Significance
An abnormal number of chromosomes or aneuploidy accounts for most spontaneous abortions, as missegregation of a single chromosome during development is often lethal. Only individuals with trisomy 21, which causes Down syndrome, can live to adulthood but show cognitive disabilities, increased risk for leukemias, autoimmune disorders, and clinical symptoms associated with premature aging. The mechanisms by which aneuploidy affects cellular function to cause Down syndrome are not understood. Our studies revealed that aneuploidy causes several defects in cells from individuals with Down syndrome. These include increased gene and protein expression, lower viability, and increased dependency on serine to proliferate. Our studies establish a critical role of aneuploidy, independent of triplicated gene identity, in driving cellular defects associated with trisomy 21.
Keywords: Down syndrome, aneuploidy, sphingolipids, dosage compensation, trisomy 21
Abstract
An extra copy of chromosome 21 causes Down syndrome, the most common genetic disease in humans. The mechanisms contributing to aneuploidy-related pathologies in this syndrome, independent of the identity of the triplicated genes, are not well defined. To characterize aneuploidy-driven phenotypes in trisomy 21 cells, we performed global transcriptome, proteome, and phenotypic analyses of primary human fibroblasts from individuals with Patau (trisomy 13), Edwards (trisomy 18), or Down syndromes. On average, mRNA and protein levels were increased by 1.5-fold in all trisomies, with a subset of proteins enriched for subunits of macromolecular complexes showing signs of posttranscriptional regulation. These results support the lack of evidence for widespread dosage compensation or dysregulation of chromosomal domains in human autosomes. Furthermore, we show that several aneuploidy-associated phenotypes are present in trisomy 21 cells, including lower viability and increased dependency on serine-driven lipid synthesis. Our studies establish a critical role of aneuploidy, independent of triplicated gene identity, in driving cellular defects associated with trisomy 21.
An abnormal number of chromosomes or aneuploidy accounts for most spontaneous abortions, as missegregation of a single chromosome during development is often lethal (1, 2). Individuals with trisomies of chromosomes 13 or 18, which cause Patau and Edwards syndromes, respectively, are born with severe developmental defects and die soon after birth. Only individuals with trisomy 21, which causes Down syndrome, can live to adulthood. However, they display developmental abnormalities, cognitive disabilities, congenital heart defects, increased risk for leukemias and neurodegenerative disease, autoimmune disorders, and clinical symptoms associated with premature aging (3, 4). The incidence of aneuploidy is also associated with tumorigenesis and increases with age in both somatic and germline tissues in apparently healthy individuals (5, 6). The mechanisms by which aneuploidy affects cellular function to cause Down syndrome, promotes tumor formation, or promotes aging are not well understood.
The phenotypes associated with trisomy 21 at the organismal level are complex. Indeed, individuals with Down syndrome are born with varying severities and higher risks for several human diseases (4). Two main gene-centric hypotheses have been proposed to explain the biological consequences of trisomy 21. One is that increased expression of a particular gene and its downstream effectors cause the pathophysiology in Down syndrome. For example, increased expression of the amyloid-beta precursor protein (APP), the High Mobility Group Nucleosome Binding Domain 1 (HMGN1), or the transcription factor RUNX1 are associated with the increased incidence of early onset of Alzheimer’s disease, B cell acute lymphoblastic leukemia, or transient myeloproliferative disorder in individuals with Down syndrome, respectively (7–9). A second hypothesis is that abnormalities arise due to the indirect effects of increased gene activity on chromosome 21 that alter specific cellular processes. Examples include genes that regulate splicing (U2AF1L5, RBM1, and U2AF1), chromatin regulators (HMGN1 and BRWD1), secretory-endosomal functions (DOPEY2, CSTB, and SYNJ1), protein turnover (USP25), or metabolism (SOD1). Nonetheless, it has been challenging to prove that a third wild-type copy of a given gene on chromosome 21 is the sole driver of a particular disease (3, 4). Indeed, analysis of human segmental trisomies of chromosome 21 provides evidence against the existence of a single chromosomal region contributing to Down syndrome phenotypes (10).
A third, less well-explored consequence of trisomy 21 is the disruption of cellular physiology due to the presence of an extra chromosome independent of the identity of the triplicated genes (i.e., the aneuploid status of the cell). Several studies have revealed aneuploidy-driven phenotypes shared among different organisms that are independent of the abnormal karyotype (11, 12). These include defects in cell proliferation, signs of proteotoxic stress, genomic instability, and altered metabolism. We recently showed in both yeast and human cells that the presence of an extra copy of a chromosome disrupts nuclear morphology (13). This phenotype is associated with dysregulation of the synthesis of lipids that constitute the nuclear membrane. In particular, we found that increasing the levels of long-chain bases suppresses nuclear abnormalities and improves the fitness of aneuploid cells, including human fibroblasts with trisomy 21 (13, 14).
