Significance
Honey bees pollinate the majority of crops, but their survival is under threat. The bee parasitic mite Varroa destructor is a major cause of honey bee decline. The successful control of Varroa mites depends on a small number of in-hive acaricides. Species-specific activation of the chemical coumaphos is a powerful “pro-drug” approach to attain this selective toxicity and protect bees. We identified a coumaphos-resistant Varroa population that escapes toxicity by down-regulating the activating enzyme, a P450 monooxygenase. Decreased activation is a rare but evolutionarily powerful solution to achieve resistance. It demonstrates that the current proinsecticide approach is under threat, as it can be circumvented, and highlights the need to restart the pipeline to develop selective pest control agents.
Keywords: pesticide, honey bee, selectivity, resistance management, CYPome
Abstract
Varroa destructor is one of the main problems in modern beekeeping. Highly selective acaricides with low toxicity to bees are used internationally to control this mite. One of the key acaricides is the organophosphorus (OP) proinsecticide coumaphos, that becomes toxic after enzymatic activation inside Varroa. We show here that mites from the island Andros (AN-CR) exhibit high levels of coumaphos resistance. Resistance is not mediated by decreased coumaphos uptake, target-site resistance, or increased detoxification. Reduced proinsecticide activation by a cytochrome P450 enzyme was the main resistance mechanism, a powerful and rarely encountered evolutionary solution to insecticide selection pressure. After treatment with sublethal doses of [14C] coumaphos, susceptible mite extracts had substantial amounts of coroxon, the activated metabolite of coumaphos, while resistant mites had only trace amounts. This indicates a suppression of the P450 (CYP)-mediated activation step in the AN-CR mites. Bioassays with coroxon to bypass the activation step showed that resistance was dramatically reduced. There are 26 CYPs present in the V. destructor genome. Transcriptome analysis revealed overexpression in resistant mites of CYP4DP24 and underexpression of CYP3012A6 and CYP4EP4. RNA interference of CYP4EP4 in the susceptible population, to mimic underexpression seen in the resistant mites, prevented coumaphos activation and decreased coumaphos toxicity.
Beekeeping plays an important role in providing nutritional, economic, and ecological security globally, with direct contribution to the economy valued at >230 billion dollars per year (1). Bee colony collapse disorder (CCD) is a major problem internationally, leading to a loss of >10 million beehives in North America alone and causing dramatic economic losses (1). Several possible causes for CCD have been proposed, such as the impact and side effects of pesticides used in agriculture, although this is currently still under considerable debate (2). Other suggested causes include damage by pests and diseases, malnutrition, genetic factors, immunodeficiencies, loss of habitat, and changing beekeeping practices. In reality, a combination of factors are likely to be involved.
The major bee ectoparasitic mite Varroa destructor is the most important pest for apiculture (3, 4), and it has been directly implicated with major economic losses (5). In addition to its direct parasitism effects on bee physiology and sociability, Varroa transmits viruses and pathogens, likely associated with CCD (6, 7). Commercial beekeepers primarily use selective chemical acaricides with low bee toxicity to control Varroa infestation (8, 9). Given the need for selectivity, very few acaricides can be used in-hive, and common commercial use is restricted to the pyrethroids tau-fluvalinate and flumethrin, the formamidine amitraz, and the organophosphorus (OP) coumaphos (10).
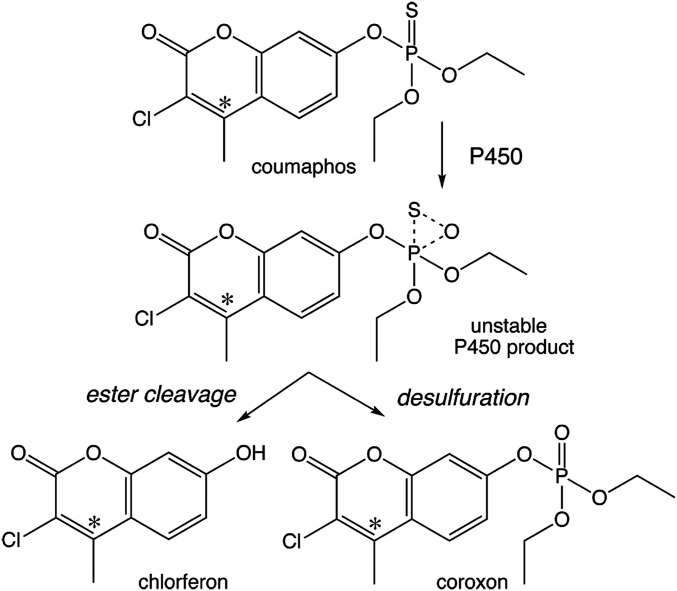
Coumaphos was synthesized 70 y ago for use in livestock to control insect ectoparasites, such as ticks and helminths. Coumaphos is a phosphorothioate proinsecticide (11) that requires in vivo bioactivation by cytochrome P450 (CYP) monooxygenases (MOs) to its active phosphate metabolite coroxon (coumaphos-oxon), which is an irreversible inhibitor of acetylcholinesterase (AChE) (Fig. 1).
Fig. 1.
Metabolism of [14C]coumaphos in Varroa. Radiolabeled coumaphos (* indicates the position of the label) is converted to two main metabolites, chlorferon and coroxon. Both compounds result from P450 monooxygenation of the P = S moiety of coumaphos that initially leads to an unstable product in the active site. Ester cleavage to the nontoxic chlorferon and desulfuration to the AChE inhibitor coroxon are the two outcomes of P450 metabolism. The ratio of the two outcomes is specific to each P450. See text for details.
The extensive use of acaricides against Varroa has selected resistant populations in several countries (12): pyrethroid (tau-fluvalinate and flumethrin) resistance has been reported in Europe and many other parts of the world (13–18); formamidine (amitraz) resistance has been detected in Mexico (19) and Argentina (20); and coumaphos resistance has been recorded in the United States and Argentina (12, 21).
Insecticide and acaricide resistance mechanisms are typically mediated either by changes in the target site, making it insensitive, or by changes in penetration, activation, or detoxification, that alters the amount of insecticide that reaches the target site (22). Mutations, such as the L925V, L925I, and L925M, in the voltage-gated sodium channel (VGSC), the target site for pyrethroids, have been associated with pyrethroid resistance in Varroa (13–18), but the resistance mechanisms for amitraz and coumaphos are unknown. However, amitraz target-site resistance mutations in the octopamine receptor (23) and detoxification-based resistance of coumaphos due to enhanced metabolism of coroxon (Fig. 1) have been reported for the tick Rhipicephalus (Boophilus) microplus (24, 25).
