Significance
Although previous studies highlight the importance of ecological variation in driving divergence in brain morphology, whether these changes accumulate after ecological transitions or play a significant role in facilitating them is unclear. We show that divergent selection associated with microhabitat partitioning drove heritable divergence in brain composition and neural gene expression between sister clades of Heliconius butterflies. Neuroanatomical divergence is restricted to visual structures and reflect adaptations to contrasting sensory niches. We show how divergent neural traits can act as barriers to gene flow by demonstrating that hybrids have intermediate traits and by presenting evidence that genes with adaptive patterns of expression divergence are more likely to occur in regions of the genome that are resistant to interspecific gene flow.
Keywords: brain evolution, ecological speciation, neuroecology, niche partitioning, reproductive isolation
Abstract
The importance of behavioral evolution during speciation is well established, but we know little about how this is manifest in sensory and neural systems. A handful of studies have linked specific neural changes to divergence in host or mate preferences associated with speciation. However, the degree to which brains are adapted to local environmental conditions, and whether this contributes to reproductive isolation between close relatives that have diverged in ecology, remains unknown. Here, we examine divergence in brain morphology and neural gene expression between closely related, but ecologically distinct, Heliconius butterflies. Despite ongoing gene flow, sympatric species pairs within the melpomene–cydno complex are consistently separated across a gradient of open to closed forest and decreasing light intensity. By generating quantitative neuroanatomical data for 107 butterflies, we show that Heliconius melpomene and Heliconius cydno clades have substantial shifts in brain morphology across their geographic range, with divergent structures clustered in the visual system. These neuroanatomical differences are mirrored by extensive divergence in neural gene expression. Differences in both neural morphology and gene expression are heritable, exceed expected rates of neutral divergence, and result in intermediate traits in first-generation hybrid offspring. Strong evidence of divergent selection implies local adaptation to distinct selective optima in each parental microhabitat, suggesting the intermediate traits of hybrids are poorly matched to either condition. Neural traits may therefore contribute to coincident barriers to gene flow, thereby helping to facilitate speciation.
Ecological adaptation is a major force driving the evolution of new species (1, 2). Although it is well established that divergent selection can influence behavioral traits and promote speciation (3), there are few empirical examples of how divergent selection acts on the underlying sensory and neural systems. For example, existing studies on adaptation across divergent light regimes have largely focused on the peripheral sensory systems, often in the context of divergent mate preference (4, 5). However, sensory perception is only the first of many mechanisms within the nervous system that may experience divergent selection, and mating preferences are only one of many behaviors that can be affected by the environment, and only one of many behaviors that may contribute to reproductive isolation. Behavioral challenges imposed by novel environmental conditions can instead be met by changes in how sensory information is processed, often reflected in differential investment in brain components that refine the sensitivity, acuity to, or integration of, different stimuli.
The intimate relationship between brain structure and ecology is apparent in many comparative studies of neuroanatomy. For example, the expansion of visual pathways in primates (6), the cerebellar expansion and refinement of the exterolateral nucleus in electric fish (7–9), the contrasting adaptations of diurnal and nocturnal lifestyles in hawk moths (10), and changes in sensory brain investment during independent colonizations of cave systems that underlie the radiation in Mexican cavefish (11), all indicate the importance of neuroanatomical adaptations to contrasting ecological needs. However, with some exceptions (SI Appendix, Table S1), comparative studies of brain evolution generally focus on phylogenetic comparisons across relatively distantly related species that encompass a large range of morphological and ecological variation. More recently, several studies have also investigated intraspecific variation in neural traits across populations, or between eco-morphs, and highlight the potential for plasticity in brain development to optimize brain structure and function to local conditions (12–14).
Between these population and phylogenetic levels there is a scarcity of information about the role brains play in facilitating speciation across environmental gradients, either through developmental plasticity or the accumulation of heritable changes during ecological divergence (SI Appendix, Table S1). Hence, whether evolutionary changes in neural systems contribute to the evolution of reproductive isolation during ecological divergence (15), or accumulate over longer time frames after reproductive isolation is complete, is unknown. A handful of insect studies have linked specific changes in neural processing to the evolution of reproductive isolation among close relatives; however, these specifically focus on divergent host preferences and the detection of host cues (16–22). Whether brains respond to changes in broader features of the environment, such as luminance or habitat structure, at a similar timescale is yet to be established. Recently, studies of closely related populations on the path to speciation have begun to address this question (11, 13, 23, 24) (summarized in SI Appendix, Table S1). Importantly, however, these studies are often unable to disentangle the effects of drift and selection, and have not determined whether hybrids between ecologically distinct populations show intermediate brain morphologies that may reflect major fitness deficits, which would support a more causative role for divergence in neural systems during the incipient stages of speciation.
Here, we investigate the role of heritable divergence in neuroanatomy and gene expression in a clade of closely related Heliconius butterflies. Heliconius are well known for their bright warning patterns and Müllerian mimicry (25, 26). Speciation within Heliconius is also often associated with ecological transitions (27–30), and habitat partitioning among sister taxa is generally required for complete speciation (28, 31). In particular, within the melpomene–cydno clades, coexisting species are often found in “mosaic sympatry,” with sister taxa inhabiting relatively open forest edge, or closed canopy forest, respectively (31–33). These species are ecologically distinct and mate assortatively, but at a population level continue to exchange alleles, implying speciation is incomplete (34). These environmental differences are associated with changes in light environment, with cydno clade taxa occupying forest with lower light levels (34). Wild caught Heliconius melpomene/cydno show evidence of divergence in peripheral eye structure and light sensitivity (35, 36), suggesting a potential response to the contrasting light environments. Indeed, across a range of taxa, transitions between dark/light conditions are consistently associated with changes in brain morphology that are assumed to be adaptive (6, 10, 11). We therefore hypothesized that differences in habitat use between melpomene and cydno clade taxa have imposed distinct sensory challenges throughout the evolution of reproductive isolation, leading to consistent, divergent changes in brain structure and function.
To test this hypothesis, we collected data to examine the evolution of brain morphology at two levels of divergence: population-level variation among melpomene and cydno clade taxa, and divergence between these two clades. Using these data, we test the following predictions that are expected under adaptive divergence: 1) there will be a consistent signal of divergent brain morphologies between melpomene and cydno clade taxa; 2) these differences should be heritable, not induced by the environment; and 3) divergence should exceed rates expected by neutral evolution. We then complement these data with neural transcriptomes from common-garden–reared individuals to test whether divergence has also occurred in processes that cannot be captured by volumetric data, and whether this can also be explained by divergent selection. Finally, we replicated these data in first-generation hybrids to test the hypothesis that hybrid offspring show intermediate traits and expression profiles. Combined with evidence of divergent selection, which would suggest that melpomene and cydno brains are adapted to distinct fitness optima, intermediacy in hybrid neural traits would imply that they fall outside both parental optima, likely being maladaptive.
Results and Discussion
Divergence in Neuroanatomy in the H. Melpomene–Cydno Complex.
To investigate the effects of ecological divergence on brain morphology within the melpomene–cydno complex, we sampled butterflies from Costa Rica, Panama, Peru, and French Guiana (Fig. 1). Where members of the melpomene and cydno clades are sympatric, the species boundary is maintained by ecological divergence and disruptive selection against hybrids, which now occur at low frequencies (37).
