Significance
Citrus Huanglongbing (HLB) is the most destructive citrus disease worldwide and has caused billions of dollars in annual production losses, threatening the entire citrus industry. Despite extensive research efforts, there are still no effective management tools to treat HLB-positive trees or to prevent new infections. Current HLB management strategies include chemical application of insecticides and traditional heat-sensitive antibiotics, which pose threats to humans, animal health, and the environment, and likely generate drug resistant insects and microbes. Here, we identified a novel class of stable antimicrobial peptides (SAMPs) from Australian finger lime and other HLB-tolerant citrus close relatives, which has dual functions of inhibiting CLas growth in HLB-positive trees and activating host immunity to prevent new infections.
Keywords: citrus Huanglongbing, antimicrobial peptide, Candidatus Liberibacter, plant immunity, Microcitrus
Abstract
Citrus Huanglongbing (HLB), caused by a vector-transmitted phloem-limited bacterium Candidatus Liberibacter asiaticus (CLas), is the most devastating citrus disease worldwide. Currently, there are no effective strategies to prevent infection or to cure HLB-positive trees. Here, using comparative analysis between HLB-sensitive citrus cultivars and HLB-tolerant citrus hybrids and relatives, we identified a novel class of stable antimicrobial peptides (SAMPs). The SAMP from Microcitrus australiasica can rapidly kill Liberibacter crescens (Lcr), a culturable Liberibacter strain, and inhibit infections of CLas and CL. solanacearum in plants. In controlled greenhouse trials, SAMP not only effectively reduced CLas titer and disease symptoms in HLB-positive trees but also induced innate immunity to prevent and inhibit infections. Importantly, unlike antibiotics, SAMP is heat stable, making it better suited for field applications. Spray-applied SAMP was taken up by citrus leaves, stayed stable inside the plants for at least a week, and moved systemically through the vascular system where CLas is located. We further demonstrate that SAMP is most effective on α-proteobacteria and causes rapid cytosol leakage and cell lysis. The α-helix-2 domain of SAMP is sufficient to kill Lcr. Future field trials will help determine the efficacy of SAMP in controlling HLB and the ideal mode of application.
Citrus Huanglongbing (HLB), also known as citrus greening, is caused by the vector-transmitted phloem-limited bacterium Candidatus Liberibacter asiaticus (CLas). It is the most destructive disease threatening citrus industries worldwide (1, 2), and thus far, no cure has been discovered. Current management strategies include insecticide application to control the transmission vector Asian citrus psyllids (ACP) and antibiotics treatment to inhibit CLas (3), but neither of these could control HLB effectively. Since the first report of HLB in Florida in 2005, citrus acreage and production in Florida decreased by 38% and 74%, respectively (2, 4). The disease has spread to most citrus-producing states, including Texas and California. Along with drastic losses in fruit production, increasing chemical applications to control the vector and the bacteria have raised costs significantly, making citrus production for growers unsustainable. In severely affected areas, such as Florida, effective therapy is demanded because disease eradication is impractical. In recently impacted areas, such as California, the focus remains on preventing new infections. Hence, innovative therapeutic and preventive strategies to combat this lethal citrus disease are urgently needed to ensure the survival of the citrus industry.
One of the most effective and ecofriendly strategies to combat pathogen infection is to utilize existing plant innate immunity–related genes from disease resistant or tolerant varieties for plant protection. Upon pathogen infection, plant defense response genes undergo expression reprogramming to trigger plant innate immunity. Plant endogenous small RNAs play a pivotal role in this regulatory process (5, 6). In addition, primary pathogen infection or application of some phytohormone analogs and chemicals, such as salicylic acid (SA) analogs, could induce systemic acquired resistance or defense priming in plants, which can promote faster and stronger host immune responses upon subsequent pathogen challenges (7, 8).
Although all commercially important citrus varieties are susceptible to HLB (9, 10), HLB tolerance has been observed in some hybrids [e.g., US-942 and Sydney hybrid 72 (11, 12)] and close citrus relatives (e.g., Microcitrus australiasica, Eremocitrus glauca, and Poncirus trifoliata) (13). By comparative expression analysis of small RNAs and messenger RNAs (mRNAs) between HLB-sensitive cultivars and HLB-tolerant citrus hybrids and relatives (11, 12), we identified a list of candidate natural defense genes potentially responsible for HLB tolerance (14). One of the candidate regulators is a novel antimicrobial peptide (AMP), which we named “stable antimicrobial peptide” (SAMP). Here, we demonstrate that SAMP not only has the antimicrobial activity but also has the priming activity and can induce citrus systemic defense responses. This dual-functional SAMP can reduce CLas titer and suppress disease symptoms in HLB-positive trees and activate plant systemic defense responses against new infection.
Results
Identification and Characterization of a Novel Class of AMPs from HLB-Tolerant Citrus Relatives.
Through the comparative expression analysis of small RNAs and mRNAs between HLB-sensitive cultivars, HLB-tolerant citrus hybrid US-942 (Poncirus trifoliata × Citrus reticulata), and microcitrus Sydney hybrid 72 (Syd 72, Microcitrus virgate from M. australis × M. australasica) (14), we identified a list of candidate plant immune response genes that are potentially responsible for HLB tolerance. One candidate gene encodes a 67 amino acid (aa) peptide containing two predicted α-helix domains (15), which is homologous to a 109-aa Arabidopsis heat-stable protein HS1 with antimicrobial and antifungal activity (16). Here, we named this peptide SAMP.
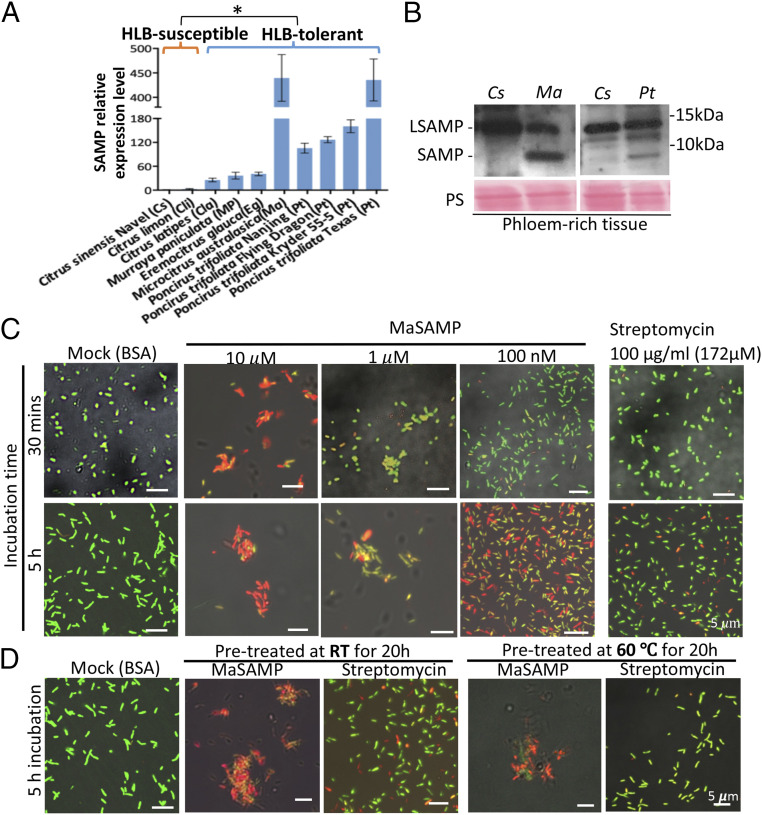
To determine whether SAMP is associated with HLB tolerance, we cloned SAMP genes from HLB-tolerant citrus relatives, including the Australian finger lime (M. australasica), Australian desert lime (E. glauca), Hawaiian mock orange (Murraya paniculata), Khasi papeda (Citrus latipes), and seven trifoliate oranges (P. trifoliate). All of these citrus relatives have a long (109-aa) and at least one short (67-aa) version of SAMPs (SI Appendix, Figs. S1B and S2), whereas HLB-susceptible citrus varieties Citrus clementine (Cc) and Citrus sinensis (Cs) have only the long SAMP (LSAMP) with 118 aa and 109 aa in length, respectively, based on the Citrus Genome Database (https://www.citrusgenomedb.org) (SI Appendix, Fig. S2). SAMP has significantly higher mRNA levels in both HLB-tolerant hybrids US-942 and Syd 72 (SI Appendix, Fig. S1A) than LSAMP in the HLB-susceptible control trees. The SAMPs share high sequence similarity to the C-terminal domain of the LSAMPs. In finger lime (Ma), desert lime (Eg), trifoliate (Pt) Flying dragon, and Pt Florida, we identified two closely related SAMPs. Since the genome sequences of these citrus relatives are currently unavailable, there may be more SAMPs encoded in these genomes. We found that SAMP transcripts have a significantly higher mRNA expression level in HLB-tolerant varieties compared to LSAMP in the HLB-susceptible varieties (Fig. 1A).