Characterization of the resistance mechanisms in Varroa has been hampered by their biological cycle inside hives (26), but the recent publication of the Varroa genome sequence has provided the needed impetus to better study the molecular biology of resistance (27). The development of robust reverse genetic (RNA interference [RNAi]) tools (28) also facilitates functional studies for the implication of specific genes in the resistant phenotypes. Using these tools and classical approaches, we have now identified the underlying cause of coumaphos resistance in a Varroa population from a Greek Aegean island.
Results
Coumaphos Resistance in Varroa.
Among six populations tested, the Andros coumaphos resistant (AN-CR) population, from the Aegean island Andros, exhibited substantially reduced mortality rates (7%) (SI Appendix, Table S1) in bioassays at a diagnostic dose of 200 mg/L. This population was sampled from a beekeeper who used coumaphos almost exclusively for many years. Dose–response bioassays were subsequently conducted to determine the strength of the phenotype. Resistance ratio (RR) scaled up to 217-fold in the AN-CR compared to the coumaphos-susceptible Athens susceptible (ATH-S) population (Table 1).
Table 1.
Concentration probit mortality bioassay data of coumaphos and coroxon (coumaphos oxon) against V. destructor mites
| Treatment | n* | LC50 (95% fiducial limits)† | Slope ± SE | χ2‡ | df | RR50 § |
| ATH-S | ||||||
| Coumaphos | 276 | 14.35 (4.25 to 35.54) | 0.86 ± 0.09 | 7.09 | 3 | — |
| Coroxon | 195 | 0.021 (0.005 to 0.070) | 0.91 ± 0.12 | 5.08 | 3 | — |
| AN-CR | ||||||
| Coumaphos | 152 | 3,112.2 (1,291.4 to 10,832.5) | 1.7 ± 0.26 | 6.16 | 3 | 217 (117.61 to 399.79) |
| Coroxon | 160 | 0.51 (0.02 to 1.001) | 0.73 ± 0.12 | 1.16 | 2 | 25 (8.68 to 71.00) |
Numbers of mites tested.
LC, lethal concentration, expressed in mg/L.
χ2 testing linearity.
RR, resistance ratio: LCAN-CR (coumaphos)/LC ATH-S (coumaphos); LCAN-CR (coroxon)/LC ATH-S (coroxon).
Penetration, Detoxification, and Altered Target Site Are Not Involved in Resistance.
Acaricide uptake (penetration) was not significantly different between resistant and susceptible mites (SI Appendix, Table S2). Analysis of detoxification enzyme activities with different substrates—more specifically, carboxyl/choline esterase (CCE) activity measured with the substrates p-nitrophenol (PNPA), 1-naphthyl acetate (1-NA), and 2-naphthyl acetate (2-NA); glutathione S-transferase (GST) activity measured with the substrate 1-chloro-2,4-dinitrobenzene (CDNB); and MO activity measured with the substrate 7-ethoxycoumarin (7-EC)—did not show any significant differences between the AN-CR and the ATH-S mites (Table 2). In contrast, a significant reduction of GST activity measured with the substrate monochlorobimane (MCB) was recorded (Table 2). The isoelectric focusing (IEF) profile of CCEs was identical between the ATH-S and AN-CR populations (SI Appendix, Fig. S1). No significant differences in the total AChE activity or in the inhibition of AChE by coroxon or malaoxon were observed between the ATH-S and AN-CR populations (Table 2).
Table 2.
Detoxification enzyme activities and AChE inhibition by organophosphate oxon analogs in the ATH-S and AN-CR V. destructor populations
| Population | CCEs | MO | GST | AChE | ||||||
| PNPA | 1-NA | 2-NA | 7-EC | CDNB | MCB | Activity | IC50 | |||
| Malaoxon | Coroxon | |||||||||
| ATH-S | 16.43 ± 1.9 | 17.66 ± 1.1 | 24.31 ± 1.7 | 8.53 ± 3.3 | 0.013 ± 0.002 | 537.18 ± 54.9 | 75.01 ± 5.35 | 2.37 ± 0.12 x 10−5 | 6.90 ± 0.5 x 10−7 | |
| AN-CR | 15.84 ± 1.7 | 18,81 ± 0.6 | 23.55 ± 1.5 | 10.46 ± 4.7 | 0.019 ± 0.002 | 74.14 ± 8.32* | 73.89 ± 4.72 | 3.23 ± 0.16 x 10−5 | 8.08 ± 0.7 x 10−7 | |
CCEs, carboxyl/choline esterases: PNPA nmol/min/mg; 1-NA/2-NA, nmol a- or b-naphthol/min/mg; MO, monooxygenases: pmol 7OH/min/mg; GSTs, glutathione S-transferases: CDNB μmol CDNB conjugated/min/mg; MCB: relative fluorescence units (RFU)/μg; AChE, acetylcholinesterases: nmol/min/mg protein; IC50: μg/mL. Values are mean ± SE. Statistical analysis was performed with the independent samples t test. Statistical significance, *P < 0.05.
Decreased Coumaphos Activation Causes Resistance.
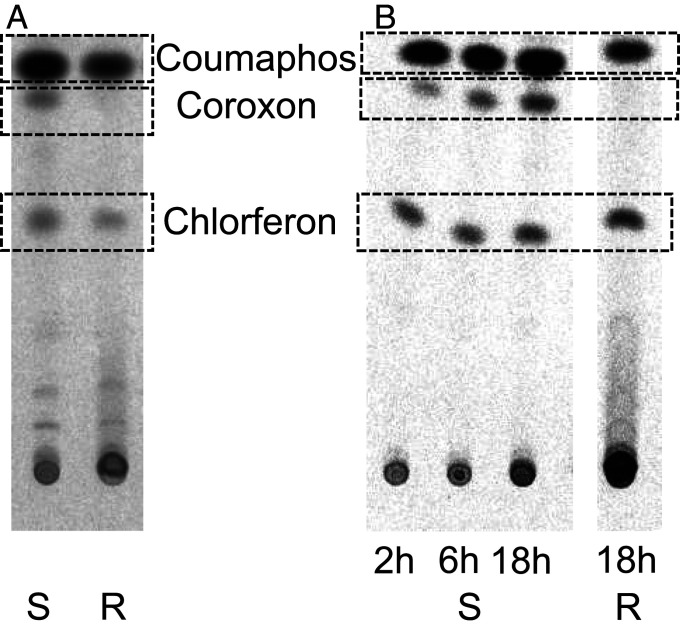
Analysis of whole body extracts of ATH-S and AN-CR Varroa mites after treatment with [14C] coumaphos, using normal phase thin layer chromatography (TLC), revealed besides the parental substrate (retention factor [Rf] = 0.83), two main known metabolites and some putative unknown secondary metabolites: the activated phosphate metabolite coroxon (Rf = 0.75); the nontoxic metabolite chlorferon (Rf = 0.50), and some more polar compounds at around Rf = 0.25 and lower (Fig. 2). The formation (detoxification rate) of the main coumaphos detoxification product (inactive metabolite) chlorferon was similar between the AN-CR and the ATH-S populations, as was the formation of the unknown metabolites. However, a significant difference in the formation of coroxon, the activated toxic organophosphate acaricide, was observed. Coroxon was readily formed in the ATH-S population, in a time-dependent manner (Fig. 2). However, this metabolite was barely detected in the resistant AN-CR mites, even 18 h posttreatment with 200 mg active ingredient/L [14C] coumaphos (Fig. 2).
