Significance
Our study has identified a minimalist nanovaccine with C1 lipid nanoparticle (LNP) that effectively promotes mRNA delivery and antigen presentation with a self-adjuvant feature through activating TLR4 signaling. The C1 LNP mRNA nanovaccine exhibited significant in vivo efficacy in both tumor prevention and therapeutic vaccine settings. Therefore, our work presents a mRNA nanovaccine platform for developing cancer immunotherapy for a wide range of tumor types.
Keywords: mRNA nanovaccine, TLR4, self-adjuvant, cancer immunotherapy, lipid-like material
Abstract
Intracellular delivery of messenger RNA (mRNA)-based cancer vaccine has shown great potential to elicit antitumor immunity. To achieve robust antitumor efficacy, mRNA encoding tumor antigens needs to be efficiently delivered and translated in dendritic cells with concurrent innate immune stimulation to promote antigen presentation. Here, by screening a group of cationic lipid-like materials, we developed a minimalist nanovaccine with C1 lipid nanoparticle (LNP) that could efficiently deliver mRNA in antigen presenting cells with simultaneous Toll-like receptor 4 (TLR4) activation and induced robust T cell activation. The C1 nanovaccine entered cells via phagocytosis and showed efficient mRNA-encoded antigen expression and presentation. Furthermore, the C1 lipid nanoparticle itself induced the expression of inflammatory cytokines such as IL-12 via stimulating TLR4 signal pathway in dendritic cells. Importantly, the C1 mRNA nanovaccine exhibited significant antitumor efficacy in both tumor prevention and therapeutic vaccine settings. Overall, our work presents a C1 LNP-based mRNA cancer nanovaccine with efficient antigen expression as well as self-adjuvant property, which may provide a platform for developing cancer immunotherapy for a wide range of tumor types.
In the past decades, cancer immunotherapy has made major breakthroughs that significantly advanced our understanding of cancer biology and improved therapeutic outcome for cancer patients (1). Although the therapeutic cancer vaccine against metastatic prostate cancer was approved by the Food and Drug Administration (FDA) in 2010, the clinical efficacy was limited, likely due to the single cancer antigen used, weak antigen presenting potency of in vitro prepared dendritic cells (DCs), and immunosuppressive tumor microenvironment (2, 3). How to induce potent antitumor immune response in vivo is still a major challenge in the area of therapeutic cancer vaccine development.
The principle of cancer vaccine is to induce potent antitumor responses by vaccinating patients with cancer-specific antigen(s) and immunostimulating adjuvant in either soluble or particulate formulation (3, 4). The cancer-specific antigen is then processed and presented to T cells by antigen presenting cells (APCs), mainly DCs in vivo. Recent findings suggest that patient-specific neoantigens may induce more specific and robust antitumor responses compared to cancer-associated antigen, but precise prediction of neoantigen is still in its infancy. As such, multiple predicted neoantigens are often used for broad immunogenicity (5). Traditionally, cancer antigens are often delivered in forms of either whole protein, antigen peptides, or DNA plasmids encoding specific cancer antigens (2, 3, 6). However, DNA plasmids may bring a potential risk of chromosome integration, and peptide synthesis of multiple antigens could be costly and time consuming. Recently, messenger RNA (mRNA)-based vaccine has emerged as an increasingly popular means to deliver vaccine for treating infectious diseases or cancer (7, 8). Notably, mRNA can provide rapid protein expression of multiple antigens in nondividing and hard-to-transfect cells such as DCs without the need of nuclear translocation and transcription. Other advantages of mRNA vaccine include high potency, high potential for rapid development, low-cost manufacture, and safe administration (7).
Efficient mRNA delivery and translation in vivo is critical in achieving therapeutic efficiency for cancer treatment. Due to the special characteristics of mRNA such as large size, highly negative charge, and susceptibility to degradation, the delivery of mRNA vaccine presents several potential challenges (7, 9). Naked mRNA injection only produced weak and transient protein expression in vivo (10). The early approach for in vivo mRNA administration was via protamine-mRNA formulation, which was later used in a prostate cancer vaccine and is currently under clinical trial (11). Over the past few years, nanoparticle-delivered vaccines (nanovaccines) start to show remarkable advances in cancer immunotherapy (12–16). As antigen and adjuvant need to be codelivered to antigen presenting cells for efficient antigen presentation, biomimetic nanoparticles with large carrying capacity are well suitable for vaccine formulation (17, 18). Importantly, nanoparticles can be targeted to DCs in vivo by passive deposit or active targeting moiety-facilitated delivery (19). Multiple factors including the size, charge, and shape may affect the DC-targeting and immune-activating efficacy of nanovaccines (20). By changing the surface charge from positive to slightly positive or neutral, Kranz et al. recently reported a lipoplex-based mRNA vaccine that efficiently targeted dendritic cells in vivo and induced specific immune responses by systematic injection (21). Nevertheless, neutral or negative charge may present a challenge for efficient mRNA loading into LNP formulation.
Due to easy synthesis and low toxicity, cationic lipid-like materials have been employed for siRNA and immunostimulatory RNA adjuvant delivery in the past few years, but their use in mRNA vaccine has been less well studied (22–25). Previous work using such a lipid nanoparticle for mRNA vaccine delivery achieved impressive in vitro and in vivo immunization efficacy (26). However, such vaccine still contained bacteria cell wall-derived lipopolysaccharide (LPS) as an adjuvant, and thus would increase the manufacturing difficulty and potential toxicity. To maintain innate immune-stimulating effect while avoiding systemic toxicity, recent “minimalist vaccine” approaches employed heterocyclic lipids as an mRNA vaccine carrier as well as a “self-adjuvant” by stimulating STING-mediated type I interferon innate immune response (16, 27). Whether other innate immune signaling responses also contribute to LNP-formulated mRNA nanovaccine has not been tested. To explore more versatile self-adjuvant nanocarrier for mRNA vaccine delivery, we developed a library of cationic lipid-like compounds through efficient ring opening of epoxides by generation 0 of poly(amidoamine) (PAMAM) dendrimers and screened them based on an in vitro antigen presentation assay. In this library, D2 was known as G0-C14 and was previously used for gene delivery (23–25). In the antigen presentation assay, the top candidate we identified was the C1 LNP with a 12-carbon tail that effectively delivered antigen-encoding mRNA into DCs, produced efficient mRNA translation into antigen, and induced a robust T cell response. Importantly, C1 itself could stimulate the expression of inflammatory cytokines such as IL-12 in DCs via activating TLR4 signaling. By testing C1 nanovaccine formulated with specific cancer antigen-encoding mRNAs on different tumor models, we observed robust immune responses and antitumor efficacy in both tumor prevention and therapeutic settings with no obvious toxicity. Thus, such C1 LNP-formulated mRNA vaccine would provide a feasible approach for cancer immunotherapy with a good safety profile.
Results
Design and Screening of Lipid-Like Material-Based Nanoparticles.