Fig. 1.
A heat-stable antimicrobial peptide, SAMP, identified from HLB-tolerant citrus relatives has bactericidal activity. (A) The expression level of SAMPs in different citrus and citrus relatives was analyzed by qRT-PCR and normalized to Actin. The significant difference is indicated by *P < 0.05 analyzed by t test. (B) SAMPs were detected by Western blot analysis using an anti-MaSAMP antibody in phloem-rich tissue bark peels of Cs, Ma, and Pt. Ponceas staining (PS) was used as the loading control. (C and D) The bactericidal activity of different concentrations (C) or heat pretreatment (D) of MaSAMP solution was examined by Lcr viability/cytotoxicity assays. Streptomycin was used for comparative analysis. The buffer with 10 μM BSA was used as the control. The DMAO (green dye) and EthD-III (red dye) stains the live and dead bacteria, respectively.
Next, to probe if SAMP is present in phloem, where CLas is located, we generated a native antibody against SAMP and detected the 6.7-kD short version in the phloem-rich tissue-bark peels of HLB-tolerant Ma and Pt but not in the susceptible Cs (Fig. 1B). These results further support that the SAMPs are likely associated with the HLB-tolerance trait.
SAMP Has Bactericidal Activity and Is Heat Stable.
To test the ability of SAMPs to treat Liberibacter diseases, and to identify the most effective SAMP variant, we developed a rapid functional screening method, using a C. Liberibacter solanacearum (CLso)/potato psyllid/Nicotiana benthamiana interaction system to mimic the natural transmission and infection circuit of the HLB complex (14). We screened SAMPs from several citrus relatives and found that the SAMP from M. australasica Australian finger lime (MaSAMP) had the strongest effect on both suppressing CLso disease and inhibiting bacterial growth in plants (SI Appendix, Fig. S3), whereas the LSAMP has no or very weak effect. Thus, MaSAMP was used for the subsequent analyses.
To directly determine the bactericidal activity of MaSAMP on Liberbacter spp., we developed a viability/cytotoxicity assay of Liberibacter crescens (Lcr), a close culturable relative of the CLas and CLso (17–19). Using this assay, we found that 10 μM MaSAMP can kill the bacterial cells and induce aggregation as rapidly as 30 min after treatment (Fig. 1C). Lower MaSAMP concentrations of 1 μM or 100 nM can still kill the bacterium within 5 h after treatment (Fig. 1C). Furthermore, we found that MaSAMP, at a concentration as low as 100 nM, is more efficient at killing bacteria than the bactericidal antibiotic Streptomycin at a concentration as high as 172 μM (100 μg/mL) after 5 h of treatment (Fig. 1C). Streptomycin commonly needs longer time to kill the bacteria.
While the heat sensitivity of antibiotics is a major drawback for controlling CLas in citrus fields, we found that SAMPs are surprisingly heat stable. A prolonged exposure to extreme temperatures of 60 °C for 20 h had minimal effect on MaSAMP, which still retained most of its bactericidal activity (Fig. 1D), whereas Streptomycin completely lost its antibacterial activity following the same temperature incubation (Fig. 1D). Thus, SAMP is a heat stable plant-derived AMP that can directly kill Lcr and suppress CLso in plants.
SAMP Suppresses CLas in HLB-Positive Trees.
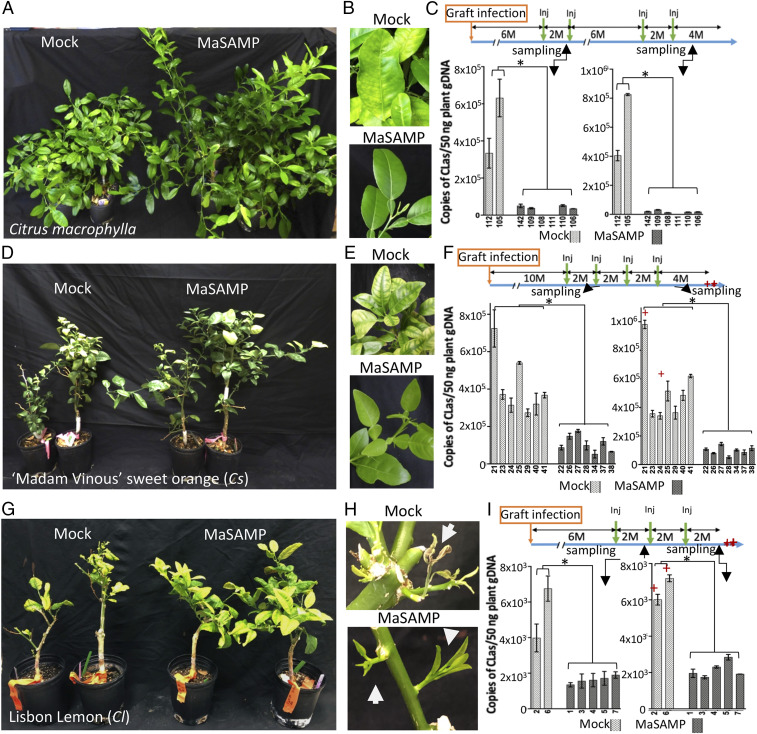
To determine whether MaSAMP can also suppress CLas in citrus trees, we used the pneumatic trunk injection method to deliver the MaSAMP solution into the HLB-positive citrus trees (SI Appendix, Fig. S4). In our first experiment, we obtained eight CLas-positive Citrus macrophylla with similar bacterial titer and disease symptoms for the treatment. Six trees were injected with MaSAMP (10 μM) and two trees were injected with the mock solution (Fig. 2). Eight wk after the first dose of MaSAMP injection, leaves were collected from the new flush on each tree for CLas titer analysis. The disease symptoms and the bacterial titer in all six treated trees were drastically reduced in comparison to the mock-treated plants (Fig. 2 A–C). Furthermore, CLas was undetectable in one of the MaSAMP-treated trees (number 111, Fig. 2C). After the first two initial injections with 2 mo between the treatment, we monitored CLas titer on the trees without additional treatment for 6 mo and observed that the CLas titer started to increase after 5 mo, although the new leaves still looked healthy. CLas remained undetectable in tree 111. We then injected MaSAMP solution two more times (with 2 mo between treatments) and observed a rapid decrease in CLas titer (Fig. 2C). Moreover, new flush from the MaSAMP-treated plants displayed no HLB symptoms, whereas new flush from the mock-treated plants continued to display yellow striping symptoms (Fig. 2B). These results suggest that MaSAMP treatments at 2-mo intervals are effective to control HLB.