Fig. 2.
Analysis of metabolites after in vivo treatment with [14C] coumaphos. Normal phase TLC in chloroform/methanol (100/7) of whole body extracts of ATH-S and AN-CR mites (V. destructor) treated with [14C] coumaphos. Coumaphos (Rf = 0.83); Coroxon (Rf = 0.75); Chlorferon (Rf = 0.50); unknown metabolites (Rf = 0.1). (A) Reduced activation of coumaphos to coroxon associated with resistance: ATH-S and AN-CR mite homogenates 2 h after treatment with [14C] coumaphos. (B) Time-dependent formation of coroxon in ATH-S but not in AN-CR Varroa mites. ATH-S mites after 2 h, 6 h, and 18 h in vivo treatment and AN-CR after 18 h treatment with [14C] coumaphos (vials coated with 200 mg active ingredient/L). S: susceptible, R: resistant.
To confirm that the failure to activate the proinsecticide was the cause of resistance, we conducted bioassays directly with coroxon, thus bypassing the activation step inside the mite. The bioassays showed that the resistance phenotype was dramatically alleviated (RR dropped from 217-fold for coumaphos, to 25-fold for coroxon) (Table 1).
Cytochrome P450 Monooxygenases Are Associated with Coumaphos Resistance.
To investigate the transcriptional basis of coumaphos resistance in Varroa, we performed RNA-seq on ATH-S, AN-CR, and AN-CR pretreated with coumaphos (tAN-CR). After filtering the Illumina RNA-seq dataset (SI Appendix, Table S3) for the presence of viral sequences (SI Appendix, Table S4), a differential gene expression analysis was performed between the AN-CR and the ATH-S population and between the tAN-CR population and the AN-CR population. A total of 270 differentially expressed genes (DEGs) were identified in AN-CR compared to ATH-S (fold change [FC] > 2 and a Benjamini–Hochberg adjusted P value < 0.05) (SI Appendix, Dataset S2). Among them, only 12 encoded typical detoxification enzymes (P450s [InterPro domain IPR001128], CCEs [IPR002018], and GSTs [IPR036282]) or proteins involved in xenobiotic transport (ABC transporters [IPR003439]) (SI Appendix, Dataset S2). Three genes encoded CYPs: two were underexpressed (CYP4EP4: with trimmed mean of M values [TMM] normalized counts per million [CPM] between 288 and 594 in ATH-S vs. 94 to 127 for AN-CR: i.e., relatively highly expressed; and CYP3012A6: TMM normalized CPM between 14 and 25 in ATH-S vs. 4 to 10 in AN-CR: i.e., lowly expressed), and one was overexpressed (CYP4DP24: TMM normalized CPM between 7 and 21 in ATH-S vs. 27 to 59 in AN-CR). The coding sequence (CDS) of all three CYP genes was identical in the transcriptome shotgun assembly (TSA) data of AN-CR and ATH-S.
Only one gene was differentially expressed in the pairwise comparison between the tAN-CR and AN-CR populations (SI Appendix, Dataset S2); hence, in line with principal component analysis (PCA) (SI Appendix, Fig. S2), the DEG profiles of the AN-CR and the tAN-CR populations were highly similar, indicating that very limited gene expression changes are associated with exposure (induction) of the AN-CR to the apparently nonactivated coumaphos, inside these Varroa mites. However, the possibility that the relatively low dose used (200 parts per million [ppm]; lethal concentration, expressed in mg/L [LC5] for this population) was not sufficient to induce changes in genes expression cannot be excluded. We mined the RNA-seq data of the ATH-S and AN-CR population for the presence of mutations in the V. destructor Ace genes. The protein encoded by the V. destructor gene LOC111243720 showed the best BLASTp hit with Tetranychus urticae AChE (E-value 0, 54% identity). This gene is the ortholog of the tick R. microplus AChE 1 (29). The protein encoded by LOC111246636 was the second best BLASTp hit to T. urticae AChE (E-value of E−101, 34% identity). This gene is the ortholog of R. microplus AChE 3 (29); 82.2 and 99.8% of the CDS of LOC111243720 and 100% of the CDS of LOC111246636 were covered by the RNA-seq consensus sequence of the ATH-S and AN-CR population, respectively. Single nucleotide polymorphisms were not identified compared to the reference genomic sequence. Thus, in agreement with our biochemical data, we have no evidence of AChE target site resistance in Varroa.
Cytochromes P450 of Varroa Destructor.
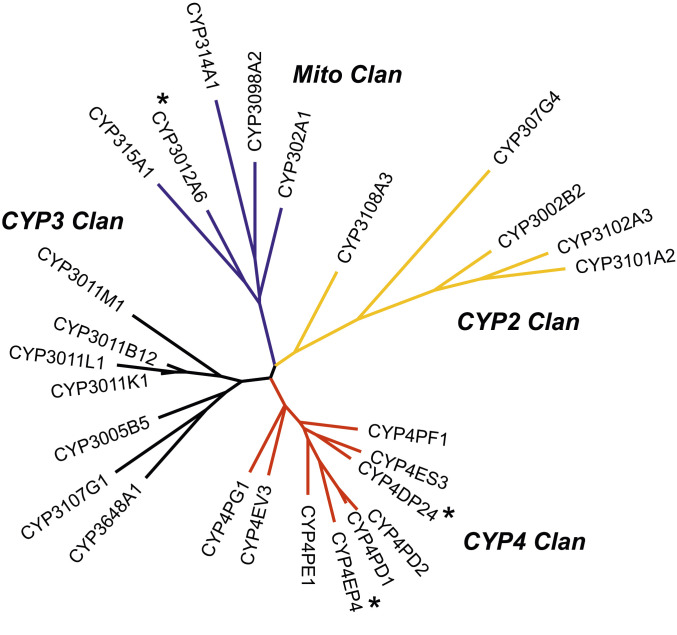
The curated genome complement of CYP genes (CYPome) of V. destructor consists of 26 full-length sequences. This is one of the smallest CYPomes of arthropods. It is much smaller than the 63 CYPs of the western orchard mite, Metaseiulus occidentalis and only half the size of the CYPome of the honey bee, the Varroa host (30). The 26 Varroa CYPs form four CYP clans (Fig. 3 and SI Appendix, Dataset S1), as in the spider mite and most insects. CYP307G4 of the CYP2 clan and CYP302A1, CYP314A1, and CYP315A1 in the mitochondrial CYP clan are involved in ecdysteroid biosynthesis and are highly conserved. There was no CYP18 or CYP306 ortholog in Varroa, a situation similar to the spider mite T. urticae.
