Significance
Alu elements, comprising more than 10% of the human genome, propagate via retrotransposition. This genomic expansion requires enzymatic activity of L1 that reverse transcribes Alu RNA into Alu cDNA in the nucleus. We report Alu also undergoes L1-mediated reverse transcription via self-priming in the cytoplasm independent of retrotransposition, providing evidence of human DNA synthesis in this cellular compartment. This newly discovered shunt molecule in the Alu replication cycle also induces death of the retinal pigmented epithelium, a hallmark of atrophic age-related macular degeneration. A Big Data Archeology analysis of multiple health insurance databases reveals that use of FDA-approved nucleoside reverse transcriptase inhibitors is associated with protection against macular degeneration, identifying a repurposing candidate for this blinding disease.
Keywords: Alu, retrotransposon, macular degeneration, retina, health insurance databases
Abstract
Alu retroelements propagate via retrotransposition by hijacking long interspersed nuclear element-1 (L1) reverse transcriptase (RT) and endonuclease activities. Reverse transcription of Alu RNA into complementary DNA (cDNA) is presumed to occur exclusively in the nucleus at the genomic integration site. Whether Alu cDNA is synthesized independently of genomic integration is unknown. Alu RNA promotes retinal pigmented epithelium (RPE) death in geographic atrophy, an untreatable type of age-related macular degeneration. We report that Alu RNA-induced RPE degeneration is mediated via cytoplasmic L1–reverse-transcribed Alu cDNA independently of retrotransposition. Alu RNA did not induce cDNA production or RPE degeneration in L1-inhibited animals or human cells. Alu reverse transcription can be initiated in the cytoplasm via self-priming of Alu RNA. In four health insurance databases, use of nucleoside RT inhibitors was associated with reduced risk of developing atrophic macular degeneration (pooled adjusted hazard ratio, 0.616; 95% confidence interval, 0.493–0.770), thus identifying inhibitors of this Alu replication cycle shunt as potential therapies for a major cause of blindness.
Reverse transcription of RNA into DNA occurs as part of the replication cycle of retroelements, genetic elements that reproduce via a copy-and-paste mechanism using a retrotransposon-encoded reverse transcriptase (RT). Retroelements have multiplied to occupy ∼42% of the human genome (1), yet the fate of retroelement-derived cDNA not integrated into the genome is poorly understood.
Age-related macular degeneration (AMD) is a blinding disease that affects 180 million people (2). In geographic atrophy, an advanced vision-threatening form of AMD without effective therapies (3), Alu RNA expressed from endogenous Alu retrotransposons by RNA polymerase III accumulates in the retinal pigmented epithelium (RPE) (4, 5). Alu RNA induces RPE cytotoxicity in human cells and mice; surprisingly, numerous RNA sensors are dispensable for this toxicity, and several other structurally similar RNAs are not toxic to the RPE (4, 6, 7). Therefore, we explored the replication cycle of the nonautonomous retrotransposon Alu, which includes reverse transcription of the Alu RNA by the L1-encoded RT in trans at the nuclear genomic insertion site—termed target-primed reverse transcription (TPRT)—and integration of the Alu cDNA into the genome (8–10).
Here, we demonstrate the existence of endogenous reverse-transcribed Alu cDNA synthesized in the cytoplasm of human cells independently of TPRT and provide evidence that Alu RNA can undergo self-priming to form Alu cDNA in the cytoplasm. We also present evidence from four independent patient health records databases that nucleoside reverse transcriptase inhibitor (NRTI) use is associated with reduced development of atrophic AMD; thus, these clinically approved drugs potentially could be repurposed for this disease.
Results
L1 Is Required for Alu RNA Toxicity.
Previously, we demonstrated NRTIs have two distinct inhibitory targets: RT and the NLRP3 inflammasome (11). While the RT-inhibitory function was dispensable for the anti-inflammatory effects of NRTIs, whether reverse transcription of Alu RNA is required for its toxicity was not tested. Thus, we examined whether endogenous L1-encoded RT mediated Alu RNA toxicity because L1-encoded ORF2p harboring RT and endonuclease (EN) activities can use Alu RNA as a template for reverse transcription in trans (12, 13).