Fig. 2.
MaSAMP suppresses CLas in different HLB-positive citrus varieties. Citrus macrophylla (A–C, after four injection doses), 'Madam Vinous' sweet orange (D–F, after four injection doses), and 'Lisbon' Lemon (G–I, after three injection doses) were injected with buffer (mock) or MaSAMP (10 μM). B, E, and H show new leaves or flushes in trees shown in A, D, and G, respectively. The treatment programs for A, D, and G were indicated in a timeline with months (M) at the upper of C, F, and I, respectively. The time points of injection (Inj, green arrow) and time of tree death (red star) are indicated. The CLas titer of individual trees in test A, D, and G at the indicated sampling time was measured by qPCR on CLas 16S rDNA using the USDA standard protocol (C, F, and I, Lower). Significant difference is indicated by *P < 0.01 analyzed by t test.
In our next round of testing, we used 14 HLB-positive 'Madam Vinous' sweet oranges (Cs) 10 mo after grafting inoculation. This set of trees had severe HLB declining symptoms with similar and high CLas titer. After MaSAMP treatments, the trees had increased growth and developed symptomless new flushes, while mock trees continued to exhibit symptomatic flushes (Fig. 2 D and E). Two mo after the first and fourth treatment, the CLas titer was examined. The titer was reduced after the first treatment and remained low after the fourth treatment in MaSAMP-treated trees, while it continued to increase in the mock-treated trees, two of which eventually died from HLB 4 mo after the last mock injection (Fig. 2F). In our third round of testing, we used seven HLB-positive 'Lisbon' Lemon trees with similar CLas titer. The two mock-treated trees were unable to produce new leaves and died 6 mo after the first injection, whereas the MaSAMP-treated trees had enhanced growth and exhibited healthy new flush (Fig. 2 G and H). Comparing the CLas titer 2 mo after first and third treatment, we found that the MaSAMP treatment could suppress the CLas growing in the lemon trees compared to the mock-treated trees (Fig. 2I). Taken together, these results demonstrate across three trials that SAMP injection can suppress CLas titer in three different HLB-susceptible citrus varieties and can cause trees in declining health to recover.
SAMP Treatment Safeguards Healthy Citrus Trees from CLas Infection.
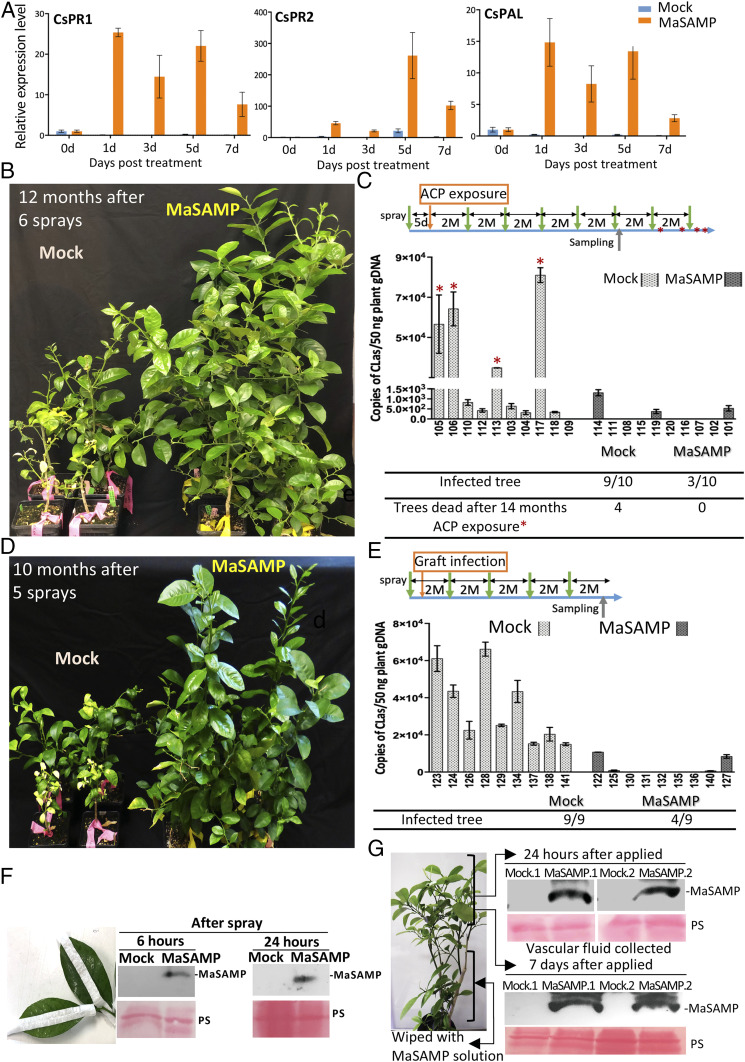
Protecting HLB-negative citrus trees and saplings from CLas infection is critical for managing HLB. Establishment of defense priming in plants can promote faster and/or stronger host immune responses upon pathogen challenges (7, 8). To determine whether MaSAMP has priming activity, we applied it by foliar spray to Nb, tomato, and citrus plants. We found that MaSAMP application can induce the expression of a set of defense genes and activate systemic defense responses in Nb and tomato (SI Appendix, Fig. S5). Similarly, MaSAMP clearly triggered prolonged induction of defense response genes such as pathogenesis-related proteins PR1 and PR2 and an enzyme of SA biosynthesis and phenyl propanoid pathways, phenylalanine ammonia-lyase1 (PAL), in citrus trees (Fig. 3A) (7). Thus, SAMP can potentially “vaccinate” uninfected citrus trees and induce defense responses to combat against HLB and maybe other pathogen threats.
Fig. 3.
MaSAMP protects the healthy citrus trees from CLas infection. (A) The expression level of defense marker genes in the MaSAMP-treated 'Valencia' sweet orange trees was highly induced over a prolonged time course. The relative expression level was analyzed by qRT-PCR and normalized to Actin. (B–E) Two sets of the 'Madam Vinous' sweet orange trees were foliar-sprayed with buffer (mock) or MaSAMP solution (10 μM) before ACP exposure (B) or graft infection (D). The treatment programs for B and D were indicated in a timeline with months (M) at the upper of C and E, respectively. The spray time points (green arrow) and the time when trees died (red star) were indicated. The CLas titer of individual trees in test B and D at the indicated sampling time was shown in the middle of C and E, respectively. The tables of the lower panel of C and E represent the number of the infected and dead trees. (F) MaSAMP was detected with Western blot by anti-MaSAMP antibody in the vascular fluid collected from MaSAMP-sprayed leaves. The midveins were tap-protected before spraying to avoid direct contact of MaSAMP with midveins. (G) Systemic leaves of trees that have lower leaves wiped with MaSAMP solution using a cotton ball were collected at 24 h or 7 d after MaSAMP application. The MaSAMP was detected by Western blot in the vascular fluid of the midvein from systemic untreated leaves. Ponceas staining (PS) was used as the loading control.