Fig. 3.
Maximum likelihood phylogeny of the 26 CYP sequences of V. destructor. The four CYP clans are marked with a different color: yellow, CYP2 clan; red, CYP4 clan; black, CYP3 clan; blue, mitochondrial clan. The three differentially expressed CYPs are marked with an asterisk.
Silencing of the CYP4EP4 Prevents Coumaphos Activation and Induces Tolerance in Susceptible Varroa.
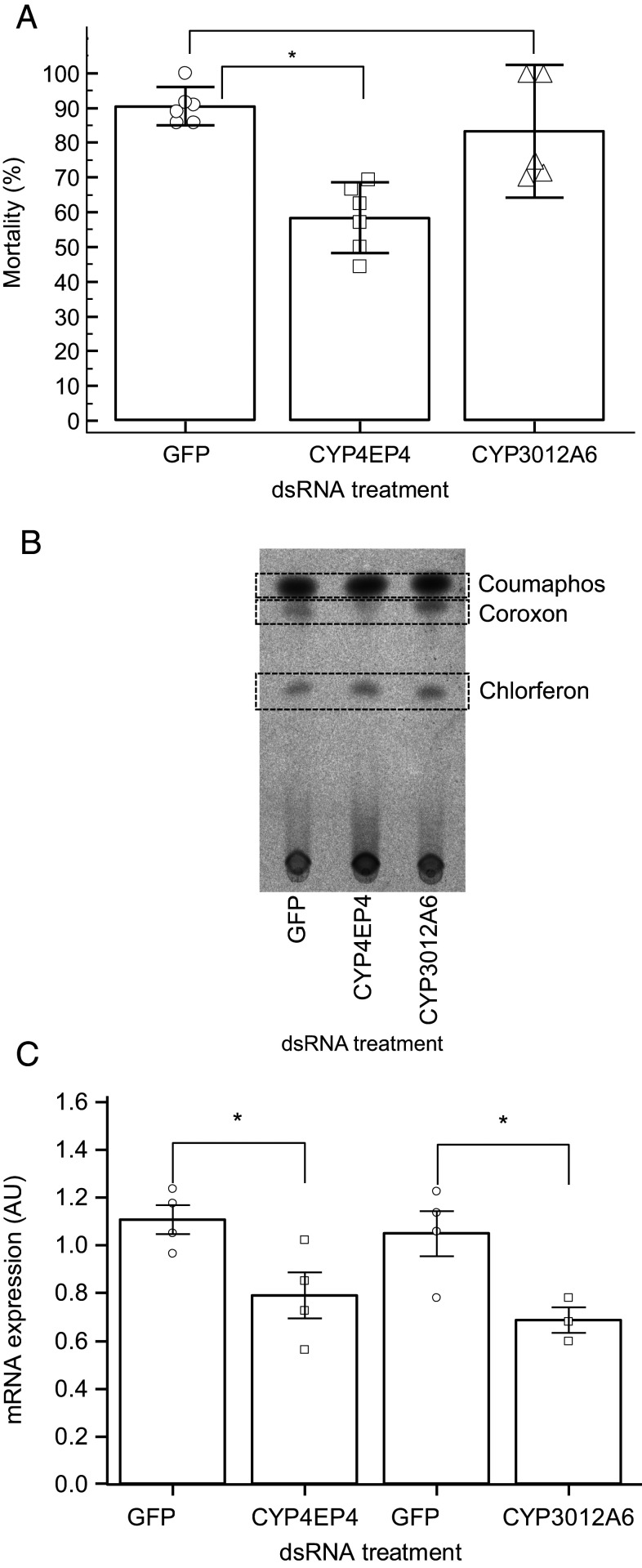
Having established that the underexpression of two CYPs (CYP3012A6 and CYP4EP4) in AN-CR Varroa mites is associated with resistance, we silenced these CYPs, using RNAi, in the ATH-S–susceptible population, aiming to mimic the underexpression in resistant mites. Silencing of CYP4EP4 significantly reduced mortality at the diagnostic dose by 35%: i.e., from 90.48% (95% CI = 84.92 to 96.04%) in the GFP-injected control mites to 58.33% (95% CI = 48.16 to 68.50%) in the dsCYP4EP4-treated mites (Fig. 4A). In contrast, silencing of the CYP3012A6 in ATH-S mites did not significantly reduce mortality at the diagnostic dose compared to the GFP control mites (Fig. 4A). Silencing of CYP4EP4 but not of GFP or CYP3012A6 in the susceptible ATH-S mites was also associated with a substantial reduction of the activation of coumaphos to its active metabolite coroxon (Fig. 4B). The silencing efficiency was verified by the statistically significant reductions in CYP4EP4 (28.7% SE = 10.3%, P = 0.032) and CYP3012A6 (34.59% SE = 11.6%, P = 0.031) messenger RNA (mRNA) levels, respectively, compared to the GFP control (Fig. 4C).
Fig. 4.
Silencing of the CYP4EP4 prevents coumaphos activation and induces tolerance in susceptible V. destructor. (A) Mortality rates against coumaphos of ATH-S immersed for 6 h in dsRNA (GFP, CYP4EP4, and CYP3012A6) and tested with diagnostic coumaphos concentration (200 ppm) after 72 h. (B) Normal Phase TLC of whole body extracts of ATH-S mites immersed for 6 h in dsRNA (GFP, CYP4EP4, and CYP3012A6) and treated with [14C] coumaphos as in A. (C) Validation of gene silencing efficacy by RNAi. Relative expression levels in dsRNA‐submersed Varroa (GFP control, CYP4EP4, and CYP3012A6). mRNA expression was calculated using the 2-ΔCT formula and was normalized to housekeeping genes; * indicates statistical significance at the P < 0.05 level, calculated using independent samples t test; error bars represent SE of mean. AU, arbitrary units (×20 for CYP3012A6 expression to harmonize the scale).
Discussion
An unusual case of CYP-mediated reduced proinsecticide activation conferring very high levels of acaricide resistance in the major bee ectoparasitic mite V. destructor was identified and elucidated at the molecular level. Many OP compounds, which dominated the insecticide market in the 1970s and 1980s, are phosphorothioates (with a P = S moiety). These are proinsecticides, with a 4 to 5 order of magnitude lower AChE inhibition than that of their activated metabolite, the corresponding phosphates (P = O). The activation step can lead to selective toxicity. For instance, malathion is rapidly inactivated by mammalian carboxylesterases while insects preferentially activate it to malaoxon. The P = S to P = O activation step is catalyzed by CYPs: for instance, house fly CYP6A1 and CYP12A1 (31, 32).