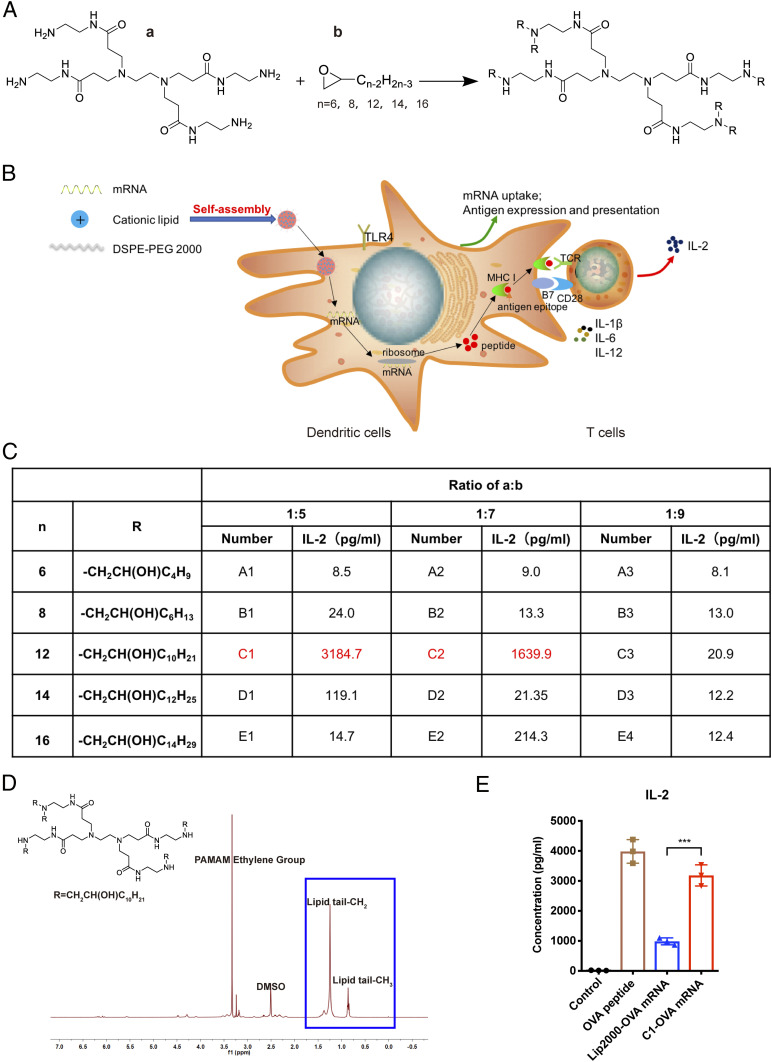
To facilitate mRNA encapsulation, cationic lipid-like materials were synthesized through the ring opening of 1,2-epoxytetradecane by generation 0 of PAMAM dendrimers with a different length of carbon tails (Fig. 1A). A polyethylene glycol containing lipid, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG 2000) was formulated with the cationic lipid-like material (A1-A3, B1-B3, C1-C3, D1-D3, and E1-E3) into LNP structure (SI Appendix, Fig. S1A). The lipid-like material compound was synthesized with cationic head groups that could efficiently condense mRNA via electrostatic interactions. Then the lipid-like material with mRNA could self-assemble with DSPE-PEG 2000 to form stable NPs.
Fig. 1.
T cell activation-based screening of mRNA-delivering lipid-like materials. (A) Synthesis of 15 cationic lipid-like compounds through efficient ring-open epoxides by generation 0 of PAMAM dendrimers for mRNA delivery. “a” represents G0 of PAMAM, “b” represents epoxide. R= -CH2CH(OH)Cn-2H2n-3. (B) Schematic illustration showing the in vitro antigen presentation assay. Antigen-encoding mRNA formulated with lipid nanoparticles was taken and expressed into antigen by dendritic cells, then presented to T cells, which secreted IL-2 upon antigen stimulation. (C) T cell-secreted IL-2 levels measured by ELISA after being induced by DCs treated with different material-formulated OVA mRNA. (D) H-NMR spectra for lipid-like material C1. (E) T cell-secreted IL-2 levels induced by DCs treated with OVA mRNA formulated with C1 or Lipofectamine 2000 (Lip2000). OVA peptide served as a positive control. Control: medium only. Data represent the mean ± SD of three independent experiments. ***P < 0.001.
Next, we applied LNPs as a nanovaccine to deliver mRNA-encoding antigen. In Fig. 1B, the schematic illustration showed the screening experimental process including the antigen presentation and T cell activation assay in vitro. LNP-formulated antigen-encoding mRNA was taken up by DCs, translated, processed, and presented to T cells, which secrete interleukin-2 (IL-2) upon activation from antigen-TCR binding. We used mRNA encoding the CD8 epitope (encoding OVA257–264) of a model antigen, ovalbumin (OVA), to test the effects of different formulations on T cell activation. OVA mRNA was encapsulated into LNPs using 15 kinds of lipid-like materials to form the mRNA nanovaccine. Mouse dendritic cells DC2.4 were treated with mRNA nanovaccine, then coincubated with B3Z cells (an OVA-specific mouse CD8 T cell hybridoma), which secrete the cytokine IL-2 upon OVA antigen stimulation. ELISA results showed that the lipid-like material C1-formulated mRNA vaccine induced the highest level of IL-2 secretion (Fig. 1C). The results demonstrated that C1 could deliver mRNA to DCs with efficient translation and antigen presentation to T cells. Fig. 1D showed the H-NMR (H-NMR spectroscopy) result of C1. OVA mRNA-loaded C1 (C1-OVA) yielded an approximately three times higher level of IL-2 than that from mRNA formulated with Lipofectamine 2000 (Lip2000), a commercial liposome reagent often used for nucleic acid transfection (Fig. 1E). To further validate such results in primary mouse immune cells, we coincubated mouse bone marrow-derived dendritic cells (BMDCs) with C1-OVA mRNA and then cocultured them with primary mouse OT-1 cells (mouse CD8 T cells specifically recognize OVA antigen epitope). The secretion level of IL-2 and IFN-γ by T cells in the C1-mRNA group was much higher than that of the Lip2000 group (SI Appendix, Fig. S1B). Importantly, C1-OVA mRNA also induced a much higher expression level of MHC-I-OVA complex (measured by an antibody recognizing H2Kb-OVA257–264 complex) on DCs than that of the Lip2000 group (SI Appendix, Fig. S1C). These results showed that C1-mRNA nanovaccine effectively delivered antigen-encoding mRNA to DCs and promoted efficient mRNA translation and antigen presentation for T cell activation.
Characterization and Structure of the C1-mRNA Nanovaccine.
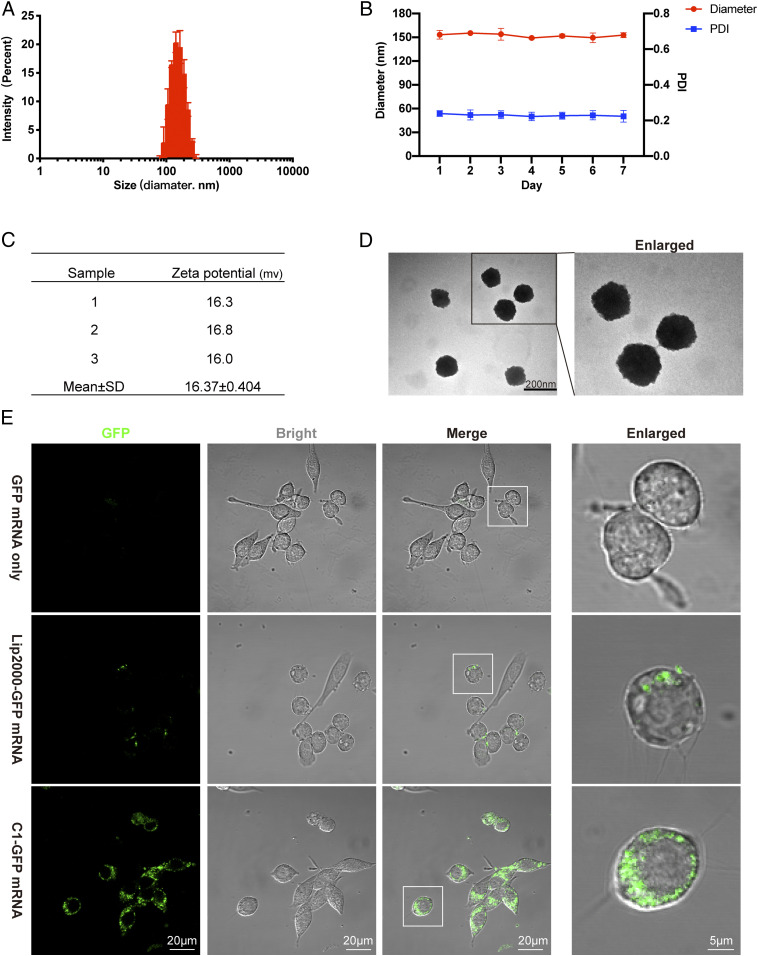
We next optimized the C1/mRNA ratio for the highest IL-2 induction, and the ratio of 160:1 was chosen to prepare the C1-mRNA nanovaccine in the rest of the study (SI Appendix, Fig. S1D). We then characterized the physical and chemical properties of the C1-mRNA vaccine. The results showed that the C1-mRNA complex was nanoparticles with mean diameter of about 150 nm (Fig. 2A). The polydispersity index (PDI) of C1-mRNA particles remained at around 0.23 for 7 d and the diameter remained stable (Fig. 2B). The average surface potential of C1-mRNA particles was near +16 mV (Fig. 2C). Such features may contribute to the efficiency of C1 carrying mRNA into cells. Transmission electron microscopy (TEM) images showed C1-mRNA particles as round-shaped nanoparticles (Fig. 2D).
