Significance
Opsins are photosensitive receptors capturing specific wavelengths of incoming light to convey color vision across animals. Lack of reliable expression systems to study invertebrate Gq opsins has limited our ability to tease apart genotype–phenotype relationships underlying spectral tuning and visual adaptations in insects compared to homologous yet phylogenetically distant vertebrate Gt opsin lineages. We developed a robust method to express invertebrate opsin proteins in vitro, which we apply to study the visual system of a lycaenid butterfly. Our detailed molecular characterization of red-shifted long-wavelength and duplicate short-wavelength Gq insect opsins, together with a broad mutagenesis approach for exploring invertebrate opsin sequence-visual pigment functions, begins pinpointing the proximate evolutionary mechanisms underlying the diversification of color vision systems across animal life.
Keywords: molecular evolution, ecological adaptation, visual system, spectral sensitivity, insects
Abstract
Color vision has evolved multiple times in both vertebrates and invertebrates and is largely determined by the number and variation in spectral sensitivities of distinct opsin subclasses. However, because of the difficulty of expressing long-wavelength (LW) invertebrate opsins in vitro, our understanding of the molecular basis of functional shifts in opsin spectral sensitivities has been biased toward research primarily in vertebrates. This has restricted our ability to address whether invertebrate Gq protein-coupled opsins function in a novel or convergent way compared to vertebrate Gt opsins. Here we develop a robust heterologous expression system to purify invertebrate rhodopsins, identify specific amino acid changes responsible for adaptive spectral tuning, and pinpoint how molecular variation in invertebrate opsins underlie wavelength sensitivity shifts that enhance visual perception. By combining functional and optophysiological approaches, we disentangle the relative contributions of lateral filtering pigments from red-shifted LW and blue short-wavelength opsins expressed in distinct photoreceptor cells of individual ommatidia. We use in situ hybridization to visualize six ommatidial classes in the compound eye of a lycaenid butterfly with a four-opsin visual system. We show experimentally that certain key tuning residues underlying green spectral shifts in blue opsin paralogs have evolved repeatedly among short-wavelength opsin lineages. Taken together, our results demonstrate the interplay between regulatory and adaptive evolution at multiple Gq opsin loci, as well as how coordinated spectral shifts in LW and blue opsins can act together to enhance insect spectral sensitivity at blue and red wavelengths for visual performance adaptation.
Opsins belong to a diverse multigene family of G protein-coupled receptors that bind to a small nonprotein retinal moiety to form photosensitive rhodopsins and enable vision across animals (1–4). The tight relationship between opsin genotypes and spectral sensitivity phenotypes offers an ideal framework to analyze how specific molecular changes give rise to adaptations in visual behaviors (5). Notably, independent opsin gene gains and losses (6–13), genetic variation across opsins (14–16), spectral tuning mutations within opsins (17–21), and alterations in visual regulatory networks (22, 23) have contributed to opsin adaptation. Yet, the molecular and structural changes underlying the remarkable diversification of spectral sensitivity phenotypes identified in some invertebrates, including crustaceans and insects (24–27), are far less understood than those in vertebrate lineages (28–32).
The diversity of opsin-based photoreceptors observed across animal visual systems is produced by distinct ciliary vertebrate c-opsin and invertebrate rhabdomeric based r-opsin subfamilies that mediate separate phototransduction cascades (31, 33–35). Vertebrate c-opsins function through the G protein transducing (Gt) signaling pathway, which activates cyclic nucleotide phosphodiesterase, ultimately resulting in a hyperpolarization response in photoreceptor cells through the opening of selective K+ channels (31, 36). By contrast, insect opsins transmit light stimuli through a Gq-type G protein (33, 37) with phosphoinositol (PLCβ) acting as an effector enzyme to achieve TRP channel depolarization in the invertebrate photoreceptor cell (34, 38).
All vertebrate visual cone opsins derive from four gene families: short-wavelength-sensitive opsins SWS1 (or ultraviolet [UV]) with λmax 344 to 445 nm and SWS2 with λmax 400 to 470 nm, and longer-wavelength-sensitive opsins that specify the green MWS (or Rh2) pigments with λmax 480 to 530 nm and red-sensitive LWS pigments with λmax 500 to 570 nm (5, 30). Most birds and fish have retained the four ancestral opsin genes (39), with notable opsin expansions in cichlid fish opsins (23, 40), whereas SWS1 is extinct in monotremes, and SWS2 and M opsins are lost in marsupials and eutherian mammals (41). In primates, trichromatic vision is conferred through SWS1 (λmax = 414 nm) and recent duplicate MWS (λmax = 530 nm) and LWS opsins (λmax = 560 nm) (42–44). In vertebrates, molecular evolutionary approaches and well-established in vitro opsin purification have identified the complex interplay between opsin duplications, regulatory and protein-coding mutations controlling opsin gene tuning, and spectral phenotypes notably in birds, fish, and mammals (45–47).
Insect opsins are phylogenetically distinct but functionally analogous to those of vertebrates, and the ancestral opsin repertoire consists of three types of light-absorbing rhabdomeric Gq-type opsin specifying UV (350 nm), short-wavelength (blue, 440 nm) and long-wavelength pigments (LW, 530 nm) (48). Given the importance of color-guided behaviors and the remarkable photoreceptor spectral diversity observed in insects (26, 27), the dynamic opsin gene diversification found across lineages (Fig. 1) highlights their potentially central role in adaptation (27, 49, 50), yet the molecular basis of opsin functionality of rhabdomeric invertebrate Gq opsins remains understudied.
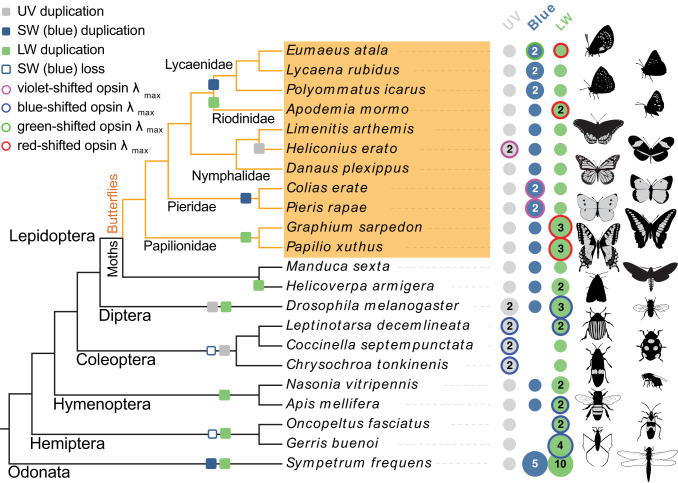
Fig. 1.
Visual opsin gene evolution and spectral tuning mechanisms in insects. Visual opsin genes of the Atala hairstreak (E. atala, Lepidoptera, Lycaenidae) in comparison with those encoded in the genomes of diverse insects. The opsin types are highlighted in gray for UV, in blue for short wavelength (SW), and in green for long wavelength (LW). Numbers indicate multiple opsins, whereas no dot indicates gene loss. Colored circles indicate instances of shifted spectral sensitivities in at least one of the encoded opsins. The direction of shift is inferred from the opsin lambda max that departs from the typical range of absorbance in the opsin subfamily using wavelength boundaries for the various colors: UV <380 nm, violet 380 to 435 nm, blue 435 to 492 nm, green 492 to 530 nm, and red shifted >530 nm. Coleopteran lineages, and some hemipterans, lost the blue opsin locus and compensated for the loss of blue sensitivity via UV and/or LW gene duplications across lineages (11, 12). In butterflies, extended photosensitivity at short wavelengths is observed in Heliconius erato with two UV opsins at λmax = 355 nm and 398 nm (10) and in P. rapae with two blue opsins with λmax = 420 and 450 nm (17). A blue opsin duplication occurred independently in lycaenid butterflies (61). LW opsin duplications occurred independently in most major insect lineages (6, 16, 55) and confer a variable range of LW sensitivities with or without additional contributions from lateral filtering. In order to extend spectral sensitivity at longer wavelengths while sharpening blue acuity, some lycaenid butterflies have evolved a new color vision mechanism combining spectral shifts at a duplicate blue opsin and at the LW opsin. Images credit: Christopher Adams (illustrator).
The recurrent evolution of red receptors in insects in particular suggests that perception of longer wavelengths can play an important role in the context of foraging, oviposition, and/or conspecific recognition (6, 27, 51–54). In butterflies, several mechanisms are likely to have provided extended spectral sensitivity to longer wavelengths. LW opsin duplications along with the evolution of lateral filtering between ommatidia has been demonstrated in two papilionids, Papilio xuthus (27) and Graphium sarpedon (55), as well as in a riodinid (Apodemia mormo) (6, 54). Lateral filtering pigments are relatively widespread across butterfly lineages, e.g., Heliconius (56), Pieris (57), Colias erate (58), and some moths [Adoxophyes orana (59) and Paysandisia archon (60)]. These pigments absorb short wavelengths and aid in shifting the sensitivity peak of green LW photoreceptors to longer wavelengths (27, 51, 56, 57, 61, 62). Despite creating distinct spectral types that can contribute to color vision, as identified in nymphalid (56), pierid (57), and lycaenid (62) species, all of which lack duplicated LW opsins (61, 63), lateral filtering alone cannot extend photoreceptor sensitivity toward the far red (700 to 750 nm) beyond the exponentially decaying long-wavelength rhodopsin absorbance spectrum (51). Thus, molecular variation of ancestral LW opsin genes is likely to have contributed an as yet underexplored mechanism to the diversification of long-wavelength photoreceptor spectral sensitivity. However, disentangling the relative contributions of lateral filtering and pure LW opsin properties has remained technically challenging using classical electrophysiological approaches (14, 64, although see, e.g., refs. 65, 66, 67) and has been limited by the lack of in vitro expression systems suitable for LW opsins.
While opsin duplicates have been identified in numerous organisms, the spectral tuning mechanisms and interplay between new opsin photoreceptors in invertebrate visual system evolution are less well understood. Here we combine physiological, molecular, and heterologous approaches to start closing this gap in our knowledge of invertebrate Gq opsin evolution by investigating the functions, spectral tuning, and implications of evolving new combinations of short- and long-wavelength opsin types in lycaenid species. This butterfly group, comprising the famous blues, coppers, and hairstreaks, is the second largest family with about 5,200 (28%) of the some 18,770 described butterfly species (68). In light of their remarkable behavioral, ecological, and morphological diversity (69, 70), as well as pioneer studies in the Lycaena and Polyommatus genera supporting the rapid evolution of color vision in certain lineages (56, 61, 62), lycaenids provide an ideal candidate system for investigating opsin evolution and visual adaptations. Using the Atala hairstreak, Eumaeus atala, as a molecular and ecological model, we find coordinated spectral shifts at short- and long-wavelength Gq opsin loci and demonstrate that the combination of six ommatidial classes of photoreceptors in the compound eye uniquely extend spectral sensitivity at long wavelengths toward the far-red while concurrently sharpening acuity of multiple blue wavelengths. Together, these findings link the evolution of four-opsin visual systems to adaptation in the context of finely tuned color perception critical to the behavior of these butterflies.
Results
Red Rhodopsin Receptors Drive Spectral Sensitivity at Long Wavelengths in Lycaenid Butterflies.
To identify red-sensitive receptors, we performed analyses of in vivo photochemical rhodopsin measurements in adult lycaenid butterflies. A remarkable red eyeshine coloration was observed in the dorsoequatorial region of two species in the Theclinae, the Japanese Oakblue, Arhopala japonica, and the Atala hairstreak, E. atala (Fig. 2 A and B and SI Appendix, Fig. S1 A and B). These observations raised the question of whether these butterflies might have enhanced red photoreceptor sensitivity due to lateral filtering and/or the properties of opsin proteins bound to ommatidial photoreceptor cell membranes.