To identify the key signaling components involved in the SAMP-induced immune responses and to elucidate the underlying mechanism, we selected several master regulators of plant immune responses, including nonexpressor of pathogenesis-related gene 1 (NPR1) (20), suppressor of G2 allele of skp1 (SGT1) (21), and the coreceptor of several receptor-like kinases involved in plant defense, BRI1-associated receptor kinase1 (BAK1)/somatic embryogenesis receptor kinase3 (SERK3) (22), to assess their role in SAMP-triggered immunity. We performed virus-induced gene silencing (VIGS) (23) to knock down these immune regulators in Nb plants and then examined the PR gene expression after MaSAMP treatment (SI Appendix, Fig. S6). The results showed that silencing of NPR1 and SGT1 largely abolished SAMP-induced PR gene expression (SI Appendix, Fig. S6A), indicating that SAMP-triggered plant immunity is NPR1- and SGT1-dependent. However, VIGS of both BAK1 homologs SERK3A and SERK3B in Nb didn’t have an effect, suggesting that BAK1/SERK3 are not required for SAMP recognition and signaling.
To test the protection ability of SAMP on citrus trees, we applied the MaSAMP solution or buffer as mock treatment by foliar spray onto 20 young healthy 'Madam Vinous' sweet orange trees. At 5 d after treatment, the trees were exposed to ACP carrying CLas under the “no choice feeding” condition for 21 d. We subsequently treated trees with MaSAMP solution by foliar spray every 2 mo. At 12 mo after inoculation, the trees sprayed with MaSAMP exhibited enhanced growth compared to the mock-treated trees (Fig. 3B). At 14 mo after inoculation, 9 of 10 mock-treated trees tested CLas positive and 4 died (Fig. 3C). In the MaSAMP-treated trees, only three tested positive, each with a very low CLas titer (Fig. 3C). For the next trial, we grafted HLB-positive budwood as inoculum onto nine MaSAMP-treated and nine mock-treated HLB-negative rootstock trees. Similar to the ACP-mediated inoculation, the MaSAMP-treated trees had enhanced growth compared to the mock treatment (Fig. 3D). At 10 mo post grafting, all of the mock-treated trees were HLB positive, whereas only four of nine MaSAMP-treated trees were HLB positive, with significantly low CLas titer (Fig. 3E).
Foliar-spray delivery is a common and practical method for field applications. To determine whether SAMP enters the citrus vascular system following foliar application, we cut out the covered midvein from leaves after spray application and collected the vascular fluid. We detected vascular SAMP uptake as early as 6 h post-spray (Fig. 3F). To eliminate potential mist contamination, we next manually wiped MaSAMP solution with cotton balls onto the lower leaves of the citrus tree and collected the vascular fluid from the midvein of upper untreated leaves. We detected MaSAMP was systemically transported to the upper leaves as early as 24 h post-treatment, and the transported MaSAMP remained stable in vasculature for at least 7 d (Fig. 3G). About 9.8 to 22 μM of MaSAMP was detected in the vascular fluid collected from leaf midribs at 24 h post-treatment (SI Appendix, Fig. S7A), which we found to be a sufficient concentration to rapidly kill Lcr in the viability assay (SI Appendix, Fig. S8). In addition, the amount of MaSAMP detected in the citrus leaves was negatively correlated with the CLas titer in the leaves, which further supports that MaSAMP inhibits CLas (SI Appendix, Fig. S7B). Thus, SAMP is rapidly taken up by citrus leaves and moves systemically in the trees through vasculature, where CLas is present. The existence of SAMP in the vascular system is long lasting.
SAMP Is Effective Against α-Proteobacteria and Causes Cell Leakage and Lysis.
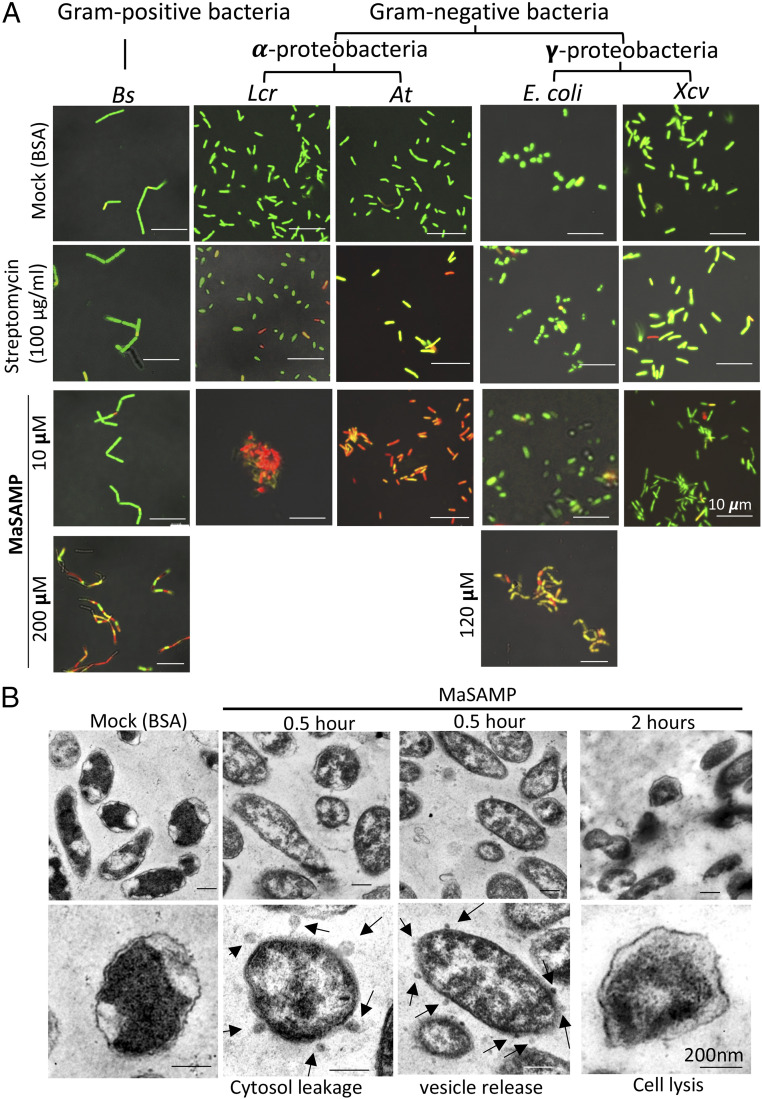
Many AMPs exhibit antimicrobial activity across an array of microorganisms (24). To better understand the range of SAMP’s antimicrobial activity, we performed viability assays on a variety of bacteria including gram-positive Bacillus subtillis (Bs), gram-negative α-proteobacteria including Lcr and Agrobacterium tumefaciens (At), and γ-proteobacteria including Escherichia coli and Xanthomonas campestris pv. Vesicatoria (Xcv). MaSAMP has strong antibacterial activity against Lcr and At at 10 μM but not against Bs, E. coli, or Xcv (Fig. 4A). The minimum inhibitory concentration of MaSAMP to inhibit Lcr and At is about 10 μM (SI Appendix, Fig. S8). High MaSAMP concentrations of 200 μM and 120 μM were needed to significantly inhibit Bs and E. coli, respectively (Fig. 4A). With the limited number of bacteria strains tested, we found that SAMP may be more effective on α-proteobacteria.