To further characterize aneuploidy-associated phenotypes in human cells, we performed a global transcriptome and proteome quantification in primary fibroblasts isolated from individuals with Down, Edwards, or Patau syndromes. We found that several aneuploidy-associated phenotypes are present in primary fibroblasts with trisomy 21. These include lower viability, altered lipid metabolism, and an increased dependency on serine to proliferate. Altogether, these studies demonstrate that, in addition to the gene-driven pathologies associated with Down syndrome, aneuploidy-related cellular defects that are independent of chromosome 21 gene identity can contribute to disease.
Results
Transcriptome Analyses of Primary Fibroblasts Show a Proportional Relationship between Copy Number and Transcript Levels.
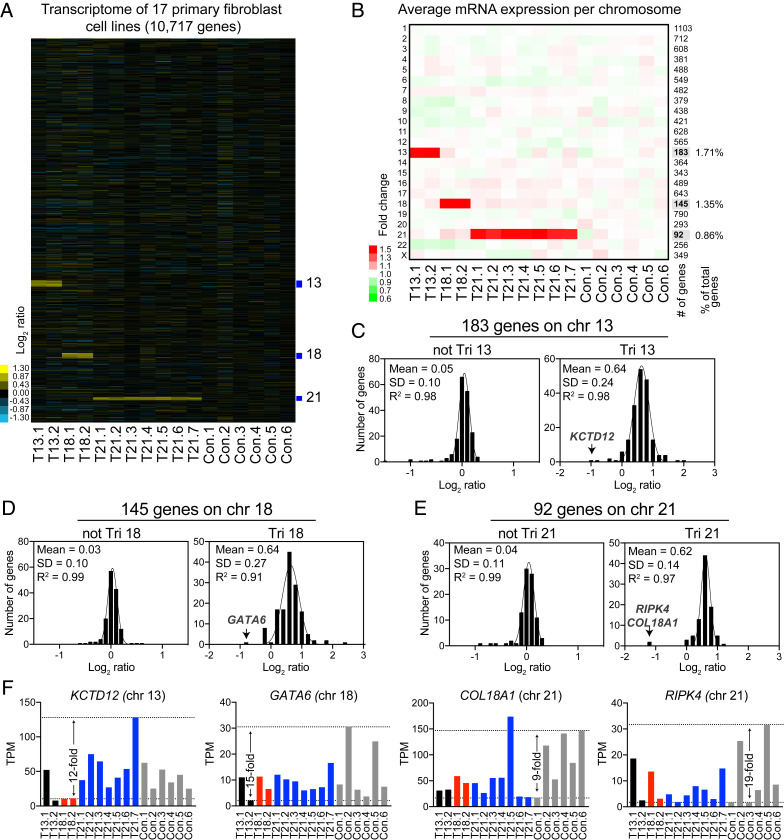
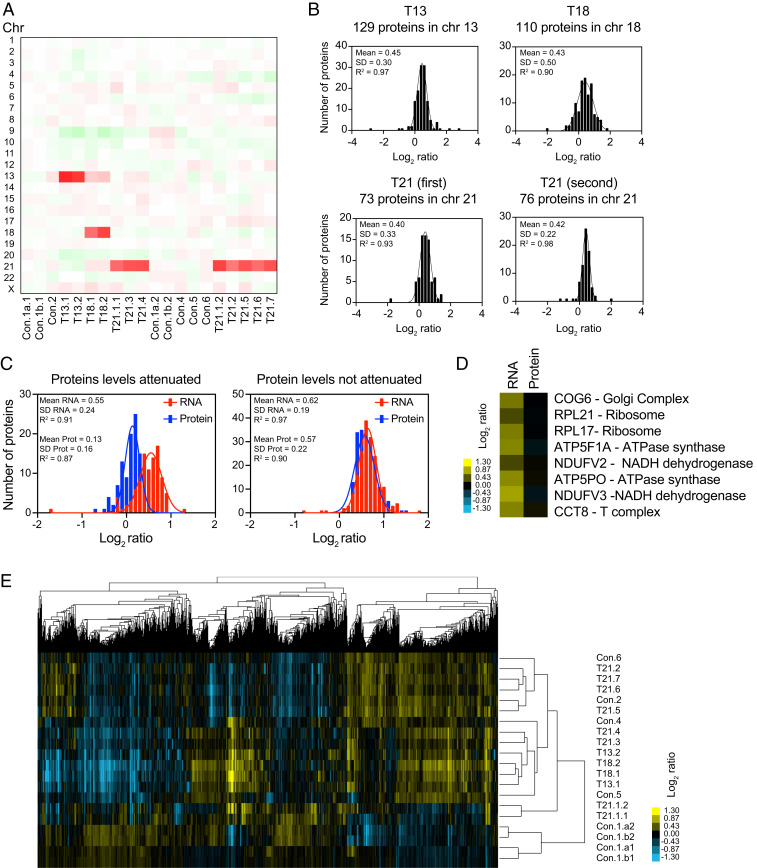
Several studies have suggested a complicated relationship between copy number, messenger RNA (mRNA), and protein levels of genes located on chromosome 21 (15–18). It has been proposed that gene dosage compensation can ameliorate the expression levels of genes located on triplicated chromosomes, yet the mechanisms for this process remain unclear (19). Furthermore, it is thought that an extra copy of chromosome 21 has a significant impact on the rest of the genome, causing dysregulation of expression of defined chromosomal domains (16). To test these hypotheses, we initially focused on quantifying the mRNA changes of the genes expressed on trisomic human chromosomes in primary fibroblasts. We obtained from the Coriell Institute 2 cell lines with trisomy 13, 2 with trisomy 18, 7 with trisomy 21, and 6 euploid controls, resulting in a total of 17 cell lines (SI Appendix, Fig. S1A). All cell lines were grown in the standard cell culture growth conditions, and samples were split into two for transcriptome and proteome quantifications. RNA sequencing (RNAseq) of the transcriptomes provided quantitative information for 10,717 genes across all 17 samples (Fig. 1A and Dataset S1). We calculated the log2 of the fold changes (FC) for all 17 samples using the average expression of the six controls as the reference genome. We obtained log2 ratios for 92, 145, and 183 genes on chromosomes 21, 18, and 13, respectively (Fig. 1B). We found that, on average, the triplicated chromosomes in all 11 analyzed trisomic cell lines displayed a 1.5-fold increase (3/2 copies, log2 FC = 0.58) in gene expression, consistent with an absence of dosage compensation (Fig. 1B and SI Appendix, Fig. S1B).
Fig. 1.
Transcript levels increase proportionally with gene copy number in trisomic primary fibroblasts. (A) The gene expression of 17 primary fibroblast cell lines ordered by chromosome position. The experiments (columns) for each cell line are shown. T13 = trisomy 13, T18 = trisomy 18, T21 = trisomy 21, and Con = euploid control; refer to SI Appendix, Fig. S1A for detailed cell line nomenclature. (B) The average gene expression per chromosome was calculated for each cell line. The number of genes quantified per chromosome is shown in the right of the heat map. Chromosomes 21, 18, and 13 represent 0.86%, 1.35%, and 1.71% of the total transcriptome, respectively. (C) A histogram of the average log2 ratios of the RNA copy number of genes located on euploid chromosomes (Left) and genes present on trisomic chromosome 13 (Right) in cell lines GM00526 and GM02948, relative to euploid controls, are shown. (D) A histogram of the average log2 ratios of the RNA copy number of genes located on euploid chromosomes (Left) and genes present on trisomic chromosome 18 (Right) in cell lines GM00734 and GM03538, relative to euploid controls are shown. (E) A histogram of the average log2 ratios of the RNA copy number of genes located on euploid chromosomes (Left) and genes present on trisomic chromosome 21 (Right) in cell lines GM04616, GM04592, AG05397, AG06922, GM02767, AG08941, and AG08942, relative to euploid controls are shown. In C–E, the bin size for all histograms is log2 ratio of 0.2 and medians are identical to means. Fits to a normal distribution (black line), means, SD, and goodness of fit (R2) are shown for each distribution. (F) Examples of the normalized TPM of a couple of triplicated genes that are outliers in the fits of the normal distributions. Black, red, blue, and gray bars correspond to expression in trisomy 13, 18, 21, and controls, respectively.
The values of the average log2 ratios of the triplicated genes fit a normal distribution with means of 0.6 and SDs of 0.24, 0.27, and 0.14 for trisomies 13, 18, and 21, respectively (Tri 13, 18, and 21, Fig. 1 C–E). The average log2 ratios of the same genes in the cell lines that harbor two copies also fit a normal distribution with means of 0.0 and SDs of 0.10 to 0.11 (not Tri 13, 18, or 21, Fig. 1 C–E). Although the log2 ratios of the triplicated genes also fit a normal distribution quite well (R2 between 0.91 and 0.98), few genes fall outside the Gaussian fits (outlier genes), as their expression does not seem to increase with copy number. Closer inspection of the normalized transcript counts (transcript per million, TPM) of the outliers in each distribution revealed that their expression levels were highly variable across individual cell lines, including the six euploid control cell lines (Fig. 1F and SI Appendix, Fig. S1C). For example, one outlier on chromosome 13 is KCTD12, a gene whose expression varies up to 12-fold among cell lines and up to sevenfold in the two different trisomies 13. The levels of the transcription factor GATA6 located on chromosome 18 vary up to 15-fold among cell lines not trisomic for chromosome 18. COL18A1 and RIPK4 are the main outliers in trisomy 21 and show, respectively, ninefold and 19-fold variability among control cell lines. Indeed, the few outliers of the distributions of triplicated genes, including below or above the fits, show high variability in expression among different individuals (SI Appendix, Fig. S1C). Our analysis indicates that, on average, most genes located on the trisomic chromosomes increase 1.5-fold in expression and that interindividual variability in gene expression accounts for a couple of outliers in the dataset.