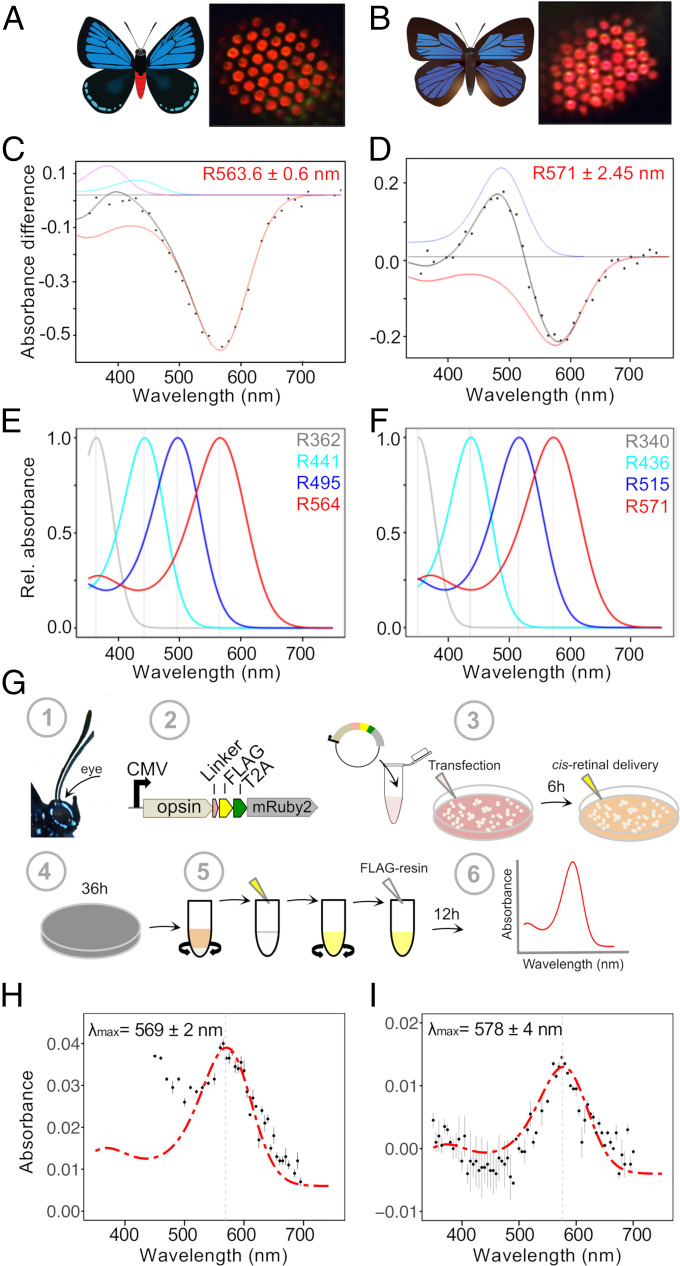
Fig. 2.
Red-shifted long-wavelength opsins extend spectral sensitivity toward the far-red in lycaenid butterflies. (A and B) Illustrations of E. atala (A) and A. japonica (B). Butterfly eyeshine photographs on the Right show a series of adjacent ommatidial units filled primarily with red rhodopsins in the dorsal retina (SI Appendix, Fig. S1). (C and D) Photochemical difference spectra (DS, black dots) were obtained following partial bleaches of long-wavelength rhodopsins from dorsal retina of intact butterflies and represent absorbance differences between amounts of rhodopsin (R, red) and metarhodopsin photoproducts when present (M, blue) after a dark period that followed photoconversion. Each black curve represents a computed difference spectrum for least square fits estimates at (C) R564 of E. atala and (D) R571 of A. japonica. In E. atala, the difference spectrum was acquired upon complete degradation of the M photoproduct and with small contributions from R440 (cyan) and retinal binding protein (RBP395, magenta). (E and F) Normalized eye spectral sensitivity of E. atala (E) and A. japonica (F) computed from a retinal densitometry analysis (SI Appendix, Fig. S2 I and J) reveals the contribution of four rhodopsins: UV, two blue opsins, and one long-wavelength opsin. (G) Schematics of the in vitro opsin purification assay. Opsin open reading frames (ORFs) were amplified from eye cDNA (1) and subcloned in an expression cassette derived from the pcDNA5 vector and containing a C-terminal FLAG epitope flanked by a peptide linker, a T2A cleavage site, and the cytoplasmic fluorescent mRuby2 marker for visualizing cell transfection efficiency (2). DNA–lipid complexes were transfected into HEK293T cells followed by 11-cis-retinal delivery. All subsequent steps were performed under dim red-light illumination (3). Culture plates were wrapped in aluminum foil and incubated for 36 h (4). Cells were harvested, active membrane-bound rhodopsins were extracted via membrane solubilization, and rhodopsin–chromophore complexes were nutated as detailed in Materials and Methods and SI Appendix (5). After an overnight incubation with FLAG resin, rhodopsin complexes were purified via FLAG resin affinity (6). All fractions were analyzed by Western blot, and the absorbance properties of the eluate fractions containing purified rhodopsins were measured via UV-VIS spectroscopy. (H and I) Functional characterization of red-shifted long-wavelength lycaenid butterfly opsins. Dark absorbance spectra of long-wavelength rhodopsin (LWRh) expressed and purified using the HEK293T transient cell culture system. LWRh absorbance spectra are indicated with black dots, and a rhodopsin template (136) was computed to obtain the best estimates of λmax fitting the data. (H) E. atala purified LW opsin with λmax = 569 nm, n = 2 protein eluate aliquots. (I) A. japonica purified LW opsin with λmax = 578 nm; n = 2 protein eluate aliquots. Bars represent ±SEs of the mean.
To investigate this question, we first used repeated white-light flashes to partially bleach the eye. This analysis revealed that only a small fraction of ommatidia are surrounded by non-light-sensitive lateral cherry-red granules (SI Appendix, Fig. S1C). The remaining ommatidia in the illuminated compound eye region lost the red eyeshine when subjected to repeated flashes of bright light. Photochemical difference spectra obtained from fully dark-adapted eyes identified LW spectral sensitivities in dark-adapted eyes of E. atala with λmax ± SE at 563.6 ± 0.6 nm (Fig. 2C), and with λmax at 571 ± 2.5 nm in A. japonica (Fig. 2D and Dataset S1A). These photochemical results support the hypothesis that elevated sensitivity at long wavelengths in E. atala and A. japonica is primarily due to opsin-based photoreceptors.
Motivated by these findings, we further examined in detail the in vivo contributions of all other rhodopsins in both species. Epi-microspectrophotometric (epi-MSP) difference spectra obtained after ommatidial flashing revealed a blue rhodopsin at 441.3 ± 4.7 nm in E. atala (SI Appendix, Fig. S2A and Dataset S1B) and at 435.8 ± 0.8 nm in A. japonica (SI Appendix, Fig. S2B and Dataset S1C). Maximal sensitivity at short wavelengths was found to be accounted for by UV-receptors absorbing maximally at 361.7 ± 2.9 nm in E. atala (SI Appendix, Fig. S2C and Dataset S1C) and at 340 ± 1.3 nm in A. japonica (SI Appendix, Fig. S2D and Dataset S1C). Finally, a retinal densitometric analysis indicated that these physiological data are best explained by a visual model in which a fourth rhodopsin is present in the eye with λmax values at 493.5 ± 0.6 nm for E. atala (SI Appendix, Fig. S2 E and G) and 514.7 ± 0.6 nm for A. japonica (SI Appendix, Fig. S2 F and H and Dataset S1 D and E). Thus, the eyes of E. atala (Fig. 2E) and A. japonica (Fig. 2F) likely contain a UV, two blue, and a LW rhodopsin.
Functional Variation of Long-Wavelength Opsins Accounts for Red Spectral Sensitivity.
To examine the contributions of single rhodopsins outside of the complex eye environment, we characterized their encoded genes and assessed the rhodopsin complex absorbance spectra in vitro. Eye transcriptome profiling revealed a single LW opsin gene in E. atala (SI Appendix, Supplementary Methods and Dataset S2A) and a single orthologous LW opsin gene in A. japonica, which is in line with earlier molecular evidence for a single LW opsin gene in other Lycaenidae (8, 56, 61). To functionally characterize the encoded opsin proteins, we engineered a new expression cassette to achieve heterologous expression under a strong cytomegalovirus (CMV) promoter (Fig. 2G). Combined with an optimized in vitro HEK293T purification assay (Materials and Methods and SI Appendix), we increased membrane protein expression levels to efficiently produce monomeric units of LW, UV, and blue heterologous Gq opsin proteins in vitro (SI Appendix, Fig. S3A and Dataset S2B). When purified from large-scale HEK293T cell cultures and reconstituted in vitro in the dark in the presence of 11-cis-retinal, we found that the LW rhodopsin from E. atala absorbs maximally at λmax = 569 ± 2 nm (CI95% = 565 to 573 nm) (Fig. 2H and Dataset S2C). The LW opsin from A. japonica absorbs maximally at λmax = 578 ± 4 nm (CI95% = 570 to 586 nm) (Fig. 2I and Dataset S2C). The absorbance maxima of purified LW rhodopsin measurements are within the confidence intervals of the best fit for LW linear absorbance estimates in vivo.
Our HEK293T cell culture expression system thus enabled us to assess the functionality of LW rhodopsins outside of the complex eye environment. Our findings indicate that a single red-shifted LW opsin G protein-coupled receptor gene in these lycaenid butterflies encodes the rhodopsin responsible for extending spectral sensitivity at long wavelengths above 600 nm, compared to insects equipped with ancestral green LW opsins (6, 8, 71).
Blue Opsin Duplication and Spectral Tuning Extend Photoreceptor Types.
To further examine how E. atala perceives wavelengths using four rhodopsins, we next performed a histological reconstruction of photoreceptor organization in a typical ommatidium (SI Appendix, Fig. S4). We found that E. atala hairstreaks have a straight, 480-μm long rhabdom composed of eight longitudinal photoreceptor cells and a ninth cell close to the basement membrane (SI Appendix, Fig. S4 A and B), similar to a number of butterfly species investigated to date (27). The two most distal R1-R2 photoreceptor cells contribute the majority of microvillar membranes, the structures filled with rhodopsins, from 0 to 160 μm, whereas R3-R4 distal cells contribute a majority of microvilli from 140 to 300 μm, thereby overlapping partially with R1-R2 in the distal rhabdom. The proximal R5-R8 cells contribute most microvilli in the last part of the rhabdom up to 440 μm, a depth where the longitudinal photoreceptor cells no longer bear microvilli, and the ninth cell just becomes visible (SI Appendix, Fig. S4 A and B).
Using these histological insights and double fluorescent in situ hybridization across transverse and longitudinal eye regions of males and females (Fig. 3 and SI Appendix, Fig. S5), we examined the photoreceptor cell expression patterns of all E. atala visual opsin-like transcripts generated in our eye transcriptomics analysis. We found that the red-shifted LW opsin mRNA is expressed in six photoreceptor cells across all ommatidia in both sexes (R3 to R8, Fig. 3 E and F), which is typical of many butterfly species (15, 22) but not all (63). No fluorescent signal was detected for the long-wavelength opsin in R1-R2 cells (Fig. 3G).