Fig. 4.
MaSAMP is most effective on α-proteobacteria. (A) Bacteria viability/cytotoxicity assays of MaSAMP were performed on Bacillus subtilis (Bs), Liberibacter crescens (Lcr), Agrobacterium tumefaciens (At), Escherichia coli (E. coli), and Xanthomonas campestris pv. Vesicatoria (Xcv). The green and red cells indicate the live and dead cells, respectively. Pictures were taken at 5 h post-treatment. (B) TEM image of Lcr cells treated with 10 μM MaSAMP or BSA (mock) showed cytosol leakage and vesicle releasing 0.5 h post-treatment. Cell lysis was observed at 2 h post-treatment. Cytosol leakage or vesicle release are indicated by the black arrows.
To understand the mechanism of MaSAMP bactericidal activity, morphological changes of Lcr after MaSAMP treatment were observed using transmission electron microscopy. Application of 10 μM MaSAMP to Lcr caused cytosol leakage and the release of small extracellular vesicles after 30 min of incubation (Fig. 4B). The Lcr cells were lysed within 2 h of incubation. Vesicle release was potentially caused by compromised maintenance of membrane lipid asymmetry, induced lipopolysaccharide modifications, or accumulation of misfolded proteins in the outer membrane (25). We isolated the membrane fraction from the MaSAMP-treated Lcr and detected the enrichment of MaSAMP in the outer membrane fraction as compared with the inner membrane fraction (SI Appendix, Fig. S9). Thus, MaSAMP likely disrupts mainly the outer membrane of Lcr and breaks the bacterial cells, which leads to cell lysis.
The Second α-Helix of SAMP Is the Major Bactericidal Motif.
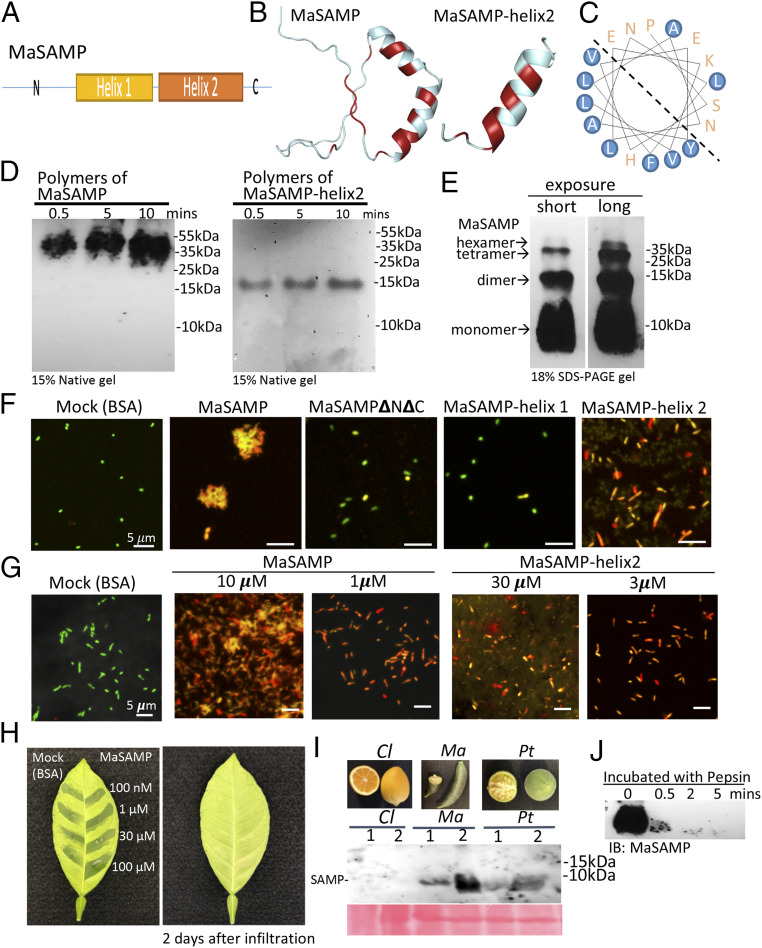
To understand the mechanism of action of SAMP, we modeled its structure, which contains two short α-helical fragments connected by a proline hinge region with a loose N and C terminus (Fig. 5 A and B) (26). The amphipathic helix2 has the hydrophobic residues facing one side (Fig. 5C). We detected that MaSAMP forms polymers (probably hexamers based on the molecular weight) in the native gel (Fig. 5D), suggesting that this peptide likely forms a pore-like structure. Different sodium dodecyl sufate (SDS)-resistant MaSAMP oligomers were observed in the SDS denaturing gel (Fig. 5E), indicating that the oligomers of SAMP are rather stable. To determine the critical domain of SAMP for its function, we generated a series of truncated versions of MaSAMP, including the double-helix hairpin (MaSAMP▵N▵C), α-helix1 (MaSAMP-helix1), and α-helix2 only (MaSAMP-helix 2), to test their bactericidal activity. The results indicate that α-helix2 is largely responsible for the antibacterial activity, though full-length MaSAMP activity is slightly higher (Fig. 5 F and G). By tripling the amount, MaSAMP-helix2 can reach up to 90% activity of the full-length MaSAMP (Fig. 5G and SI Appendix, Fig. S10). Furthermore, we also detected the polymers (again most likely hexamers based on the molecular weight) using only the helix2 domain (MaSAMP-helix2) (Fig. 5D), further suggesting that this peptide forms oligomers using its helix2 domain.
Fig. 5.
The α-helix2 domain of MaSAMP is the key bactericidal motif, and SAMP is present in fruits and rapidly degraded by pepsin. (A) The diagram of the SAMP structure. (B) The predicted structure of SAMP by the SWISS-MODEL. The hydrophobic residues are marked in red. (C) The helical wheel diagram of the α-helix2 domain was predicted. The hydrophobic residues are circled in blue. (D) MaSAMP and MaSAMP-helix2 domain form only polymers (likely hexamers) in the native PAGE gel. (E) MaSAMP forms SDS-resistant oligomers. (F and G) The bactericidal activity of various truncated MaSAMPs was examined using Lcr viability/cytotoxic assay. The green and red cells indicate the live and dead cells, respectively. (H) MaSAMP phytotoxicity was assessed by infiltrating different concentrations of MaSAMP or BSA solution into the leaf of sweet orange. (I) MaSAMP was detected by Western blot using the anti-MaSAMP antibody in the fruit tissue of Australian finger lime (Ma) and trifoliate orange (Pt) but not Lemon (Cl). The corresponding fruit pictures are shown in the upper panel. (J) MaSAMP was rapidly degraded after incubation with human pepsin over a time course.
Toxicity Assessment of SAMP.
Because SAMP is internalized by citrus, it is important to test its phytotoxicity. We injected different concentrations of MaSAMP solution directly into citrus leaves and found that MaSAMP has little phytotoxicity even at a concentration as high as 100 μM (Fig. 5H). Furthermore, we found that MaSAMP can be detected in fruit tissue of both HLB-tolerant Australian finger lime and trifoliate orange by Western blot analysis (Fig. 5I). MaSAMP is very sensitive to human endopeptidase Pepsin, a major gastric enzyme produced by stomach chief cells (Fig. 5J). Thus, MaSAMP in Australian finger lime has already been consumed by humans for hundreds of years and can be easily digested. These results suggest a low possibility of toxicity of SAMP on citrus and humans, although additional safety assessment tests are necessary for regulatory approval.