Fig. 2.
Physicochemical characterization and mRNA expression efficiency of C1-mRNA nanoparticles. (A) The average size of C1-mRNA nanoparticles. (B) The size and PDI of C1-mRNA nanoparticles measured for 7 d. (C) The Zeta potential of C1-mRNA nanoparticles. (D) The TEM picture showing the shape of the C1-mRNA complex. (E) GFP mRNA expression in DC2.4 cells by naked mRNA, Lip2000, or C1 delivery. Data represent the mean ± SD from three independent experiments.
Cellular Uptake Route and Intracellular Localization of the C1-mRNA Nanovaccine.
Nanoparticles with a size over 100 nm often enter cells through a macropinocytosis or phagocytosis route (20). Pretreatment of DC 2.4 cells with cytochalasin D (an inhibitor for phagocytosis), but not amiloride (an inhibitor for macropinocytosis), significantly reduced the cellular uptake of the C1-mRNA-coumarin6 nanovaccine (SI Appendix, Fig. S2 A and B), suggesting that phagocytosis is the major cellular uptake route for the C1-mRNA nanovaccine.
We next examined the uptake and intracellular localization of the C1-mRNA nanovaccine by DCs. To facilitate tracking the intracellular localization of C1-mRNA, the mRNA was labeled with the fluorescence dye Fluorescein 12 (F12) by replacing the UTP with F12-UTP during mRNA synthesis. DC2.4 cells were incubated with F12-mRNA only, Lip2000-F12-mRNA, and the C1-F12-mRNA nanovaccine. Confocal microscopy revealed that the fluorescence intensity of the C1-F12-mRNA group was significantly stronger than F12-mRNA only and Lip2000-F12-mRNA groups (SI Appendix, Fig. S2 C and D), indicating an effective intracellular mRNA delivery by C1 LNP. To ensure that mRNA can be efficiently translated into protein/peptide, we prepared a C1-GFP mRNA encoding green fluorescent protein (GFP). Confocal microscopy revealed that about 50% of cells transfected with Lip2000-GFP mRNA showed GFP signal at 24 h after transfection, but almost 100% of cells expressed GFP protein with strong fluorescence intensity in the C1-GFP mRNA transfected group (Fig. 2E and SI Appendix, Fig. S2E). These results indicated that the GFP mRNA delivered by C1 LNP was efficiently translated into protein. We also compared protein expression levels of GFP mRNA formulated with different lipid-like materials by flow cytometry, and found that the C1-GFP mRNA groups showed markedly higher levels of GFP protein expression than other groups (SI Appendix, Fig. S1E).
The C1-mRNA Nanovaccine Induced the Immune Activation of BMDCs.
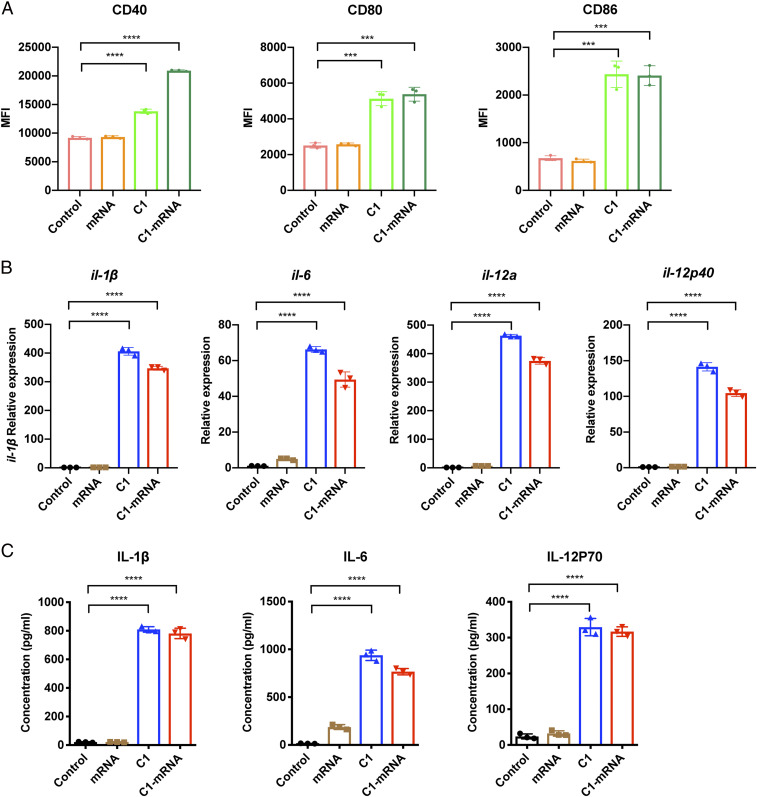
We next investigated how the C1-mRNA nanovaccine would influence the activation status of DCs. C1 and C1-mRNA markedly up-regulated the surface expression of costimulatory molecules such as CD80, CD86, and CD40 on DCs (Fig. 3A), indicating DC maturation and activation. As innate immune signaling often promotes inflammatory cytokine gene expression for DC activation, we next examined cytokine gene expression in primary mouse BMDCs. The results showed that C1 and C1-mRNA treatment significantly induced the expression of proinflammatory cytokine genes IL-1β, IL-6, IL-12a, and IL-12p40 in BMDCs (Fig. 3B), and type I interferon genes including IFN-α1, IFN-α4, and IFN-β1, in a milder magnitude (SI Appendix, Fig. S3A). ELISA results confirmed the up-regulation of protein levels of IL-1β, IL-6, and IL-12p70 cytokines in culture supernatant from cells treated with C1 and C1-mRNA (Fig. 3C). Therefore, we concluded that the C1 LNP induced proinflammatory cytokine production by BMDCs.
Fig. 3.
Innate immune activation of BMDCs by the C1-mRNA nanovaccine. (A) C1-mRNA nanovaccine promoted the maturation of BMDCs. The expression of CD40, CD80, and CD86 were up-regulated by C1-mRNA treatment. BMDCs were gated for CD11c+ cells for analysis on the DC population. Control: medium only. Concentration used: mRNA: 120 ng/mL, C1: 19.2 μg/mL; treatment time: 24 h. MFI, mean fluorescence intensity. (B) The mRNA levels of cytokines including il-1β, il-6, il-12a, and il-12p40 in BMDCs by different treatments as in A, measured by qPCR. Treatment time: 12 h. (C) Protein levels of IL-1β, IL-6, and IL-12p70 in culture media of BMDCs by different treatments as in A, measured by ELISA. Treatment time: 24 h. Data represent the mean ± SD of three independent experiments. ***P < 0.001; ****P < 0.0001.
Recent studies identified a critical role of STING-mediated type I inferferon signaling in cyclic lipid nanoparticle-mediated mRNA vaccine efficacy (16, 27). To address whether the immune activation effect of C1 was STING dependent, we tested the immune stimulatory effect of C1 in wild-type (WT) BMDCs and STING-deficient BMDCs derived from a dendritic cell-specific Sting knockout mouse line (designated as Sting cKO here). The result showed that C1-induced levels of T cell activation, expression of costimulatory molecules, and type I interferon genes were comparable between WT and STING-deficient cells (SI Appendix, Fig. S3 B–D). Therefore, we conclude that C1 LNP-induced immune activation is largely STING independent.