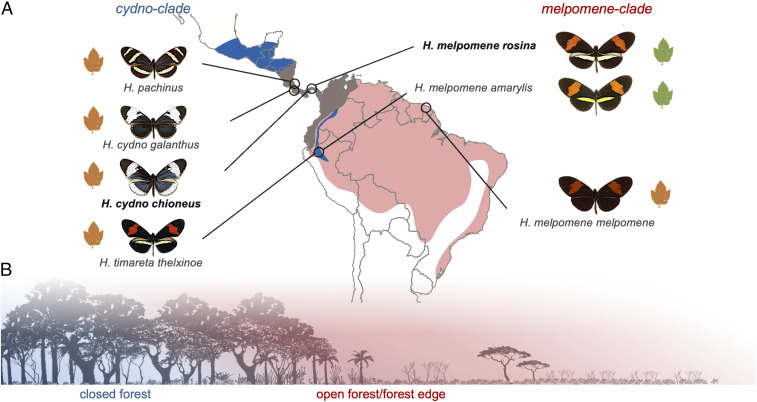
Fig. 1.
Population sampling and ecological divergence. (A) Outline map of Central and South America showing the range of the cydno clade (blue), the melpomene clade (red), and their overlap (brown). Circles indicate sampled populations in Costa Rica, Panama, Peru, and French Guiana. In the Andes, the cydno clade species, H. timareta, is restricted to high elevations but overlaps with H. melpomene at its lower margins. Green Passiflora sp. leaves indicate oligophagous populations that are host-plant specialists; orange leaves indicate polyphagous populations that lay on multiple Passiflora species. Populations included in the common-garden experiments are shown in bold. (B) Illustration of niche partitioning between melpomene (red; open forest, forest edge) and cydno (blue; closed forest).
Across the two clades, while controlling for population effects, the average volume of the combined optic lobe neuropils (OL) is significantly larger in cydno clade species (including H. cydno, Heliconius pachinus, and Heliconius timareta) than H. melpomene (n = 77, Χ2 = 17.354, P < 0.001). This difference is not explained by allometric scaling (y axis shift in OL ∼ unsegmented central brain neuropil [rCBR]: Χ2 = 12.260, P < 0.001; Fig. 2C). Five of the six optic lobe neuropils are significantly larger in the cydno clade (Fig. 2 C, D, and I and SI Appendix, Table S4), with the sole exception being the lobula. For a given brain size, these five neuropils are between 13 and 27% larger in cydno, suggesting altered patterns of investment are unequal across structures. However, in each case, the increase is associated with grade shifts in allometric scaling (SI Appendix, Table S5). Across insects, these structures are vital for summation and parallelization of photoreceptor signals (38–40), and a diverse range of visual processes including color vision (41–43), shape and motion detection, maneuverability in flight (44, 45), and circadian rhythms (46). The ventral lobula (vLOB), which is only present in some butterflies (47–50), also acts as a relay center sending visual information to the mushroom body (50), a major site of insect learning and memory.
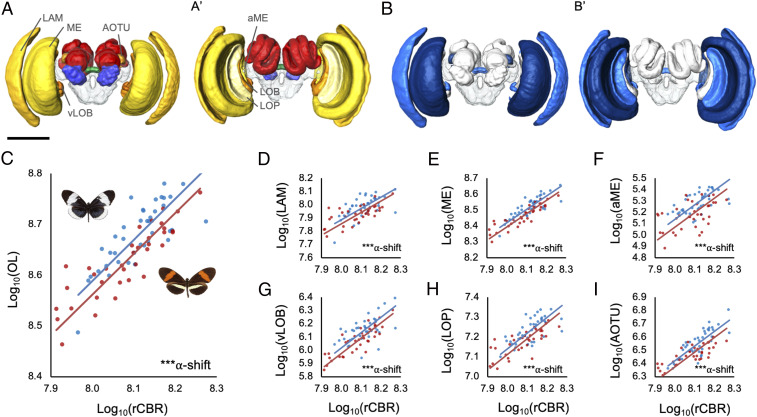
Fig. 2.
Divergence in brain morphology between H. melpomene and H. cydno clades. (A) Three-dimensional volumetric models of a Heliconius brain showing segmented neuropils from anterior (A) and posterior (A′) views; visual neuropils in yellows–oranges, antennal lobe in blue, the central complex in green, the mushroom bodies in red, and the unsegmented rCBR is transparent. Visual neuropils discussed in the main text are labeled as follows: aME, accessory medulla; AOTU, anterior optic tubercle; LAM, lamina; LOB, lobula; LOP, lobula plate; ME, medulla; vLOB, ventral lobula. (B) Three-dimensional volumetric models of a Heliconius brain showing segmented neuropils from anterior (A) and posterior (A′) views where blue neuropils are significantly different in size between H. melpomene and H. cydno clades, based on the lme4 results including all populations, with darker neuropils indicating higher significance. (Scale: in A/B, 500 μm.) (C) Grade shift in the scaling relationship between optic lobe (OL) and central brain (rCBR) volume between melpomene (red points) and cydno (blue points) clade taxa. (D–I) Nonallometric shifts in the size of individual visual neuropils between melpomene and cydno clade taxa; lamina (LAM), medulla (ME), accessory medulla (aME), ventral lobula (vLOB), lobula plate (LOP), and anterior optic tubercule (AOTu). C–I show results of the SMATR analysis, including all populations. α-shift indicates the significance of a grade shift in neuropil scaling, where ***P < 0.001.
The anterior optic tubercle (AOTU) is also 23% larger in the cydno-clade populations (Χ2 = 10.050, P < 0.001). The AOTU is the most prominent optic glomerulus in the central brain and is involved in processing sky light and spectral cues, as well as polarized light, which is less abundant in inner forests (51–53). The finding that this structure is expanded in the cydno clade is consistent with forest-type–dependent variation in light regimes affecting visual processing pathways. Contrary to claims that there is a trade-off between investment in major insect visual and olfactory neuropils (54), we find no evidence of volumetric shifts in the antennal lobe (Χ2 = 0.615, P = 0.615). Excluding the AOTU, no other central brain neuropil shows robust evidence for nonallometric expansion (SI Appendix, Tables S4 and S5). Divergence in brain structure is therefore restricted to neuropils associated with visual processing. As melpomene and cydno clade taxa occupy forest of different light intensities and physical structure (25, 31–33), differential investment in these neuropils likely reflects contrasting demands on visual processing. Consistent with this interpretation, previous data suggest H. cydno responds to lower intensities of light than H. melpomene (35).
Distinct Patterns of Intraclade Variation Reveal a Consistent Role of Ecology in Shaping Brain Morphology.
To further understand the origins of differential investment in visual neuropil, we next considered variation within the H. cydno and H. melpomene clades. Despite evidence of genetic substructuring (55), brain morphology was highly consistent across the four cydno-clade populations we sampled, with no neuropil showing significant geographic variation (SI Appendix, Table S4B). In contrast, we do find evidence of variation across populations of H. melpomene, both in total optic lobe volume (Χ2 = 9.917, P = 0.007) and for several of the individual visual neuropils that differentiate the H. cydno and H. melpomene clades (SI Appendix, Tables S4B and S5B). These include the largest visual neuropil, the medulla (Χ2 = 11.161, P = 0.004), and the AOTU (Χ2 = 9.647, P = 0.008) (SI Appendix, Fig. S1). Post hoc analysis reveals that these results are not driven solely by a single divergent population (SI Appendix, Table S5C), raising the possibility that H. melpomene may occupy more visually heterogeneous habitats than H. cydno, and may be exposed to a greater range of local sensory conditions.