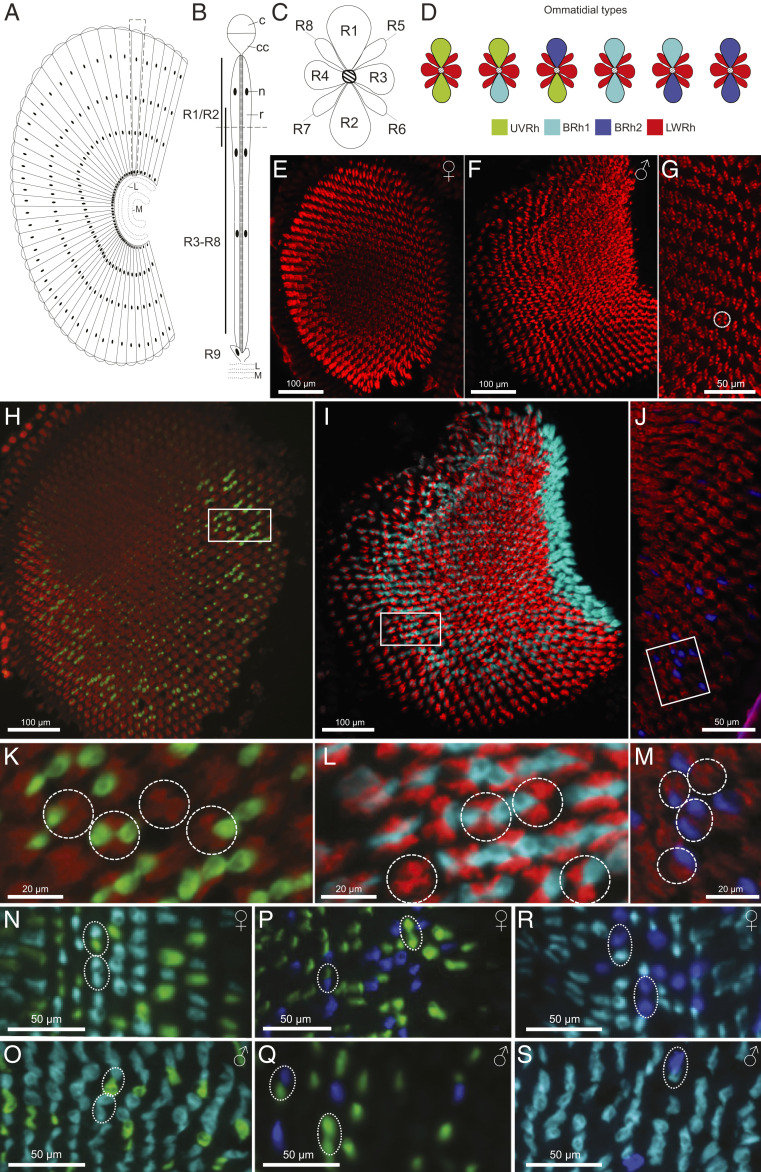
Fig. 3.
Adjacent photoreceptor localization of duplicate blue opsin mRNAs drives retinal mosaic expansion. (A) Schematic diagram of a longitudinal section of the compound eye and optic lobe proximal to the retina. L, lamina; M, medulla. (B) Longitudinal view of a E. atala ommatidium (see Eye Histology and SI Appendix, Fig. S4); c, cornea; cc, crystalline cone; n, photoreceptor cell nucleus; r, rhabdom; R1-R9, photoreceptor cells. (C) Transverse section of a dorsal ommatidium showing anatomical R1-R8 photoreceptor cells whose photosensitive membranes form the fused rhabdom. (D) R1-R2 cells in males and females express short visual pigments (UVRh, BRh1, and BRh2) forming six ommatidial classes namely UVRh-UVRh, UVRh-BRh1, UVRh-BRh2, BRh1-BRh1, BRh1-BRh2, and BRh2-BRh2 as shown in double stained ommatidia (N–S). (E and F) Fluorescent in situ hybridization of male and female dorsal eyes with a LW opsin cRNA probe shows that R3-R8 cells express LWRh in all ommatidia. (G) Higher magnification of E with one ommatidium circled. (H–J) Double fluorescent mRNA hybridization of transversal dorsal eye sections (H and J, female; I, male) with cRNA opsin probes for (H) UVRh (green) and LWRh (red), (I) BRh1 (cyan) and LWRh (red), and (J) BRh2 (blue) and LWRh (red). The boxed areas are shown under higher magnification in K–M, with dashed circles around individual ommatidia showing differences in R1-R2 expression patterns (i.e., no expression, expression in R1 or R2, expression in R1 and R2) of UVRh (K), BRh1 (L), and BRh2 (M) opsin mRNAs. (N–S) Transversal dorsal female (N, P, and R) and male (O, Q, and S) eye sections with double probe labeling for UVRh-BRh1 (N and O), UVRh-BRh2 (P and Q), and BRh1-BRh2 (R and S) indicate that short opsin mRNAs are not coexpressed in photoreceptors R1 and R2, thus revealing an exclusive one-cell one-mRNA expression pattern in single ommatidia. The dashed ovals correspond to individual classes of ommatidia (as shown in D). The BRh2-BRh2 ommatidial type was not observed in males. Longitudinal views of the eye show that ommatidia expressing BRh2 are more abundant in the upper dorsal eye in males compared to females (SI Appendix, Fig. S5).
We next examined the cellular localization of a short UV and two blue opsin-like mRNAs in transverse sections in the dorsal eye (Fig. 3 H–M). Using probes targeting LWRh in combination with cRNA probes for UVRh (Fig. 3 H and K), BRh1 (Fig. 3 I and L) or BRh2 mRNAs (Fig. 3 J and M), our data provided evidence that the three latter rhodopsin mRNAs are expressed in either or both R1-R2 receptor cells forming single ommatidia (Fig. 3 K–M), suggesting a stochastic cell fate patterning in individual ommatidia (72).
We also considered the possibility that these opsins are coexpressed in R1 and R2 cells using cRNA probes for UVRh and BRh1 (Fig. 3 N and O), UVRh and BRh2 (Fig. 3 P and Q), or BRh1 and BRh2 (Fig. 3 R and S). We found mutually exclusive expression of UVRh, BRh1, and BRh2 mRNAs in R1 and R2 photoreceptors (Fig. 3 N–S), indicating that in both sexes opsin gene expression follows a one-cell one-opsin regulation pattern (72) in these two cells. To further characterize the abundance of photoreceptor cells, we counted 1,182 female ommatidia and 1,504 male ommatidia from high-quality transversal in situ tissue sections. Female R1 and R2 cells expressed on average 25.8% UV cells, and ∼75% of blue cells, split between BRh1 (62.7%) and BRh2 (11.5%), whereas males have on average 28.4% UV photoreceptors, 63.3% BRh1, and 8.3% BRh2 photoreceptors. The relative abundance of UVRh:BRh1:BRh2:LWRh cell types is thus similar between females (0.06:0.16:0.03:0.75) and males (0.07:0.16:0.02:0.75). Females, however exhibit a dorsoventral expression gradient in which BRh2-expressing cells are sparse in the dorsal eye region compared to males, but similarly abundant in the ventral eye of both sexes (SI Appendix, Fig. S5 A, C, and E). Additional structural features of the eye including lateral filtering cherry-red granules likely contribute to spectral sensitivity in some of the LW photoreceptor cells. Nonetheless, these cellular expression patterns show that overall, the retina forms an expanded stochastic mosaic of six opsin-based ommatidial classes: UV-UV-LW, B1-B1-LW, B2-B2-LW, UV-B1-LW, UV-B2-LW, and B1-B2-LW (Fig. 3).
Finally, we examined the activity of heterologously expressed blue mRNA opsin transcripts in E. atala. To do so, we reconstituted EatBRh1 and EatBRh2 rhodopsin complexes in vitro and measured their spectral sensitivity maxima via UV-visible (UV-VIS) spectroscopy. These analyses determined that EatBRh1 absorbs maximally at λmax = 435 ± 2 nm and EatBRh2 at λmax = 500 ± 2 nm (Fig. 4 A and B and SI Appendix, Fig. S3C and Dataset S2C), thereby confirming that they encode the rhodopsins conferring the blue to green spectral sensitivity in E. atala. Finally, the UV-like transcript was found to absorb UV wavelengths in vitro at λmax = 352 ± 3.5 nm (SI Appendix, Fig. S3D), which is in close range with optophysiological measurements (SI Appendix, Fig. S2C and Dataset S1A), notwithstanding the near absorbance spectra of residual cis-retinal at λmax = 380 nm.
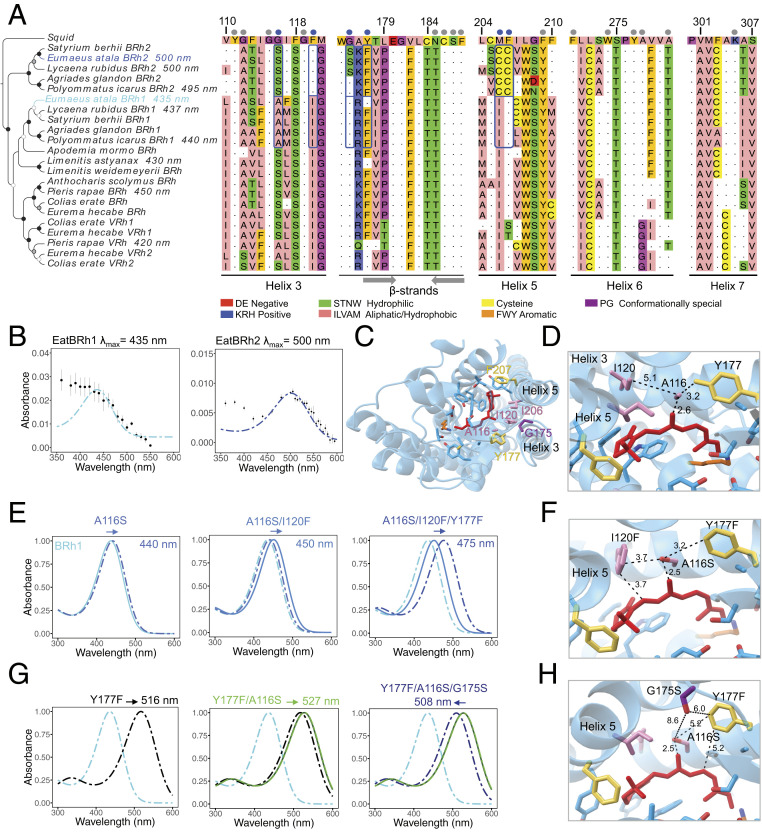
Fig. 4.
Four residues contribute to spectral tuning shifts between duplicate blue opsins. (A) Neighbor-Joining tree of selected lepidopteran blue opsin amino acid sequences. The squid opsin (Todarodes pacificus, accession no. CAA49906) is used as outgroup. Bootstrap node support is as follows: 50 to 74%, white circle; 75 to 94%, gray circle; ≥95%, black circle. Dots above the partial multiple sequence alignment show the 21 amino acid residues residing within 5 Å of any carbon atom in the retinal polyene chain. Blue dots identify the six positions where amino acid residues differ between E. atala EatBRh1 and EatBRh2; blue rectangles highlight variants at these positions in lycaenids. Residue numbering is based on residue position in the squid opsin. Residues are colored according to their physicochemical properties in Jalview v2. Gray arrows indicate β-strands forming the binding pocket. (B) Blue rhodopsin absorbance spectra (dots) fitted to the visual template (cyan and blue line functions), respectively (n = 4 protein eluate aliquots, errors bars represent ±SEM). (C) Predicted structure for EatBRh1 based on homology modeling with the squid rhodopsin with variant sites A116S, I120F, G175S, Y177F, I206C, and Y207C. (D) Model of the EatBRh1 rhodopsin bearing A116, I120, and Y177. (E) Spectral tuning trajectory when substituting residues A116S, I120F, and Y177F show a 40-nm partial blue shift in rhodopsin absorbance λmax in the triple mutant (SI Appendix, Supplementary Methods and Fig. S6). This tuning shift may be mediated by several possible mechanisms, including additive effects caused by novel hydrogen bond formation at the coevolving adjacent sites A116 and I120 (F) and with nearby conserved residues G115 and G121. (G) Spectral tuning trajectory substituting G175S, which partially compensates the green tuning shift of double mutant Y177F/A116S (SI Appendix, Fig. S6) and tunes the absorbance spectrum near 500 nm. The data underlying the alternative trajectories in E and G are presented in SI Appendix, Fig. S6 and highlight additive and epistatic interactions at four variant residues (F and H) in the acquisition of the green-shifted EatBRh2 opsin function.
Together, the combination of in situ and in vitro analyses demonstrates that the ommatidial colocalization of a blue, a green-shifted opsin, and the ubiquitous red LW opsin receptor in E. atala confers robust eye spectral sensitivity at medium and long wavelengths. This finding raised the question as to which molecular changes conferred the necessary functional variation for the evolution of green sensitivity by the duplicate blue opsin.
Four Critical Spectral Tuning Sites Confer Green Sensitivity to a Blue Duplicate Opsin.