We then tested whether the side chain length of R groups and chemical structures would affect the stimulation effects of the LNPs. Among the 15 lipids tested, only C1 and C2 activated cytokine production in BMDCs (SI Appendix, Fig. S3F). Intriguingly, C3 did not stimulate cytokine production, indicating minor structure change/difference may cause different biological function (chemical structures of C1, C2, and C3 were shown in SI Appendix, Fig. S3G). These results suggest that a length of 12-carbon is optimal for BMDC activation, and meanwhile exclude the possibility of endotoxin (LPS) contamination during nanoparticle synthesis.
The C1-mRNA Nanovaccine Activated BMDCs through TLR4 Signaling.
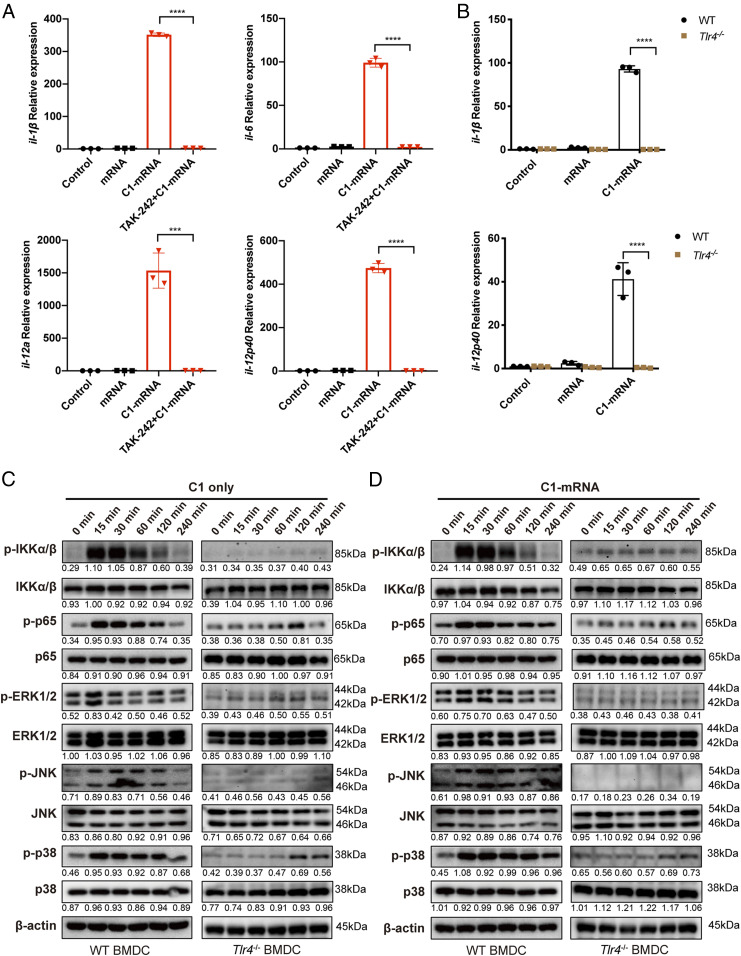
The inflammatory cytokine production in BMDCs is mainly regulated by innate immune receptor activation-induced NF-κB signaling (28). To determine if the activation of BMDCs by C1-mRNA nanovaccine was dependent on any known Toll-like receptor (TLR) signaling pathway, we pretreated BMDCs with small molecule inhibitors for TLR3, TLR4, TLR7/8, and TLR9, and then analyzed the cytokine gene expression in C1- or C1-mRNA-treated BMDCs. Our results showed that TAK-242, a TLR4-specific inhibitor, blocked the ability of C1 or C1-mRNA-induced cytokine production in BMDCs (Fig. 4A). In contrast, pretreatment of TLR3 inhibitor (CU CPT4a), TLR7/8 inhibitor (ODN2088 control), or TLR9 inhibitor (ODN2088) only partially inhibited the cytokine production (SI Appendix, Fig. S3H). Consistent with the effect of TLR4 inhibitor, cytokine activation induced by C1 and C1-mRNA in WT BMDCs was almost completely absent in TLR4-deficient BMDCs derived from Tlr4−/− mice (Fig. 4B). At the molecular level, C1 or C1-mRNA treatment markedly induced the phosphorylation level of protein kinases such as IKKα/β and mitogen-activated protein kinases (MAPKs), including ERK, JNK, and p38 in WT BMDCs, but such phosphorylation was largely abolished in Tlr4−/− BMDCs (Fig. 4 C and D). C1 LNP, but not C3, also induced NF-κB reporter activation in DC2.4 cells, and such activation was also inhibited by TAK-242 (SI Appendix, Fig. S3E). Thus, we concluded that C1 LNP could strongly induce phosphorylation of MAPKs and activate the NF-κB pathway in dendritic cells in a TLR4-dependent manner. Together, these data show that phagocytosis of C1-mRNA nanovaccine by BMDCs activated the TLR4-dependent NF-κB signaling pathway, thereby promoting a cytokine response and DC activation for efficient antigen presentation.
Fig. 4.
C1-mRNA induced cytokine expression in BMDCs through TLR4 signaling. (A) Expression levels of il-1β, il-6, il-12a, and il-12p40 in BMDCs after incubation with control, mRNA, C1-mRNA, and C1-mRNA with TAK-242 (5 μM, pretreatment for 1 h), measured by qPCR. Control: medium only. Concentration used: mRNA: 120 ng/mL, C1: 19.2 μg/mL; treatment time: 12 h. (B) Expression levels of il-1β, il-6, il-12a, and il-12p40 in WT or Tlr4−/− BMDCs after incubation with control, mRNA, and C1-mRNA, measured by qPCR. Control: medium only. Treatment time: 12 h. (C and D) Phosphorylated and total protein levels of innate immune signaling molecules (IKKα/β, p65, ERK, JNK, and p38) in BMDCs after incubation with C1 (C) or C1-mRNA (D). The number below each band is the quantified density value relative to β-actin. Data represent the mean ± SD of three independent experiments in A and B. ***P < 0.001; ****P < 0.0001.
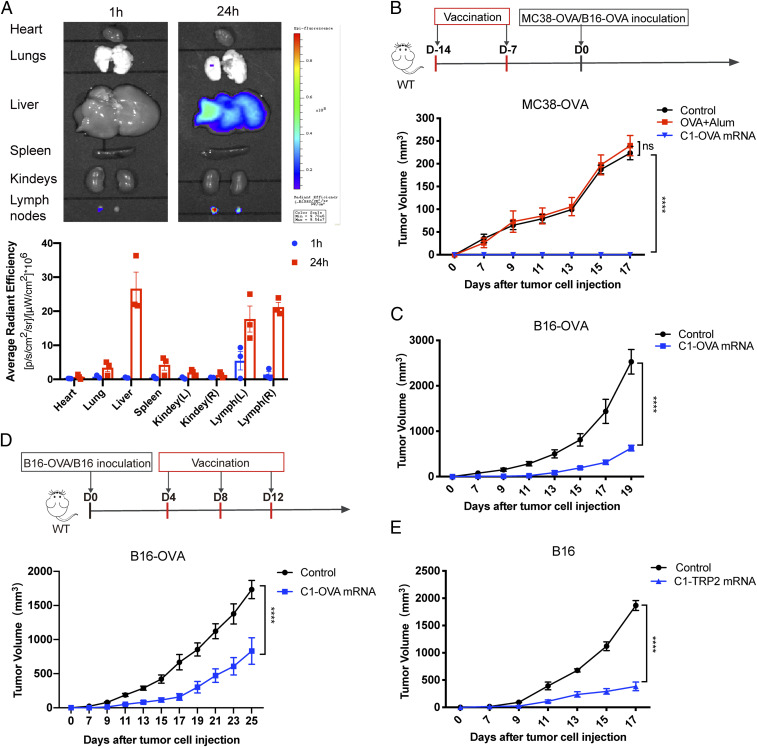
In Vivo Biodistribution and Antitumor Efficacy of the C1-mRNA Nanovaccine.