Despite greater variability within H. melpomene, comparisons between sympatric species pairs suggest a consistent pattern of divergent investment between melpomene and cydno clade populations. In Panama, H. m. rosina and H. c. chioneus are differentiated by total optic lobe volume (Χ2 = 12.708, P < 0.001), with five of seven visual neuropils having larger volumes in H. cydno (SI Appendix, Table S4A). Similarly, in Peru, H. m. amaryllis and H. timareta thelxinoe vary in total optic lobe volume (Χ2 = 6.773, P = 0.009) and the two largest visual neuropils, the medulla and lamina (SI Appendix, Table S4A). Given H. m. amaryllis and H. t. thelxinoe are comimics and do not appear to distinguish conspecifics using visual cues (56), the shift in visual investment is unlikely to be related to mate choice. In contrast, H. c. galanthus and H. pachinus, which are ecologically equivalent but geographically isolated across Costa Rica’s central valley (31, 57, 58), show no evidence of neuroanatomical divergence despite strong visual mate preferences (58), supporting the causative role of divergent ecologies (SI Appendix, Table S4A). Comparisons between H. m. melpomene, which is allopatric with respect to H. cydno, to all cydno populations also detect evidence of divergence in OL volume (Χ2 = 4.974, P = 0.026) with levels of phenotypic divergence comparable to other melpomene populations (SI Appendix, Table S4A). This suggests an absence of strong character displacement for this trait.
Neuroanatomical Differences Are Heritable.
We next reared H. melpomene rosina and H. cydno chioneus under common-garden conditions to determine whether the variation we observe is heritable. As in our comparisons between wild-caught individuals, we observed a nonallometric expansion of the optic lobe in insectary-reared H. cydno relative to insectary-reared H. melpomene (33%; n = 20, Χ2 = 11.363, P = 0.001; SI Appendix, Table S6). This was driven by volumetric increases ranging from 24 to 57% across specific visual neuropils in H. cydno, including five of the six structures that differed between wild-caught individuals (SI Appendix, Table S6). The most pronounced shifts were found in the lamina (57% larger, Χ2 = 13.702, P < 0.001), vLOB (49%, Χ2 = 6.359, P < 0.001), and AOTU (40%, Χ2 = 21.749, P < 0.001). We found no evidence that the extent of divergence between H. cydno and H. melpomene was higher in wild-caught than common-garden individuals for any individual neuropil (SI Appendix, Table S6C). Differences in brain morphology therefore appear to have a substantial heritable component and are not the product of environmentally induced plasticity during development.
Neuroanatomical Divergence Is Likely Driven by Natural Selection.
Heritable trait divergence could be explained by neutral genetic drift or divergent selection. Allometric scaling among traits, where component sizes vary consistently with total size, is evidence for constraint on trait evolution (59–62), including on brain structure (63, 64). This suggests populations evolving under genetic drift should follow conserved allometric scaling relationships, as is typical among recently diverged taxa (61). In contrast, our observation of nonallometric variation of brain components, among both wild-caught and common-garden–reared individuals, strongly implicates divergent natural selection.
To explicitly test for selection, we calculated PST for variation in neuropil volumes in Panamanian H. m. rosina and H. c. chioneus raised under common-garden conditions. PST is a direct phenotypic analog of FST, which measures population differentiation relative to the total variance across populations (65). Comparisons between PST and FST can therefore be used as a formal test of divergent selection, where PST values that exceed genome-wide FST suggest greater phenotypic divergence than expected by neutral genetic divergence (65). After accounting for allometric effects, PST significantly exceeds genomewide FST (34) for total optic lobe size (adjusted P = 0.011), lamina (adjusted P = 0.006), medulla (adjusted P = 0.020), lobula (adjusted P = 0.016), vLOB (adjusted P = 0.005), and AOTU (adjusted P = 0.005), consistent with the action of divergent natural selection. Although inferences made from PST can be vulnerable to underlying assumptions regarding trait heritability (65), our results are robust across a broad range of quantitative genetic scenarios (SI Appendix, Table S8).
As a further test for selection acting across the melpomene–cydno complex, we performed a partial-Mantel test to assess whether pairwise divergence in brain morphology between wild populations is predicted by levels of neutral genetic divergence (FST). Here, we expect that the presence of divergent selection would erode the relationship between genetic distance and phenotypic divergence (66). In contrast, if selection is absent, we expect a linear relationship between trait divergence and genetic divergence, consistent with a strong phylogenetic signal. After allometric correction, only two neuropils, the antennal lobe and lobula, show patterns of divergence consistent with neutral expectations (SI Appendix, Table S8). The lack of association for any neuropils with divergent volumes between H. melpomene and H. cydno again implies our results are not explained by drift.
Together, evidence of 1) nonallometric divergence in brain structure, 2) between-species variation that significantly exceed neutral predictions under controlled environmental conditions, and 3) a lack of an association between phenotypic and genetic divergence across the melpomene–cydno complex, strongly implicates natural selection as the driving force behind the observed differences in neuroanatomical structures.
Neuroanatomical Evolution Is Mirrored by Shifts in Neural Gene Expression.
Volumetric changes in neuroanatomy are likely the result of differences in cell number or size, which may in turn reflect replicated or divergent neural circuitry. Shifts in neural physiology or activity are also behaviorally important but may not be captured in morphometric data. These differences, however, can potentially be captured in patterns of differential gene expression between species. We therefore also examined gene expression between age-matched, adult H. m. rosina and H. c. chioneus brain tissue, from individuals raised under common-garden conditions to control for environmental effects. After accounting for the influence of tissue composition (67), we still detect significant levels of interspecific divergence in expression profiles for age and environment-matched individuals, suggesting a degree of divergence in neural gene expression that is independent of divergence in brain morphology (see Fig. 4A and SI Appendix, Figs. S3–S5). This pattern is consistent across two independently collected datasets. Differentially expressed genes are enriched for molecular functions linked to cytoskeletal and transmembrane channel activities (SI Appendix, Table S10), consistent with changes in brain physiology being achieved through alterations of neuronal wiring or activity.
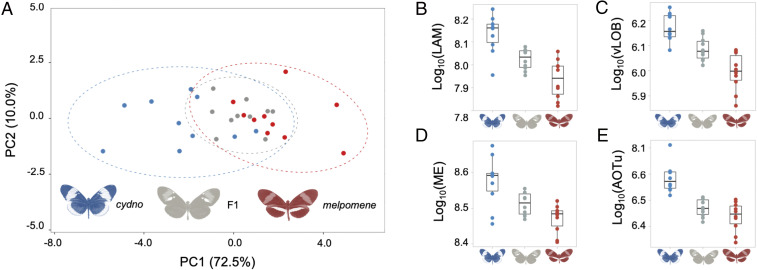
Fig. 3.
Intermediate brain morphology in H. m. rosina × H. c. chioneus F1 hybrids. (A) Variation in H. m. rosina (red), H. c. chioneus (blue), and hybrid (gray) brain morphology in a principal-component analysis of all segmented neuropils and rCBR. (B–E) Examples of neuropils with intermediate volumes in hybrids (B and C), or melpomene-like volumes (D and E) in F1 hybrids; lamina (LAM), ventral lobula (vLOB), medulla (ME), and anterior optic tubercule (AOTu).