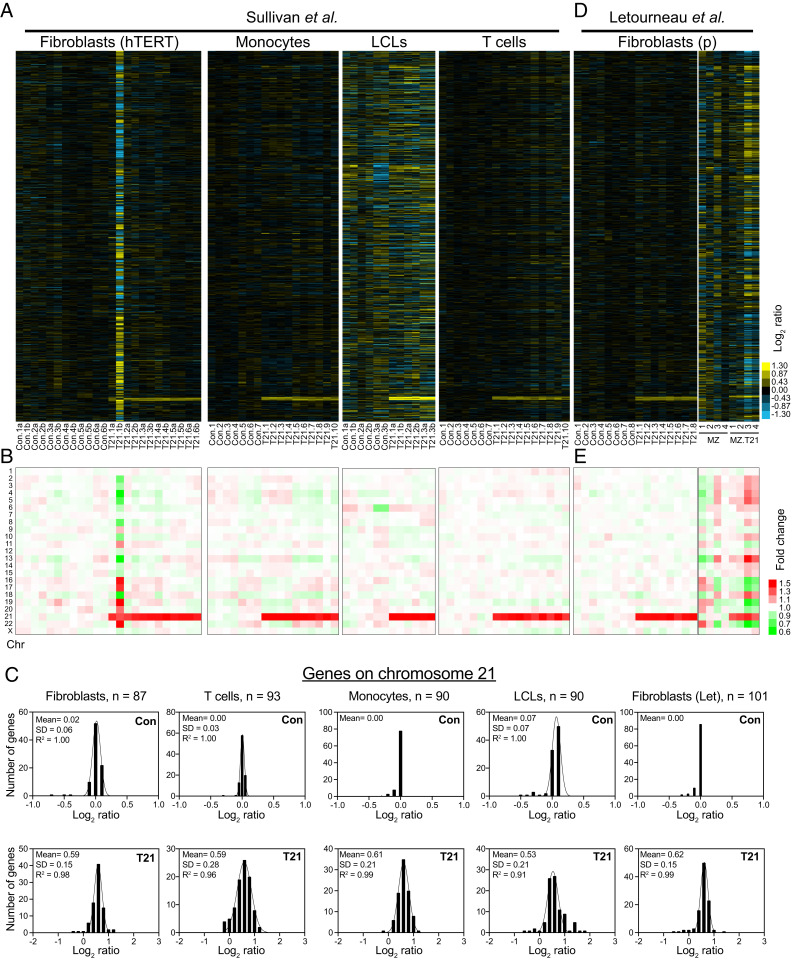
Although several recent studies quantified the transcriptome of trisomy 21 cells, they did not report a precise quantification and analyses of the mRNA levels of genes located on chromosome 21. To validate that the transcript levels of chromosome 21 genes proportionally increase with copy number, we analyzed two recently published datasets, one by Sullivan et al. (20) and the other by Letourneau et al. (16). In a tour de force approach, Sullivan et al. obtained transcriptome profiles of 24 human Telomerase reverse transcriptase (hTERT)-immortalized fibroblasts, 12 transformed B cells (lymphoblastoid cell lines, LCLs), 17 monocytes, and 17 T cell lines from different individuals with normal karyotype or with Down syndrome (SI Appendix, Fig. S2A). We calculated the log2 of the FC using the average expression of the control individuals as the reference genome for each cell type and plotted the data ordered by chromosome position (Fig. 2A and Dataset S2). Consistent with our analysis of primary fibroblasts, we found that, on average, the mRNA levels of genes located on chromosome 21 are 1.5-fold higher in all cell types and across samples from different individuals with Down syndrome relative to the controls (Fig. 2B). As observed in our data, a few outlier genes that do not fit the normal distributions can be accounted for by high variability in expression levels among control individuals (SI Appendix, Fig. S2C). Intriguingly, one fibroblast cell line (T21.1b) shows significant changes in gene expression in all chromosomes and deviates from the rest. We suspect that technical issues may account for such extreme changes in gene expression across the genome in this sample (see below). Altogether, the transcript levels of chromosome 21 genes in 37 out of 38 cell lines with trisomy 21 analyzed by Sullivan et al. fit a normal distribution with means very close to the predicted log2 ratio of 0.58 (Fig. 2C).
Fig. 2.
Transcript levels increase proportionally with gene copy number in distinct trisomy 21 cell lines. (A) The gene expression of 24 immortalized fibroblast cell lines, 17 monocytes cell lines, 12 LCLs, and 17 T cell lines obtained by Sullivan et al. (20). The experiments (columns) for each cell line are shown ordered by chromosome position. Refer to SI Appendix, Fig. S2A for detailed cell line nomenclature. (B) The average gene expression per chromosome was calculated for each cell line. (C) A histogram of the average log2 ratios of the RNA copy number of genes located on euploid chromosomes (Top) and genes present on trisomic chromosomes (Top) of the trisomic cell lines relative to euploid controls are shown. The bin size for all histograms is log2 ratio of 0.2, and medians are identical to means. Fits to a normal distribution (black line), means, SD, and goodness of fit (R2) are shown for each distribution. (D) The gene expression of 16 primary fibroblast and 4 technical replicates of each of 2 (MZ and MZ.T21) primary fibroblasts from monozygotic twins; data obtained by Letourneau et al. (16). The experiments (columns) for each cell line are shown ordered by chromosome position. Refer to SI Appendix, Fig. S2B for detailed cell line nomenclature. (E) The average gene expression per chromosome was calculated for each cell line (D).
In the second study, Letourneau et al. reported the expression profiles of primary fibroblasts from 16 unrelated individuals (8 controls and 8 with trisomy 21) and of two cell lines obtained from monozygotic twins, one with a normal karyotype and the other with trisomy 21 (SI Appendix, Fig. S2B). Consistently, we found, on average, a 1.5-fold increase in expression of chromosome 21 genes in the primary fibroblasts from individuals with Down syndrome (Fig. 2 D and E). On the other hand, the monozygotic twin with trisomy 21 shows very variable expression across the genome (Fig. 2 D and E and Dataset S2). Letourneau et al. interpreted the latter result as evidence for trisomy 21 driving the dysregulation in expression across chromosomal domains. A closer inspection of the transcript counts raises the possibility that technical issues account for this result. We found that the correlation of gene expression among biological replicates of the same cell line from the monozygotic twin with trisomy 21 is low (R2 = 0.44, SI Appendix, Fig. S2E) compared to the correlation between two different primary fibroblasts (R2 = 0.95, SI Appendix, Fig. S2D). We also found that up to 27% of the total reads in the RNAseq datasets of the monozygotic twins come from mitochondrial DNA–encoded genes (SI Appendix, Fig. S2F). The poor reproducibility of the monozygotic twins’ transcriptomes seems to be caused by low gene coverage. The total read counts of the monozygotic twins' genome are about half of the coverage of primary fibroblasts (SI Appendix, Fig. S2F). Indeed, only ∼4,000 genes show 1 read per kilobase million or greater values in the monozygotic twin dataset compared to more than 10,000 in the primary fibroblasts (SI Appendix, Fig. S2G). Unexpectedly, the average gene expression per chromosome of the outlier sample from the Sullivan et al. study (T21.1b) shows similar expression patterns but opposite values than the monozygotic twin trisomy 21 (SI Appendix, Fig. S2E). This analysis raises the possibility that similar technical issues may account for these very variable transcriptomes. Excluding the two outlier transcriptomes, our analyses support the hypothesis that, on average, the mRNA levels increase proportionally with gene copy numbers. Mechanisms to compensate and ameliorate gene expression of an extra copy of a human autosome do not seem to be engaged.
Interindividual Variability Drives Gene Expression Patterns of Human Primary Fibroblasts.
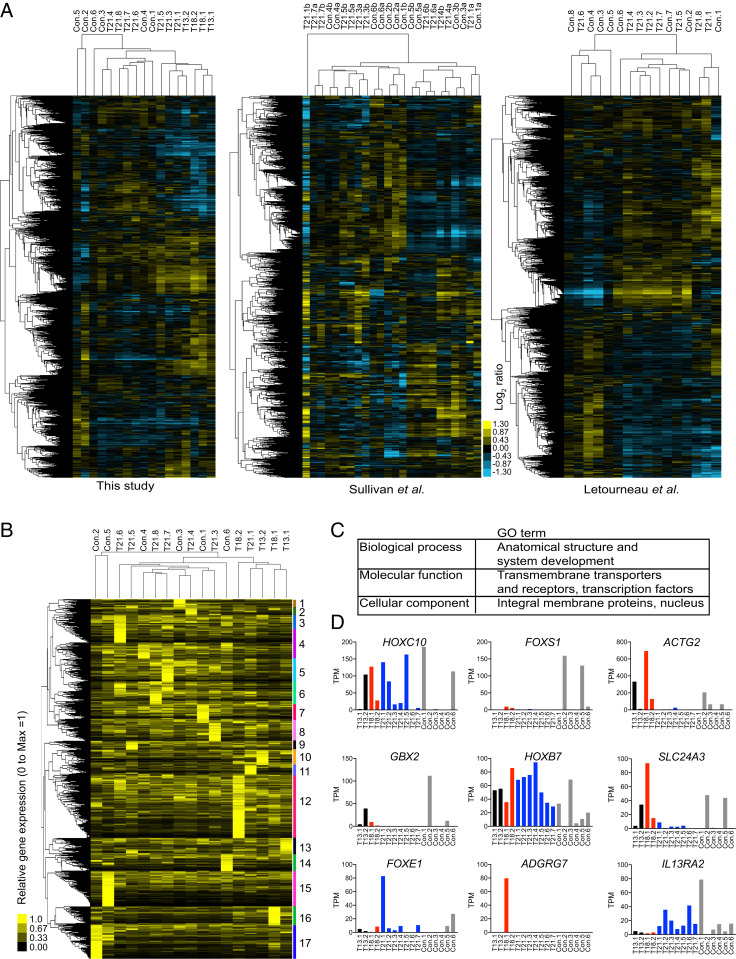
Similar to the findings reported by Sullivan et al., we found that the gene expression patterns of human fibroblasts do not cluster with karyotype, sex, or the donor's age in all three datasets (Fig. 3A). Excluding chromosome 21, we could not identify a set of genes that are commonly up- or down-regulated in all three datasets. Instead, hierarchical clustering analysis indicates that several control cell lines have gene expression patterns closer to those of a trisomic cell line than another control. These results suggest that gene expression patterns are specific to the cell’s identity coming from different individuals, thereby causing significant variability and occluding the identification of gene responses associated with aneuploidy in this context.