Here, we show that reduced CYP-mediated activation in Varroa is involved in coumaphos resistance. Bioassays with coroxon, which bypassed the CYP activation step, reduced the AN-CR resistance from 217-fold for coumaphos, to 25-fold for coroxon. The CYPome in Varroa is limited (N = 26), which is usual in parasitic organisms. The human body louse Pediculus humanus has 37 CYP genes (33), and the crustacean salmon louse Lepeophtheirus salmonis just 21 (34). There was differential expression of three CYPs, one (CYP4DP24 from the CYP4 clan) overexpressed and two underexpressed (CYP3012A6 and CYP4EP4 from the mitochondrial and CYP4 clan, respectively), in AN-CR compared to ATH-S mites. Gene expression changes, after induction by 200 ppm coumaphos in the resistant AN-CR population, were negligible (only one gene).
Changes in CYP levels in both directions could in theory lead to resistance. This is because the CYP enzymes that attack the phosphorothioate (P = S) moiety of OP compounds first generate a monooxygenated, unstable intermediate which can then result in two outcomes (see Fig. 1 for coumaphos, showing the main radiolabeled metabolites). One outcome is ester cleavage and formation of the nontoxic metabolite chlorferon, and the other outcome is desulfuration and formation of the activated, toxic acaricide coroxon. The ratio of the two products depends on the substrate and the CYP. For coumaphos, a CYP catalyzing a high coroxon/chlorferon ratio would favor activation over detoxification. Its underexpression may lead to resistance, and vice versa (see schema in SI Appendix, Fig. S3).
Analysis of coumaphos metabolism indicated that there was no difference between the ATH-S and the AN-CR strains in the formation of chlorferon or in the depletion of the parental coumaphos. However, coroxon formation was decreased in resistant mites. Thus, the underexpression of a CYP in the resistant population with a high ratio of coroxon formation is the most likely resistance mechanism.
Despite many attempts, we were not able to confirm in vitro the function of the differentially expressed CYP enzymes to directly verify our interpretation of the in vivo metabolism studies. Hence, we confirmed the resistance mechanism using reverse genetics of the two underexpressed CYPs. RNAi of the underexpressed CYP4EP4 in the susceptible strain decreased coumaphos activation and increased the tolerance to coumaphos. Thus, this reverse genetics manipulation mimics long time selection for a down-regulation mechanism of CYP4EP4, with similar outcome. Expression of CYP4EP4 is ∼20 times higher than CYP3012A6. Silencing CYP3012A6 did not change the resistance phenotype, either because this CYP does not metabolize coumaphos or because its expression level is too low to affect the toxic outcome.
Most studies on OP resistance have documented the overexpression of CYP genes which generate a high ester cleavage (detoxification)/desulfuration (activation) ratio, such as CYP6A1 in diazinon-resistant house flies (ratio 2.66) (31). Examples of the reverse, i.e., decreased levels of enzymes with higher desulfuration/ester cleavage ratios, have not been reported. However, methyl parathion resistance in a field strain of Heliothis virescens was associated with lower microsomal activation to methyl paraoxon, and this activation product had a significantly lower resistance factor (35). This situation is similar to the mechanism described here for coumaphos resistance in Varroa.
The possibility that additional mechanisms may also play a role in the coumaphos resistance cannot be excluded as toxicological assays with coroxon still allow an RR of 25. For example, the reduced GST activity with the substrate MCB and the underexpression of GSTs (Table 3) in the AN-CR may have a direct or supportive role in the resistance phenotype.
Table 3.
Differentially expressed transcripts between the coumaphos resistant (AN-CR) and susceptible V. destructor population (ATH-S), encoding detoxification/transporter (P450, CCE, GST, and ABC) genes
| Gene ID | Enzyme/Transporter family | Gene name (InterPro domain) | log2FC RNA-seq |
| LOC111248731 | P450 | CYP4EP4 (IPR036396) | −1.84 |
| LOC111246534 | P450 | CYP3012A6 (IPR036396) | −1.62 |
| LOC111246379 | P450 | CYP4DP24 (IPR036396) | 1.64 |
| LOC111254737 | CCE | (IPR002018) | −1.22 |
| LOC111248568 | CCE | (IPR002018) | −1.34 |
| LOC111250396 | CCE | (IPR002018) | −1.05 |
| LOC111251676 | CCE | (IPR002018) | −1.23 |
| LOC111250776 | CCE | (IPR002018) | −2.06 |
| LOC111244895 | GST | (IPR040079) | −1.00 |
| LOC111253932 | GST | (IPR040079) | −1.10 |
| LOC111244969 | ABCC | (IPR003439) | −1.22 |
| LOC111253112 | ABCA | (IPR003439, IPR026082) | −1.37 |
Varroa infestations are associated with severe economic losses in apiculture, and coumaphos is one of the few acaricides available for its control worldwide. The presence and frequency of this coumaphos resistance mechanism can now be tracked, to assess if it is driving coumaphos resistance in other places in the world. If it is common, but not restricted to the Greek Aegean islands, the Janus face of CYP metabolism, with different activation vs. detoxification profiles for different insecticides catalyzed by the same CYP, could potentially open up new options for insecticide resistance management (IRM). For example, Adolfi et al. (2019) (36) showed that the overexpression of the Anopheles AgCYP6M2 CYP confers permethrin resistance via detoxification and increases the susceptibility to malathion, via bioactivating it to its more toxic metabolite malaoxon. Similarly pyrethroid-resistant H. virescens is more susceptible to the proinsecticide chlorfenapyr (37). This negative cross-resistance between different insecticide/proinsecticide classes, due to differential regulation of CYPs in resistant insects, could be exploited in a push–pull strategy of IRM.
Materials and Methods
Chemicals and [14C]-Labeled Coumaphos.
The following technical grade chemicals were used in the bioassays: coumaphos 98% purity (Sigma Aldrich), malaoxon, coumaphos-oxon 96% (LGC Standards), and 3-chloro-7-hydroxy-4-methylcoumarin (chlorferon) 97% purity (Sigma Aldrich). All the other reagents were obtained from Sigma Aldrich, unless otherwise stated.
Radiolabeled [coumarin-4-14C] coumaphos (Fig. 1) of high chemical and radiochemical purity (>99%) was purchased from Institute of Isotopes Co., Ltd. (Izotop, Hungary). The specific activity of the [14C] coumaphos was 2.024 MBq/mg (1 MBq = 60 × 106 disintegrations per minute [dpm]).
Mite Populations.