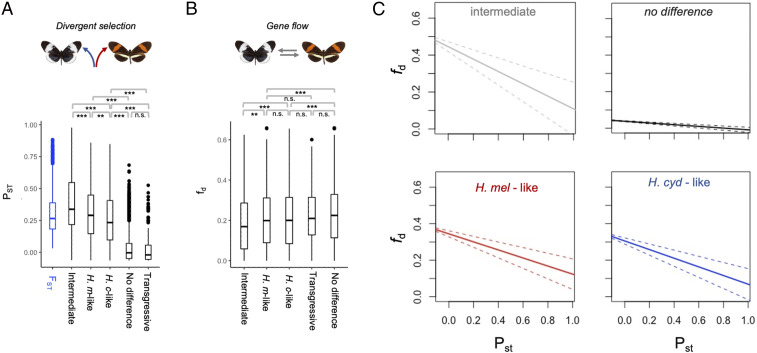
Differential expression between species could, again, be explained by genetic drift, rather than divergent selection. We therefore applied the same approach used to test for divergent selection in morphological traits. Estimated PST exceeds genome-wide FST for 18.5% (305 of 1,647) of differentially expressed genes, strongly implicating divergent selection in driving at least some shifts in neural gene expression. Divergent selection between melpomene and cydno implies that these species occupy distinct selective optima. This could create a reproductive barrier by selecting against individuals that possess allelic variation poorly matched to the local environment, thereby reducing gene flow between species around loci under divergent selection. Consistent with this hypothesis, fd, a measure of shared allelic variation that is used to infer barriers to gene flow (68), is negatively correlated with values of PST for neural gene expression, even after accounting for variation in local recombination rate (Χ2 = 179.0, P < < 0.001). Previous genome-wide analyses have highlighted a highly heterogeneous pattern of genetic divergence between H. m. rosina and H. c. chioneus, with selection against gene flow acting across the genome (34, 68). This suggests that the species barrier is determined by multiple, polygenic traits. Because fd, and by extension PST, is not clustered across the genome (68), our data are consistent with this inference. We therefore suggest that divergence in neural traits is, at least in part, shaping the landscape of genetic differentiation between H. m. rosina and H. c. chioneus.
Hybrids Show Evidence of Intermediate Traits that Deviate from Both Parental Conditions.
Reproductive isolation can result from a mismatch between intermediate hybrid phenotypes and the environment, such that hybrids suffer lower fitness in either parental environment (1, 2). To explore whether divergent brain structures might contribute to fitness deficits in hybrids, we produced multiple crosses between H. m. rosina and H. c. chioneus. We focus on first generation (F1) individuals, which account for a major proportion of natural Heliconius hybrids (37). In F1 hybrids, strong dominance patterns at genes under divergent selection could mean that individuals predominantly appear like one parental species, suggesting they may have similar fitness. In contrast, if polygenic traits have incomplete or mixed dominance patterns, F1s may have intermediate traits. This could disrupt behavioral performance leading to fitness deficits, potentially contributing to the evolution of reproductive isolation. Multivariate analysis of the seven visual neuropils reveals that hybrids indeed show intermediate brain morphologies (Fig. 3A and SI Appendix, Table S7). This intermediate state is the product of variable dominance effects on specific neuropil (Fig. 3 B–E and SI Appendix, Table S7). Four of the seven neuropils are significantly larger in H. cydno than F1 hybrids, but are not significantly different between F1s and H. melpomene (SI Appendix, Table S7B), suggesting that these are largely influenced by loci with melpomene-dominant alleles. In contrast, two neuropils, the lamina and vLOB, are significantly different between F1s and both parental species (Fig. 3 B and C and SI Appendix, Table S7B), implying incomplete or mixed dominance across multiple loci. Importantly, this mosaic pattern also leads to disrupted scaling relationships between some visual neuropil, which may affect the flow and integration of visual information in the brain (SI Appendix, Table S7 C and D and Fig. S2).
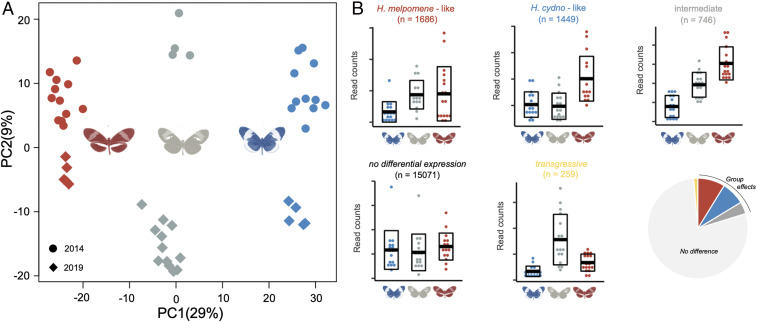
We observed a similar pattern in our neural transcriptomic data, where hybrids show intermediate patterns of gene expression (Fig. 4 and SI Appendix, Figs. S4 and S5). Focusing on genes that are differentially expressed between H. cydno and H. melpomene, F1 hybrids cluster outside the range of both parental species (Fig. 4A). As inferred for the visual neuropils, the expression of individual genes shows variable patterns of dominance (SI Appendix, Fig. S6): 36% of differentially expressed genes are “melpomene-like” in F1 hybrids, 21% are “cydno-like,” and 43% are statistically intermediate. Again, consistent with divergent selection playing a role in gene expression evolution, genes with intermediate expression in F1 hybrids show increased levels of PST (Χ2 = 5825.9, P < <0.001), with a greater proportion (23%) of intermediate genes showing PST values in excess of genome-wide FST, compared to genes with melpomene-like (9%) or cydno-like expression (7%). In contrast, only 0.01% of genes with consistent expression between H. cydno and H. melpomene, and no genes with transgressive expression in hybrids, show such signatures of selection (Fig. 5A). Again, these results are robust across a broad range of quantitative genetic scenarios (SI Appendix, Fig. S7). In addition, as expected given their enrichment for high PST values, genes with intermediate hybrid expression are more likely to coincide with regions of reduced gene flow than other differentially expressed genes (Χ2 = 116.1, P < <0.001; Fig. 5C). In sum, genes with intermediate expression in hybrids are more likely to have evolved under divergent selection in H. melpomene and H. cydno, and are more likely to be associated with selection that removes alleles that introgress between species.
Fig. 4.
Divergence in gene expression between H. m. rosina and H. c. chioneus. (A) Principal-component analysis of neural gene expression for differentially expressed genes. H. c. chioneus samples are colored in blue, F1 hybrids in gray, and H. m. rosina in red. Sequencing year is denoted by dot shape: circular (2014); rhomboid (2019). Note that the batch effect of year may be due to technical aspects associated with library preparation and sequencing, rather than biological variation, but is orthogonal to the main group effect. Sequencing batch was included in statistical models testing for differential expression. (B) Examples of expression profiles for genes that fall into different gene categories defined by their expression in F1 hybrids relative to both parental species, with horizontal bars indicating the mean and boxplots delineating ±SD of the normalized gene counts (n indicates quantity, considering all genes), with a pie chart depicting the proportion of genes that fall into each category (red, H. melpomene-like; blue, H. cydno-like; dark gray, intermediate; light gray, not differentially expressed; yellow, transgressive).
Fig. 5.
Gene expression, selection, and gene flow between H. m. rosina and H. c. chioneus. (A) Medians, interquartile ranges, and distributions of FST and PST values for genes assigned to different categories based on their expression profiles in F1 hybrids (see also Fig. 4). (B) Median, interquartile range, and distributions of admixture proportions (fd), estimated in 100-kb windows, between H. cydno and H. melpomene, for different gene categories. **P < 0.001; *P < 0.01; n.s., not significant, Kruskal–Wallis test with post hoc Dunn test, with Bonferroni correction (see SI Appendix, Fig. S8 for full comparisons). (C) Regression lines between fd and PST for the four key categories, showing significant negative associations that imply loci under selection for gene expression level overlap with regions of low gene flow. This association is most pronounced for intermediate genes (slope = −0.336 [95% CIs, −0.219 to −0.453], P << 0.001; see main text). The relationships are also significant (P << 0.001), and similar between melpomene (slope = −0.221 [95% CIs −0.154 to −0.288]) and cydno-like genes (slope = −0.239 [95% CIs, −0.169 to −0.305]), but shallower for genes with no group differences (slope = −0.047 [95% CIs, −0.036 to −0.058]).