Discussion
HLB is the largest threat to the world citrus industry. Current methods to prevent infections and maintain productivity of HLB-infected trees include insecticidal control of the vector (27), antibacterial treatments (28–31), and nutrient supplements (32, 33). The overuse of insecticides and antibiotics is known to pose threats to human and animal health and to select for resistance in the target insect population (34). Furthermore, current bactericidal or bacteriostatic treatments mostly involve the spray of antibiotics, such as streptomycin and oxytetracycline, which are likely to select for antibiotic-resistant bacteria strains and can disrupt the citrus microbiome, ecosystem, and may further affect the effectiveness of these antibiotics for medical antibacterial treatment in humans and animals. On the contrary, SAMPs have a distinct mode of action and tend to disrupt the bacterial cell membrane through nonspecific mechanisms, making the emergence of resistant bacteria less likely (35, 36). Moreover, SAMP kills bacteria faster than antibiotics, which reduces bacterial generations and further lowers the possibility of evolved resistance (37). Most importantly, the heat stability of SAMP can provide a prolonged and durable effect in the field compared to heat-sensitive antibiotics.
The major bactericidal activity of SAMP is from the amphipathic α-helix2, which is different from most of the plant-derived cysteine-rich AMPs (38). Most known plant cystine-rich AMPs rely on proper disulfide bridges to form specific structures for their function (13). SAMP, however, is not a cystine-rich peptide—it only contains two cystine residues at its N terminus. The mode of action of SAMP is to form pore-like oligomers (likely hexamers) through its amphipathic α-helix2 domain, which can insert into the bacterial membrane and cause cytosol leakage and cell lysis. Pore-forming proteins often have amphipathic α-helices for membrane insertion or spanning (39–41). For example, the N-terminal α-helix of plant intracellular nucleotide-binding domain leucine-rich repeat-containing receptor (NLR) protein ZAR1 is oligomerized to form a funnel-like structure that associates with the plasma membrane and leads to plant cell death (42). In mammalian systems, gasdermin proteins trigger pyroptosis by lipid binding through its N-terminal amphipathic α-helix and protein oligomerization (43, 44). These α-helix domains function mostly within a big protein. The HR4Fei-0 in plants, which mediates the oligomerization of NLR RPP7, could form SDS-resistant oligomers with cytotoxicity when expressed in E. coli (45). The 22-aa amphipathic α-helix2 domain from SAMP shares no sequence similarity to any known animal or plant pore-forming proteins and can oligomerize and function by itself as a bactericide to attack bacterial membranes. Our findings revealed that SAMP is mainly associated with the outer membrane of Lcr. The outer membrane of gram-negative bacteria is primarily composed of a coat of lipopolysaccharides (glycolipids in the outer leaflet and phospholipids in the inner leaflet of membrane), which can be the target of SAMP (46). Different structures and modifications of lipopolysaccharides from α-proteobacteria and γ-proteobacteria could affect the antimicrobial efficiency of AMPs (47). The diversity and complexity of the membrane lipids in different classes of bacteria could affect the membrane integration and impact the activity of AMPs (48, 49). Here, we found SAMP is most effective on targeting cell membranes of gram-negative α-proteobacteria and causes cytosol leakage and cell lysis. The difference of membrane lipid composition and structures of α- and γ-proteobacteria and the major SAMP-binding molecules still needs further investigation.
SAMP not only kills α-proteobacteria cells—it can also prime plant immune responses to prevent/reduce infection. This activity is largely dependent on the master regulators of plant immunity, NPR1 and SGT1. NPR1 plays a central role in salicylic acid-dependent systemic acquired resistance (7) and activates the expression of downstream defense response genes (20). SGT1 is a cochaperone of heat shock protein 90 and a cofactor of the E3 ubiquitin ligase complex, and it is required for the signal transduction of many NLR protein–mediated effector-triggered immunity (ETI) (21, 50). We speculate that SAMP may act as a peptide ligand that is recognized by a receptor-like protein or a receptor-like kinase and activates defense responses. Although there are hundreds of receptor-like proteins or receptor-like kinases encoded in the plant genomes, some of them share common coreceptors, such as BAK1/SERK3, for their signal transduction and function. Arabidopsis BAK1 is not only a coreceptor for the pattern recognition receptor-like kinases flagellin-sensing 2 (FLS2), elongation factor-Tu receptor (EFR), and pep receptor 1 (PEPR1)/PEPR2—those activate pathogen-associated molecular pattern-triggered immunity (PTI)—but also a coreceptor for the brassinosteroid receptor BRI1 (22). Our results indicate that BAK1/SERK3 are not required for SAMP recognition and signaling. Future studies will help identify the receptor-like proteins that recognize SAMP to better understand the molecular mechanism of downstream defense signaling pathways.
In our greenhouse trials, SAMP has been shown to both treat HLB-positive trees and inhibit the emergence of new HLB infection in healthy trees. Field trials, which can take several years, are currently being initiated in Florida to confirm the efficacy of SAMP in controlling HLB. Field trials also include testing multiple peptide application methods for citrus growers to prevent and treat HLB.
Materials and Methods
Citrus Material, CLas Infection, and Sample Collection.
The citrus trees used in the experiments were Citrus macrophylla, 'Madam Vinous' sweet oranges (Citrus sinensis L.), and 'Lisbon' lemon scion (Citrus limon; Limonoira 8A; California Citrus Clonal Protection Program) on Carrizo rootstock. The plants were grown in round one-gallon pots in a greenhouse at 27 °C (± 1.5 °C) with supplemental lighting (high-pressure sodium lights; 16-h light/8-h dark photoperiod) at the University of California Davis’ Contained Research Facility. The plants were ∼6 mo old when they were graft inoculated or treated. For graft inoculation, one branch was selected to receive three T-bud grafts. The CLas (California Hacienda Heights isolate (51)-positive budwood for the grafts were buds taken from Citrus macrophylla, 'Madam Vinous' sweet oranges, and Lisbon lemon on Carrizo tested positive (cycle threshold [Ct] value of 21) using qPCR for the detection of CLas 16S ribosomal DNA using the US Department of Agriculture (USDA) standard protocol (52). The graft-infected HLB-positive Citrus macrophylla, 'Madam Vinous' sweet oranges, and Lisbon lemon have an average Ct value of 24, 23, and 30, respectively. All citrus experiments were conducted in accordance with state and federal guidelines regulating the culture, transport, and disposal of ACP and plant material associated with the plant bacterial pathogen CLas.
The citrus and citrus relatives collected from the Citrus Variety Collection at the University of California, Riverside, included the Australian finger lime (Microcitrus australasica, CRC3670), Australian desert lime (Eremocitrus glauca, CRC3463), Hawaiian mock orange (Murraya paniculata, CRC3171), trifoliate oranges (Poncirus trifoliate, CRC 2861, CRC3215, CRC3330A, CRC3345, CRC3888, CRC2862), and Khasi papeda(Citrus latipes, CRC3052).
CLas Detection in Citrus Trees.
New leaves from each branch (six to eight leaves in total from each tree) were collected at approximately two month intervals after inoculation, and DNA was extracted from dissected midribs following the cetyltrimethyl ammonium bromide protocol from the midvein of leaves (53). The extracted DNA was analyzed using qPCR for the presence of CLas, using the USDA standard protocol for CLas detection with primers and TaqMan probe designed against CLas 16S rDNA (52).