To understand the proximate mechanisms driving the large 65-nm bathochromic spectral shift (a shift in absorption toward longer wavelengths) between the duplicate blue opsins in E. atala, we turned to site-directed mutagenesis (Materials and Methods and SI Appendix) followed by in vitro assays using mutants of EatBRh1 proteins bearing specific EatBRh2 substitutions. We modeled the predicted 380-aa-long EatBRh1 and EatBRh2 opsin protein structures and found that these opsins exhibit 101 amino acid residue differences, of which six variant sites were identified within 5 Å of any carbon forming the cis-retinal binding pocket (Fig. 4 A, C, and D). Interestingly, among these variants, A116S and Y177F are shared with a blue/violet opsin duplication that occurred independently in pierid butterflies and led to a UV-shifted (VRh λmax = 420 nm) and a blue rhodopsin (BRh λmax = 450 nm) in Pieris rapae (17). The four remaining variant sites, namely I120F, G175S, I206C, and F207C, are unique to duplicated lycaenid blue opsins (Fig. 4A). This led us to investigate whether E. atala blue opsin bathochromic tuning involves new residues or partially conserved molecular convergence with similar tuning residues.
We first mutated individual EatBRh1 residues A116S, I120F, G175S, and Y177F and measured their spectral tuning effect in vitro (SI Appendix, Fig. S6 and Dataset S2D). Mutant EatBRh1 opsins showing bathochromic shifts were used sequentially to substitute additional adjacent variant sites following two candidate evolutionary trajectories (Fig. 4 E and G). We observed an additive bathochromic shift totaling 40 nm (λmax = 475 nm) by substituting A116S together with I120F and Y177F (Fig. 4 E and F), whereas in a second tuning trajectory, we observed that Y177F alone conferred a 81-nm bathochromic shift (λmax = 516 nm), which could then be compensated for by an 8-nm hypsochromic shift (λmax = 508 nm) by combining G175S and A116S (Fig. 4 G and H). A third evolutionary trajectory explored the contribution of two lycaenid-specific cysteine substitutions (I106C and F207C) in helix 5 (SI Appendix, Fig. S6). These changes caused a strong green-wave spectral shift that was not compensated for by additional candidate tuning residues in a quintuple mutant, EatBRh1/A116S/I120F/Y177F/I206C/F207C (SI Appendix, Fig. S6 and Dataset S2D). G175S tested alone or in various double mutant combinations caused bathochromic shifts (SI Appendix, Fig. S6), but we did not obtain the sextuple chimeric construct bearing G175S and cannot therefore exclude the possibility that it may play a hypsochromic role in this particular case. Our results from variants EatBRh1 bearing I106C and F207C, however, suggest that the cysteine residues on helix 5 are not likely to contribute to blue spectral shifts in EatBRh2.
Coordinated Spectral Sensitivity Shifts and Coincidental Wing Coloration Trait Evolution.
The spectral sensitivities of visual photoreceptors may be mere by-products of evolution or may represent adaptions to specific color signals (73, 74). For a butterfly to interpret colors, it must 1) possess at least two spectral types of receptors sensitive to the reflectance spectrum of incident visible light illuminating colored objects, and 2) be able to compare individual receptor responses neuronally to create an output chromatic signal (75, 76). To begin investigating the evolutionary consequences of molecular changes in butterfly opsins in the broader context of behaviors requiring color vision, such as finding oviposition sites and intraspecific recognition (49, 77–80), we derived a four-opsin color vision model (Fig. 5) and evaluated the overlap between receptor spectral sensitivities against the spectral composition of the reflected light produced by host plant foliage and all major colored patches on conspecific wings (Fig. 5 and SI Appendix, Figs. S7–S9 and Dataset S3 A–M).
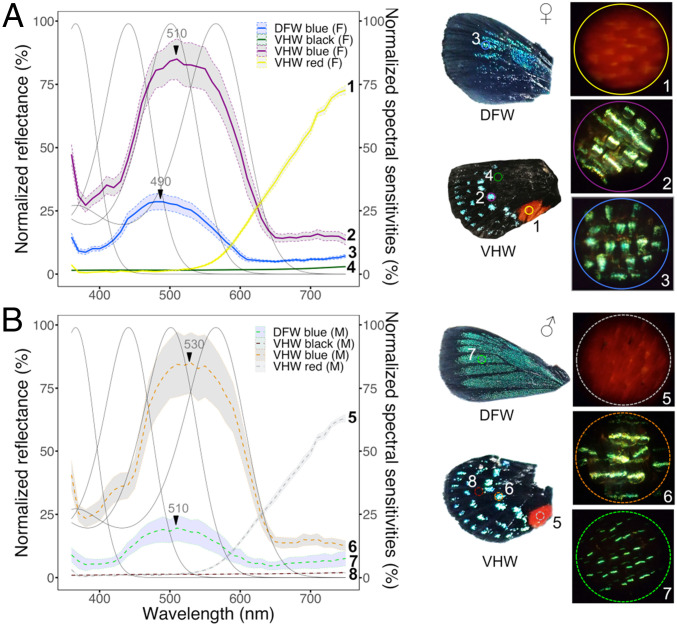
Fig. 5.
Reflectance spectra of wing and body coloration correlate with photoreceptor spectral sensitivity. Discriminability analysis of female (A) or male (B) wing colors overlaid with visual spectral sensitivities. Graphs of mean reflectance spectra of wing scale patches ± SEs of the mean (n = 3 to 5 individuals, with duplicate measurements). Gray lines indicate normalized photoreceptor spectral sensitivity functions. Reflectance spectra are normalized against total brightness. Numbers next to each reflectance curve correspond to numbers on each wing photograph (DFW, dorsal forewing; VHW, ventral hindwing). Magnified views of representative wing scales measured by epi-MSP are shown on the Right. The field of view for each scale photograph is 210 μm in diameter. The reflectance curves correspond with the absorbing wavelengths of at least two rhodopsins, which may allow the butterfly to efficiently perceive colors associated with foliage and conspecific wing patterns (SI Appendix, Figs. S7 and S9). Analyses from black areas of ventral hindwings reveal that brightness in cyan regions increases 64-fold compared to black scales in both sexes, which improves contrast with adjacent colored scales.
In E. atala males, dorsal forewings are bright iridescent blue in summer, whereas scales appear more generally green/teal in winter generations (81). Female dorsal wings, on the other hand, display a darker royal blue color along the edge of their upper forewings. Both sexes also have conserved wing and body patterns, including regularly spaced rows of blue spots visible on closed and open hindwings, and a bright red abdomen enhanced by a large red spot on the midcaudal hindwing area that falls precisely along the abdomen when the wings are closed (Fig. 2A).
We used epi-microspectrophotometry to measure the reflectance spectra associated with leaf surfaces of the butterfly’s primary host cycad, Zamia integrifolia, and noted a peak of reflectance at 550 nm and a red edge inflection point around 700 nm with high reflectance in the far-red region (SI Appendix, Fig. S7A and Dataset S3A). We analyzed leaf reflectance against spectral sensitivity functions in three- and four-opsin vision systems. Our analyses show that the overlap in spectral sensitivity between BRh2/LWRh (R495/R564) is expected to improve dichromatic discrimination in the Eumaeus eye (SI Appendix, Fig. S8A) at long wavelengths by 25 nm and up to 90 nm compared to intermediate visual systems bearing R495/R530, R475/R530, R440/R530, or R440/R564 receptors (SI Appendix, Fig. S8 C–F).
Next, we examined colored patches from males and females including blue scales on the abdomen and thorax, and black, blue, and red scales on forewings and hindwings (Fig. 5 and SI Appendix, Fig. S7 B and C and Dataset S3). Measurements from male and female ventral hindwings showed that black scales reflect only about 1.5% incident light in both sexes (Fig. 5 A and B and Dataset S3 B and C) compared to light reflected by adjacent colored scales in the blue/green band at 450 to 520 nm (Fig. 5 A and B and Dataset S3 D and E), indicating a 64-fold increase in the relative brightness of wing colored patches (Fig. 5 A and B). Blue scales on the dorsal forewings have distinct reflectance maxima at 490 nm in females (F) and 510 nm in males (M) (Fig. 5 A and B and Dataset S3 F and G) with female scales reflecting 1.3 to 1.9 times more light overall in the 400- to 500-nm wavelength range. Blue scales on the ventral hindwings have a maximal reflectance peak at 510 nm (F) and 530 nm (M) (Fig. 5 A and B) and under our experimental conditions, reflect similar light levels across 450- to 650-nm wavelengths between sexes. Light reflected from blue scales on the thorax is 1.9 times greater than in blue regions on dorsal forewings in females and 2.9 times higher in males (SI Appendix, Fig. S7B and Dataset S3 H and I); however, thorax reflectance spectra overlap tightly in both sexes, which suggests that males and females are dimorphic only on the dorsal wings. Finally, red scales on male and female hindwings reflect maximally in the far red (750 nm) (Fig. 5 and Dataset S3 J and K), similar to red abdominal scales (SI Appendix, Fig. S7C and Dataset S3 L and M). These results show the extent of overlap between E. atala photoreceptor spectral sensitivities and wing color traits.
Discussion
Long-Wavelength Red Opsins Contribute to Far-Red Spectral Sensitivity.
Opsins are the first elements in visual transduction, and they primarily mediate variation in light detection, image formation, and visual spectral sensitivity (3, 5, 27, 46, 82). Recent research has also uncovered multiple cases where they have been coopted for novel functions (83–86). As insect visual opsin proteins belong to a dynamically evolving multigene family (Fig. 1), they represent not only a robust system to link molecular genetic variation to phenotypic changes in color vision (5), but more generally offer insights into the consequences of molecular variation contributing to adaptive phenotypes across distant animal groups (87–89).
Here, we identify molecular patterns of evolution and functional mutations underlying concomitant spectral changes to longer wavelengths in Gq opsin gene lineages driving lycaenid butterfly visual phenotypes, and the partial convergence in blue opsin tuning residues between the Gt and Gq opsin families. By optimizing a cell culture assay that allows the efficient purification and characterization of insect rhodopsin complexes in vitro, we identified a type of red-shifted LW opsin that absorbs significantly longer wavelengths. This is due to a remarkable 35- to 40-nm bathochromic shift in λmax compared to ancestral green insect LW rhodopsins (15, 90) and produces photoreceptors sensitive to 560 to 700 nm of red light (Fig. 2). The ability to obtain recombinant insect opsin proteins provides an opportunity to uncover important functional insights underlying the independent evolution of vertebrate and invertebrate LW opsins involved in red sensing and enables exploration of the diversity of genetic mechanisms underlying functional variation of insect LW opsins, which are thus far largely unknown except in P. xuthus (91). Our functional approach together with in-depth in vivo MSP and optophysiological data (Fig. 2 and SI Appendix, Figs. S1 and S2) isolated the contribution of the pure LW rhodopsin from red-filtering effects that often affect light absorbance properties in insect eye photoreceptors, demonstrating that the LW opsin proteins alone can dramatically increase the photoreceptor response at longer wavelengths (Fig. 2 and SI Appendix, Fig. S8). Consequently, compared to butterflies and other insect species whose photoreceptors express typical LW rhodopsins (λmax 520 to 530 nm) (8, 26), our findings demonstrate that lycaenid species expressing red-absorbing rhodopsins have a significantly extended spectral sensitivity toward the far red (700 to 750 nm) directly controlled by molecular variation underlying the absorbance properties of their long-wavelength opsin. In Papilio butterflies, which express combinations of five rhodopsin proteins (UV, B, and PxRh1-3), green sensitivity is achieved via tuning residues in helix 3 of two LW opsins (22, 91, 92). Lycaenid LW opsins instead possess a highly conserved helix 3 lacking those spectral substitutions, supporting the hypothesis that distinct spectral tuning mechanisms have evolved independently to achieve red sensitivity in lycaenids.
Visual adaptations toward red sensitivity in vertebrate cones have been shown to facilitate long-wavelength light perception in primates (42), birds (93, 94), and in multiple fish groups, notably in threespine sticklebacks following repeated evolution of a red-shifted L opsin (95), via L opsin duplication and spectral tuning in far-red bioluminescent deep-sea dragonfish (96, 97) or in L opsin sensitivity shifts in cichlids that inhabit red-light-dominated waters (98). In insects, discriminating green leaves is difficult for herbivorous insects without host-specific chemosensory cues because the foliar spectral characteristics of chlorophyll reflectance (500 to 580 nm) are highly similar among the vast majority of plants (99). Hence, many insects that have a green receptor can only compare dichromatic signals from green and blue (and/or UV) receptors, resulting in yellow color preference (26). In contrast, papilionid and pierid butterflies with red-sensitive photoreceptors have behaviorally been shown to use green-red color vision for green leaf color discrimination (53). The unique combination of spectral shifts in green and red receptor sensitivity (Fig. 2 and SI Appendix, Fig. S8) raises the intriguing possibility that ovipositing Eumaeus females, and other lycaenids with red receptors, may as well extract more information from reflectance slope differences and leaf water content in the red range (>600 nm) compared to insects lacking red receptors (53, 100).