On the island of Andros, a population was collected from a site where coumaphos had been used almost exclusively for the last 14 y and where control failure was reported (Andros coumaphos resistant [AN-CR]). A Varroa population collected from beehives at the Agricultural University of Athens was used as a susceptible strain as the bee colonies have been maintained without chemical treatments for the last 20 y (Athens susceptible [ATH-S]).
Bioassays.
Chemicals dissolved in acetone were freshly prepared, and 0.5 mL of solution was placed into individual 12-mL glass vials. Vials were rolled until the acetone evaporated. Batches of 10 adult female mites were introduced into the coated vials, closed with holed parafilm, and kept at 25 °C for 20 h. Dose–response bioassays were conducted. For gene expression analysis (described in Cytochrome P450 Monooxygenases Are Associated with Coumaphos Resistance), induction bioassays were conducted at the coumaphos LC5 (200 mg/L) for the AN-CR, and the treated mites were collected at the end of the bioassay (tAN-CR). After the treatment period, mites were considered dead in the absence of leg movement when prodded with a fine paintbrush. Toxicity data were analyzed using PoloPC (LeOrA Software, Berkeley, CA). RRs were calculated by dividing the LC50 of a population with the LC50 of the ATH-S–susceptible/reference population.
Detoxification Enzyme Activities and AChE Inhibition.
The enzymatic activity of CCEs, P450 monooxygenases, GSTs, and AChE was determined according to Van Leeuwen et al. (38) with slight modifications. Briefly, mass homogenates were prepared by crushing 1 to 10 freshly sampled adult mites in sodium phosphate buffer (0.1 M, pH 7.2, 200 μL) with a Teflon pestle. The homogenate was centrifuged at 5,000 × g and 4 °C for 5 min, and the resulting supernatant was used as an enzyme source. Approximately 10 to 20 μg of crude protein extract was included in each assay, with at least three to five replicates for each population and assay. Fluorescence and absorbance were measured with a TECAN SpectraFluor microplate reader.
For the CCE assays, the substrates PNPA, 1-NA, and 2-NA were used. For the GST assays, the substrates CDNB (kinetically, for 10 min) and MCB (end point after 20 min) were used. For MO assays, the substrate 7-EC was used to measure the O-deethylation activity kinetically.
For AChE activity and inhibition rates with the OP oxon analogs malaoxon and coroxon, the reaction was conducted in 0.2 mL of substrate–reagent solution containing 0.5 mM 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) and 1.25 mM acetylthiocholine (AcSCh). Residual AChE activities, after 10-min incubation with malaoxon and coroxon (at concentrations 10−8 M to 10−4 M), were measured kinetically at 405 nm. The half maximal inhibitory concentration (IC50) was determined using GraFit Version 3.0 (Erithacus Software Ltd., Staines, UK) (39).
Analysis of Coumaphos Metabolism.
Mites exposed to [14C] coumaphos vials, as described above, were washed and homogenized in 200 µL of methanol. Supernatant was collected after centrifugation (5 min at 5,000 rpm), and the pellet was resuspended in methanol. The supernatant was reduced with airflow to 10 μL. TLC was prepared by saturating the chamber with chloroform/methanol (100:7) and allowing it to equilibrate for 1 h. Then, 10 μL of each sample was spotted onto a silica gel precoated plate. After chromatography in chloroform/methanol (100:7), the position of cold coumaphos, coroxon, and chlorferon was marked, and the plate was wrapped in plastic foil. Radioactivity was recorded with a phosphorimager (Typhoon FLA 7000; GE Healthcare), followed by calculation of the Rf as the ratio of migration distance of the compound of interest to that of the solvent front. CorelDRAW Home and Student X7 software was used for image processing. Cold chemistry unlabeled ultraviolet (UV)-active controls were used to localize the [14C]-determined spots and link retention time.
Illumina RNA-seq.
Varroa mites were introduced in 12-mL glass vials coated with either acetone (ATH-S and AN-CR) or 200 mg active ingredient coumaphos/L acetone (tAN-CR) and kept at 25 °C for 20 h. Glass vials were coated as described in Bioassays. After 20 h, alive mites were collected in an Eppendorf tube (10 to 20 mites per tube), snap frozen with liquid nitrogen, and stored at −80 °C until RNA extraction. RNA was extracted, using the TRI reagent protocol (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions and treated with TURBO DNase I (ThermoFisher Scientific) to remove genomic DNA contamination. Illumina libraries were constructed from the RNA samples with the TruSeq Stranded mRNA Library Preparation Kit with polyA selection (Illumina) and sequenced on an Illumina HiSeq 2500 to generate strand-specific paired reads of 2 × 125 base pairs (bp) (library construction and sequencing were performed at the High-Throughput Genomics Core of the Huntsman Cancer Institute, University of Utah). All RNA-seq data that were generated during the current study are available at the Gene Expression Omnibus (GEO) repository with accession number GSE153472. The quality of the reads was verified using FASTQC version 0.11.5 (40), and, subsequently, viral contamination was removed (SI Appendix, Table S3). A PCA was created as described by Love et al. (41) (SI Appendix, Fig. S1). A differential expression (DE) analysis was performed using voom (version 3.34.9) (42), and the read count data were obtained by HTSeq 0.6.0. DEGs (log2FC > 1 and Benjamini–Hochberg adjusted P value < 0.05) were determined for the following comparisons: AN-CR vs. ATH-S, tAN-CR vs. ATH-S, and tAN-CR vs. AN-CR).
The RNA-seq data of both ATH-S and AN-CR were also used to search for mutations in Ace, the gene(s) encoding the target site of coumaphos. First, Varroa Ace sequences were identified by a BLASTp search against the Varroa proteome (version of October 2017) and using T. urticae Ace as query (tetur19g00850, https://bioinformatics.psb.ugent.be/orcae/overview/Tetur). Next, a BAM file for the four samples of the ATH-S population and for the eight samples of the AN-CR population (both AN-CR and tAN-CR) was generated by merging the individual BAM files of the ATH-S and tAN-CR samples that were created during the STAR alignment (see paragraph above), respectively. Subsequently, this merged BAM file was used to generate ATH-S and AN-CR Ace consensus sequences using the “mpileup” and “call” command of the samtools 1.4.1 and bcftools 1.3.1 software (43), respectively, and the Varroa genome as reference (GCF_002443255.1). The resulting FASTQ file was then converted to a FASTA file using the seqtk 1.2 software (using default Illumina settings: “-aQ 64 -q 20” as suggested on https://github.com/lh3/seqtk). Last, we assembled a de novo transcriptome using CLC Genomics workbench 11 (Qiagen) with default settings, and using 24 million random subsampled, bee virus filtered reads (8 million per sample) from ATH-S2-ATH-S4 and 24 million random, bee virus filtered reads (3.42 million per sample) from AN-CR1-AN-CR4 and tAN-CR1-tAN-CR3 (ATH-S1 and tAN-CR4 were not used for de novo assembly as these samples had less than eight million reads) (SI Appendix, Dataset S2).
