez-Parra E., Fründt C., Gutierrez C., A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J. 33, 801–811 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Takahashi N., et al., The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet. 6, e1000817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi N., et al., The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 27, 1840–1851 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandepoele K., et al., Genome-wide identification of potential plant E2F target genes. Plant Physiol. 139, 316–328 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naouar N., et al., Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J. 57, 184–194 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Haga N., et al., Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 157, 706–717 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haga N., et al., R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134, 1101–1110 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Ito M., et al., G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13, 1891–1905 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato K., et al., Preferential up-regulation of G2/M phase-specific genes by overexpression of the hyperactive form of NtmybA2 lacking its negative regulation domain in tobacco BY-2 cells. Plant Physiol. 149, 1945–1957 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubos C., et al., MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi K., et al., Transcriptional repression by MYB3R proteins regulates plant organ growth. EMBO J. 34, 1992–2007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menges M., Murray J. A., Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203–212 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Trolet A., et al., Cell cycle-dependent regulation and function of ARGONAUTE1 in plants. Plant Cell 31, 1734–1750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubeda-Tomás S., et al., Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19, 1194–1199 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Iyer-Pascuzzi A. S., Benfey P. N., Fluorescence-activated cell sorting in plant developmental biology. Methods Mol. Biol. 655, 313–319 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Schnittger A., De Veylder L., The dual face of cyclin B1. Trends Plant Sci. 23, 475–478 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Yin K., et al., A dual-color marker system for in vivo visualization of cell cycle progression in Arabidopsis. Plant J. 80, 541–552 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Bulankova P., Akimcheva S., Fellner N., Riha K., Identification of Arabidopsis meiotic cyclins reveals functional diversification among plant cyclin genes. PLoS Genet. 9, e1003508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandepoele K., et al., Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menges M., de Jager S. M., Gruissem W., Murray J. A., Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41, 546–566 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Willems A., et al., The cyclin CYCA3;4 is a postprophase target of the APC/CCCS52A2 E3-ligase controlling formative cell divisions in Arabidopsis. Plant Cell 32, 2979–2996 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brady S. M., et al., A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Cools T., Iantcheva A., Maes S., Van den Daele H., De Veylder L., A replication stress-induced synchronization method for Arabidopsis thaliana root meristems. Plant J. 64, 705–714 (2010). [DOI] [PubMed] [Google Scholar]
- 39.O’Malley R. C., et al., Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166, 1598 (2016). [DOI] [PubMed] [Google Scholar]
- 40.von Wangenheim D., et al., Live tracking of moving samples in confocal microscopy for vertically grown roots. eLife 6, e26792 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M., Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Hayashi K., Hasegawa J., Matsunaga S., The boundary of the meristematic and elongation zones in roots: Endoreduplication precedes rapid cell expansion. Sci. Rep. 3, 2723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otero S., Desvoyes B., Peiró R., Gutierrez C., Histone H3 dynamics reveal domains with distinct proliferation potential in the Arabidopsis root. Plant Cell 28, 1361–1371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mähönen A. P., et al., PLETHORA gradient formation mechanism separates auxin responses. Nature 515, 125–129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez R. E., et al., MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell 27, 3354–3366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coego A.et al.; TRANSPLANTA Consortium , The TRANSPLANTA collection of Arabidopsis lines: A resource for functional analysis of transcription factors based on their conditional overexpression. Plant J. 77, 944–953 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Müller S., Plant cell division - defining and finding the sweet spot for cell plate insertion. Curr. Opin. Cell Biol. 60, 9–18 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Petricka J. J., et al., The protein expression landscape of the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 109, 6811–6818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawakatsu T., et al., Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants 2, 16058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersson S. V., et al., An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21, 1659–1668 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito M., Conservation and diversification of three-repeat Myb transcription factors in plants. J. Plant Res. 118, 61–69 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Fischer M., Müller G. A., Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit. Rev. Biochem. Mol. Biol. 52, 638–662 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Nakano T., Suzuki K., Fujimura T., Shinshi H., Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van den Broeck L., et al., From network to phenotype: The dynamic wiring of an Arabidopsis transcriptional network induced by osmotic stress. Mol. Syst. Biol. 13, 961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W., et al., Identification and characterization of dwarf 62, a loss-of-function mutation in DLT/OsGRAS-32 affecting gibberellin metabolism in rice. Planta 232, 1383–1396 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Tong H., et al., DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58, 803–816 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Achard P., et al., Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188–1193 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Moubayidin L., et al., The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 20, 1138–1143 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Csikász-Nagy A., Battogtokh D., Chen K. C., Novák B., Tyson J. J., Analysis of a generic model of eukaryotic cell-cycle regulation. Biophys. J. 90, 4361–4379 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ortiz-Gutiérrez E., et al., A dynamic gene regulatory network model that recovers the cyclic behavior of Arabidopsis thaliana cell cycle. PLOS Comput. Biol. 11, e1004486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi N., et al., A regulatory module controlling stress-induced cell cycle arrest in Arabidopsis. eLife 8, e43944 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Őszi E., et al., E2FB interacts with RETINOBLASTOMA RELATED and regulates cell proliferation during leaf development. Plant Physiol. 182, 518–533 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez P., Luo A., Sylvester A., Rasmussen C. G., Proper division plane orientation and mitotic progression together allow normal growth of maize. Proc. Natl. Acad. Sci. U.S.A. 114, 2759–2764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y., Iakovidis M., Costa S., Control of patterns of symmetric cell division in the epidermal and cortical tissues of the Arabidopsis root. Development 143, 978–982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Costa S., Are division plane determination and cell-cycle progression coordinated? New Phytol. 213, 16–21 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Sparks E. E., Benfey P. N., Tissue-specific transcriptome profiling in Arabidopsis roots. Methods Mol. Biol. 1610, 107–122 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Schmid M., et al., A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Irizarry R. A., Ooi S. L., Wu Z., Boeke J. D., Use of mixture models in a microarray-based screening procedure for detecting differentially represented yeast mutants. Stat. Appl. Genet. Mol. Biol. 2, Article1 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Lemon W. J., Liyanarachchi S., You M., A high performance test of differential gene expression for oligonucleotide arrays. Genome Biol. 4, R67 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J., qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, R19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schindelin J., et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ercoli M. F., Vena R., Goldy C., Palatnik J. F., Rodriguez R. E., Analysis of expression gradients of developmental regulators in Arabidopsis thaliana roots. Methods Mol. Biol. 1863, 3–17 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Dello Ioio R., et al., A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Van Damme D., et al., In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 136, 3956–3967 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marc J., et al., A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10, 1927–1940 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pedroza-Garcia J. A., et al., Role of the Polymerase ε sub-unit DPB2 in DNA replication, cell cycle regulation and DNA damage response in Arabidopsis. Nucleic Acids Res. 44, 7251–7266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Notredame C., Higgins D. G., Heringa J., T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
- 78.Rodriguez R. E., Breakfield N., Benfey P. N., Palatnik J., Transcriptome of plants cells in the G2/M phase of the cell cycle. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144842. Deposited 6 February 6 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers (Arabidopsis Genome Initiative locus identifiers) for the genes described here are provided in SI Appendix, Table S3. Microarray data have been deposited in the Gene Expression Omnibus database (78).