Three Photoreceptors Derived from Ommatidial Expression Pattern and Green Spectral Tuning of a Blue Opsin Duplicate.
Opsin evolution has undergone recurrent events of gene duplication and loss across animals, including insects (Fig. 1). Gene duplication followed by functional divergence among duplicate copies is known to increase transcriptional and functional diversity across lineages (101, 102), and in some instances favor the evolution of lineage-specific phenotypic traits (103, 104). Whereas structural, functional, or gene network constraints can lead to evolutionary trade-offs for biochemically stable alternatives among duplicated genes (105, 106), alternative mechanisms may arise to alleviate constraints of retaining redundant gene copies in existing eukaryotic gene networks (105). These include the repeated allelic fixation of segmental duplications (107), cooperation between encoded products of duplication (88), or specialization of ancestral functions (48, 108, 109).
Several bug and beetle lineages that have lost blue rhodopsins independently, for instance, recruited duplicate UV or LW gene(s) to restore blue sensitivity (12, 110). Among butterflies, the Heliconius, Pieris, and Papilio lineages have respectively recruited additional UV, blue, or LW opsins, which confer acute spectral sensitivity in the range of violet to green visible light (10, 92, 111, 112). Our functional results in E. atala demonstrate that two short wavelength blue-like opsin loci encode a typical blue opsin (λmax 435 to 440 nm) and a significantly green-shifted blue opsin (λmax 495 to 500 nm) (Fig. 4). This increases overlapping sensitivity between two classes of blue receptors, suggesting finer wavelength discrimination and potentially improved perception of subtle color differences in shades of blue (SI Appendix, Fig. S10), in a manner similar to that found in species of Pieris in the Pieridae (111, 113) and species of Lycaena and Polyommatus in the Lycaenidae (61–63).
One interesting insight in the evolution of blue spectral tuning in E. atala comes from chimeric BRh1 variants bearing mutations A116S, G175S, and Y177F. Together, these mutations confer a 73-nm bathochromic shift (λmax = 508 nm) most closely recapitulating the spectral properties of EatBRh2 (λmax = 500 nm) compared to other tested variants (Fig. 4G and SI Appendix, Fig. S6). Intermediate adaptive phenotypes were also revealed, suggesting that the acquisition of green sensitivity may occur via gradual evolutionary steps and epistatic interactions between sites. For instance, EatBRh1 variant A116S causes a +5-nm shift alone, but together with I120F, it shifts maximal absorbance by an additional +10 nm to λmax = 450 nm. As these two residue substitutions are conserved across all characterized lycaenid BRh1 loci (Fig. 4A), these results indicate that adjacent sites in helix 3 may contribute to intermediate blue absorbance spectra accounting for variable visual ecologies across lycaenids (62).
A third residue, Y177F, is a key spectral tuning mutation in E. atala, as the triple BRh1 variant (A116S/I120F/Y177F) displays a 30-nm bathochromic shift (λmax = 475 nm) compared to the wild-type rhodopsin. These findings illustrate the multiple ways that gradual spectral tuning may have evolved, at least in this insect species, and point out the importance of spectral residues lying on the ionone ring portion of the chromophore binding pocket (31, 42). In agreement with these findings, two of the reverse tuning substitutions at the same sites are responsible for hypsochromic spectral shifts both in a blue-shifted blue rhodopsin of a Limenitis butterfly (F177Y, −5 nm) (82) and in a violet-shifted blue rhodopsin of a Pieris butterfly (S116A, −13 nm; F177Y −4 nm) (17). Spectral tuning modulation in blue rhodopsin duplicate genes has therefore involved parallel evolutionary trajectories through convergent biochemical changes along multiple insect lineages. Reverse mutations are not necessarily functionally equivalent in their absolute magnitudes (Δλmax), underscoring the role of epistatic interactions at neighboring sites, resulting in distinct λmax shifts across distinct lineages. A homologous tyrosine residue in the ionone ring portion of the chromophore-binding pocket (Y262) in the human blue cone opsin (SWS2 λmax = 414 nm) is responsible for a 10-nm bathochromic spectral tuning when mutated to tryptophan (Trp) (114). However, Y177 is unique to E. atala; other BRh1 loci at this position often have the F177 seen in EatBRh2 (Fig. 4A). Not all BRh2 loci have S175 but instead possess G175 in both blue opsin loci, potentially suggesting an additional tuning role for adjacent residues in positions R176K and I178V. These two residues do not differ much in hydrophobicity but may still provide the essential molecular interactions with the ionine ring portion of the chromophore necessary to modulate distinct BRh2 spectral sensitivity maxima. Human ancestors achieved blue sensitivity gradually and almost exclusively via epistasis among seven amino acid residues (46). Therefore, testing the nonadditive interactions at coevolving BRh2 adjacent sites 175 to 178 in lycaenids will further help to recapitulate intermediate phenotypes across derived green-shifted insect blue rhodopsins.
Additionally, molecular and phylogenetic studies of SWS2 opsin evolution in vertebrates, which are functionally analogous to insect blue opsins, with the ancestral vertebrate SWS2 pigment absorbing at ∼440 nm (115), have shown that opsins shifted to longer wavelengths recurrently involve substituting polar residues with a hydroxyl side chain (e.g., Ser, Thr, Tyr). For example, A94S causes a 14-nm change between zebrafish (λmax 416 nm) and goldfish (λmax 443 nm) blue pigments (116), and A269T is one of several important replacements in the tuning of avian SWS2 pigments (115), a site also involved in spectral tuning of bovine Rh1 (117). It is worth stressing that the EatBRh2 short-wavelength blue opsin duplicate achieves a remarkable green absorbance shift not found in SWS2 opsins but encoded by other opsin lineages, Rh2 in fish (93) or birds (118), and M in primates (42, 44), where polar residue changes are also observed to cause additive tuning effects in long-wavelength shifted pigments (115). Our mutagenesis findings not only illustrate the partial sharing of genetic mechanisms of spectral tuning between independent blue opsin duplications across butterfly lineages, but reveal a partial conservation of structural and biochemical tuning constraints between invertebrate Gq blue opsin lineages and those observed in the evolution of SWS2 vision genes in vertebrates (5, 106).
Extended Blue and Red Spectral Sensitivity Enhance Visual Signal Perception.
Variation in peripheral photoreceptor cell sensitivity is a visual tuning strategy that can maximize the extraction of complex color information in natural environments and incidentally affect subsequent higher-order processing controlling behavior (119), as shown in marine habitat species (120), and some insects (113, 121, 122). In the lycaenid species examined in this study, spectral sensitivity evolved sharply in response to coordinated peripheral tuning relying on complementary changes in two opsin proteins (BRh2 with λmax 495 to 515 nm and LW with λmax 564 to 574 nm), and the acquisition of three additional photoreceptor classes (BRh2-UVRh, BRh2-BRh2, and BRh2-BRh1). The combined retinal densitometry and in situ hybridization evidence are consistent with the presence of two blue rhodopsins in separate photoreceptors in the same region in the eye and in sufficient abundance for color vision. In total, the six-ommatidial stochastic rhodopsin mosaic offers remarkable spectral properties, which would have been unlikely to achieve with the limited sensitivity of a single blue opsin or the sensitivity maxima of an ancestral green opsin. Given the high color richness and behavioral diversity of lycaenids, we may expect that photoreceptor and color vision evolution has played an important role in adaptation. Whereas shifts in opsin absorbance spectra may evolve recurrently or over longer evolutionary times, photoreceptor expression pattern differences such as observed in LW photoreceptors between males of E. atala and Lycaena rubidus, albeit sharing similar opsin spectral sensitivities (63), may reflect more rapidly evolving, lineage-specific ecological differences (95, 96).
Adaptations in visual sensitivity in E. atala are likely relevant in conveying spectral signals underlying distinct color-guided behaviors. The derived green-shifted BRh2 photoreceptors, despite a lower density across the eye compared to BRh1 photoreceptors, could further be beneficial in decreasing the minimal wavelength difference (Δλ) that the butterfly can in theory discriminate (46, 76, 123). Similarly to duplicate UV opsins that allow Heliconius butterflies to more finely discriminate UV signals (63), E. atala could effectively tune blue wavelength discrimination and chromatic resolution in the blue-green spectrum (56, 61–63), since these additional BRh2 photoreceptors predictably increase the number of possible opponency channels available for color processing (123–125).
As daylight is reflected in the butterfly scales, the peak positions of the reflected light spectrum may differ, and color discrimination will be maximized at the steepest points of overlap (420 to 480 nm) between BRh1/BRh2 receptor sensitivity functions compared to lineages that did not undergo blue opsin diversification (Fig. 5 and SI Appendix, Figs. S9 and S10). Taking into account color-opponent postreceptor neuronal perception (75), and in keeping with our analyses of spectral sensitivities, this raises the intriguing possibility that wing color differences may function as recognition signals, with adjacent black scales in E. atala wings further enhancing the perceived brightness of contiguous blue color signals, similar to the superblack plumage coloration found in birds of paradise (126), peacock spiders (127), and recent examples in papilionid and nymphalid butterflies (128, 129).
Members of the genus Eumaeus are additionally characterized by red abdomens as well as a bright red spot at the bottom of the hindwings, and this is thought to advertise the toxins that the butterflies sequester as larvae from their Zamia cycad host plants (130). Although the use of red aposematic coloration has been coopted as an additional intraspecific recognition signal, e.g., in Heliconius butterflies (79, 131), the red scale reflectance spectra are similar between the sexes in E. atala and shared among all Eumaeus species. We show that Eumaeus can perceive these long wavelengths, but the orange-red coloration may have evolved primarily as an aposematic signal toward predators (132). The bright red abdominal/hindwing reflectance in the far-red spectrum would very likely stimulate the L cones of many bird species with red sensitivity from about 550 nm to 700 nm (39, 73), and they would readily associate toxicity with a color close to pure red.
Trichromatic systems are capable of recording most spectral information from natural scenes (121); yet a color vision system with higher dimensionality generally benefits color vision in bright light compared to a system based upon fewer receptor types (133). Although we could not record responses of different individual receptors, it is reasonable to assume that tetrachromaticity in E. atala is an adaptation for the discrimination of specific colors, such as body coloration or oviposition sites. Whether this is a specific adaptation for signaling via body coloration remains to be determined. Color vision theory further suggests that a system reaches its full potential when the width of the visual range is increased (133), which is in line with the red-shifted receptor in E. atala and suggests that the butterfly is able to robustly detect color differences across an extended range of wavelengths and filter environmental reflectance cues in parallel. In fact, the combined shifts in blue and red receptor physiological differences are likely to contribute a much larger effect to the efficacy of photoreceptor spectral sensitivity for maximizing stimuli detection than would any individual receptor shift (121). These concomitant peripheral molecular changes thus offer mechanistic insights into the diversification of insect visual spectral sensitivities, supporting the scenario of a sensory system where gene duplication generates new blue opsin paralogs, regulatory changes specify new spectral subtypes as reflected by evidence from in situ hybridization of the composition of photoreceptors making up individual ommatidia, and coding mutations modify the blue and LW rhodopsin spectral sensitivity maxima, providing expanded spectral sensitivity at longer wavelengths as well as a finely tuned ability to discriminate wavelengths of incoming light in the blue band.
By developing a Gq opsin heterologous expression system and by demonstrating the fine tuning between the evolution of blue and red opsin functions for long-wavelength spectral sensitivity our study provides a promising invertebrate molecular toolkit to disentangle the molecular–functional interplay between peripheral sensory genes and adaptations in invertebrate color vision performance.
Materials and Methods
Butterflies.