Our results therefore reveal both divergence of neural phenotypes between ecologically distinct populations, and the potential disruption of these phenotypes in F1 hybrids. We consider it highly unlikely that hybrids experience “hybrid vigor” due to 1) the lack of a distinct “intermediate” forest habitat where intermediate investment in visual processing could be advantageous; 2) evidence that introgressed alleles are removed from parental populations, which implies selection against immigrants and hybrids (that occur at very low frequencies in the wild); and 3) high levels of heterozygosity in both parental populations meaning inbreeding effects are unlikely. Instead, we suggest the intermediate neural phenotypes are likely to act as further barriers to gene flow. Divergence in gene expression is a major source of genetic incompatibilities between species (69–71), and causes abnormal development and reduces survival (72). Hybrid disruption of expression profiles has been reported in diverging species pairs (72–77), and some evidence points to the importance of divergence in neural gene expression during ecological divergence (78, 79). In Heliconius, phylogenetic comparisons of neural gene expression also provide some evidence of selection at deeper timescales (80). Our data add clear support for this hypothesis. More broadly, disruption in the scaling relationships between components of the sensory systems that coevolve within species, but are under divergent selection between species, likely alters the way in which environmental stimuli are perceived and processed. This occurs at anatomical and molecular levels and may lead to a mismatch between the visual system of hybrids and their sensory environment.
In summary, using a large sample of multiple, geographically disparate populations, we provide strong evidence that divergent selection has acted on brain composition and gene expression during the evolution of habitat partitioning between melpomene and cydno clades of Heliconius butterflies. These changes are heritable, significantly exceed expected rates of neutral divergence, and result in intermediate traits in F1 hybrids. Neuroanatomical divergence is restricted to the visual neuropils, strongly suggesting that adaptation to contrasting sensory niches contributes to hybrid fitness deficits. These data are consistent with known differences in ecology between the two clades (25, 31–33, 35, 81). We therefore hypothesize that the divergent traits we identify reflect adaptations to contrasting light levels and conditions, including differences in the availability of polarized light and sun cues. These changes would impact multiple behaviors that contribute to fitness, from foraging efficiency to mating behavior, implying neural evolution has an underappreciated role during ecological speciation. Although we have not directly demonstrated the fitness effects of divergent neural traits, our hypothesis is consistent with previous evidence that H. melpomene and H. cydno have different visual sensitivities (35, 36), and the evidence for divergent selection itself implies these fitness differences must be present.
While disruptive selection on color pattern has a major role in maintaining reproductive isolation between species (82–84), habitat divergence is thought to be critical to “complete” speciation in Heliconius (28, 31). Our results suggest environmentally dependent selection on neural traits contributes to this process. Whether local adaptation in neural traits is a primary driver of reproductive isolation, whether shifts in color pattern initiate speciation with other traits diverging only secondarily, or if speciation only occurs when divergence in these traits is coincident in time, is unclear. However, given that the quality of the aposematic signal is environment and community dependent (85–87), changes in microhabitat preference, and the corresponding neurobiological adaptations to local environmental conditions, likely occur at the early stages of divergence. Our results are consistent with this early role, as differences between melpomene and cydno clades are comparable across populations, implying they originated as these clades diverged. Together, divergent ecological selection on behavior, and their neural bases, in addition to disruptive selection on mimetic warning patterns, would ultimately provide strong, coincident barriers to gene flow (88), thereby facilitating continued divergence.
At a macroevolutionary scale, diverse studies, ranging from recent adaptive radiations in cichlid fish (89) to more ancient diversification of mammals (90), highlight the importance of ecological transitions in driving divergence in sensory regions of the brain. However, whether these changes in brain composition accumulate after ecological transitions or play a significant role in facilitating them is unclear. Our data provide evidence that brain evolution has a facultative role in ecological transitions. Our results mirror a previous analysis of divergence in brain morphology between Heliconius himera and Heliconius erato cyrbia (23), which are isolated across a steep ecological transition between dense lowland wet forest and more open higher altitude dry forest (30, 91). In this case, heritable shifts in investment are again most notable in sensory neuropils, including visual neuropils (23). Similar conclusions can be drawn from the evolution of several fish ecotypes (13, 14, 92–94); however, here environment-dependent plasticity often plays a dominant role in producing population differences (13, 14, 95). By demonstrating heritable divergence in brain composition, rates of neural gene expression that exceed neutral expectations, and hybrid disruption at both an anatomical and molecular level, our data provide a robust case for adaptive neural evolution. Given the prevalent role of niche separation and environmental gradients in many adaptive radiations, we suggest that local adaptation in brain and sensory systems may have an underappreciated role during ecological speciation.
Methods
Animals.
We sampled three pairs of species in Costa Rica, Panama, and Peru, and a population of H. m. melpomene from French Guiana (Fig. 1) with permission from local authorities (SI Appendix). All wild individuals (n = 77) were hand netted, and brain tissue was fixed in situ in a ZnCl2-formalin solution (96) within a few hours of collection. Common-garden samples of H. c. chioneus and H. m. rosina were reared at the Smithsonian Tropical Research Institute’s Gamboa insectaries. Hybrids were produced from multiple H. c. chioneus × H. m. rosina crosses in 2013 and 2019. Insectary individuals were dissected at 2 to 3 wk for neuroanatomical (n = 30), and 9 to 15 d for gene expression samples (n = 49) (SI Appendix, Tables S2 and S3).
Immunohistochemistry and Imaging.
Brain structure was revealed using immunofluorescence staining against a vesicle-associated protein at presynaptic sites, synapsin (anti-SYNORF1; obtained from the Developmental Studies Hybridoma Bank, Department of Biological Sciences, University of Iowa, Iowa City, IA; RRID: AB_2315424) and Cy2-conjugated affinity-purified polyclonal goat anti-mouse IgG (H+L) antibody (Jackson ImmunoResearch Laboratories), obtained from Stratech Scientific Ltd. (Jackson ImmunoResearch; catalog no. 115-225-146; RRID: AB_2307343). All imaging was performed on a confocal laser-scanning microscope (Leica TCS SP5 or SP8; Leica Microsystem) using a 10× dry objective with a numerical aperture of 0.4 (Leica Material No. 11506511), a mechanical z-step of 2 μm and an x–y resolution of 512 × 512 pixels. Confocal scans were segmented using Amira 5.5 (Thermo Fisher Scientific) to produce estimates of neuropil volumes.
RNA Extraction and Sequencing.
Brains were dissected out of the head capsule in cold (4 °C) 0.01 M PBS. In 2019, total RNA was extracted using TRIzol Reagent (Thermo Fisher) and a PureLink RNA Mini Kit, with PureLink DNase digestion on column (Thermo Fisher). Illumina 150-bp paired-end RNA-seq libraries were prepared and sequenced at Novogene. In 2014, RNA was extracted using TRIzol Reagent and a RNeasy kit (Qiagen), samples were treated with DNase I (Ambion), and libraries were prepared and sequenced with 100-bp paired-end reads at Edinburgh Genomics. After trimming adaptor and low-quality bases from raw reads using TrimGalore v.0.4.4 (http://www.bioinformatics.babraham.ac.uk/projects), Illumina reads were mapped to the H. melpomene 2 genome (97)/H. melpomene 2.5 annotation (98) using STAR v.2.4.2a in two-pass mode (99). We kept only reads that mapped in “proper pairs,” using Samtools (100). The number of reads mapping to each gene was estimated with HTseq v. 0.9.1 (model = union) (101).
Statistical Analyses of Neuropil Volumes.