Cytochrome P450 (CYP) Gene Annotation.
The predicted V. destructor CYP sequences from the National Center for Biotechnology Information (NCBI) annotation release 100 were manually curated for accuracy and completeness. Seven were corrected, including an artifactual fusion of three CYPs. In addition, one full-length CYP was found in our transcriptome assembly, and, while not predicted in the NCBI RefSeq models, it was present in the genome as an intronless gene. This resulted in a CYPome of 26 full-length genes, for which official CYP names were obtained from Dr. D. R. Nelson (University of Tennessee, Memphis, TN). All CYP genes were found in the de novo transcriptome assembly (Dataset S1). Six partial CYP sequences and CYP pseudogenes were also found and were excluded from further analysis. Full-length CYP sequences were aligned by MUSCLE (44), a phylogeny obtained by PhyML at http://www.phylogeny.fr, and the tree was drawn and annotated with FigTree v1.4.4. (http://tree.bio.ed.ac.uk/).
RNAi Functional Validation Assays.
Reverse genetic experiments using submersion in double-stranded RNA (dsRNA) were performed according to Campbell et al. (28). Briefly, PCR was performed on complementary DNA (cDNA) from AN-CR mites using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) following the manufacturer’s instructions, with specific primer pairs, designed using NCBI Primer BLAST to produce a product with a length of 400 to 600 bp and a guanine-cytosine (GC) content of 20 to 50%, that also introduce a T7 promoter sequence: dsGFP (control), dsCYP4EP4, and dsCYP3012A6 (SI Appendix, Table S2). The dsRNA fragments were carefully designed based on the analysis of the Varroa CYPome to ensure maximum specificity against target genes, but not against other CYP clan 4 members. dsRNA was created by using the Megascript Kit from Ambion and the T7 RNA polymerase with a 16-h 37 °C incubation, following the manufacturer’s instructions. The dsRNA was cleaned using a MegaClear Transcription Clean-Up kit (Ambion). The resultant dsRNA product was analyzed using a Nanodrop spectrometer (Nanodrop Technologies). Batches of 5 female Varroa mites were immersed into dsRNA (2.5 μg/μL) for 6 h. Eight biological experiments were performed with each dsRNA. As a control, nonendogenous GFP dsRNA was used at the same concentration.
Quantitative Real-Time PCR.
The efficiency of the dsRNA assays was validated by quantitative PCR (qPCR). RNA was extracted in a similar way as for Illumina RNA-seq experiments and subsequently used for cDNA synthesis, using SuperScript III reverse transcriptase and Oligo-dT 20 primers (Invitrogen, Athens, Greece). Amplification reactions of 10 μL final volume were performed on a CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA) and Kapa SYBR FAST qPCR Master Mix (Kapa-Biosystems), using the primers listed in SI Appendix, Table S5. The thermal protocol consisted of a polymerase activation step at 95 °C for 3 min and 40 cycles of denaturation and annealing/extension steps at 95 °C for 10 s and 60 °C for 45 s, followed by a melting curve analysis step. Two housekeeping genes, NADH (LOC111249888) and 18S ribosomal RNA (rRNA) (LOC111253957), were used as reference genes for normalization (45). A 10-fold dilution series of pooled cDNA was used to assess the efficiency of the qPCR reaction for each gene-specific primer pair. A no template control (NTC) was included to detect possible contamination, and a melting curve analysis was done in order to check the presence of a unique PCR product. Experiments were performed with four biological replicates and two technical replicates for each reaction.
Supplementary Material
Acknowledgments
We thank Robert Greenhalgh and Prof. Richard M. Clark (University of Utah) for help with sequencing; and David R. Nelson (University of Tennessee) for naming the CYP genes. We thank beekeepers John Rerras (Andros, Greece), Dimitris Lazarakis (Sparti, Greece), and Ariadni Kritikou (Naxos, Greece) for providing Varroa samples from their beehives for the bioassays; and Dr. Aris Ilias (Institute of Molecular Biology and Biotechnology of the Foundation for Research and Technology Hellas) for helping with the analysis of bioassay data. This research has been financed by Greek national funds through the Public Investments Program of the General Secretariat for Research and Technology, under the Emblematic Αction “The Bee Road” (project code: 2018ΣE01300000). S.V. was supported by Greece and the European Union (European Social Fund) through the Operational Programme “Human Resources Development, Education and Lifelong Learning” in the context of the project “Strengthening Human Resources Research Potential via Doctorate Research” (MIS-5000432), implemented by the State Scholarships Foundation. During this study, W.D. was a postdoctoral research fellow of the Research Foundation Flanders (Grant 1274917N). This work was in part supported by Research Foundation Flanders Grant G053815N and the European Research Council under the European Union's Horizon 2020 research and innovation program, Grant 772026-POLYADAPT.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020380118/-/DCSupplemental.
Data Availability.
Transcriptomics data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE153472). All other study data are included in the article and/or SI Appendix.