Fig. 3.
Interindividual variability drives gene expression patterns of human primary fibroblasts. (A) The hierarchical clustering analysis of the expression patterns of primary fibroblasts analyzed in this study and in the studies by Sullivan et al. (20) and Letourneau et al. (16) are shown. The expression patterns cluster independent of the karyotype of the cell line. (B) The relative gene expression of genes uniquely expressed in some cell lines. The hierarchical clustering analysis revealed 17 clusters specific for each cell line. (C) The gene ontology enrichment analysis shows that the uniquely expressed genes are enriched in master transcription factors that regulate development and cell surface markers, including transmembrane transporters and receptors proteins. (D) Examples of the normalized TPM of a couple of genes highly expressed in some cell lines but poorly expressed in others. Black, red, blue, and gray bars correspond to expression in trisomy 13, 18, 21, and controls, respectively.
To better define the source of variability in gene expression among primary fibroblasts, we analyzed the identity of genes uniquely expressed in individual cell lines. Initially, genes whose expression level was not detected (zero reads) in several cell lines were omitted as part of our cutoff to calculate FC among all samples. We found a total of 2,543 that were significantly expressed in specific cell lines but not detected in others (Dataset S3). Rather than calculating ratios, we normalized the relative TPM across genes from no reads to maximum reads on a range from 0 to 1 (0 is in black, 1 is in yellow, Fig. 3B). Hierarchical clustering revealed a set of genes specifically expressed in each cell line. We identified 17 clusters representing 17 cell lines. Remarkably, the clustering pattern among the uniquely expressed genes in the cell lines is similar to the clustering pattern of their whole transcriptome (Fig. 3 A and B). Gene ontology enrichment analyses revealed that this set of genes includes several master transcription factors that determine the anatomical structure and regulate development, including homeobox and forkhead box genes (Fig. 3 C and D). Several membrane transporters and receptors are present in this set of genes. These results indicate that cell identity, including anatomical and developmental properties specific to each individual, governs the gene expression patterns of primary fibroblasts in both trisomic and euploid cell lines.
Other sources of variability in gene expression come from culture conditions. We found that biological replicates of the same cell line cultured in parallel show good reproducibility (linear regression fit, slope = 1.0 and R2 = 0.97, SI Appendix, Fig. S3B). However, when we compared the gene expression of the same cell lines grown months apart using independent medium preparations, hierarchical clustering patterns were influenced by the time the cultures were performed (SI Appendix, Fig. S3 C and D). These results indicate that, in addition to cell identity, distinct growth conditions can significantly contribute to physiological noise in gene expression, perhaps fueling the discrepancies between different research groups.
Despite interindividual variability, we sought to determine whether a set of genes change expression in response to aneuploidy or trisomy 21 in our dataset. Using hierarchical clustering analyses, we identified a set of commonly down-regulated genes in 7 out of 11 trisomic cell lines (SI Appendix, Fig. S3A). This cluster is enriched for genes that regulate metabolic processes, including amino acid metabolism (P = 6.8 × 10−6) and response to nutrient levels (P = 7.1 × 10−7). These results support the hypothesis that aneuploidy disrupts cellular metabolism. Next, by sorting the average change in gene expression between aneuploids compared to euploids, we also identify a set of up-regulated genes in trisomic fibroblasts. The up-regulated cluster is enriched for genes in the DNA repair pathway (P = 6.2 × 10−21), consistent with aneuploidy-causing replication stress and genomic instability. It is noteworthy that Sullivan et al. found that trisomy 21 fibroblasts display a gene expression pattern associated with an activated immune response driven by interferon signaling (20), a result not observed in the analysis of our dataset. Furthering these discrepancies, Letourneau et al. reported another distinct signature associated with reduced expression of secreted proteins involved in cytokine–cytokine receptor pathways and inflammatory response (16). Altogether, the three studies revealed different sets of genes that might be regulated by trisomy 21 without reaching a consensus on a conserved cellular response. Several other studies have reported different cellular pathways to be affected by trisomy 21 (21–23). We conclude that interindividual variability and experimental conditions mask gene expression patterns that might be associated with cellular responses associated with aneuploidy.
Protein Levels Increase Proportionally with Gene Copy Number and mRNA Levels.
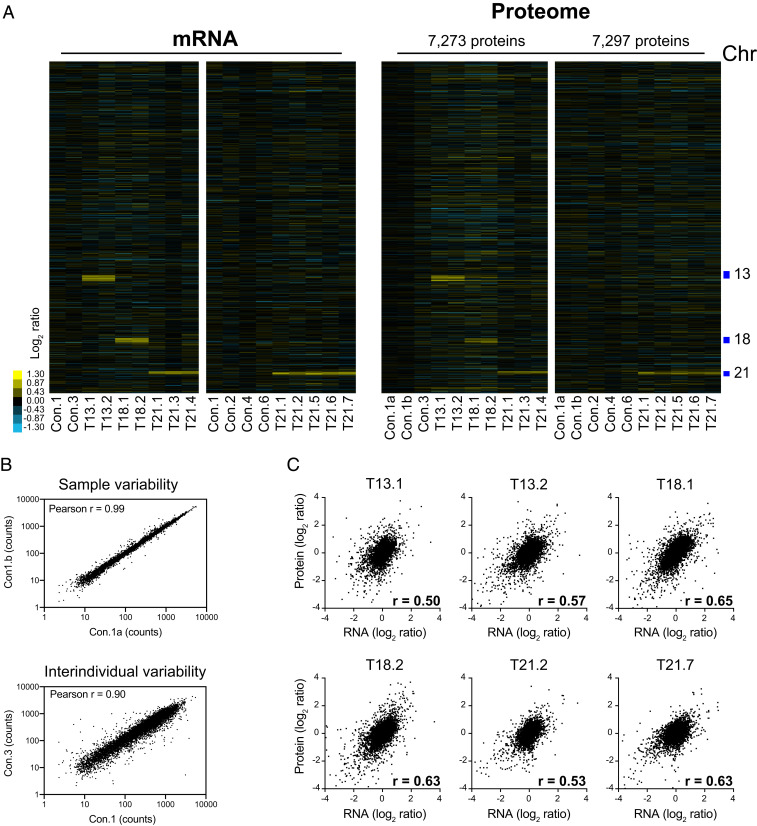
Next, we analyzed how changes in mRNA levels modify the proteome. We used isobaric tandem mass tag (TMT)-based quantitative mass spectrometry (MS) to quantify protein levels in primary fibroblasts. We analyzed two sets of 10 samples using a TMT 10plex protocol. One control (Con.1) was analyzed in duplicate in both sets of samples to monitor our approach's technical variability. Also, samples for one trisomy 21 cell line (T21.1) were included in both sets of quantifications to test reproducibility. We obtained quantitative information for 7,273 proteins and 7,297 proteins in the first and second sets of analysis, respectively, with 6,486 proteins overlapping (Fig. 4A and Dataset S4). A total of five controls, two trisomies 13, two trisomies 18, and seven trisomies 21 proteomes were obtained in our analysis. A comparison of the peptide counts of the two control samples (Con.1a and Con.1b) shows good reproducibility with a linear correlation of slope equal to 1 (R2 = 0.99), while the comparison of the proteomes of two unrelated individuals shows more variability (R2 = 0.81, Fig. 4B).