To examine the biodistribution of C1-mRNA in vivo, we injected a DiR (a fluorescent dye)-labeled C1-mRNA (C1-mRNA-DiR) subcutaneously at the right inguinal region of B6 mice. Twenty-four hours after injection, we observed a strong fluorescence signal in liver and lung tissues. Importantly, the C1-mRNA-DiR nanovaccine also exhibited a strong accumulation in draining lymph nodes and contralateral lymph nodes (Fig. 5A). This result prompted us to further investigate the in vivo immune effect of the C1-mRNA nanovaccine. When vaccinated with C1-OVA mRNA, the mice showed the highest level of OVA-specific CD8+ T cells reflected by OVA-tetramer staining (SI Appendix, Fig. S4 A and B), and lymph node T cells produced much higher levels of IFN-γ than that of mice vaccinated with OVA protein formulated with the common vaccine adjuvant aluminum salt (OVA+Alum group) (SI Appendix, Fig. S4C). These results demonstrated that the C1-mRNA nanovaccine induced a robust antigen-specific T cell response in vivo.
Fig. 5.
Antitumor effect of the C1-mRNA nanovaccine as a preventative or therapeutic vaccine. (A) In vivo biodistribution of the C1-mRNA nanovaccine (Upper). Fluorescence measured at 1 h or 24 h after subcutaneous injection of C1-mRNA-DiR (Lower). C1-mRNA-DiR dose per mouse: mRNA, 10 μg; C1, 1.6 mg; DiR:16 μg; n = 3 mice per group. (B and C) The antitumor efficacy of the C1-mRNA nanovaccine as a preventative cancer vaccine. Upper: The vaccination schedule; Lower: the preventative effect of C1-OVA mRNA nanovaccine in the MC38-OVA tumor model (B) and the B16-OVA tumor model (C). Alum served as a control adjuvant. Control: PBS injection group. C1-OVA mRNA dose per mouse: mRNA encoding OVA257–264,10 μg; C1, 1.6 mg; OVA+Alum per mouse: 200 μg peptide per mouse; n = 5 mice per group. (D and E) The antitumor efficacy of the C1-mRNA nanovaccine as a therapeutic cancer vaccine. (D) Upper: The vaccination schedule; Lower: the therapeutic effect of the C1-OVA mRNA nanovaccine in the B16-OVA tumor model. (E) The therapeutic effect of the C1-Trp2 mRNA nanovaccine in the B16 tumor model. Control: PBS injection group; C1-OVA mRNA dose per mouse: mRNA encoding OVA257–264, 10 μg; C1,1.6 mg. C1-TRP2 mRNA dose per mouse: mRNA encoding TRP2, 10 μg; C1, 1.6 mg; n = 5 mice per group. Data represent the mean ± SEM. ****P < 0.0001.
Next, we examined the antitumor efficacy of the C1-mRNA nanovaccine in both cancer prevention and therapeutic settings. We first constructed an OVA-expressing colorectal cancer cell line MC38-OVA and melanoma cell line B16-OVA for the tumor experiment (SI Appendix, Fig. S4D). B6 mice were randomly assigned into three different groups: a control group (with phosphate-buffered saline [PBS] injection), an OVA+Alum group, and a C1-OVA mRNA nanovaccine group. All vaccines were injected subcutaneously at the inguinal region of mice, and MC38-OVA tumor cells were subcutaneously inoculated 7 d later (Fig. 5B). The C1-OVA mRNA nanovaccine group showed the best tumor growth inhibition compared to that from the OVA+Alum vaccine group and the control group (Fig. 5B and SI Appendix, Fig. S5A). A similar tumor inhibition effect was observed in the B16-OVA tumor model (Fig. 5C and SI Appendix, Fig. S5B). These results indicated that the C1-OVA mRNA nanovaccine could serve as an effective vaccine to prevent tumor development from OVA-expressing tumor cell inoculation. When the C1-mRNA nanovaccine was used as a therapeutic vaccine after tumor inoculation, it also effectively inhibited the growth of B16-OVA tumors (Fig. 5D and SI Appendix, Fig. S5C).
In previous experiments, mRNA encoding the exogenous antigen OVA257–264 was designed in the C1-mRNA nanovaccine as a surrogate tumor antigen. However, many tumor antigens are self-antigens and the host is more tolerant to such antigens. To further determine if the C1-mRNA vaccine could break self-tolerance and induce antitumor immunity against self-tumor antigen, we synthesized a new C1 nanovaccine with mRNA encoding TRP2, a self-antigen with mouse melanoma, to treat the B16 mouse melanoma model. Tumor growth curves revealed that the C1-TRP2 mRNA nanovaccine also effectively inhibited B16 tumor growth as a therapeutic vaccine (Fig. 5E and SI Appendix, Fig. S5D), suggesting that the vaccine efficacy of the C1 mRNA vaccine with a self-antigen was as good as the model antigen.
C1-mRNA Nanovaccine Efficacy Is Dependent on TLR4 Signaling.
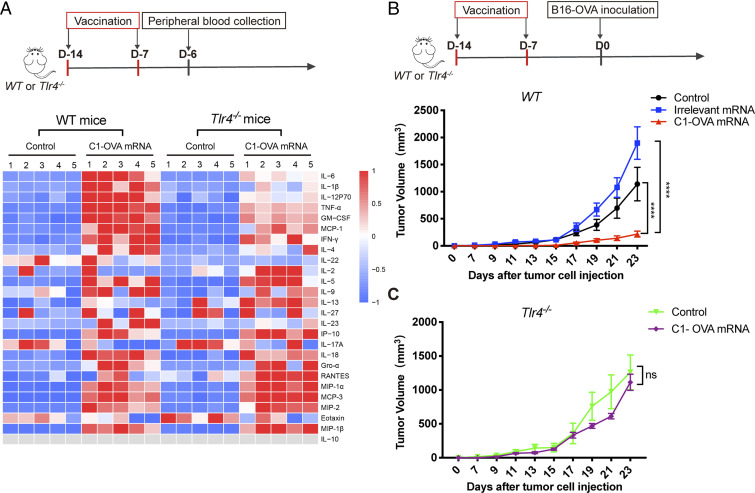
We next tested the in vivo role of TLR4 on the antitumor effect of the C1-mRNA nanovaccine using WT and Tlr4−/− mice. We analyzed the levels of 26 different cytokines/chemokines in mouse serum samples after C1-mRNA nanovaccine treatment (Fig. 6A). Multifactor immunoassay results showed that the levels of six cytokines (IL-6, IL-β, IL-12P70, TNF-α, GM-CSF, and MCP-1) were significantly different between WT and Tlr4−/− mice after two nanovaccine immunizations (Fig. 6B). We reasoned that such levels of cytokine release might contribute to immune activation and antitumor immunity, while not causing systemic inflammation. We then tested how TLR4 deficiency altered the antitumor activity of the C1-mRNA nanovaccine. Consistent with previous results, C1-OVA mRNA, but not C1 with mRNA encoding an irrelevant peptide, effectively inhibited B16-OVA tumor growth on WT mice, confirming that such an effect was tumor antigen specific (Fig. 6B and SI Appendix, Fig. S6A). More importantly, the tumor inhibition effect of the C1-OVA mRNA vaccine was almost completely abolished in Tlr4−/− mice, confirming the essential role of TLR4 in the C1-mRNA vaccine-induced antitumor effect (Fig. 6C and SI Appendix, Fig. S6B). In addition, the OVA mRNA vaccine formulated by nonimmune stimulatory C3 and D1 LNPs did not inhibit B16-OVA tumor growth (SI Appendix, Fig. S6 C–E). All these data strongly suggested that the antitumor efficacy of the C1-mRNA vaccination required both efficient antigen expression and TLR4 signaling activation.