Nonallometric differences in brain component sizes were estimated using nested linear models in lme4 R (102). Linear models included each brain component as the dependent variable, the volume of rCBR, and taxonomic/experimental grouping as independent variables, with sex and country (where relevant) included as random factors. Including country as a random factor controls for the fact that populations within each clade are nonindependent; however, we also test the hypothesis that phylogenetic structure explains our results using Mantel tests (see below). The log-likelihoods of nested models were compared using likelihood ratio tests and a Χ2 distribution, with 1 degree of freedom between nested models including/excluding focal factors, and sequential Bonferroni correction (103). For neuropils showing a significant clade/species effect, we subsequently explored the scaling parameters responsible for group differences using SMATR v.3.4–3 (104). Partial-Mantel tests were performed between pairwise differences in neuropil volumes and FST (55), controlling for rCBR, using ECODIST (105) with Pearson correlations and 1,000 permutations. We calculated PST using the PSTAT package (106) with a c/h2 ratio of 1, and allometric correction with the res() function. The significance of PST was calculated as the proportion of the FST distribution (34) that was above each PST value. Finally, to identify intermediate traits in hybrids we also performed principal-component analysis and ANOVAs among parental and hybrid individuals, with post hoc Tukey tests to compare group means, using base R packages (107).
Statistical Analyses of Gene Expression Data.
Differential gene expression analyses were conducted in DESeq2 (108), including sex and sequencing batch as random factors, with a minimum fold change in expression of 2 to counter effects of tissue composition (67). We conducted a principal-component analysis on rlog-transformed gene count data (as implemented in DESeq2) to inspect clustering of expression profiles. ANOVAs on normalized gene expression counts of species and hybrids, with post hoc Tukey tests, were performed using base R packages (107). PST from normalized gene counts in H. m. rosina and H. c. chioneus was calculated following Uebbing et al. (109), with h2 set to 0.5 and c to 1.0. Estimated admixture proportions (fd) between H. m. rosina and H. c. chioneus, and population recombination rates (rho) were taken from Martin et al. (68). To test for an association between low gene flow and high PST, we fitted a linear mixed model: fd ∼ rho + PST + (1|chromosome), with a Gaussian distribution, using 100-kb nonoverlapping windows of fd. We used InterProScan v.5 (110) to retrieve gene ontology (GO) terms for the Hmel2.5 gene set and performed GO enrichment tests with the TopGO package in R (111), using the “elim” algorithm, which corrects for nonindependence among GO terms.
Supplementary Material
Acknowledgments
We are indebted to the environmental agencies in Costa Rica, Panama, Peru, and French Guiana for permissions to carry out this work. We thank Neil Rosser, Ronald Mori Pezo, and the Dasmahapatra group for assistance in Peru; the Organization for Tropical Studies at Las Cruces and La Selva, and Le Leona Eco Lodge for assistance in Costa Rica; Adriana Tapia, Moises Abanto, Oscar Paneso, Cruz Batista Saez, Chi-Yun Kuo, Morgan Oberweiser, the W.O.M., Chris Jiggins, and Evolution of Brains and Behaviour laboratories, and Smithsonian Tropical Research Institute (STRI) for support at the Gamboa insectaries, Panama. We also thank the University College London Confocal Imaging facility, and Matt Wayland and the Department of Zoology Imaging Facility, University of Cambridge, for assistance. This work was funded by a Royal Commission for the Great Exhibition Research Fellowship, a Leverhulme Trust Early Career Fellowship, a short-term STRI Fellowship, British Ecological Society Research Grant (3066), and a Natural Environment Research Council Independent Research Fellowship (NE/N014936/1) (to S.H.M.) and a Deutsche Forschungsgemeisnchaft Emmy Noether fellowship and research grant (GZ: ME 4845/1-1) (to R.M.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015102118/-/DCSupplemental.
Data Availability.
Full descriptions of the methodology and the neuroanatomical dataset are available in SI Appendix and have been deposited on DataDryad, along with R code (https://doi.org/10.5061/dryad.7wm37pvs3). The majority of the raw reads from the genetic dataset have been already available in The European Nucleotide Archive (accession no. PRJEB39935), and the remaining have been deposited on The European Nucleotide Archive (accession no. PRJEB42387).
All study data are included in the article and/or supporting information.
References
- 1.Rundle H. D., Nosil P., Ecological speciation: Ecological speciation. Ecol. Lett. 8, 336–352 (2005). [Google Scholar]
- 2.Schluter D., Evidence for ecological speciation and its alternative. Science 323, 737–741 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Coyne J. A., Orr H.A., Speciation (Oxford University Press, Oxford, 2004). [Google Scholar]
- 4.Cummings M. E., Endler J. A.; Fuller R. C., Handling editor. 25 years of sensory drive: The evidence and its watery bias. Curr. Zool. 64, 471–484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price T. D., Sensory drive, color, and color vision. Am. Nat. 190, 157–170 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Barton R. A., Visual specialization and brain evolution in primates. Proc. Biol. Sci. 265, 1933–1937 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson B. A., Arnegard M. E., Neural innovations and the diversification of African weakly electric fishes. Commun. Integr. Biol. 4, 720–725 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vélez A., Kohashi T., Lu A., Carlson B. A., The cellular and circuit basis for evolutionary change in sensory perception in mormyrid fishes. Sci. Rep. 7, 3783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sukhum K. V., Shen J., Carlson B. A., Extreme enlargement of the cerebellum in a clade of teleost fishes that evolved a novel active sensory system. Curr. Biol. 28, 3857–3863.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Stöckl A., et al., Differential investment in visual and olfactory brain areas reflects behavioural choices in hawk moths. Sci. Rep. 6, 26041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loomis C., et al., An adult brain atlas reveals broad neuroanatomical changes in independently evolved populations of Mexican cavefish. Front. Neuroanat. 13, 88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snell-Rood E. C., An overview of the evolutionary causes and consequences of behavioural plasticity. Anim. Behav. 85, 1004–1011 (2013). [Google Scholar]
- 13.Gonda A., Herczeg G., Merilä J., Population variation in brain size of nine-spined sticklebacks (Pungitius pungitius)—local adaptation or environmentally induced variation? BMC Evol. Biol. 11, 75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eifert C., et al., Brain size variation in extremophile fish: Local adaptation versus phenotypic plasticity: Brain size variation in extremophile fish. J. Zool. (Lond.) 295, 143–153 (2015). [Google Scholar]
- 15.Wcislo W. T., Behavioral environments and evolutionary change. Annu. Rev. Ecol. Syst. 20, 137–169 (1989). [Google Scholar]
- 16.Dekker T., Ibba I., Siju K. P., Stensmyr M. C., Hansson B. S., Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr. Biol. 16, 101–109 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Linn C. Jr, et al., Fruit odor discrimination and sympatric host race formation in Rhagoletis. Proc. Natl. Acad. Sci. U.S.A. 100, 11490–11493 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linz J., et al., Host plant-driven sensory specialization in Drosophila erecta. Proc. Biol. Sci. 280, 20130626 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride C. S., et al., Evolution of mosquito preference for humans linked to an odorant receptor. Nature 515, 222–227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prieto-Godino L. L., et al., Evolution of acid-sensing olfactory circuits in drosophilids. Neuron 93, 661–676.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Tait C., Batra S., Ramaswamy S. S., Feder J. L., Olsson S. B., Sensory specificity and speciation: A potential neuronal pathway for host fruit odour discrimination in Rhagoletis pomonella. Proc. Biol. Sci. 283, 20162101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seeholzer L. F., Seppo M., Stern D. L., Ruta V., Evolution of a central neural circuit underlies Drosophila mate preferences. Nature 559, 564–569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery S. H., Merrill R. M., Divergence in brain composition during the early stages of ecological specialization in Heliconius butterflies. J. Evol. Biol. 30, 571–582 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Keesey I. W., Grabe V., Knaden M., Hansson B. S., Divergent sensory investment mirrors potential speciation via niche partitioning across Drosophila. Elife. 30, e57008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown K. S., The biology of Heliconius and related genera. Annu. Rev. Entomol. 26, 427–457 (1981). [Google Scholar]
- 26.Merrill R. M., et al., The diversification of Heliconius butterflies: What have we learned in 150 years? J. Evol. Biol. 28, 1417–1438 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Jiggins C. D., Ecological speciation in mimetic butterflies. Bioscience 58, 541–548 (2008). [Google Scholar]
- 28.Mallet J., McMillan W. O., Jiggins C. D., “Mimicry and warning color at the boundary between races and species” in Endless Forms: Species and Speciation, Howard D. J., Berlocher S. H., Eds. (Oxford University Press, Oxford, 1998), pp. 390–403. [Google Scholar]
- 29.Jiggins C. D., Mallet J., Bimodal hybrid zones and speciation. Trends Ecol. Evol. 15, 250–255 (2000). [DOI] [PubMed] [Google Scholar]
- 30.McMillan W. O., Jiggins C. D., Mallet J., What initiates speciation in passion-vine butterflies? Proc. Natl. Acad. Sci. U.S.A. 94, 8628–8633 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mérot C., Salazar C., Merrill R. M., Jiggins C. D., Joron M., What shapes the continuum of reproductive isolation? Lessons from Heliconius butterflies. Proc. Biol. Sci. 284, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smiley J., “The host plant ecology of Heliconius butterflies in Northeastern Costa Rica,” PhD thesis, University of Texas at Austin, Austin, TX (1978).