Fig. 4.
Protein levels proportionally increase with copy number in trisomic fibroblasts. (A) A comparison of the mRNA (Left) and protein levels (Right) of human primary fibroblasts. The genes are ordered by chromosome position in each experiment (columns). (B) A linear regression analysis of protein counts in technical replicates of a control cell line and between two cell lines from different individuals. (C) The Pearson correlation r was calculated for the mRNA and protein levels for six representative trisomic cell lines.
We then calculated the log2 of the FC using the average counts of the control samples as the reference proteome in each dataset. Increases in protein levels readily reveal the identities of the triplicated chromosomes by plotting the proteome profiles for each cell line ordered by chromosome position (Fig. 4A). Changes in RNA and protein levels correlate with coefficients (Pearson r) ranging from 0.4 to 0.7 among all samples analyzed (examples for six pairs of transcriptomes and proteomes of trisomic cells are shown in Fig. 4C). Proteome profiles could easily be matched to their corresponding RNA profiles using correlation analysis between log2 ratios of RNA and protein in all samples (SI Appendix, Fig. S4A). A total of 129, 110, and 73 proteins on chromosomes 13, 18, and 21, respectively, were quantified in the first dataset. A total of 76 proteins on chromosome 21 were quantified in the second set. On average, the protein levels of the triplicated chromosomes increase by log2 ratios of 0.45, 0.43, and 0.40 in trisomies 13, 18, and 21, respectively (SD between 0.3 to 0.5, Fig. 5 A and B). These results imply that protein levels proportionally increase with copy number but are slightly lower (1.3- to 1.4-fold) than the theoretical 1.5-fold increase observed in transcript levels. The distributions of the log2 ratios fit quite well a normal distribution (R2 between 0.90 and 0.98) with a couple of outliers both below and above the curve. As observed in the transcript levels, closer inspection of the outliers indicates that interindividual variability accounts for their apparent dysregulation (a few examples are shown in SI Appendix, Fig. S4B).
Fig. 5.
Protein levels of subunits of macromolecular complexes are attenuated in trisomic primary fibroblasts. (A) A heatmap of the average protein levels per chromosome in primary fibroblasts. The experiments (columns) for each cell line are ordered by chromosome position. (B) A histogram of the average log2 ratios of the protein levels of genes located on chromosome 13 in trisomy 13 cell lines on chromosome 18 in trisomy 18 cell lines, on chromosome 21 in trisomy 21 cell lines in the first and second datasets, relative to euploid controls are shown. The bin size for all histograms is log2 ratio of 0.2. Fits to a normal distribution (black line), means, SD, and goodness of fit (R2) are shown for each distribution. (C) A histogram of the average log2 ratios of the RNA (red) and protein (blue) levels of triplicated genes that show protein levels lower than predicted (Left) and 1.5-fold changes in trisomic fibroblasts. The bin size for all histograms is log2 ratio of 0.1. Fits to a normal distribution (solid lines), means, SD, and goodness of fit (R2) are shown for each distribution. (D) A few examples of individual subunits located on triplicated chromosomes that show attenuation at the protein levels compared to RNA. COG6 and RPL21 are in chromosome 13. RPL17, ATP5F1A, and NDUFV2 are in chromosome18. ATP5PO, NDUFV3, and CCT8 are in chromosome 21. (E) The hierarchical clustering analyses of proteome profiles do not cluster by karyotype of the cell lines.
Subunits of Macromolecular Complexes Are Regulated Posttranscriptionally.
To better understand why the fold increase of the average protein levels of triplicated chromosomes is slightly lower than the predicted 1.5-fold, we analyzed the identity of a set of proteins encoded on the triplicated chromosomes with lower levels than expected in the trisomic cell lines. Excluding the outliers caused by interindividual variability, we found that about one-third of triplicated genes (97 out of 281) have protein changes of log2 ratios less than 0.3 and fit a normal distribution with a mean of 0.13 (SD = 0.16, Fig. 5C). Remarkably, the average change in mRNA levels of this set of proteins is 1.5-fold (mean log2 ratio = 0.55, Fig. 5C). These results indicate that the levels of this set of proteins are attenuated via posttranscriptional mechanisms. The other 184 triplicated genes show the predicted average increase of 1.5-fold in both mRNA and protein levels (Fig. 5C).
Several studies showed that a set of attenuated proteins in aneuploid yeast and mammalian cell lines include subunits of macromolecular complexes (24–27). We hypothesized that protein instability of individual subunits in the absence of the up-regulation of entire complexes is the cause of this effect (26). Here, we found that 39% (38 of 97) of attenuated proteins are subunits of macromolecular complexes. Notably, two out of two ribosomal subunits in our dataset show attenuation, consistent with the finding that ribosomal subunits are rapidly degraded unless they assemble into a stable complex (26, 28, 29). Other examples include two subunits of the ATPase synthase and two subunits of NADH dehydrogenase (Fig. 5D). We also found that several membrane proteins and secreted proteins account for the attenuation in protein levels (see Dataset S5 for a complete list of attenuated proteins). Consistent with previous studies in aneuploid yeast cells, our findings in human cells indicate that protein levels increase in abundance proportionally with mRNA levels, with a set of proteins being attenuated by posttranscriptional mechanisms most likely mediated by protein degradation pathways. Therefore, a conserved consequence of aneuploidy in trisomy 21 cells is increased protein synthesis leading to an imbalanced proteome and higher activity of the protein quality control pathways.
Interindividual Variability Determines Proteome Profiles of Human Primary Fibroblasts.
As seen in the transcriptomes, hierarchical clustering analysis demonstrates that the proteome profiles cluster independent of karyotype (Fig. 5E). Several proteome profiles of controls are more similar to trisomic proteomes than to other controls. For example, the proteome profile of Con.2 is more similar to T21.6, T21.7, and T21.2 than to the proteome of Con.1. Importantly, we found a significant correlation between the proteome profiles of the replicates within each dataset (Con.1a versus Con.1b) and in between datasets (T21.1.1 versus T21.1.2), indicating good reproducibility of our analysis.
Hierarchical clustering did not reveal a clear pattern of commonly up- or down-regulated proteins in the aneuploid cell lines. Since there is a strong correlation between RNA and protein changes, we analyzed the protein levels of the down- and up-regulated gene clusters identified in the transcriptome analyses (SI Appendix, Fig. S5). Indeed, we found that these clusters also show specific down- and up-regulation at the protein levels. However, analysis of the average changes indicates that these responses are slightly attenuated. Importantly, this set of proteins remains enriched for factors that regulate amino acid biosynthesis in the down-regulated cluster and DNA replication in the up-regulated cluster.
Trisomic Fibroblasts Rely on Serine-Driven Lipid Synthesis to Proliferate.