Fig. 6.
The C1-mRNA nanovaccine induced a TLR4-dependent immune response in vivo. (A) Cytokine and chemokine profiles in serum samples from WT and Tlr4−/− mice after C1-mRNA vaccination. Upper: The schedule of vaccination and peripheral blood collection. Lower: Multiplex immunoassay measuring 26 cytokines/chemokines from serum samples. Control: PBS injection group. C1-OVA mRNA dose per mouse: mRNA encoding OVA257–264,10 μg; C1, 1.6 mg; n = 5 mice per group. (B and C) Antitumor efficacy of the C1-mRNA nanovaccine on WT and Tlr4−/− mice. Irrelevant mRNA that does not encode antigen serve as a negative control. Control: PBS injection group. The mRNA nanovaccine dose per mouse: mRNA, 10 μg; C1, 1.6 mg; n = 5 mice per group. Data represent the mean ± SEM ****P < 0.001; ns, not significant.
In Vivo Safety Profile of the C1-mRNA Nanovaccine.
To evaluate the safety profile of the C1-mRNA nanovaccines in vivo, main organs and blood serum of mice were collected on day 20 after immunization. Immunohistochemical staining revealed no major histological abnormality in the heart, liver, spleen, lung, and kidney tissues from mice with the C1-mRNA nanovaccine (SI Appendix, Fig. S7A). Hematological analysis also showed normal liver function (assessed by aspartate aminotransferase [AST] and alanine aminotransferase [ALT] activities), kidney function (assessed by uric acid [UA], creatinine [CR], and blood urea nitrogen [BUN] levels), and cardiac function (assessed by creatine kinase [CK] and lactate dehydrogenase [LDH] activities) of mice treated with nanovaccine, suggesting that the C1-mRNA nanovaccine did not cause notable toxicity in vivo (SI Appendix, Fig. S7B).
Discussion
Since the first report about lipid-like materials, synthesis, and screening for siRNA delivery, such materials have been widely used for siRNA, chemotherapeutic drugs, immunostimulating RNA, and mRNA delivery (22–25). Interestingly, the structures of the most efficient lipid-like materials screened for siRNA and mRNA delivery are quite different. This may reflect the different chemistry and binding affinity of double-stranded short siRNA versus single-stranded mRNA.
Previous study on antigen presentation revealed that antigen and innate immune-stimulating adjuvant needed to be codelivered into single antigen presenting cells to promote antigen presentation (18). As nanoparticles may serve as delivery vectors as well as self-adjuvants, they are particularly suitable for vaccine formulation (15, 29). Early work showed RNA molecules themselves could be immune stimulatory via engaging TLR3/7/8 signaling when delivered with specific lipid nanoparticles, and a recent study also reported such signaling contributed to the efficacy of the mRNA nanovaccine the authors developed (21, 24). Recently, Luo et al. reported a polymeric nanoparticle with a cyclic side chain that could activate STING-dependent type I interferon signaling and boost antitumor immunity, thereby acting as an effective minimalist nanovaccine (16). More recently, a screening of ionizable lipid-like materials identified a type of heterocyclic lipid with efficient mRNA delivery and intrinsic STING activation property for mRNA vaccine formulation (27). In comparison, the C1 lipid-like material we identified had a PAMAM ethylene group with a lipid tail of 12-carbon length, and activated TLR4-mediated proinflammatory signaling in DCs to potentiate antitumor immunity. The immune effect of C1-mRNA is largely STING independent, as we observed comparable levels of DC stimulation and T cell activation by C1-mRNA in WT and STING-deficient BMDCs. By contrast, TLR4 is essential for C1-mRNA-induced immune activation and in vivo antitumor efficacy. It is possible that C1-mRNA could activate other innate immune receptors besides TLR4, as TLR3/7/8 inhibitors partially inhibited C1-mRNA-induced inflammatory gene expression, and C1-mRNA-induced change of serum cytokine levels was not completely abolished in Tlr4−/− mice. However, the complete absence of antitumor efficacy of C1-mRNA on Tlr4−/− mice strongly argues the pivotal role of TLR4 signaling in C1-mRNA vaccine-induced antitumor immunity. TLR4 activation induces both NF-κB signaling and type I interferon signaling, which are both critical for antigen presentation function of DCs (28). As C1 induced expression of both inflammatory cytokine genes and type I interferon genes, further work is warranted to delineate which downstream signaling of TLR4 contribute to the efficacy of the C1-mRNA vaccine.
It is well documented that either excess type I interferon response or TLR4 activation-induced proinflammatory cytokine could be systemically toxic (4), while the above-mentioned cyclic lipids or C1 nanovaccine-induced innate immune activation did not show obvious toxicity. The exact reason is currently unknown. It is possible that cyclic lipids or C1 lipid are mild agonists for these innate immune receptors, and thus eliciting a response sufficient for dendritic cell activation while not causing systemic inflammation. Monophosphoril lipid A, a vaccine adjuvant currently under test in several clinical trials, is a detoxified component extracted from LPS and acts as a milder TLR4 agonist compared to LPS (30). We indeed observed a lower magnitude of cytokine induction by C1-mRNA than that of LPS, but further characterization is warranted to delineate the underlying mechanisms.
Notably, C1-mRNA exhibited the highest antigen expression level compared to that of mRNA complexed with other lipid-like materials tested, and efficient expression of mRNA-encoded antigen is often a prerequisite for a successful mRNA vaccine. Because C1-mRNA also induced innate immune activation to promote antigen presentation and T cell activation, we could not convincingly distinguish the contributions of efficient mRNA translation and innate immune activation to the therapeutic efficacy of the C1-mRNA vaccine. Based on the available data, it is most likely that efficient mRNA translation and innate immune activation may both contribute to the therapeutic efficacy of C1-mRNA. As such, the relative contributions of these two factors merit future study.
In summary, we have identified a lipid-like material-based minimalist mRNA nanovaccine with self-adjuvant activity through TLR4 signaling. The C1 nanovaccine exhibited potent antitumor immunity when loaded with tumor antigen-encoding mRNA without obvious toxicity. Considering that neoantigens from cancer patients are highly personalized, the C1-mRNA nanovaccine could offer a versatile platform to prepare personalized and effective vaccine for cancer treatment. Further optimization of large-scale nanoparticle production and mRNA encoding predicted neoantigens from human cancer patients need to be tested to advance the future clinical application of the C1-mRNA nanovaccine to cancer immunotherapy.
Materials and Methods
The data authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit platform (https://www.researchdata.org.cn/default.aspx), and inspected/approved by Sun Yat-sen University Cancer Center Data Access/Ethics Committee with the approval number RDDB2021001065.
Cell Culture.
B16 murine melanoma cell line and HEK293 cells were obtained from ATCC. MC38 murine colon adenocarcinoma cell line was kindly provided by Xuanming Yang, Shanghai Jiaotong University, Shanghai, China. Xuanming Yang, Shanghai Jiaotong University, Shanghai, China. B16-OVA and MC38-OVA cells were constructed by stably expressing ovalbumin cDNA. DC2.4 is a murine dendritic cell line kindly provided by Kenneth Rock, University of Massachusetts Medical School, Worcester, MA. B3Z hybridoma cells were kindly gifted by Nilabh Shastri, University of California, Berkeley, CA. Cells were maintained either in DMEM or RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin antibiotic (Gibco). Cell culture and all biological experiments were performed at 37 °C in 5% CO2 conditions in a cell culture incubator. All the cell lines were tested to be mycoplasma free.
Six- to eight-week-old female mice (C57BL/6J mice, Tlr4−/− or STING cKO mice) were used to isolate BMDCs. Briefly, the femur and tibia from mice hind legs were collected and bone marrow cells were flushed out with 1% FBS-containing PBS using a syringe. Cells were briefly treated with ACK lysis buffer to remove red blood cells and then resuspended into RPMI1640 medium with 10% FBS, antibiotics, and β-mercaptoethanol (55 μM, Gibco). Cells were grown with a supplement of recombinant murine granulocyte macrophage–colony stimulating factor (GM-CSF) (20 ng/mL, Peprotech) and IL-4 (20 ng/mL, Peprotech). Cell culture medium was refreshed every other day. Semiattached cells were harvested as immature BMDCs on day 6.