- 33.Estrada C., Jiggins C. D., Patterns of pollen feeding and habitat preference among Heliconius species. Ecol. Entomol. 27, 448–456 (2002). [Google Scholar]
- 34.Martin S. H., et al., Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23, 1817–1828 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seymoure B. M., “Heliconius in a new light: The effects of light environments on mimetic coloration, behavior, and visual systems,” PhD dissertation, Arizona State University, Tempe, AZ (2016).
- 36.Seymoure B. M., Mcmillan W. O., Rutowski R., Peripheral eye dimensions in Longwing (Heliconius) butterflies vary with body size and sex but not light environment nor mimicry ring. J. Res. Lepidoptera 48, 83–92 (2015). [Google Scholar]
- 37.Mallet J., Beltrán M., Neukirchen W., Linares M., Natural hybridization in heliconiine butterflies: The species boundary as a continuum. BMC Evol. Biol. 7, 28 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stöckl A. L., Ribi W. A., Warrant E. J., Adaptations for nocturnal and diurnal vision in the hawkmoth lamina. J. Comp. Neurol. 524, 160–175 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Stöckl A. L., O’Carroll D. C., Warrant E. J., Hawkmoth lamina monopolar cells act as dynamic spatial filters to optimize vision at different light levels. Sci. Adv. 6, eaaz8645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borst A., Drosophila’s view on insect vision. Curr. Biol. 19, R36–R47 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Morante J., Desplan C., Building a projection map for photoreceptor neurons in the Drosophila optic lobes. Semin. Cell Dev. Biol. 15, 137–143 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Rister J., et al., Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron 56, 155–170 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Paulk A. C., Dacks A. M., Phillips-Portillo J., Fellous J.-M., Gronenberg W., Visual processing in the central bee brain. J. Neurosci. 29, 9987–9999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hausen K., “The lobula-complex of the fly: Structure, function and significance in visual behaviour” in Photoreception and Vision in Invertebrates (NATO ASI Series, Series A: Life Sciences; M. A. Ali, Ed. (Springer, Boston, MA, 1984), pp. 523–559.
- 45.Buschbeck E. K., Strausfeld N. J., The relevance of neural architecture to visual performance: Phylogenetic conservation and variation in Dipteran visual systems. J. Comp. Neurol. 383, 282–304 (1997). [PubMed] [Google Scholar]
- 46.Homberg U., Würden S., Movement-sensitive, polarization-sensitive, and light-sensitive neurons of the medulla and accessory medulla of the locust, Schistocerca gregaria. J. Comp. Neurol. 386, 329–346 (1997). [PubMed] [Google Scholar]
- 47.Heinze S., Reppert S. M., Anatomical basis of sun compass navigation I: The general layout of the monarch butterfly brain. J. Comp. Neurol. 520, 1599–1628 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Montgomery S. H., Ott S. R., Brain composition in Godyris zavaleta, a diurnal butterfly, reflects an increased reliance on olfactory information. J. Comp. Neurol. 523, 869–891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montgomery S. H., Merrill R. M., Ott S. R., Brain composition in Heliconius butterflies, posteclosion growth and experience-dependent neuropil plasticity. J. Comp. Neurol. 524, 1747–1769 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Kinoshita M., Shimohigasshi M., Tominaga Y., Arikawa K., Homberg U., Topographically distinct visual and olfactory inputs to the mushroom body in the swallowtail butterfly, Papilio xuthus. J. Comp. Neurol. 523, 162–182 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Pfeiffer K., Kinoshita M., Homberg U., Polarization-sensitive and light-sensitive neurons in two parallel pathways passing through the anterior optic tubercle in the locust brain. J. Neurophysiol. 94, 3903–3915 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Mappes M., Homberg U., Surgical lesion of the anterior optic tract abolishes polarotaxis in tethered flying locusts, Schistocerca gregaria. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 193, 43–50 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Heinze S., “Polarized-light processing in insect brains: Recent insights from the desert locust, the monarch butterfly, the cricket, and the fruit fly” in Polarized Light and Polarization Vision in Animal Sciences, Hováth G., Ed. (Springer, Berlin, 2014), pp. 61–111. [Google Scholar]
- 54.Keesey I. W., et al., Inverse resource allocation between vision and olfaction across the genus Drosophila. Nat. Commun. 10, 1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arias C. F., et al., Phylogeography of Heliconius cydno and its closest relatives: Disentangling their origin and diversification. Mol. Ecol. 23, 4137–4152 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Mérot C., Frérot B., Leppik E., Joron M., Beyond magic traits: Multimodal mating cues in Heliconius butterflies. Evolution 69, 2891–2904 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Kronforst M. R., Young L. G., Blume L. M., Gilbert L. E., Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution 60, 1254–1268 (2006). [PubMed] [Google Scholar]
- 58.Kronforst M. R., et al., Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc. Natl. Acad. Sci. U.S.A. 103, 6575–6580 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huxley J. S., Problems of Relative Growth (Methuen, London, 1932). [Google Scholar]
- 60.Gould S. J., Geometric similarity in allometric growth: A contribution to the problem of scaling in the evolution of size. Am. Nat. 105, 113–136 (1971). [Google Scholar]
- 61.Voje K. L., Hansen T. F., Egset C. K., Bolstad G. H., Pélabon C., Allometric constraints and the evolution of allometry. Evolution 68, 866–885 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Pélabon C., et al., Evolution of morphological allometry. Ann. N. Y. Acad. Sci. 1320, 58–75 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Montgomery S. H., The human frontal lobes: Not relatively large but still disproportionately important? A commentary on Barton and Venditti. Brain Behav. Evol. 82, 147–149 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Montgomery S. H., Mundy N. I., Barton R. A., Brain evolution and development: Adaptation, allometry and constraint. Proc. Biol. Sci. 283, 20160433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brommer J. E., Whither Pst? The approximation of Qst by Pst in evolutionary and conservation biology. J. Evol. Biol. 24, 1160–1168 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Whitehead A., Crawford D. L., Neutral and adaptive variation in gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 5425–5430 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montgomery S. H., Mank J. E., Inferring regulatory change from gene expression: The confounding effects of tissue scaling. Mol. Ecol. 25, 5114–5128 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Martin S. H., Davey J. W., Salazar C., Jiggins C. D., Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17, e2006288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haerty W., Singh R. S., Gene regulation divergence is a major contributor to the evolution of Dobzhansky-Muller incompatibilities between species of Drosophila. Mol. Biol. Evol. 23, 1707–1714 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Ortíz-Barrientos D., Counterman B. A., Noor M. A. F., Gene expression divergence and the origin of hybrid dysfunctions. Genetica 129, 71–81 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Landry C. R., Hartl D. L., Ranz J. M., Genome clashes in hybrids: Insights from gene expression. Heredity 99, 483–493 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Renaut