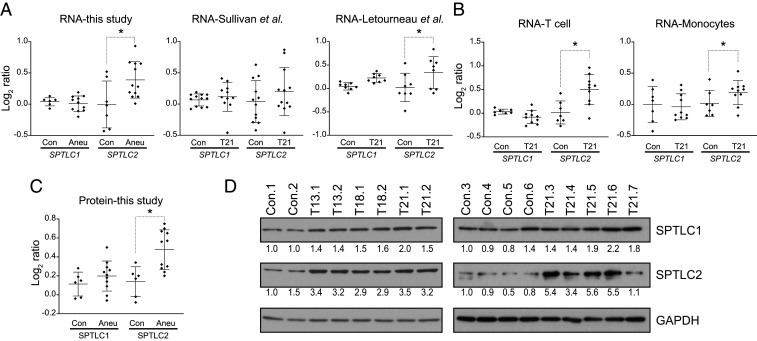
Altered sphingolipid metabolism is a conserved aneuploid-associated phenotype of aneuploid yeast and mammalian cells (13, 14, 30). Sphingolipids are essential molecules that are integral structural components of cellular membranes. We recently showed that altered sphingolipid levels are associated with abnormalities in the morphology of the nucleus of aneuploid cells, including human primary fibroblasts trisomic for chromosomes 13, 18, or 21 (13). Furthermore, we found that sphingolipid levels in human trisomic fibroblasts are elevated compared to euploid cells. The pathway of the synthesis of sphingolipids is conserved in yeast and humans. The initial step involves the condensation of the amino acid serine to palmitoyl-CoA by serine palmitoyltransferase (SPT) to generate long-chain bases (SI Appendix, Fig. S6). Analysis of the mRNA levels of trisomic cell lines revealed that the expression of one of the central subunits of SPT, SPTLC2, is up-regulated in several trisomic fibroblasts cell lines compared to controls in our data set (Fig. 6A). Notably, the SPTLC2 mRNA up-regulation is also detected in several trisomy 21 cell lines analyzed in the Sullivan et al. and Letourneau et al. studies (Fig. 6 A and B). The SPTLC2 up-regulation could lead to increased SPT activity as this subunit is essential for the stability of the enzyme (31). Consistently, our proteome analyses indicate that the levels of SPTLC2 increase in trisomic fibroblasts compared to controls (Fig. 6C). We performed Western blot analysis of SPTLC1 and SPTLC2 in 6 euploids and all 11 trisomic cell lines to validate these results. We found that, indeed, the SPT protein levels increase in all trisomic cell lines (Fig. 6D). These results provide a mechanism by which sphingolipid synthesis is up-regulated in aneuploid cells (SI Appendix, Fig. S6). Furthermore, they suggest that the regulation of the mRNA levels of one of the main subunits can stabilize the complex and increase the cells' enzyme levels.
Fig. 6.
Trisomic primary fibroblasts up-regulate the de novo synthesis of sphingolipids. (A) The expression levels of subunits SPTLC1 and SPTLC2 of the serine palmitoyltransferase enzyme in fibroblasts in this study and in the Sullivan et al. (20) and Letourneau et al. (16) studies. (B) The expression levels of subunits SPTLC1 and SPTLC2 in T cells and monocytes (20). (C) The protein levels of SPTLC1 and SPTLC2 in primary fibroblasts. (D) The Western blots of SPTLC1 and SPTLC2 in primary fibroblasts. GADPH was used as a loading control.
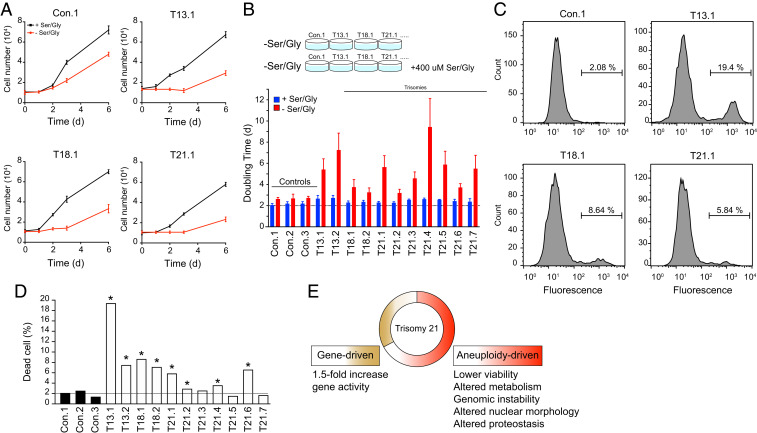
Sphingolipid synthesis and levels are dependent on the availability of intracellular serine, and aneuploid yeast cells strictly rely on serine synthesis to proliferate (14, 32). Here, we tested whether trisomy fibroblasts rely on serine to proliferate. Remarkably, we found that the proliferation rates of all 11 trisomic cell lines were significantly more affected than controls when cells were cultured in a medium depleted of serine (Fig. 7 A and B). On average, euploids slowed down and increased their doubling times by 20% (2.1 to 2.6 d). Trisomic cell lines slowed down by changing their doubling times from 40% in Tri21.2 up to 360% in Tri21.4. These results indicate that serine, an amino acid utilized for protein, lipid, and nucleotide biosynthesis, is essential for the proliferation of trisomy 21 cells. They also support the hypothesis that aneuploidy disrupts cellular metabolism due to a higher metabolic demand driven by increased nucleotide, protein, and lipid synthesis.
Fig. 7.
Trisomic primary fibroblasts rely on serine-driven lipid biosynthesis and show lower viability compared to euploid controls. (A) The proliferation of control fibroblasts and trisomic fibroblasts with or without serine and glycine. The glycine was depleted because it can be used to generate serine by the SHMT1/2 enzymes. The growth curves of four representative cell lines are shown. (B) Doubling times of primary fibroblasts were calculated in medium with (blue bars) or without serine (red bars). A schematic of the experimental procedure is show above. The error bars represent SD, n = 3. (C, D) The viability assay shows that there is an increase in cell death in the trisomic fibroblasts. Fluouresence-activated cell sorting is shown for four representative measurements, n = 5,000 cells (see SI Appendix, Fig. S7 for assay schematic and control experiment). In D, *P < 0.05 between trisome and average control, Student’s t test. (E) The consequences of trisomy 21 in human cells are driven by gene- and aneuploidy-driven phenotypes.
Aneuploidy Lowers the Viability of Human Primary Fibroblasts.
Previous studies have shown that mammalian trisomic cells proliferate slower than their euploid counterparts (13, 25, 27, 33, 34), an observation reported almost 50 y ago for trisomy 21 cells (35). However, the mechanisms behind the slow proliferation phenotype remain unknown. Early passage trisomic cells show no signs of cell cycle arrest or senescence as assayed by beta-galactosidase staining. As reported in trisomic mouse embryonic fibroblasts (MEFs) (33), we could not detect significant cell cycle delays in human trisomic fibroblasts. Since cell cycle delays or senescence do not account for lower proliferation rates of trisomic cells, and because aneuploidy lowers viability in yeast (27), we tested whether cell viability is affected in trisomic fibroblasts using the LIVE/DEAD Fixable Green Dead Cell Stain Kit. This kit uses a dye that reacts with free amines yielding intense fluorescent staining. In viable cells, the dye’s reactivity is restricted to the cell surface, resulting in less-intense fluorescence than in nonviable cells where the dye diffuses inside the cell (SI Appendix, Fig. S7). Remarkably, we found that human trisomic fibroblasts have a significant increase in the population of nonviable cells. While control cell lines show up 2% dead cells in their populations, the trisomic cell lines show between 2 to 20% nonviable cells, depending on their karyotype (Fig. 7 C and D). Such decreases in viability can translate into a significant difference in cell numbers during exponential growth. Phenotypic variation in cell viability is noted among the seven trisomic 21 cell lines, as would be expected from previous studies (36). Nevertheless, the lower viability phenotype is significantly more pronounced in trisomies 13 and 18, consistent with the hypothesis that larger chromosomes are more deleterious to cell physiology than smaller ones.
Discussion
The acquisition of an extra autosome has detrimental consequences to organismal development. Studies in yeast at the cellular level indicate that the deleterious effects of aneuploidy increase with the size of the extra chromosome (11, 27). In humans, three copies of chromosomes 21, 18, or 13, which encode the least number of genes, in that order, are the only viable trisomies. Notably, the effects on human development in trisomies 13 and 18 are more deleterious than in trisomy 21. This fact is consistent with a correlation between the number of genes on the triplicated chromosome and the degree by which aneuploidy disrupts cellular physiology. In the context of Down syndrome, a gene-centric view poses that increased expression of the triplicated genes located on chromosome 21 causes cellular defects (3, 4). Our studies show that trisomy 21 cells share several phenotypes with other mammalian aneuploid cells, including trisomic MEFs, human trisomies 13 and 18, and even aneuploid yeast cells (13, 25, 27, 33, 37). These aneuploidy-associated phenotypes across organisms and independent of the triplicated chromosome identity include hampered proliferation, genomic instability, abnormal nuclear morphology, and altered metabolism and proteostasis. Therefore, one must consider the aneuploidy effects on cell physiology to better understand the causes of the pathophysiology associated with Down syndrome (Fig. 7E).