Materials.
Cationic ethylenediamine core PAMAM dendrimer generation 0 (G0), DSPE-PEG 2000, and coumarin6 were purchased from Sigma-Aldrich. DiR iodide [1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide] was also from Sigma-Aldrich. Lipofectamine 2000 was purchased from Invitrogen. LPS, ODN2088 (TLR9 inhibitor), and ODN2088 control (TLR7/8 inhibitor) were purchased from Invivogen, Inc. Fluorescein12-labeled UTP (F12-UTP), amiloride (macropinocytosis inhibitor), cytochalasin D (phagocytosis inhibitor), CU CPT 4a (TLR3 inhibitor) and TAK-242 (TLR4 inhibitor) were purchased from Apexbio, Inc.
Mice.
Six- to eight-week-old female C57BL/6J mice were purchased from Vital River Laboratory (Beijing). OT-I mice and Tlr4−/− mice were from The Jackson Laboratory. Stingflox/floxCD11c-CRE (designated as STING cKO in the text) mice were from Shanghai Model Organisms, Inc. All mice were maintained under specific pathogen-free conditions and used in accordance with the animal experimental guidelines. All the animal work was approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University.
Synthesis of Cationic Lipid-Like Materials.
The cationic lipid-like materials were synthesized from PAMAM dendrimer G0 using a ring-opening reaction by reacting with 1,2 epoxytetradecane according to the following described procedures. First, PAMAM dendrimer G0 was mixed with 1,2-alkylene oxide of different segment length (including 1,2-epoxyhexane, 1,2-epoxyoctane, 1,2-epoxydodecane, 1,2-epoxytetradecane, and 1,2-epoxyhexadecane1) at different molar ratios of 1:5, 1:7, and 1:9. Then, 10 mL methanol was added into the mixture in a round-bottom flask. Ring-opening reaction was set to 90 °C in an oil bath for 48 h. At last, the crude mixtures were separated by steaming to obtain an oily mixture after the reaction. Fifteen different kinds of lipid-like materials were obtained and grouped as A1-A3, B1-B3, C1-C3, D1-D3, and E1-E3.
Preparation of Modified mRNA.
Plasmid vectors including pcDNA3.1-SIINFEKL (OVA257–264) and pcDNA3.1-VYDFFVWL (Trp2180–188) were constructed. We also modified the different sites of mRNA including 5′ end modification and 3′ end poly(A) tailing according to previous reports (31, 32). Briefly, different linker sequences and signal peptides to link epitopes were added to enhance transcription in vitro. The modification of 3′ end includes a 100-bp poly(A) tail and two serial fragments (2× β-globin UTR) in front of the poly(A) tail. The 3′ β-globin UTR was amplified from 3′ β-globin UTR sense, 5′-ttactcgagagctcgctttcttgctgtccaatttct-3′, 3′ β-globin UTR antisense, 5′-ttagtcgacgcagcaatgaaaataaatgttttttattaggca-3′. Synthetic DNA fragments connected by nonimmunogenic glycine/serine linkers (start linker: GGSGGGGSGG and end linker: GGSLGGGGSG) were cloned into a starting vector containing MITD-domains (MITD, IVGIVAGLAVLAVVVIGAVVATVMCRRKSSGGKGGSYSQAASSDSAQGSDVSLTA) for optimized routing to HLA class I and II pathways. In addition, a 78-bp signal peptide derived from an MHC class I molecule (Sec) was amplified (primers: Sec sense, 5′-aagcttagcggccgcaccatgcgggtcacggcgccccgaacc-3′; Sec antisense, 5′-ctgcagggagccggcccaggtctcggtcag-3′) and inserted in front of the antigen peptide sequence.
In vitro mRNA transcription (IVT) was performed with HiScribe T7 ARCA mRNA kit (with tailing) (NEB). The plasmid vector was linearized by BsmBI enzyme and purified, and in vitro mRNA transcription was performed according to the manufacturer’s instructions. F12-mRNA was synthesized by adding 20% F12-UTP into the 2× ARCA/NTP mix when conducting IVT reactions.
Preparation and Characterization of Lipid-Like Material-Based Nanovaccine.
A self-assembly method was used to prepare mRNA-encapsulated polymer-lipid-like material nanoparticle. In brief, lipid-like materials and DSPE-PEG 2000 were dissolved separately in dimethyl sulfoxide at concentrations of 1 mg/mL and 2 mg/mL, respectively. First, lipid-like material and synthesized epitope-encoding mRNA in aqueous solution were mixed at a weight ratio of 160:1 in a small glass vial to form cationic lipid-like material-mRNA complexes. Then, DSPE-PEG 2000 was added followed by vortex mixing. The weight ratio of lipid-like material to DSPE-PEG 2000 was 5:1. Next, the mixture was slowly dropped into the corresponding volume of sterile water with vortex mixing. Upon nanoprecipitation, nanoparticles formed instantly for 10 min standing still at room temperature. The nanoparticles were then washed with sterile water using Amicon Ultra Centrifugal Filters (molecular weight cutoff, 100 kDa; Millipore) to remove the organic solvent and free compounds and finally concentrated into a suitable volume of water. The nanoparticles were freshly prepared for in vitro and in vivo experiments. The nanoparticles prepared with lipid-like material C1, antigen-encoding mRNA, and DSPE-PEG 2000 were termed C1-mRNA nanovaccine. In some experiments, coumarin6 was used to label the C1-mRNA nanovaccine. The weight ratio of coumarin6 to C1 was 1:100, and the product was termed C1-mRNA-coumarin6.
The particle size and zeta potential of the C1-mRNA nanovaccine complex were characterized by dynamic light scattering (DLS) using a Zetasizer NanoZS90 (Malvern Instruments) at room temperature. The morphology and shape of the C1-mRNA nanovaccine complex were observed under the JEM-14000PLUS TEM (JEOL Ltd, Japan). Phosphotungstic acid (1%) was used for the negative staining of the nanovaccine complex to enhance the electron contrast.
In Vitro Antigen Presentation Assay.
An in vitro antigen presentation assay was performed as following protocol. DC2.4 cells or mature BMDCs from WT mice, Tlr4−/− mice, or Stingflox/floxCD11c-CRE (designated as STING cKO in the text) mice were seeded in 24-well, flat-bottom culture plates at a density of 2 × 105 cells/well and cultured for 12 h at 37 °C. Each well was washed once with PBS and then the cells were pulsed with soluble OVA257–264 peptide (SIINFEKL), Lipfectamine 2000-mRNA encoding OVA257–264, lipid-like material-mRNA encoding OVA257–264 for indicated times. The concentration of mRNA and lipid were 120 ng/mL and 19.2 μg/mL, respectively. Then, the cells were cocultured with 1 × 106 B3Z cells or OT-I cells isolated from OT-I mice for 16 h at 37 °C. The activation of stimulated B3Z or OT-I cells was detected by determining IL-2 concentration in the culture supernatant using a murine IL-2 ELISA kit (eBioscience).
For lacZ activity measurement, B3Z T cells in the cell culture plate were lysed by 50 μL LacZ lysis buffer and freeze thawed, followed by adding of 50 μL PBS containing 0.5% bovine serum albumin and 100 μL substrate solution (1 mg/mL chlorophenol red β-D-galactopyranoside) dissolved in β-galactosidase buffer. The plate was incubated at 37 °C for 5 to 10 h until color development reached a proper level, followed by color intensity reading at 590 nm using a microtiter plate reader.
In Vitro Transfection of Dendritic Cells by C1-mRNA.