S., Bernatchez L., Transcriptome-wide signature of hybrid breakdown associated with intrinsic reproductive isolation in lake whitefish species pairs (Coregonus spp. Salmonidae). Heredity 106, 1003–1011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Michalak P., Noor M. A., Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol. Biol. Evol. 20, 1070–1076 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Ranz J. M., Namgyal K., Gibson G., Hartl D. L., Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 14, 373–379 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mavarez J., Audet C., Bernatchez L., Major disruption of gene expression in hybrids between young sympatric anadromous and resident populations of brook charr (Salvelinus fontinalis Mitchill). J. Evol. Biol. 22, 1708–1720 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Wurmser F., et al., Population transcriptomics: Insights from Drosophila simulans, Drosophila sechellia and their hybrids. Genetica 139, 465–477 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Ometto L., Ross K. G., Shoemaker D., Keller L., Disruption of gene expression in hybrids of the fire ants Solenopsis invicta and Solenopsis richteri. Mol. Ecol. 21, 2488–2501 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Whiteley A. R., et al., The phenomics and expression quantitative trait locus mapping of brain transcriptomes regulating adaptive divergence in lake whitefish species pairs (Coregonus sp.). Genetics 180, 147–164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Filteau M., Pavey S. A., St-Cyr J., Bernatchez L., Gene coexpression networks reveal key drivers of phenotypic divergence in lake whitefish. Mol. Biol. Evol. 30, 1384–1396 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Catalán A., Briscoe A. D., Höhna S., Drfit and directional selection are the evolutionary forces driving gene expression divergence in eye and brain tissue of Heliconius butterflies. Genetics 213, 581–594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Merrill R. M., Naisbit R. E., Mallet J., Jiggins C. D., Ecological and genetic factors influencing the transition between host-use strategies in sympatric Heliconius butterflies. J. Evol. Biol. 26, 1959–1967 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Merrill R. M., et al., Disruptive ecological selection on a mating cue. Proc. Biol. Sci. 279, 4907–4913 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Naisbit R. E., Jiggins C. D., Mallet J., Disruptive sexual selection against hybrids contributes to speciation between Heliconius cydno and Heliconius melpomene. Proc. Biol. Sci. 268, 1849–1854 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiggins C. D., Naisbit R. E., Coe R. L., Mallet J., Reproductive isolation caused by colour pattern mimicry. Nature 411, 302–305 (2001). [DOI] [PubMed] [Google Scholar]
- 85.Chouteau M., Arias M., Joron M., Warning signals are under positive frequency-dependent selection in nature. Proc. Natl. Acad. Sci. U.S.A. 113, 2164–2169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dell’Aglio D. D., Troscianko J., McMillan W. O., Stevens M., Jiggins C. D., The appearance of mimetic Heliconius butterflies to predators and conspecifics. Evolution 72, 2156–2166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seymoure B. M., Raymundo A., McGraw K. J., Owen McMillan W., Rutowski R. L., Environment-dependent attack rates of cryptic and aposematic butterflies. Curr. Zool. 64, 663–669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Butlin R. K., Smadja C. M., Coupling, reinforcement, and speciation. Am. Nat. 191, 155–172 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Huber R., van Staaden M. J., Kaufman L. S., Liem K. F., Microhabitat use, trophic patterns, and the evolution of brain structure in African cichlids. Brain Behav. Evol. 50, 167–182 (1997). [DOI] [PubMed] [Google Scholar]
- 90.Barton R. A., Purvis A., Harvey P. H., Evolutionary radiation of visual and olfactory brain systems in primates, bats and insectivores. Philos. Trans. R. Soc. Lond. B Biol. Sci. 348, 381–392 (1995). [DOI] [PubMed] [Google Scholar]
- 91.Jiggins C. D., McMillan W. O., Neukirchen W., Mallet J., What can hybrid zones tell us about speciation? The case of Heliconius erato and H. himera (Lepidoptera: Nymphalidae). Biol. J. Linn. Soc. Lond. 59, 221–242 (1996). [Google Scholar]
- 92.Gonda A., Herczeg G., Merilä J., Adaptive brain size divergence in nine-spined sticklebacks (Pungitius pungitius)? J. Evol. Biol. 22, 1721–1726 (2009). [DOI] [PubMed] [Google Scholar]
- 93.Park P. J., Bell M. A., Variation of telencephalon morphology of the threespine stickleback (Gasterosteus aculeatus) in relation to inferred ecology. J. Evol. Biol. 23, 1261–1277 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Keagy J., Braithwaite V. A., Boughman J. W.; Riesch R., Handling editor, Brain differences in ecologically differentiated sticklebacks. Curr. Zool. 64, 243–250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Park P. J., Chase I., Bell M. A., Phenotypic plasticity of the threespine stickleback Gasterosteus aculeatus telencephalon in response to experience in captivity. Curr. Zool. 58, 189–210 (2012). [Google Scholar]
- 96.Ott S. R., Confocal microscopy in large insect brains: Zinc-formaldehyde fixation improves synapsin immunostaining and preservation of morphology in whole-mounts. J. Neurosci. Methods 172, 220–230 (2008). [DOI] [PubMed] [Google Scholar]
- 97.Davey J. W., et al., Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3 (Bethesda) 6, 695–708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pinharanda A., et al., Sexually dimorphic gene expression and transcriptome evolution provide mixed evidence for a fast-Z effect in Heliconius. J. Evol. Biol. 32, 194–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anders S., Pyl P. T., Huber W., HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bates D., et al., “lme4.” Version 1.1-23. https://github.com/lme4/lme4/. Accessed 6 March 2020.
- 103.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995). [Google Scholar]
- 104.Warton D. I., Duursma R. A., Falster D. S., Taskinen S., Smatr 3—an R package for estimation and inference about allometric lines. Methods Ecol. Evol. 3, 257–259 (2012). [Google Scholar]
- 105.Goslee S. C., Urban D. L., The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22, 1–19 (2007). [Google Scholar]
- 106.Da Silva S. B., Da Silva A., Pstat: An R package to assess population differentiation in phenotypic traits. R J. 10, 447–454 (2018). [Google Scholar]
- 107.R Core Team , A Language and Environement for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2014). https://www.r-project.org/. Accessed 4 August 2019.
- 108.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Uebbing S., et al., Divergence in gene expression within and between two closely related flycatcher species. Mol. Ecol. 25, 2015–2028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jones P., et al., InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alexa A., Rahnenfuhrer J., topGO: Enrichment Analysis for Gene Ontology. R package version 2.42.0 (2020). https://bioconductor.org/packages/release/bioc/html/topGO.html. Accessed 18 May 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full descriptions of the methodology and the neuroanatomical dataset are available in SI Appendix and have been deposited on DataDryad, along with R code (https://doi.org/10.5061/dryad.7wm37pvs3). The majority of the raw reads from the genetic dataset have been already available in The European Nucleotide Archive (accession no. PRJEB39935), and the remaining have been deposited on The European Nucleotide Archive (accession no. PRJEB42387).
All study data are included in the article and/or supporting information.