Trisomies associated with sex chromosomes, such as the triple X and XYY syndromes, are not as deleterious to human development as autosomal trisomies. In triple X, the noncoding RNA Xist silences the expression of the third copy of the X chromosomes. Because the X chromosome is larger in sequence than chromosomes 21, 18, or 13, its trisomy indicates that the mere presence and maintenance of extra DNA are not significant drivers of aneuploidy-associated phenotypes. In support of this, yeast-harboring yeast artificial chromosomes with heterochromatic human DNA comparable in size to the yeast chromosomes do not show aneuploidy-associated phenotypes (14, 27). In the case of an extra copy of the Y chromosome, the number of genes expressed and encoded on the Y chromosome is significantly smaller than that of chromosome 21. Altogether, these observations support the hypothesis that increases in gene expression are the main cause for cellular imbalance and disruption of cellular homeostasis.
Several studies have suggested that dosage compensation could ameliorate the RNA levels of the genes present on an extra copy of an autosome (19, 38). However, mechanisms for dosage compensation of an entire extra copy of an autosome are not known in human cells. Here we show that, on average, mRNA levels increase 1.5-fold in human cells trisomic for chromosomes 13, 18, or 21, supporting a lack of dosage compensation of human autosomes. Furthermore, quantitative proteomics indicates that increases in transcript levels lead to proportional increases in protein levels, except for a subset of proteins that encode subunits of macromolecular complexes. As evidenced in several studies, individual subunits of macromolecular complexes are unstable unless they assemble into a stable complex (24–26, 39). Excess subunits are either degraded or aggregate since protein aggregation is also an effective mechanism to lower extra proteins (39). Importantly, evolutionary mechanisms are in place to synthesize and assemble subunits of a particular complex in equimolar amounts, presumably sparing the cell from the wasteful synthesis of individual subunits (40, 41). Secreted and membrane proteins are other classes of proteins that show lower levels than the predicted 1.5-fold. The secreted proteins will not accumulate in the cell but can lead to physiological changes by modifying the extracellular environment. Membrane protein levels may depend on mechanisms that control protein trafficking and endocytosis. Increased protein synthesis, folding, and degradation may affect protein quality control pathways and alter the metabolic demands of trisomy 21 cells, independent of the identity and function of the triplicated genes.
Analysis of different cell types from several individuals with or without trisomy 21 indicates that transcriptome profiles cluster independent of karyotype, sex, or age of the donor. Our analyses of the transcriptomes of primary fibroblasts from several studies indicate that each cell line uniquely expresses a set of transcription factors associated with anatomical structure and development. To circumvent this issue, Letourneau et al. analyzed the expression pattern of two fibroblast cell lines isolated from monozygotic twins, one of which has trisomy for chromosome 21. Their studies concluded that trisomy 21 leads to genome-wide disruption of chromosomal domains or territories. Two previous publications questioned the findings by Letourneau et al. (42, 43). Our analysis indicates that the monozygotic cell lines display noisy and nonreproducible gene expression profiles due to insufficient sequence coverage. Lastly, gene expression analyses of trisomic MEFs and human aneuploid cell lines generated by different approaches do not converge to a unique gene expression pattern in response to aneuploidy (25, 33, 44). In isogenic aneuploid yeast strains, we showed that gene expression changes are associated with slow proliferation (27). The fact that cell cycle delays are not present in primary human cells is consistent with the lack of a transcriptional response associated with slow proliferation.
Other aneuploidy-associated phenotypes in human trisomy cells include increased replication stress measured by foci positive for the p53 Binding Protein 1 (13, 45). Mutations in genes involved in DNA replication and repair cause premature aging (46). Thus, aneuploidy-driven genomic instability may play an active role in the premature aging characteristics of individuals with Down syndrome. Importantly, trisomic MEFs and human fibroblasts do not show significant defects in cell cycle progression or increased senescence. Here we found that reduced proliferation measured in cell accumulation assays is mainly due to increased cell death in tissue culture. Interestingly, we did not detect increases in caspase-3 cleavage, indicating cell death occurs through noncanonical apoptosis or other cell death pathways. A crucial question is whether lower cellular viability contributes to abnormal human development associated with Down syndrome, including reduced organ size (47).
The expression of the triplicated chromosome creates an increased metabolic demand for the amino acid serine. As observed in aneuploid yeast cells, we found that human trisomic cells rely on serine to proliferate. Serine is a crucial amino acid used to make glycine, and together, they account for a large proportion of the codon usage in protein synthesis. Serine is also essential for nucleotide and lipid biosynthesis (48). Several studies showed that aneuploid yeasts, trisomic primary fibroblasts, and trisomic MEFs have elevated levels of long-chain bases, the direct product of SPT (13, 14, 30). Here, we found that trisomic human fibroblasts have increased expression of SPT subunits. Long-chain bases are essential components of the nuclear membrane, and cells harboring an extra chromosome show abnormal nuclear morphologies. A recent study of the metabolome profiles of plasma samples from individuals with Down syndrome revealed serine and sphingosine-1-phosphate, the main form of long-chain base in the blood, as the top metabolites with lower abundance compared to control individuals (49). Because dietary serine can have significant consequences on human physiology (50, 51), future research will determine whether a serine-enriched diet could have beneficial effects on the health of individuals with Down syndrome.
Experimental Procedures
Culture of Human Cell Lines.
Primary human fibroblasts from euploid individuals and trisomy fibroblasts (SI Appendix, Fig. S1A) were purchased from Coriell Cell Repositories. They were maintained in Minimum Essential Medium (MEM, with nonessential amino acid; Gibco 10370088) supplemented with 15% fetal bovine serum and 2 mM l-glutamine. To make serine/glycine-depleted medium, MEM (Gibco 11095080) was supplemented with alanine, asparagine, aspartic acid, glutamic acid, proline, and dialyzed serum (3-k molecular weight cutoff).
RNASeq.
Cells were grown for 48 h and 2 × 105 cells were harvested between 50 to 70% confluency. The RNAeasy Kit from Qiagen (catalog number 74104) was used to purify the RNA, and a nanoDrop was used to measure concentration. Samples were shipped on dried ice for transcriptome sequencing to BGI Americas (https://www.bgi.com/). Paired-end reads were aligned to human primary genome hg38, with star_2.5.3a 1, annotated with GENECODE GRCh38.p12 annotation release 292. Aligned exon fragments with mapping quality higher than 20 were counted toward gene expression with featureCounts_1.5.2 3. Expression normalization was performed using the TPM method (52–54). The parameters are as follows: genome, hg38.primary.fa; GTF, gencode.v29.primary_assembly.annotation.gtf. The log2 ratios are presented in Dataset S1. The sequencing data are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154418.
RNAseq Data from Two Other Studies.
Data from the Sullivan et al. study were obtained from the public available database https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79842 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84531. Data from the Letourneau et al. study were obtained from the public available database https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55426.
RNAseq Analysis.
All RNAseq reads were normalized to the total number of reads per experiments (TPM). To calculate the fold changes among all cell lines, the average TPM per gene was calculated for all the control euploid samples and the average TPM was used as a reference genome. Log2 (TPM per gene/TPM of reference) was obtained for each sample. We included FC for genes that were detected in all samples and a cutoff of 1 TPM or greater in our analysis. Hierarchical clustering was performed using the program WCluster. WCluster takes both a data table and a weight table to allow individual measurements to be differentially considered by the clustering algorithm. Gene expression data were clustered by a Pearson correlation metric with equal weighting given to all data, or with no weight given to genes on the triplicated chromosomes. The PRISM software was used to calculate frequency distributions, fit to a Gaussian curve, identify the outliers, perform linear regression analyses, and calculate Pearson r correlation coefficients.
Western Blot Assay.