Bacteria Growth Conditions.
Lcr Strain BT-1 culture was grown in BM7 medium at 28 °C for 5 d at 100 rpm (18). Bs and Xcv were grown in Tryptic Soy broth (Becton Dickinson) overnight at 30 °C and 160 rpm. E. coli and At were grown in LB Broth (Becton Dickinson) overnight at 160 rpm at 37 °C and 28 °C, respectively.
Bacterial Viability/Cytotoxicity Assay.
Staining of living and dead bacteria was performed following the manufacturer’s protocol (Viability/Cytotoxicity Assay Kit for Bacteria Live and Dead Cells, Biotium). The 5-d-cultured Lcr bacterial cells and overnight-cultured Bs, Xcv, E. coli, and At bacterial cells were used for the assay. The cultures were centrifuged (7,000 g, 10 min, 22 °C) to pellet, resuspended, and washed with 0.85% NaCl solution three times. The bacterial cells were suspended in 0.85% NaCl solution, adjusted to OD600 1.0, and diluted 100-fold for staining. A 100× MaSAMP stock (1 M, 100 μM, and 10 μM in dimethylsulfoxide [DMSO]) was prepared to dilute the bacterial suspension to create the final MaSAMP concentrations of 10 μM, 1 μM, and 100 nM. At the end of treatment, the stained bacterial cell suspensions were concentrated 100-fold and observed with Leica SP5 confocal microscopy. Alternatively, the fluorescence intensity of stained bacterial cell suspension was measured with the Promega GloMax Discover Microplate Reader.
SAMP Injection or Foliar Spray.
Trunk injections were performed using a custom pneumatic trunk injection pump. One injection port (2 mm diameter) per tree was made by drilling 2 to 3 mm into the trunk with a drill bit positioned at ∼5 to 10 cm above the root. Ports were properly sealed with injection port tips. The injection port tip was connected to a syringe containing a solution by a tube. The plungers on the syringes were pushed by a pump with a pressure of 20 psi per tree. The MaSAMP stock solution (10 mM in DMSO) was diluted 1,000-fold in 1× PBS (pH 7.3) to make 10 μM MaSAMP solution for injection. Methylated seed oil (MSO) surfactant was added to 10 μM MaSAMP solution to a final concentration of 0.5% for the foliar spray solution. Both sides of the leaves were sprayed to run-off. For the systemic MaSAMP uptake test, the 10 μM MaSAMP with 0.5% MSO was wiped onto both sides of leaves using a saturated cotton ball.
RNA Preparation and qRT-PCR.
Nb, tomato, and citrus total RNA was extracted using TRIzol Reagent (Invitrogen) and treated with DNase I (Roche). Total RNA was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) with oligo(dT) primer. For qPCR, transcripts were amplified from 2 μL 20× diluted complementary DNA and iQ SYBR Green Supermix (Bio-Rad). The PCR amplification consisted of 3 min at 94 °C, 45 cycles of 30 s at 94 °C, 30 s at 62 °C, 1 min at 72 °C, and 15 min at 72 °C, followed by the generation of a dissociation curve. The primers used are list in SI Appendix, Table S1. The generated Ct was used to calculate the transcript abundance relative to Nb Ubiquitin (NbUbi) (54), Sl elongation factor (SlEF) (55), or Citrus Actin (CsAct) (5).
Citrus Phloem-Rich Fluid or Leaf Vascular Fluid Collection.
Stems were collected from Cs, Ma, or Pt trees. The bark was stripped into pieces and manually removed from the twig. The bark strip was rinsed with deionized water, dried with Kim wipes, and cut into about 1 cm pieces using a sterile razor blade for collecting the fluid from phloem-rich tissue. For the leaf vascular fluid collection, the midveins of 10 to 20 leaves were cut out using a sterile razor blade, rinsed with deionized water, dried with Kim wipes, and then cut into about 1 cm pieces. The 1 cm bark or midvein tissues were vertically placed into a 0.5 mL Eppendorf tube. A small hole was punched at the bottom of the tube, and the tube was put into a 1.5 mL Eppendorf tube. The sample was centrifuged at 12,000 rpm for 15 min at 4 °C, and the collected fluid was stored at −80 °C until further analysis.
SAMP and SAMP Oligomer Analysis.
For SAMP and SAMP-helix2 oligomers analyses, 1 μM solutions of MaSAMP and MaSAMP-helix2 were made by diluting 1 μL 120 ng MaSAMP or 360 ng MaSAMP-helix2 peptide stock (in DMSO) in 20 μL sample buffer (100 mM Tris HCl, pH6.8, 10% glycerol, 0.1 M DTT, and 0.0006% Bromophenol blue) in a time series and resolved in a 15% PAGE gel without SDS. The oligomers of MaSAMP or MaSAMP-helix2 were detected by immunoblot or silver staining (following the manufacturer’s instructions, Bio-Rad), respectively.
Immunoblot.
For SAMP detection, the protein samples were resolved in an 18% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto nitrocellulose membranes with pore sizes of 0.1 μm in a Tris-Glycine transfer buffer. The membrane was blocked with Tris-buffered saline (TBS)/0.5% volume/volume (vol/vol) Tween 20/3% weight/volume (wt/vol) fat-free milk power and immunoblotted with the appropriate antibodies: polyclonal rabbit anti-SAMP (serum containing polyclonal antibodies was produced by rabbits immunized with 67 residues of MaSAMP produced in E. coli, Covance Inc., 1:1,000 dilution) and goat anti-rabbit IgG-HRP (Abcam, ab6721, 1:3,000 dilution). Enhanced chemiluminescence reagents (Amersham) were used for detection. The membranes after protein transfer were stained with Ponceau S staining solution (0.1% [wt/vol] Ponceau S in 5% [vol/vol] acetic acid). The membranes were incubated for up to an hour in staining solution with gentle agitation. After staining, the membranes were rinsed in distilled water until the background was clean.
Transmission Electron Microscopy.
OD600 0.01 of Lcr bacterial cells were incubated with 10 μM MaSAMP in 1× PBS pH 7.3 for 0.5 and 2 h at room temperature. Cells incubated with 10 μM bovine serum albumin for 2 h were used as mock treatment. During the incubation, Lcr cells settled to the bottom of the tube. After incubation, the extra suspension was removed, leaving 500 μL of bacteria–peptide mixture, which was then added 1:1 to the twofold fixation buffer (4% glutaraldehyde, 5 mM CaCl2, 10 mM MgCl2, and 1× PBS pH 7.3) and left for fixation overnight at 4 °C. Samples were washed three times in 1× PBS and resuspended in 1% low-melting-point agarose (Sigma). Samples were then fixed in 1% (wt/vol) osmium tetroxide following dehydration in ethanol with a graded series of concentrations and embedment in Epon 812 resin. Ultrathin sections were collected on 200 mesh nickel grids coated with Formvar and stained with uranyl acetate and lead citrate. Sections were examined with a Tecnai12 transmission electron microscope at an accelerating voltage of 80 kV.
The Protein Structure Prediction of SAMP.
The structure prediction of Ma-S1 (the SAMP used for the test in this study) was found using template form SWISS-MODEL Template Library, - 2q3p, by SWISS-MODEL (56). All structural figures were generated by PyMOL (http://www.pymol.org).