Pupae of E. atala were collected from host plants Z. integrifolia at the Montgomery Botanical Garden, Miami, FL in January 2020 and reared at 22 °C in an insectary under a 12:12 L:D cycle until emergence. Eggs and young larvae of A. narathura japonica were collected feeding on oak trees (Quercus glauca) from field sites near Ginoza, Okinawa, Japan and reared in the Museum of Comparative Zoology laboratories in Cambridge, MA (US Department of Agriculture permit P526P-18-00037) until they eclosed as adults.
Epi-Microspectrophotometry.
Compound eyes of most adult butterfly species exhibit eyeshine, a property that allows measuring rhodopsin absorbance spectra as well as spectral sensitivity of photoreceptor pupillary responses in eyes of intact butterflies. The eyes of butterflies exposed to repeated bright white flashes under incident-light microscope show a photochemical effect whereby the coloration of eyeshine changes during each flash due to changes in absorbance spectra that accompany photoisomerization of rhodopsins to their metarhodopsin photoproducts (134). At the same time, the eyes exhibit a pupillary response mediated by intracellular migration of pigment granules within photoreceptor cells, causing the intensity of eyeshine to decrease rapidly with time during each flash (135).
Quantitative epi-MSP was used to determine absorption spectra of butterfly rhodopsins by measuring eyeshine reflectance spectra after photoconversion of the rhodopsin to its metarhodopsin product (67, 134) using three MSP methods, in vivo photochemistry, optophysiology, and retinal densitometry as detailed in SI Appendix.
Functional Assays.
Cloning and protein expression.
The coding region of each opsin transcript (see SI Appendix for transcriptome analysis and opsin characterization) was amplified from eye cDNA and subcloned in a modified pFRT-TO expression vector cassette derived from pcDNA5 and containing the human CMV immediate early promoter (Invitrogen) (Fig. 2G). The expression plasmid was modified to include a C-terminal tag by the monoclonal antibody FLAG epitope sequence (DYKDDDDK), followed by a Ser-Gly-Ser linker peptide, a T2A peptide sequence (EGRGSLLTCGDVEENPG), and the fluorescent marker protein mRuby2. Plasmid DNAs were verified by Sanger sequencing and purified with the endo-free ZymoPURE II Plasmid Midiprep Kit (Zymo Research). Prior to large-scale expression, small-scale HEK293T cultures were transfected to optimize expression conditions both via mRuby2 visualization and Western blot analysis as described in SI Appendix.
Transient expression.
High-expressing clones from GPCR opsin cDNAs were transiently expressed in HEK293T cells prior to in vitro purification. For each construct, cells were seeded at a density of 1.0 × 106 cells on day 0 in 15 tissue culture dishes (10 cm diameter, ref. 25382-166, VWR) in Dulbecco’s modified Eagle medium high glucose, GlutaMAX (Life Technologies) supplemented with 10% fetal bovine serum (Seradigm Premium, VWR). Lipid complexes containing 24 μg DNA: 72 μL polyethylenimine (1 mg/mL) diluted in Opti-MEM I reduced serum (Life Technologies) were added 48 h later to cells reaching 75 to 85% confluency. At 6 h posttransfection, the culture medium was exchanged with new medium containing 5 μM 11-cis-retinal (2 mg/mL stock in 95% ethanol) and under dim red light. The 11-cis-retinal absorption peak at 380 nm was confirmed using a NanoDrop 2000/2000c UV-VIS Spectrophotometer (Thermo Fisher) prior to each experiment using a 1:100 dilution in ethanol. Culture plates supplemented with 11-cis-retinal were wrapped in aluminum foil and cells were incubated in the dark. At 48 h posttransfection, the medium was decanted under dim red light. Cells were scraped in cold filter-sterilized Hepes wash buffer (3 mM MgCl2, 140 mM NaCl, 50 mM Hepes pH 6.6 to 8.5 depending on protein isoelectric point) containing complete ethylenediaminetetraacetic acid (EDTA)-free protein inhibitors (Sigma-Aldrich), centrifuged for 10 min at 1,620 rcf at 4 °C, and resuspended in 10 mL wash buffer for two consecutive washes. After the second wash, cell pellets were gently resuspended in 10 mL cold wash buffer containing 40 μM 11-cis-retinal. Cells expressing opsin-membrane proteins were incubated in the dark during 1 h at 4 °C on a nutating mixer (VWR) to favor the formation of active rhodopsin complexes, and cells were then collected by centrifugation at 21,500 rpm for 25 min at 4 °C on a Sorvall WX Ultra 80 Series equipped with an AH-629 Swinging Bucket Rotor (Thermo Scientific).
Rhodopsin complex purification and spectroscopy.
Transmembrane proteins were gently extracted by pipetting in 10 mL of ice-cold extraction buffer (3 mM MgCl2, 140 mM NaCl, 50 mM Hepes, 20% glycerol vol/vol, 1% n-dodecyl β-D-maltoside, complete EDTA-free protein inhibitors) and incubated for 1 h at 4 °C prior to centrifugation at 21,500 rpm for 25 min at 4 °C. The 10-mL crude extract supernatant containing solubilized rhodopsin complexes was added to 1 mL Pierce Anti-DYKDDDDK Affinity Resin (Thermo Scientific) and incubated overnight at 4 °C in a 15-mL Falcon on a nutating mixer. Samples were loaded on Pierce Centrifuge Columns (Thermo Scientific) and after three washes of the resin-bound FLAG-epitope rhodopsin complexes with three-column reservoir volumes of elution buffer (3 mM MgCl2, 140 mM NaCl, 50 mM Hepes, 20% glycerol vol/vol, 0.1% n-dodecyl β-D-maltoside), the rhodopsin was eluted in 2 mL of elution buffer containing 1.25 mg (265 μM) Pierce 3× DYKDDDDK peptide (Thermo Scientific). The eluate was concentrated using an Amicon Ultra-2 centrifugal filter unit with Ultracel-10 membrane (Millipore), for 35 min at 4 °C and 3,500 rpm. The concentrated eluate (350 μL) was aliquoted in amber light-sensitive tubes (VWR) and kept on ice in the dark. UV-visible absorption spectra (200 to 800 nm) of dark-adapted purified proteins were measured in the dark from 1.5-μL aliquots using a NanoDrop 2000/2000c UV-VIS spectrophotometer (Thermo Fisher). Opsin purification yields were estimated following bovine serum albumin analysis (Dataset S2). Raw absorbance data were fitted to a visual template (136) and polynomial function analyses performed in R (v 3.6.3) (137) to determine the opsin maximal absorption peaks. The spectral mutagenesis methods are presented in SI Appendix.
Eye Histology.
Each E. atala eye was immersed for prefixation in 2.5% glutaraldehyde/2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) (Electron Microscopy Sciences) for 2 h at room temperature, then stored at 4 °C for 12 to 14 h prior to fixation, embedding, ultrathin sectioning, and mounting on copper grids for transmission electron microscopy analysis at the Harvard Medical School Electron Microscopy Facility (see method details in SI Appendix).
Preparation of RNA Probes and RNA In Situ Hybridization.
We generated in vitro transcription templates from UVRh, BRh1, BRh2, LWRh opsin cDNA cloned in ∼700-bp segments into pCRII-TOPO (Invitrogen). Antisense cRNA probes were synthesized using T7 or Sp6 polymerases using either digoxigenin (DIG) or fluorescein (FITC) labeling mix (Sigma-Aldrich) from purified PCR templates. The synthesized cRNA probes were ethanol precipitated with NH4OAc 7.5 M and 1 μL glycogen, spun down at 4 °C for 30 min, redissolved in pure water, and stored at −80 °C. These probes were first used to test mRNA expression for each opsin receptor gene. We then tested the probes by dual color in situ hybridization using combinations of DIG and FITC probes to map opsin receptor expression patterns.
For in situ hybridization, E. atala compound eyes were dissected and immersed in 1.5-mL Eppendorf tubes containing freshly made 4% formaldehyde (Fisher Scientific) in 1× phosphate-buffered saline (PBS) for 2 h at room temperature for fixation, then immersed successively in increasing sucrose gradient solutions (10%, 20%, 30% in PBS) for 30 min each, stored in 30% sucrose solution overnight at 4 °C, briefly transferred in optimal cutting temperature (OCT) compound:sucrose 1:1, embedded in OCT (Tissue-Tek), and frozen on dry ice. Tangential and longitudinal eye sections (12 μm) were obtained using a cryostat (Leica), mounted on VWR Superfrost Plus Micro slides, and used for RNA in situ hybridization following the procedure described in ref. 137. Double fluorescence in situ hybridization was performed using 100 μL hybridization solution (prehybridization buffer supplemented with 4% dextran sulfate [Sigma]) containing a combination of two opsin cRNA probes, labeled with either DIG or FITC (at 1 ng/μL for UVRh and BRh2, and 0.5 ng/μL for BRh1 and LWRh), and the signal developed using the TSA Cy3 and Cy5 Plus kits (PerkinElmer) following the manufacturer’s protocol. Tissues were mounted with Vectashield (Vector Labs). All the microscopy images were acquired using an AxioImager Z2 (Zeiss).
Wing, Body, and Leaf Reflectance.
Reflectance spectra were measured from leaves of Z. integrifolia (Zamiacae) collected at the Montgomery Botanical Garden (Miami, FL) and from discrete patches of colored scales on male and female E. atala fore- and hindwings, thorax, and abdomen (Dataset S3) from butterflies collected in January 2020 at the Montgomery Botanical Garden in a Leitz Ortholux-Pol microscope equipped with a Leitz MPV-1 photometer with epi-illumination block, fitted with a Leitz 5.6×/0.15P objective. The illuminator filled the back focal plane of the objective with axial incident light. The photometer measured reflected light from the full aperture of the objective from a spot in the front focal plane that was 210 μm in diameter. Reflectance data were corrected for stray light by subtracting data measured from the MSP objective viewing a light dump comprising a substantially out-of-focus black velvet cloth. Corrected reflectance data were normalized against the same normalization constant of 0.179 to preserve relative brightness among all measured body patches. Normalized reflectance data were analyzed in R (v3.6.3).
Supplementary Material
Acknowledgments
We thank Masaru Hojo for providing pupae of A. japonica; Sandy Koi for providing E. atala and for advice regarding rearing and ecology; Maria Eriksson and Peg Coughin for assistance with electron microscopy; Xi Shi, Sara Jones, and Jonathan Schmid-Burgk for advice on cell culture; Ian Slaymaker for advice on protein purification procedures; Hopi E. Hoekstra for access to cryostat and microscope; Rachel Gaudet and José Velilla for guidance using Pymol; Rosalie Crouch for providing the 11-cis-retinal; Jeanne M. Serb and Davide Faggionato for advice on 11-cis-retinal delivery; Kentaro Arikawa, Julien Ayroles, Gregor Belušič, Mary Caswell Stoddard, Almut Kelber, Olle Lind, Rhiannon Macrae, Samuel Aravinthan, and Mandyam Veerambudi Srinivasan for helpful discussions; Adriana Briscoe for providing helpful feedback on an earlier version of the manuscript; and Richard Belliveau, Maggie Starvish, and Rachel Hawkins for providing logistical support. The computations were run on the FASRC Odyssey cluster of the FAS Division of Science Research Computing Group at Harvard University. This work was supported by a Mind Brain Behavior Interfaculty grant (to N.E.P. and M.A.L.), an Knut and Alice Wallenberg postdoctoral fellowship at the Broad Institute of MIT and Harvard (to M.A.L.), the Royal Physiographic Society of Lund (M.A.L.), the Lars Hierta Memorial Foundation (M.A.L.), an NSF graduate research fellowship (to S. Salzman), personal funds (G.D.B.), and NSF DEB-1541560 (to N.E.P.) and PHY-1411445 (to N.E.P. and N.Y.). F.Z. is an investigator of the Howard Hughes Medical Institute and is supported by NIH Grants 1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201; the Open Philanthropy Project; the Harold G. and Leila Mathers and the Edward Mallinckrodt, Jr. Foundations; the Poitras Center for Psychiatric Disorders Research at MIT; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; and by the Phillips family and J. and P. Poitras. Publication charges supported by a grant from the Wetmore Colles Fund.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.M.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008986118/-/DCSupplemental.
Data Availability.
The E. atala eye transcriptome and opsin transcript data are available in the Sequence Read Archive Bioproject number PRJNA625881 and GenBank database under accession numbers MN831881–MN831885. Source data underlying epi-microspectrophotometry, functional expression, and reflectance spectra are available in Datasets S1–S3, respectively.