Cells were lysed in lysis buffer (25 mM Tris HCl, pH = 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), and 0.5% deoxycholate with protease inhibitors). Lysate was separated on SDS polyacrylamide gels, and then proteins were transferred onto a polyvinylidene difluoride membrane and analyzed with antibodies against SPTLC1 (Abcam, ab176706), SPTLC2 (Abcam, ab23696), and GAPDH (Millipore, AB2302). Immunoreactive signals were detected by the Super Signal reagent (Pierce).
Cell Proliferation Assay.
A total of 1 × 104 cells were plated on 24-well plates with the standard culture medium. After 8 h, cells were washed with phosphate-buffered saline (PBS) and replaced with conditional medium with or without serine/glycine (0.4 mM). Cell numbers were counted every 24 h for 6 d.
Cell Death Assay.
To distinguish between live and dead cells, cells were stained with LIVE/DEAD Fixable Dead Cell stain kit (Invitrogen, L34969). Briefly, 4 × 104 cells were plated on a 60-mM dish. After 4 d, detached cells were incubated with fluorescent reactive dye at room temperature for 30 min in the dark. Stained cells were washed with PBS and then fixed with PBS with 4% formaldehyde for 15 min. After washing cells with PBS, cells were resuspended in PBS with 1% bovine serum albumin and analyzed the fixed-cell suspension by flow cytometry. Plots in Fig. 7C were generated by FlowJo software.
Proteomics.
See SI Appendix for detailed quantitative MS methodology.
Supplementary Material
Acknowledgments
This research was supported by NIH Grant 1R01GM118481-01A1 (to E.M.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014723118/-/DCSupplemental.
Data Availability.
RNAseq data of primary fibroblasts is publicly available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154418 (61). Proteome data are available at https://massive.ucsd.edu, ID number MSV000085706, and are available for download at ftp://massive.ucsd.edu/MSV000085706.
References
- 1.Nagaoka S. I., Hassold T. J., Hunt P. A., Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassold T. J., Jacobs P. A., Trisomy in man. Annu. Rev. Genet. 18, 69–97 (1984). [DOI] [PubMed] [Google Scholar]
- 3.Antonarakis S. E., Lyle R., Dermitzakis E. T., Reymond A., Deutsch S., Chromosome 21 and down syndrome: From genomics to pathophysiology. Nat. Rev. Genet. 5, 725–738 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Antonarakis S. E., et al., Down syndrome. Nat. Rev. Dis. Primers 6, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naylor R. M., van Deursen J. M., Aneuploidy in cancer and aging. Annu. Rev. Genet. 50, 45–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland A. J., Cleveland D. W., Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10, 478–487 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldgaber D., Lerman M. I., McBride O. W., Saffiotti U., Gajdusek D. C., Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer’s disease. Science 235, 877–880 (1987). [DOI] [PubMed] [Google Scholar]
- 8.Elagib K. E., et al., RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 101, 4333–4341 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Lane A. A., et al., Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat. Genet. 46, 618–623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korbel J. O., et al., The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl. Acad. Sci. U.S.A. 106, 12031–12036 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres E. M., Williams B. R., Amon A., Aneuploidy: Cells losing their balance. Genetics 179, 737–746 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santaguida S., Amon A., Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16, 473–485 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Hwang S., et al., Suppressing aneuploidy-associated phenotypes improves the fitness of trisomy 21 cells. Cell Rep. 29, 2473–2488.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang S., et al., Serine-dependent sphingolipid synthesis is a metabolic liability of aneuploid cells. Cell Rep. 21, 3807–3818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., et al., Systematic proteome and proteostasis profiling in human Trisomy 21 fibroblast cells. Nat. Commun. 8, 1212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letourneau A., et al., Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 508, 345–350 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Stamoulis G., et al., Single cell transcriptome in aneuploidies reveals mechanisms of gene dosage imbalance. Nat. Commun. 10, 4495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonarakis S. E., Down syndrome and the complexity of genome dosage imbalance. Nat. Rev. Genet. 18, 147–163 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Kojima S., Cimini D., Aneuploidy and gene expression: Is there dosage compensation? Epigenomics 11, 1827–1837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan K. D., et al., Trisomy 21 consistently activates the interferon response. eLife 5, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zamponi E., et al., Nrf2 stabilization prevents critical oxidative damage in Down syndrome cells. Aging Cell 17, e12812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scarpato M., et al., AnaLysis of expression on human chromosome 21, ALE-HSA21: A pilot integrated web resource. Database (Oxford) 2014, bau009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzales P. K., et al., Transcriptome analysis of genetically matched human induced pluripotent stem cells disomic or trisomic for chromosome 21. PLoS One 13, e0194581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McShane E., et al., Kinetic analysis of protein stability reveals age-dependent degradation. Cell 167, 803–815.e21 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Stingele S., et al., Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 8, 608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dephoure N., et al., Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3, e03023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres E. M., et al., Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317, 916–924 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Tsay Y. F., Thompson J. R., Rotenberg M. O., Larkin J. C., Woolford J. L. Jr, Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: Balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 2, 664–676 (1988). [DOI] [PubMed] [Google Scholar]
- 29.elBaradi T. T., van der Sande C. A., Mager W. H., Raué H. A., Planta R. J., The cellular level of yeast ribosomal protein L25 is controlled principally by rapid degradation of excess protein. Curr. Genet. 10, 733–739 (1986). [DOI] [PubMed] [Google Scholar]
- 30.Tang Y. C., et al., Aneuploid cell survival relies upon sphingolipid homeostasis. Cancer Res. 77, 5272–5286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasuda S., Nishijima M., Hanada K., Localization, topology, and function of the LCB1 subunit of serine palmitoyltransferase in mammalian cells. J. Biol. Chem. 278, 4176–4183 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Cowart L. A., Hannun Y. A., Selective substrate supply in the regulation of yeast de novo sphingolipid synthesis. J. Biol. Chem. 282, 12330–12340 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Williams B. R., et al., Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torres E. M., et al., Identification of aneuploidy-tolerating mutations. Cell 143, 71–83 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal D. J., McCoy E. E., Studies on Down’s syndrome in tissue culture. I. Growth rates and protein contents of fibroblast cultures. J. Cell. Physiol. 83, 85–90 (1974). [DOI] [PubMed] [Google Scholar]
- 36.Beach R. R., et al., Aneuploidy causes non-genetic individuality. Cell 169, 229–242.e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfau S. J., Amon A., Chromosomal instability and aneuploidy in cancer: From yeast to man. EMBO Rep. 13, 515–527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Disteche C. M., Dosage compensation of the sex chromosomes and autosomes. Semin. Cell Dev. Biol. 56, 9–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennan C. M., et al., Protein aggregation mediates stoichiometry of protein complexes in aneuploid cells. Genes Dev. 33, 1031–1047 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiber A., et al., Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G. W., Burkhardt D., Gross C., Weissman J. S., Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahlfors H., et al., Gene expression dysregulation domains are not a specific feature of Down syndrome. Nat. Commun. 10, 2489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Do L. H., Mobley W. C., Singhal N., Questioned validity of gene expression dysregulated domains in down’s syndrome. F1000 Res. 4, 269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santaguida S., et al., Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell 41, 638–651.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Passerini V., et al., The presence of extra chromosomes leads to genomic instability. Nat. Commun. 7, 10754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kubben N., Misteli T., Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 18, 595–609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aldridge K., Reeves R. H., Olson L. E., Richtsmeier J. T., Differential effects of trisomy on brain shape and volume in related aneuploid mouse models. Am. J. Med. Genet. A. 143A, 1060–1070 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Locasale J. W., Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powers R. K., et al., Trisomy 21 activates the kynurenine pathway via increased dosage of interferon receptors. Nat. Commun. 10, 4766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ke W., et al., Dietary serine-microbiota interaction enhances chemotherapeutic toxicity without altering drug conversion. Nat. Commun. 11, 2587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baksh S. C., et al., Extracellular serine controls epidermal stem cell fate and tumour initiation. Nat. Cell Biol. 22, 779–790 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrow J., et al., GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y., Smyth G. K., Shi W., featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data of primary fibroblasts is publicly available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154418 (61). Proteome data are available at https://massive.ucsd.edu, ID number MSV000085706, and are available for download at ftp://massive.ucsd.edu/MSV000085706.