Supplementary Material
Acknowledgments
We thank Dr. Tracy Kahn at the University of California, Riverside, for providing the plant materials of citrus relatives and Rachael Hamby for editing the manuscript. This work was supported by grants from the USDA National Institute of Food and Agriculture (2019-70016-29067), University of California Riverside Cy Mouradick Chair Fund, the Citrus Research Board (project number 5200-195), and the California Citrus Nursery Board (Jin-19) to H.J.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019628118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Bove J. M., Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37 (2006). [Google Scholar]
- 2.Graham J., Gottwald T., Setamou M., Status of Huanglongbing (HLB) outbreaks in Florida, California and Texas. Trop. Plant Pathol. 45, 265–278 (2020). [Google Scholar]
- 3.Barnett M. J., Solow-Cordero D. E., Long S. R., A high-throughput system to identify inhibitors of Candidatus Liberibacter asiaticus transcription regulators. Proc. Natl. Acad. Sci. U.S.A. 116, 18009–18014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stokstad E., Agriculture. Dread citrus disease turns up in California, Texas. Science 336, 283–284 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Zhao H., et al., Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol. Plant 6, 301–310 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C. Y., Wang H., Hu P., Hamby R., Jin H., Small RNAs–Big players in plant-microbe interactions. Cell Host Microbe 26, 173–182 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Fu Z. Q., Dong X., Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Mauch-Mani B., Baccelli I., Luna E., Flors V., Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 68, 485–512 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Folimonova S. Y., Robertson C. J., Garnsey S. M., Gowda S., Dawson W. O., Examination of the responses of different genotypes of citrus to huanglongbing (citrus greening) under different conditions. Phytopathology 99, 1346–1354 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Gottwald T. R., Current epidemiological understanding of citrus Huanglongbing. Annu. Rev. Phytopathol. 48, 119–139 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Albrecht U., Bowman K. D., Tolerance of the trifoliate citrus hybrid US-897 (Citrus reticulata x Poncirus trifoliata) to huanglongbing. HortScience 46, 16–22 (2011). [Google Scholar]
- 12.Albrecht U., Bowman K. D., Tolerance of trifoliate citrus hybrids to Candidatus liberibacter asiaticus. Scr. Hortic. 147, 71–80 (2012a). [Google Scholar]
- 13.Ramadugu C., et al., Long-term field evaluation reveals Huanglongbing resistance in citrus relatives. Plant Dis. 100, 1858–1869 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Huang C., et al., Identification of citrus defense regulators against citrus Huanglongbing disease and establishment of an innovative rapid functional screening system. Plant Biotechnol. J., 10.1111/pbi.13502. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lytle B. L., et al., Structure of the hypothetical protein At3g17210 from Arabidopsis thaliana. J. Biomol. NMR 28, 397–400 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Park S. C., et al., Characterization of a heat-stable protein with antimicrobial activity from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 362, 562–567 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Leonard M. T., Fagen J. R., Davis-Richardson A. G., Davis M. J., Triplett E. W., Complete genome sequence of Liberibacter crescens BT-1. Stand. Genomic Sci. 7, 271–283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagen J. R., et al., Liberibacter crescens gen. nov., sp. nov., the first cultured member of the genus Liberibacter. Int. J. Syst. Evol. Microbiol. 64, 2461–2466 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Merfa M. V., et al., Progress and obstacles in culturing ‘candidatus liberibacter asiaticus’, the bacterium associated with Huanglongbing. Phytopathology 109, 1092–1101 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Yan S., Dong X., Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20, 64–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Austin M. J., et al., Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Yasuda S., Okada K., Saijo Y., A look at plant immunity through the window of the multitasking coreceptor BAK1. Curr. Opin. Plant Biol. 38, 10–18 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Ratcliff F., Martin-Hernandez A. M., Baulcombe D. C., Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Zhang L. J., Gallo R. L., Antimicrobial peptides. Curr. Biol. 26, R14–R19 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Volgers C., Savelkoul P. H. M., Stassen F. R. M., Gram-negative bacterial membrane vesicle release in response to the host-environment: Different threats, same trick? Crit. Rev. Microbiol. 44, 258–273 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Levin E. J., Kondrashov D. A., Wesenberg G. E., Phillips G. N. Jr, Ensemble refinement of protein crystal structures: Validation and application. Structure 15, 1040–1052 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stansly P. A., et al., Vector control and foliar nutrition to maintain economic sustainability of bearing citrus in Florida groves affected by huanglongbing. Pest Manag. Sci. 70, 415–426 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Gottwald T. R., Current epidemiological understanding of citrus Huanglongbing. Annu. Rev. Phytopathol. 48, 119–139 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Hu J., Jiang J., Wang N., Control of citrus Huanglongbing via trunk injection of plant defense activators and antibiotics. Phytopathology 108, 186–195 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Zhang M., et al., Effective antibiotics against ‘Candidatus Liberibacter asiaticus’ in HLB-affected citrus plants identified via the graft-based evaluation. PLoS One 9, e111032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaustein R. A., Lorca G. L., Teplitski M., Challenges for managing candidatus Liberibacter spp. (Huanglongbing disease pathogen): Current control measures and future directions. Phytopathology 108, 424–435 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Zhao H., et al., Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol. Plant 6, 301–310 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouse B. B., Rehabilitation of HLB infected citrus trees using severe pruning and nutritional sprays. Proc. Fla. State Hort. Soc. 126, 51–54 (2013). [Google Scholar]
- 34.Tiwari S., Mann R. S., Rogers M. E., Stelinski L. L., Insecticide resistance in field populations of Asian citrus psyllid in Florida. Pest Manag. Sci. 67, 1258–1268 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Rojas A., Makarova O., Rolff J., Antimicrobials, stress and mutagenesis. PLoS Pathog. 10, e1004445 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jochumsen N., et al., The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nature Commun. 7, 13002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fantner G. E., Barbero R. J., Gray D. S., Belcher A. M., Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nature Nanotechnol. 5, 280–285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tam J. P., Wang S., Wong K. H., Tan W. L., Antimicrobial peptides from plants. Pharmaceuticals (Basel) 8, 711–757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law R. H., et al., The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature 468, 447–451 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Rosado C. J., et al., The MACPF/CDC family of pore-forming toxins. Cellular Microbiol. 10, 1765–1774 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller M., Grauschopf U., Maier T., Glockshuber R., Ban N., The structure of a cytolytic alpha-helical toxin pore reveals its assembly mechanism. Nature 459, 726–730 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Wang J., et al., Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Ding J., et al., Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Liu X., et al., Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L., Habring A., Wang K., Weigel D., Atypical resistance protein RPW8/HR triggers oligomerization of the NLR immune receptor RPP7 and autoimmunity. Cell Host Microbe 27, 405–417.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Li J., et al., Membrane active antimicrobial peptides: Translating mechanistic insights to design. Front. Neurosci. 14, 73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olaitan A. O., Morand S., Rolain J. M., Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 5, 643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutcliffe I. C., A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 18, 464–470 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Sohlenkamp C., Geiger O., Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Shirasu K., The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Zheng Z., et al., Two ‘candidatus liberibacter asiaticus’ strains recently found in California Harbor different prophages. Phytopathology 107, 662–668 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Li W., Hartung J. S., Levy L., Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods 66, 104–115 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Murray M. G., Thompson W. F., Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin H., et al., NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3, 291–297 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Martinez-Medina A., et al., Shifting from priming of salicylic acid- to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 213, 1363–1377 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Waterhouse A., et al., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.