DC2.4 cells were seeded in dishes (35-mm glass bottom dishes with a 10-mm microwell, Corning) at a density of 5 × 104 cells/well and cultured overnight. Cells were transfected with F12-mRNA by different delivery vectors. The experiment groups included F12-mRNA group (F12-mRNA only), Lip2000-F12-mRNA group (Lipfectamine 2000-F12-mRNA), and the C1-F12-mRNA group. Lip2000 was used as a standard transfection reagent (according to the manufacturer’s protocol). Then the cells were incubated for 6 h followed by washing with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. Cells were then washed twice with PBS and permeabilized by incubation in 0.2% Triton X-100 in PBS for 10 min. Finally, cells were washed twice with PBS, counterstained with 500 nm DAPI (Cell Signaling Technology), and analyzed by confocal laser scanning microscopy (LSM880, Zeiss).
Real-Time PCR.
Total cell RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA was reverse transcribed using the Primer Script RT Reagent Kit with genomic DNA Eraser (Takara). Real-time PCR was performed using the SYBR Premix kit (Genstar) and analyzed using the Bio-Rad detection system. The expression of individual genes was calculated by using a ΔΔCT method and normalized to the expression of actin. The primer sequences for mouse genes are shown in SI Appendix, Table S1.
ELISA.
Cytokines (IL-1β, IL-2, IL-6, IL-12P70, and IFN-γ) released from B3Z cells, DC2.4 cells, BMDCs, or OT-I cells in cell supernatants were detected by individual cytokine ELISA kits according to the manufacturer’s instructions (eBioscience).
Biodistribution of mRNA Nanovaccine in Mice.
The C1-mRNA nanovaccine including DiR (a near infrared fluorescent) (C1-mRNA-DiR) was used for biodistribution analysis. The weight ratio of DiR to lipid-like material C1 was 1:100. Mice received a one-time injection of C1-mRNA-DiR subcutaneously at the right inguinal region at a dose of 10 µg mRNA per mouse. After 24 h, their organs were collected and imaged with an IVIS Lumina III imaging system (Perkin-Elmer) for the biodistribution analysis. The excitation wavelength and emission wavelength of DiR were 750 and 780 nm, respectively.
In Vivo Immunization and Flow Cytometry Analysis.
The spleens and draining lymph nodes of vaccinated mice were collected on day 7 after two vaccinations from the control group with PBS injection, OVA peptide plus Alum group (Sigma, OVA257–264, SIINFEKL, #7951, 200 μg/mouse; Imject Alum Adjuvant, #77162), and the C1-OVA mRNA nanovaccine group. For Alum adjuvant formulation, Alum was added dropwisely with constant mixing to the OVA peptide solution, so that the final volume ratio of Alum to OVA peptide solution was 1:2. After vaccination, spleens and lymph nodes were isolated and prepared into single-cell suspension, and cells were stimulated with OVA peptide (SIINFEKL, 1 μg/mL) for 24 h. The cells were incubated with PE conjugates of MHC tetramer specific for CD8 T cells recognizing OVA epitope SIINFEKL (MBL International, clone: TS-5001-1C, diluted 1:100 in fluorescence-activating cell sorting [FACS] buffer) for 30 min at room temperature. Subsequently, the cells were incubated with CD3e-eFlour450 (eBioscience, clone: 145-2C11, diluted 1:100 in FACS buffer) and CD8a-fluorescein isothiocyanate (eBioscience, clone: 53-6.7, diluted 1:100 in FACS buffer) for 20 min at room temperature. The cells were washed with FACS buffer, and the data were collected on a BD LSR Fortessa X-20, followed by analysis using FlowJo software version 7.6.
Statistical Analyses.
Statistical significance was determined using an independent two-tailed Student’s t test or two-way ANOVA, and P < 0.05 was considered statistically significant. Data were analyzed using GraphPad Prism 7 software.
Supplementary Material
Acknowledgments
We thank Dr. Seeruttun Sharvesh Raj for language editing. This work was supported by grants from the National Key R&D Program of China (2017YFC0908500/2017YFC0908503), the National Natural Science Foundation of China (Grants 81773051, 81803005, and 51973243), the Guangdong Innovative and Entrepreneurial Research Team Program (2016ZT06S638), and National Science and Technology Major Project of the Ministry of Science and Technology of China (2018ZX10301402).
Footnotes
Competing interest statement: A patent has been filed for materials described in this manuscript by the authors.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005191118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or SI Appendix.
References
- 1.Miller J. F., Sadelain M., The journey from discoveries in fundamental immunology to cancer immunotherapy. Cancer Cell 27, 439–449 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Kantoff P. W.et al.; IMPACT Study Investigators , Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 363, 411–422 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Melero I., et al., Therapeutic vaccines for cancer: An overview of clinical trials. Nat. Rev. Clin. Oncol. 11, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Coffman R. L., Sher A., Seder R. A., Vaccine adjuvants: Putting innate immunity to work. Immunity 33, 492–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gubin M. M., Artyomov M. N., Mardis E. R., Schreiber R. D., Tumor neoantigens: Building a framework for personalized cancer immunotherapy. J. Clin. Invest. 125, 3413–3421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartzentruber D. J., et al., gp100 peptide vaccine and interleukin-2 in patients with advanced melanoma. N. Engl. J. Med. 364, 2119–2127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines–A new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahin U., Karikó K., Türeci Ö., mRNA-based therapeutics–Developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Islam M. A., et al., Biomaterials for mRNA delivery. Biomater. Sci. 3, 1519–1533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selmi A., et al., Uptake of synthetic naked RNA by skin-resident dendritic cells via macropinocytosis allows antigen expression and induction of T-cell responses in mice. Cancer Immunol. Immunother. 65, 1075–1083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallen K. J., et al., A novel, disruptive vaccination technology: Self-adjuvanted RNActive(®) vaccines. Hum. Vaccin. Immunother. 9, 2263–2276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang R., et al., Cancer cell membrane-coated adjuvant nanoparticles with mannose modification for effective anticancer vaccination. ACS Nano 12, 5121–5129 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Liu H., et al., Structure-based programming of lymph-node targeting in molecular vaccines. Nature 507, 519–522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuai R., Ochyl L. J., Bahjat K. S., Schwendeman A., Moon J. J., Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat. Mater. 16, 489–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C., Ye Y., Hu Q., Bellotti A., Gu Z., Tailoring biomaterials for cancer immunotherapy: Emerging trends and future outlook. Adv. Mater. 29, (2017). [DOI] [PubMed] [Google Scholar]
- 16.Luo M., et al., A STING-activating nanovaccine for cancer immunotherapy. Nat. Nanotechnol. 12, 648–654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroll A. V., et al., Biomimetic nanoparticle vaccines for cancer therapy. Adv. Biosyst. 3, e1800219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blander J. M., Medzhitov R., Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature 440, 808–812 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Mehta N. K., Moynihan K. D., Irvine D. J., Engineering new approaches to cancer vaccines. Cancer Immunol. Res. 3, 836–843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachmann M. F., Jennings G. T., Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 10, 787–796 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Kranz L. M., et al., Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Akinc A., et al., A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 26, 561–569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love K. T., et al., Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. U.S.A. 107, 1864–1869 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen D. N., et al., Lipid-derived nanoparticles for immunostimulatory RNA adjuvant delivery. Proc. Natl. Acad. Sci. U.S.A. 109, E797–E803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X., et al., Enhancing tumor cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proc. Natl. Acad. Sci. U.S.A. 110, 18638–18643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oberli M. A., et al., Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 17, 1326–1335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao L., et al., Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 37, 1174–1185 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi K., Karin M., NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 18, 309–324 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Goldberg M. S., Improving cancer immunotherapy through nanotechnology. Nat. Rev. Cancer 19, 587–602 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Shetab Boushehri M. A., Lamprecht A., TLR4-Based immunotherapeutics in cancer: A review of the achievements and shortcomings. Mol. Pharm. 15, 4777–4800 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Kreiter S., et al., Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals. J. Immunol. 180, 309–318 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Holtkamp S., et al., Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 108, 4009–4017 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.