References
- 1.Terakita A., The opsins. Genome Biol. 6, 213 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama S., Evolution of dim-light and color vision pigments. Annu. Rev. Genomics Hum. Genet. 9, 259–282 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Nilsson D. E., The evolution of eyes and visually guided behaviour. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2833–2847 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briscoe A. D., Chittka L., The evolution of color vision in insects. Annu. Rev. Entomol. 46, 471–510 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Hauser F. E., Chang B. S., Insights into visual pigment adaptation and diversity from model ecological and evolutionary systems. Curr. Opin. Genet. Dev. 47, 110–120 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Frentiu F. D., Bernard G. D., Sison-Mangus M. P., Brower A. V., Briscoe A. D., Gene duplication is an evolutionary mechanism for expanding spectral diversity in the long-wavelength photopigments of butterflies. Mol. Biol. Evol. 24, 2016–2028 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Vorobyev M., Ecology and evolution of primate colour vision. Clin. Exp. Optom. 87, 230–238 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Briscoe A. D., Reconstructing the ancestral butterfly eye: Focus on the opsins. J. Exp. Biol. 211, 1805–1813 (2008). [DOI] [PubMed] [Google Scholar]
- 9.McCulloch K. J., et al., Sexual dimorphism and retinal mosaic diversification following the evolution of a violet receptor in butterflies. Mol. Biol. Evol. 34, 2271–2284 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Briscoe A. D., et al., Positive selection of a duplicated UV-sensitive visual pigment coincides with wing pigment evolution in Heliconius butterflies. Proc. Natl. Acad. Sci. U.S.A. 107, 3628–3633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armisén D., et al., The genome of the water strider Gerris buenoi reveals expansions of gene repertoires associated with adaptations to life on the water. BMC Genomics 19, 832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharkey C. R., et al., Overcoming the loss of blue sensitivity through opsin duplication in the largest animal group, beetles. Sci. Rep. 7, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D., et al., Molecular evolution of bat color vision genes. Mol. Biol. Evol. 21, 295–302 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Koshitaka H., Kinoshita M., Vorobyev M., Arikawa K., Tetrachromacy in a butterfly that has eight varieties of spectral receptors. Proc. Biol. Sci. 275, 947–954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frentiu F. D., et al., Adaptive evolution of color vision as seen through the eyes of butterflies. Proc. Natl. Acad. Sci. U.S.A. 104 (suppl. 1), 8634–8640 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Futahashi R., et al., Extraordinary diversity of visual opsin genes in dragonflies. Proc. Natl. Acad. Sci. U.S.A. 112, E1247–E1256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakakuwa M., et al., Evolution and mechanism of spectral tuning of blue-absorbing visual pigments in butterflies. PLoS One 5, e15015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neitz M., Neitz J., Jacobs G. H., Spectral tuning of pigments underlying red-green color vision. Science 252, 971–974 (1991). [DOI] [PubMed] [Google Scholar]
- 19.van Hazel I., Sabouhanian A., Day L., Endler J. A., Chang B. S., Functional characterization of spectral tuning mechanisms in the great bowerbird short-wavelength sensitive visual pigment (SWS1), and the origins of UV/violet vision in passerines and parrots. BMC Evol. Biol. 13, 250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun H., Macke J. P., Nathans J., Mechanisms of spectral tuning in the mouse green cone pigment. Proc. Natl. Acad. Sci. U.S.A. 94, 8860–8865 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauser F. E., van Hazel I., Chang B. S. W., Spectral tuning in vertebrate short wavelength-sensitive 1 (SWS1) visual pigments: Can wavelength sensitivity be inferred from sequence data? J. Exp. Zoolog. B Mol. Dev. Evol. 322, 529–539 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Perry M., et al., Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Nature 535, 280–284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parry J. W., et al., Mix and match color vision: Tuning spectral sensitivity by differential opsin gene expression in lake Malawi cichlids. Curr. Biol. 15, 1734–1739 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Porter M. L., Awata H., Bok M. J., Cronin T. W., Exceptional diversity of opsin expression patterns in Neogonodactylus oerstedii (Stomatopoda) retinas. Proc. Natl. Acad. Sci. U.S.A. 117, 8948–8957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cronin T. W., Marshall N. J., Caldwell R. L., Spectral tuning and the visual ecology of mantis shrimps. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1263–1267 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Kooi C., Stavenga D., Arikawa K., Belusic G., Kelber A., Evolution of insect colour vision–From spectral sensitivity to visual ecology. Annu. Rev. Entomol. 66, 23.1–23.28 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Arikawa K., The eyes and vision of butterflies. J. Physiol. 595, 5457–5464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs G. H., Primate photopigments and primate color vision. Proc. Natl. Acad. Sci. U.S.A. 93, 577–581 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luehrmann M., et al., Short-term colour vision plasticity on the reef: Changes in opsin expression under varying light conditions differ between ecologically distinct fish species. J. Exp. Biol. 221, jeb175281 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Jacobs G. H., Evolution of colour vision in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2957–2967 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shichida Y., Matsuyama T., Evolution of opsins and phototransduction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2881–2895 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez E. A., et al., The role of ecological factors in shaping bat cone opsin evolution. Proc. Biol. Sci. 285, 20172835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terakita A., et al., Expression and comparative characterization of Gq-coupled invertebrate visual pigments and melanopsin. J. Neurochem. 105, 883–890 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Fain G. L., Hardie R., Laughlin S. B., Phototransduction and the evolution of photoreceptors. Curr. Biol. 20, R114–R124 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plachetzki D. C., Fong C. R., Oakley T. H., The evolution of phototransduction from an ancestral cyclic nucleotide gated pathway. Proc. Biol. Sci. 277, 1963–1969 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez M. P., Nasi E., Light transduction in invertebrate hyperpolarizing photoreceptors: Possible involvement of a Go-regulated guanylate cyclase. J. Neurosci. 20, 5254–5263 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y. J., et al., The Drosophila dgq gene encodes a G alpha protein that mediates phototransduction. Neuron 13, 1143–1157 (1994). [DOI] [PubMed] [Google Scholar]
- 38.Hardie R., Phototransduction mechanisms in Drosophila microvillar photoreceptors. WIREs Membr Transp Signal 1, 162–187 (2012). [Google Scholar]
- 39.Kelber A., Bird colour vision–From cones to perception. Curr. Opin. Behav. Sci. 30, 34–40 (2019). [Google Scholar]
- 40.Carleton K. L., Kocher T. D., Cone opsin genes of african cichlid fishes: Tuning spectral sensitivity by differential gene expression. Mol. Biol. Evol. 18, 1540–1550 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Shi Y., Yokoyama S., Molecular analysis of the evolutionary significance of ultraviolet vision in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 100, 8308–8313 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowmaker J. K., Hunt D. M., Evolution of vertebrate visual pigments. Curr. Biol. 16, R484–R489 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Jacobs G. H., Losses of functional opsin genes, short-wavelength cone photopigments, and color vision–A significant trend in the evolution of mammalian vision. Vis. Neurosci. 30, 39–53 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Nathans J., Thomas D., Hogness D. S., Molecular genetics of human color vision: The genes encoding blue, green, and red pigments. Science 232, 193–202 (1986). [DOI] [PubMed] [Google Scholar]
- 45.Dulai K. S., von Dornum M., Mollon J. D., Hunt D. M., The evolution of trichromatic color vision by opsin gene duplication in New World and Old World primates. Genome Res. 9, 629–638 (1999). [PubMed] [Google Scholar]
- 46.Yokoyama S., et al., Epistatic adaptive evolution of human color vision. PLoS Genet. 10, e1004884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawamura S., Color vision diversity and significance in primates inferred from genetic and field studies. Genes Genomics 38, 779–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feuda R., Marlétaz F., Bentley M. A., Holland P. W. H., Conservation, duplication, and divergence of five opsin genes in insect evolution. Genome Biol. Evol. 8, 579–587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monteiro A., Origin, development, and evolution of butterfly eyespots. Annu. Rev. Entomol. 60, 253–271 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Dasmahapatra K.et al.; Heliconius Genome Consortium , Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernard G. D., Red-absorbing visual pigment of butterflies. Science 203, 1125–1127 (1979). [DOI] [PubMed] [Google Scholar]
- 52.Bernard G., Douglass J., Goldsmith T., Far-red sensitive visual pigment of a metalmark butterfly. Invest. Ophthalmol. 29, 350 (1988). [Google Scholar]
- 53.Kelber A., Ovipositing butterflies use a red receptor to see green. J. Exp. Biol. 202, 2619–2630 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Pratt G., Emmel J., Bernard G. D., The buckwheat metalmarks. Am Butterflies 19, 4–31 (2011). [Google Scholar]
- 55.Chen P.-J., Awata H., Matsushita A., Yang E.-C., Arikawa K., Extreme spectral richness in the eye of the common bluebottle butterfly, Graphium sarpedon. Front. Ecol. Evol. 4, 18 (2016). [Google Scholar]
- 56.Zaccardi G., Kelber A., Sison-Mangus M. P., Briscoe A. D., Color discrimination in the red range with only one long-wavelength sensitive opsin. J. Exp. Biol. 209, 1944–1955 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Wakakuwa M., Stavenga D. G., Kurasawa M., Arikawa K., A unique visual pigment expressed in green, red and deep-red receptors in the eye of the small white butterfly, Pieris rapae crucivora. J. Exp. Biol. 207, 2803–2810 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Ogawa Y., Kinoshita M., Stavenga D. G., Arikawa K., Sex-specific retinal pigmentation results in sexually dimorphic long-wavelength-sensitive photoreceptors in the eastern pale clouded yellow butterfly, Colias erate. J. Exp. Biol. 216, 1916–1923 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Satoh A., et al., Red-shift of spectral sensitivity due to screening pigment migration in the eyes of a moth, Adoxophyes orana. Zoological Lett. 3, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pirih P., et al., The giant butterfly-moth Paysandisia archon has spectrally rich apposition eyes with unique light-dependent photoreceptor dynamics. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 204, 639–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sison-Mangus M. P., Briscoe A. D., Zaccardi G., Knüttel H., Kelber A., The lycaenid butterfly Polyommatus icarus uses a duplicated blue opsin to see green. J. Exp. Biol. 211, 361–369 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Bernard G. D., Remington C. L., Color vision in Lycaena butterflies: Spectral tuning of receptor arrays in relation to behavioral ecology. Proc. Natl. Acad. Sci. U.S.A. 88, 2783–2787 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sison-Mangus M. P., Bernard G. D., Lampel J., Briscoe A. D., Beauty in the eye of the beholder: The two blue opsins of lycaenid butterflies and the opsin gene-driven evolution of sexually dimorphic eyes. J. Exp. Biol. 209, 3079–3090 (2006). [DOI] [PubMed] [Google Scholar]
- 64.McCulloch K. J., Osorio D., Briscoe A. D., Sexual dimorphism in the compound eye of Heliconius erato: A nymphalid butterfly with at least five spectral classes of photoreceptor. J. Exp. Biol. 219, 2377–2387 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Pirih P., Arikawa K., Stavenga D. G., An expanded set of photoreceptors in the Eastern Pale Clouded Yellow butterfly, Colias erate. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 196, 501–517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blake A. J., Pirih P., Qiu X., Arikawa K., Gries G., Compound eyes of the small white butterfly Pieris rapae have three distinct classes of red photoreceptors. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 205, 553–565 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Bernard G. D., Bleaching of rhabdoms in eyes of intact butterflies. Science 219, 69–71 (1983). [DOI] [PubMed] [Google Scholar]