References
- 1.Johnson B. L., Lichtveld M. Y., Environmental Policy and Public Health (CRC Press, ed. 2, 2017). [Google Scholar]
- 2.Cressey D., The bitter battle over the world’s most popular insecticides. Nature 551, 156–158 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Rosenkranz P., Aumeier P., Ziegelmann B., Biology and control of Varroa destructor. J. Invertebr. Pathol. 103, S96–S119 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Traynor K. S., et al., Varroa destructor: A complex parasite, crippling honey bees worldwide. Trends Parasitol. 36, 592–606 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Cressey D., EU states lose up to one-third of honeybees per year. Nature 15016, 10.1038/nature.2014.15016 (2014). [DOI] [Google Scholar]
- 6.Francis R. M., Nielsen S. L., Kryger P., Varroa-virus interaction in collapsing honey bee colonies. PLoS One 8, e57540 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y. P., Siede R., “Honey Bee Viruses” in Advances in Virus Research, Maramorosch K., Shatkin A. J., Murphy F. A., Eds. (Academic Press, 2007), 70, pp. 33–80. [DOI] [PubMed] [Google Scholar]
- 8.Aliano N. P., Ellis M. D., Siegfried B. D., Acute contact toxicity of oxalic acid to Varroa destructor (Acari: Varroidae) and their Apis mellifera (Hymenoptera: Apidae) hosts in laboratory bioassays. J. Econ. Entomol. 99, 1579–1582 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Damiani N., Gende L. B., Bailac P., Marcangeli J. A., Eguaras M. J., Acaricidal and insecticidal activity of essential oils on Varroa destructor (Acari: Varroidae) and Apis mellifera (Hymenoptera: Apidae). Parasitol. Res. 106, 145–152 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Vanengelsdorp D., et al., Colony collapse disorder: A descriptive study. PLoS One 4, e6481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeschke P., Propesticides and their use as agrochemicals. Pest Manag. Sci. 72, 210–225 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Maggi M. D., Ruffinengo S. R., Gende L. B., Eguaras M. J., Sardella N. H., LC50 baseline levels of amitraz, coumaphos, fluvalinate and flumethrin in populations of Varroa destructor from Buenos Aires Province, Argentina. J. Apic. Res. 47, 292–295 (2008). [Google Scholar]
- 13.González-Cabrera J., et al., Novel mutations in the voltage-gated sodium channel of pyrethroid-resistant varroa destructor populations from the Southeastern USA. PLoS One 11, e0155332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Cabrera J., Davies T. G., Field L. M., Kennedy P. J., Williamson M. S., An amino acid substitution (L925V) associated with resistance to pyrethroids in Varroa destructor. PLoS One 8, e82941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farjamfar M., Saboori A., González-Cabrera J., Hernández Rodríguez C. S., Genetic variability and pyrethroid susceptibility of the parasitic honey bee mite Varroa destructor (Acari: Varroidae) in Iran. Exp. Appl. Acarol. 76, 139–148 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Panini M., et al., Pyrethroid resistance in Italian populations of the mite Varroa destructor: A focus on the Lombardy region. Bull. Insectol. 72, 227–232 (2019). [Google Scholar]
- 17.Stara J., et al., Spatio-temporal dynamics of Varroa destructor resistance to tau-fluvalinate in Czechia, associated with L925V sodium channel point mutation. Pest Manag. Sci. 75, 1287–1294 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Hubert J., et al., Point mutations in the sodium channel gene conferring tau-fluvalinate resistance in Varroa destructor. Pest Manag. Sci. 70, 889–894 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez S., Otero-Colina G., Pardio-Sedas V., Villanueva-Jimenez J., Resistance to amitraz and flumethrin in Varroa destructor populations from Veracruz, Mexico. J. Apic. Res. 44, 124–125 (2005). [Google Scholar]
- 20.Maggi M. D., Ruffinengo S. R., Negri P., Eguaras M. J., Resistance phenomena to amitraz from populations of the ectoparasitic mite Varroa destructor of Argentina. Parasitol. Res. 107, 1189–1192 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Pettis J. S., A scientific note on Varroa destructor resistance to coumaphos in the United States. Adipologie 35, 91–92 (2004). [Google Scholar]
- 22.Van Leeuwen T., Dermauw W., The molecular evolution of xenobiotic metabolism and resistance in chelicerate mites. Annu. Rev. Entomol. 61, 475–498 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Corley S. W., et al., Mutation in the RmβAOR gene is associated with amitraz resistance in the cattle tick Rhipicephalus microplus. Proc. Natl. Acad. Sci. U.S.A. 110, 16772–16777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bull D. L., Ahrens E. H., Metabolism of coumaphos in susceptible and resistant strains of Boophilus microplus (Acari: Ixodidae). J. Med. Entomol. 25, 94–98 (1988). [DOI] [PubMed] [Google Scholar]
- 25.Li A. Y., Davey R. B., Miller R. J., George J. E., Resistance to coumaphos and diazinon in Boophilus microplus (Acari: Ixodidae) and evidence for the involvement of an oxidative detoxification mechanism. J. Med. Entomol. 40, 482–490 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Van Leeuwen T., Vontas J., Tsagkarakou A., Dermauw W., Tirry L., Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochem. Mol. Biol. 40, 563–572 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Techer M. A., et al., Divergent evolutionary trajectories following speciation in two ectoparasitic honey bee mites. Commun. Biol. 2, 357 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell E. M., Budge G. E., Bowman A. S., Gene-knockdown in the honey bee mite Varroa destructor by a non-invasive approach: Studies on a glutathione S-transferase. Parasit. Vectors 3, 73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Temeyer K. B., Schlechte K. G., McDonough W. P., Baculoviral expression of presumptive OP-resistance mutations in BmAChE1 of Rhipicephalus (boophilus) microplus (Ixodida: Ixodidae) and biochemical resistance to OP inhibition. J. Med. Entomol. 56, 1318–1323 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Dermauw W., Van Leeuwen T., Feyereisen R., Diversity and evolution of the P450 family in arthropods. Insect Biochem. Mol. Biol. 127, 103490 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Sabourault C., et al., Overproduction of a P450 that metabolizes diazinon is linked to a loss-of-function in the chromosome 2 ali-esterase (MdalphaE7) gene in resistant house flies. Insect Mol. Biol. 10, 609–618 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Guzov V. M., Unnithan G. C., Chernogolov A. A., Feyereisen R., CYP12A1, a mitochondrial cytochrome P450 from the house fly. Arch. Biochem. Biophys. 359, 231–240 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Lee S. H., et al., Decreased detoxification genes and genome size make the human body louse an efficient model to study xenobiotic metabolism. Insect Mol. Biol. 19, 599–615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humble J. L., et al., Genome-wide survey of cytochrome P450 genes in the salmon louse Lepeophtheirus salmonis (Krøyer, 1837). Parasit. Vectors 12, 563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konno T., Hodgson E., Dauterman W. C., Studies on methyl parathion resistance in Heliothis virescens. Pestic. Biochem. Physiol. 33, 189–199 (1989). [Google Scholar]
- 36.Adolfi A., et al., Functional genetic validation of key genes conferring insecticide resistance in the major African malaria vector, Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 116, 25764–25772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pimprale S. S., Besco C. L., Bryson P. K., Brown T. M., Increased susceptibility of pyrethroid-resistant tobacco budworm (Lepidoptera: Noctuidae) to chlorfenapyr. J. Econ. Entomol. 90, 49–54 (1997). [Google Scholar]
- 38.Van Leeuwen T., Van Pottelberge S., Tirry L., Comparative acaricide susceptibility and detoxifying enzyme activities in field-collected resistant and susceptible strains of Tetranychus urticae. Pest Manag. Sci. 61, 499–507 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Leatherbarrow R. J., GraFit Version 3.0. GraFit Version 3.0 (Erithacus Software Ltd., Staines, UK, 1992). [Google Scholar]
- 40.Andrews S., Fast QC, A Quality Control tool for High Throughput Sequence Data, version 0.11.5. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 41.Love M. I., Anders S., Kim V., Huber W., RNA-seq workflow: Gene-level exploratory analysis and differential expression. F1000 Res. 4, 1070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Law C. W., Chen Y., Shi W., Smyth G. K., voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell E. M., McIntosh C. H., Bowman A. S., A toolbox for quantitative gene expression in Varroa destructor: RNA degradation in field samples and Systematic analysis of reference gene Stability. PLoS One 11, e0155640 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomics data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE153472). All other study data are included in the article and/or SI Appendix.