- 68.van Nieukerken E., et al., Order Lepidoptera Linnaeus, 1758. Zootaxa 3148, 212–221 (2011). [Google Scholar]
- 69.Heikkilä M., Kaila L., Mutanen M., Peña C., Wahlberg N., Cretaceous origin and repeated tertiary diversification of the redefined butterflies. Proc. Biol. Sci. 279, 1093–1099 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Espeland M., et al., A comprehensive and dated phylogenomic analysis of butterflies. Curr. Biol. 28, 770–778.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Chang B. S. W., Ayers D., Smith W. C., Pierce N. E., Cloning of the gene encoding honeybee long-wavelength rhodopsin: A new class of insect visual pigments. Gene 173, 215–219 (1996). [DOI] [PubMed] [Google Scholar]
- 72.Chou W.-H., et al., Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron 17, 1101–1115 (1996). [DOI] [PubMed] [Google Scholar]
- 73.Osorio D., Vorobyev M., A review of the evolution of animal colour vision and visual communication signals. Vision Res. 48, 2042–2051 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Endler J. A., Basolo A. L., Sensory ecology, receiver biases and sexual selection. Trends Ecol. Evol. 13, 415–420 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Kelber A., Osorio D., From spectral information to animal colour vision: Experiments and concepts. Proc. Biol. Sci. 277, 1617–1625 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Osorio D., Vorobyev M., Photoreceptor spectral sensitivities in terrestrial animals: Adaptations for luminance and colour vision. Proc. Biol. Sci. 272, 1745–1752 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vane-Wright R., Boppre M., Visual and chemical signalling in butterflies: Functional and phylogenetic perspectives. Proc. Biol. Sci. 340, 197–205 (1993). [Google Scholar]
- 78.Fordyce J. A., Nice C. C., Forister M. L., Shapiro A. M., The significance of wing pattern diversity in the Lycaenidae: Mate discrimination by two recently diverged species. J. Evol. Biol. 15, 871–879 (2002). [Google Scholar]
- 79.Jiggins C. D., Naisbit R. E., Coe R. L., Mallet J., Reproductive isolation caused by colour pattern mimicry. Nature 411, 302–305 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Wilkins L., Osorio D., Object colours, material properties and animal signals. J. Exp. Biol., 10.1242/jeb.204487 (2019). [DOI] [PubMed]
- 81.Koi S., Hall D., “Atala butterfly, Atala hairstreak, Coontie hairstreak, Eumaeus atala Poey 1832 (Insecta: Lepidoptera: Lycaenidae)” (EENY-169, University of Florida IFAS Extension, 2016). [Google Scholar]
- 82.Frentiu F. D., et al., Opsin clines in butterflies suggest novel roles for insect photopigments. Mol. Biol. Evol. 32, 368–379 (2015). [DOI] [PubMed] [Google Scholar]
- 83.Eacock A., et al., Adaptive colour change and background choice behaviour in peppered moth caterpillars is mediated by extraocular photoreception. Commun. Biol. 2, 286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bracken-Grissom H. D., et al., Light organ photosensitivity in deep-sea shrimp may suggest a novel role in counterillumination. Sci. Rep. 10, 4485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Donohue M. W., Cohen J. H., Cronin T. W., Cerebral photoreception in mantis shrimp. Sci. Rep. 8, 9689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Porter M. L., Beyond the eye: Molecular evolution of extraocular photoreception. Integr. Comp. Biol. 56, 842–852 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Manceau M., Domingues V. S., Linnen C. R., Rosenblum E. B., Hoekstra H. E., Convergence in pigmentation at multiple levels: Mutations, genes and function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 2439–2450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chapal M., Mintzer S., Brodsky S., Carmi M., Barkai N., Resolving noise-control conflict by gene duplication. PLoS Biol. 17, e3000289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carroll S. B., Evolution at two levels: On genes and form. PLoS Biol. 3, e245 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hempel de Ibarra N., Vorobyev M., Menzel R., Mechanisms, functions and ecology of colour vision in the honeybee. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 200, 411–433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saito T., et al., Spectral tuning mediated by helix III in butterfly long wavelength-sensitive visual opsins revealed by heterologous action spectroscopy. Zoological Lett. 5, 35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arikawa K., Spectral organization of the eye of a butterfly, Papilio. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 189, 791–800 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Hart N. S., Hunt D. M., Avian visual pigments: Characteristics, spectral tuning, and evolution. Am. Nat. 169 (suppl. 1), S7–S26 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Cuthill I. C., et al., The biology of color. Science 357, eaan0221 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Marques D. A., et al., Convergent evolution of SWS2 opsin facilitates adaptive radiation of threespine stickleback into different light environments. PLoS Biol. 15, e2001627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Busserolles F., Fogg L., Cortesi F., Marshall J., The exceptional diversity of visual adaptations in deep-sea teleost fishes. Semin. Cell Dev. Biol. 106, 20–30 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Partridge J., Douglas R., Far-red sensitivity of dragon fish. Nature 375, 21–22 (1995). [Google Scholar]
- 98.Terai Y., et al., Divergent selection on opsins drives incipient speciation in Lake Victoria cichlids. PLoS Biol. 4, e433 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prokopy R. J., Owens E. D., Visual detection of plants by herbivorous insects. Annu. Rev. Entomol. 28, 337–364 (1983). [Google Scholar]
- 100.Liew O. W., Chong P. C., Li B., Asundi A. K., Signature optical cues: Emerging technologies for monitoring plant health. Sensors (Basel) 8, 3205–3239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaessmann H., Origins, evolution, and phenotypic impact of new genes. Genome Res. 20, 1313–1326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang J., Evolution by gene duplication: An update. Trends Ecol. Evol. 18, 292–298 (2003). [Google Scholar]
- 103.Lynch M., Conery J. S., The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000). [DOI] [PubMed] [Google Scholar]
- 104.Long M., Thornton K., Gene duplication and evolution. Science 293, 1551 (2001). [DOI] [PubMed] [Google Scholar]
- 105.Soskine M., Tawfik D. S., Mutational effects and the evolution of new protein functions. Nat. Rev. Genet. 11, 572–582 (2010). [DOI] [PubMed] [Google Scholar]
- 106.Castiglione G. M., Chang B. S. W., Functional trade-offs and environmental variation shaped ancient trajectories in the evolution of dim-light vision. eLife 7, e35957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang J., Zhang Y. P., Rosenberg H. F., Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30, 411–415 (2002). [DOI] [PubMed] [Google Scholar]
- 108.Liénard M. A., Wang H.-L., Lassance J.-M., Löfstedt C., Sex pheromone biosynthetic pathways are conserved between moths and the butterfly Bicyclus anynana. Nat. Commun. 5, 3957 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brand P., et al., The evolution of sexual signaling is linked to odorant receptor tuning in perfume-collecting orchid bees. Nat. Commun. 11, 244 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lord N. P., et al., A cure for the blues: Opsin duplication and subfunctionalization for short-wavelength sensitivity in jewel beetles (Coleoptera: Buprestidae). BMC Evol. Biol. 16, 107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Awata H., Wakakuwa M., Arikawa K., Evolution of color vision in pierid butterflies: Blue opsin duplication, ommatidial heterogeneity and eye regionalization in Colias erate. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 195, 401–408 (2009). [DOI] [PubMed] [Google Scholar]
- 112.Briscoe A. D., Six opsins from the butterfly Papilio glaucus: Molecular phylogenetic evidence for paralogous origins of red-sensitive visual pigments in insects. J. Mol. Evol. 51, 110–121 (2000). [DOI] [PubMed] [Google Scholar]
- 113.Arikawa K., Wakakuwa M., Qiu X., Kurasawa M., Stavenga D. G., Sexual dimorphism of short-wavelength photoreceptors in the small white butterfly, Pieris rapae crucivora. J. Neurosci. 25, 5935–5942 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fasick J. I., Lee N., Oprian D. D., Spectral tuning in the human blue cone pigment. Biochemistry 38, 11593–11596 (1999). [DOI] [PubMed] [Google Scholar]
- 115.Yokoyama S., Tada T., The spectral tuning in the short wavelength-sensitive type 2 pigments. Gene 306, 91–98 (2003). [DOI] [PubMed] [Google Scholar]
- 116.Chinen A., Matsumoto Y., Kawamura S., Spectral differentiation of blue opsins between phylogenetically close but ecologically distant goldfish and zebrafish. J. Biol. Chem. 280, 9460–9466 (2005). [DOI] [PubMed] [Google Scholar]
- 117.Chan T., Lee M., Sakmar T. P., Introduction of hydroxyl-bearing amino acids causes bathochromic spectral shifts in rhodopsin. Amino acid substitutions responsible for red-green color pigment spectral tuning. J. Biol. Chem. 267, 9478–9480 (1992). [PubMed] [Google Scholar]
- 118.Frentiu F. D., Briscoe A. D., A butterfly eye’s view of birds. BioEssays 30, 1151–1162 (2008). [DOI] [PubMed] [Google Scholar]
- 119.Ryan M., Cummings M., Conceptual biases and mate choice. Annu. Rev. Ecol. Evol. Syst. 44, 437–459 (2013). [Google Scholar]
- 120.Boughman J. W., Divergent sexual selection enhances reproductive isolation in sticklebacks. Nature 411, 944–948 (2001). [DOI] [PubMed] [Google Scholar]
- 121.Lind O., Henze M. J., Kelber A., Osorio D., Coevolution of coloration and colour vision? Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bybee S. M., et al., UV photoreceptors and UV-yellow wing pigments in Heliconius butterflies allow a color signal to serve both mimicry and intraspecific communication. Am. Nat. 179, 38–51 (2012). [DOI] [PubMed] [Google Scholar]
- 123.Kelber A., Roth L. S. V., Nocturnal colour vision–not as rare as we might think. J. Exp. Biol. 209, 781–788 (2006). [DOI] [PubMed] [Google Scholar]
- 124.Chittka L., Optimal sets of receptors and color opponent systems for coding of natural objects in insect vision. J. Theor. Biol. 181, 179–196 (1996). [Google Scholar]
- 125.Chittka L., The colour hexagon: A chromaticity diagram based on photoreceptor excitations as a generalized representation of colour opponency. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 170, 533–543 (1992). [Google Scholar]
- 126.McCoy D. E., Prum R. O., Convergent evolution of super black plumage near bright color in 15 bird families. J. Exp. Biol. 222, jeb208140 (2019). [DOI] [PubMed] [Google Scholar]
- 127.McCoy D. E., et al., Structurally assisted super black in colourful peacock spiders. Proc. Biol. Sci. 286, 20190589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Davis A. L., Nijhout H. F., Johnsen S., Diverse nanostructures underlie thin ultra-black scales in butterflies. Nat. Commun. 11, 1294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vukusic P., Sambles J. R., Lawrence C. R., Structurally assisted blackness in butterfly scales. Proc. Biol. Sci. 271 (suppl. 4), S237–S239 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Whitaker M. R. L., Salzman S., Ecology and evolution of cycad-feeding lepidoptera. Ecol. Lett. 23, 1862–1877 (2020). [DOI] [PubMed] [Google Scholar]
- 131.Rossi M., et al., Neural processing genes are linked to divergence in visual mate preference in sympatric Heliconius butterflies. Nat. Commun. 11, 4763 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Segalla R., Telles F. J., Pinheiro F., Morellato P., A review of current knowledge of Zamiaceae, with emphasis on Zamia from South America. Trop. Conserv. Sci. 12, 1–21 (2019). [Google Scholar]
- 133.Taddei-Ferretti C., Ed., “Costs and benefits of increasing the dimensionality of color vision system” in Biophysics of Photoreception: Molecular and Phototransductive Events (World Scientist, 1997), pp. 280–289. [Google Scholar]
- 134.Bernard G. D., Dark-processes following photoconversion of butterfly rhodopsins. Biophys. Struct. Mech. 9, 277–286 (1983). [Google Scholar]
- 135.Stavenga D. G., Numan J., Tinbergen J., Kuiper J., Insect pupil mechanisms. II. Pigment migration in retinula cells of butterflies. J Comp Phys A 113, 73–93 (1977). [Google Scholar]
- 136.Palacios A. G., Goldsmith T. H., Bernard G. D., Sensitivity of cones from a cyprinid fish (Danio aequipinnatus) to ultraviolet and visible light. Vis. Neurosci. 13, 411–421 (1996). [DOI] [PubMed] [Google Scholar]
- 137.Isogai Y., et al., Molecular organization of vomeronasal chemoreception. Nature 478, 241–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The E. atala eye transcriptome and opsin transcript data are available in the Sequence Read Archive Bioproject number PRJNA625881 and GenBank database under accession numbers MN831881–MN831885. Source data underlying epi-microspectrophotometry, functional expression, and reflectance spectra are available in Datasets S1–S3, respectively.