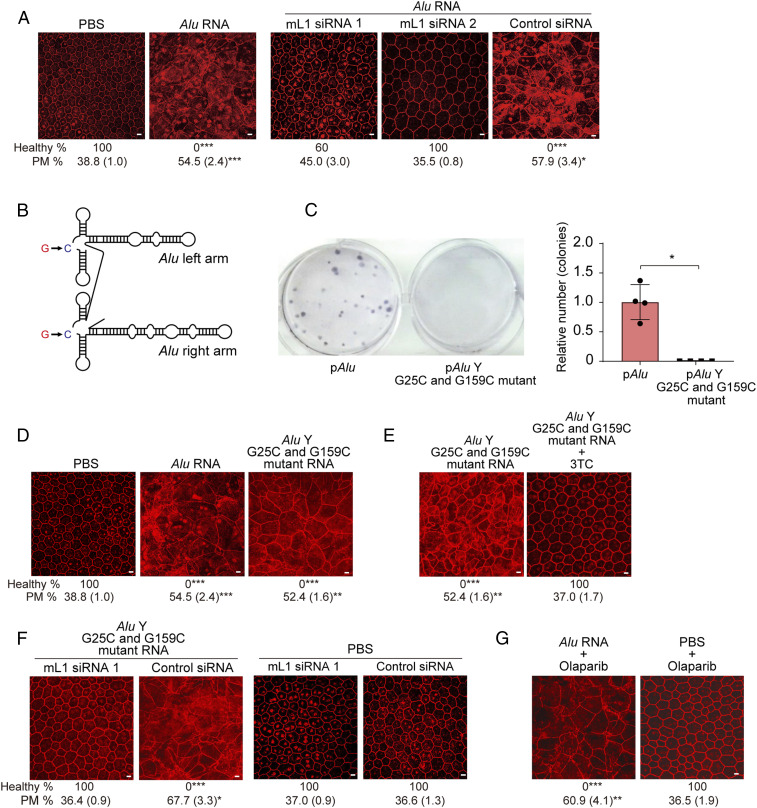
We identified two mouse L1 (mL1) small interfering RNAs (siRNAs) that reduced endogenous L1 ORF2p abundance in mouse RPE cells (SI Appendix, Fig. S1 A and B). Subretinal delivery of either mL1 siRNA, synthesized in a cell-permeable, nonimmunogenic format (14, 15), prevented Alu RNA-induced RPE degeneration in wild-type (WT) mice (Fig. 1A and SI Appendix, Figs. S1C and S2A). These similar outcomes argue against off-target siRNA activity; nevertheless, several mRNAs contain embedded sequences corresponding to varying L1 segments. However, mouse mRNAs containing sequences complementary to mL1 siRNA were not down-regulated following mL1 siRNA treatment in mouse RPE cells (SI Appendix, Fig. S1D). Next, we performed in vivo functional rescue of endogenous L1 RT via either of two expression plasmids: one encoding a synthetic and codon-optimized full-length human L1 element [pORFeus-Hs (16)] and the other encoding rat L1 ORF2p [pORF2-Rn (17)] (SI Appendix, Fig. S1E). Both these siRNA-refractory L1 expression constructs restored Alu RNA toxicity in mice despite coadministering mL1 siRNAs (SI Appendix, Fig. S1 F and G). These data suggest L1 activity is necessary and sufficient for Alu RNA toxicity in mice.
Fig. 1.
Endogenous L1 is required for Alu RNA-induced RPE toxicity. (A) RPE sheet micrographs of mice. RPE cellular boundaries were visualized by immunostaining flat mounts with zonula occludens-1 (ZO-1, red) antibody. Loss of regular hexagonal cellular boundaries in flat mounts represents degenerated RPE. (Scale bars, 10 μm.) Binary and morphometric quantification of RPE degeneration are shown. (*P < 0.05; **P < 0.01; ***P < 0.001, Fisher’s exact test for binary; two-tailed t test for morphometry.) PM, polymegethism [mean (SEM)]. RPE morphology in wild-type (WT) mice administered with Alu RNA or PBS, and Alu RNA with either of two L1-targeted siRNAs or control siRNA. n = 6–15. (B) Schematic of secondary structure of an Alu RNA harboring G25C/G159C mutations. (C) Retrotransposition frequency of Alu G25C/G159C double mutant RNA compared to Alu RNA in a cellular Alu retrotransposition reporter assay (described in SI Appendix, Supplementary Methods). *P < 0.05, Mann–Whitney U test. The error bars represent the mean ± SEM. (D and E) RPE morphology in WT mice following administration of Alu RNA or Alu G25C/G159C double-mutant RNA. (Scale bars, 10 μm.) n = 6. (E) Alu G25C/G159C double-mutant RNA-induced RPE degeneration in WT mice was blocked by 3TC. (Scale bars, 10 μm.) n = 5–6. (F) Alu G25C/G159C double-mutant RNA or PBS subretinal injection into WT mice with mL1 siRNA or control siRNA. (Scale bars, 10 μm.) n = 6–11. (G) RPE degeneration (ZO-1 flat mounts) in WT mice treated with Alu RNA and olaparib, a chemical inhibitor of L1 retrotransposition. (Scale bars, 10 μm.) n = 4–7.
Next, we studied whether retrotransposition, i.e., genomic insertion of reverse-transcribed Alu cDNA, which occurs infrequently in somatic cells, was responsible for Alu RNA toxicity. We synthesized a mutant Alu element with two mutations (G25C/G159C) (Fig. 1B), which has diminished retrotransposition ability (18). Despite retrotransposition deficiency, reconfirmed in a transmobilization assay (Fig. 1C), Alu G25C/G159C RNA induced RPE degeneration in WT mice (Fig. 1D and SI Appendix, Fig. S2B) in a dose-dependent manner alike WT Alu RNA (SI Appendix, Fig. S3 A and B). Alu G25C/G159C toxicity was prevented by the NRTI lamivudine (3TC) (Fig. 1E and SI Appendix, Fig. S2C) or mL1 siRNA (Fig. 1F and SI Appendix, Fig. S2D). Olaparib, a chemical inhibitor of L1 retrotransposition (19), inhibited Alu retrotransposition (SI Appendix, Fig. S4A) but did not block reverse transcription of Alu RNA into Alu cDNA in primary human RPE cells (SI Appendix, Fig. S4B) or Alu RNA-induced RPE degeneration in WT mice (Fig. 1G and SI Appendix, Fig. S2E). These findings suggest L1-mediated reverse transcription, but not retrotransposition, is essential for Alu RNA toxicity. Hence, we hypothesized that Alu cDNA reverse transcribed by L1 RT but not inserted into the genome is a key intermediate in Alu RNA toxicity.
Reverse-Transcribed Alu cDNA in Cells and Mice.
We generated a strand-specific probe to detect endogenous single-stranded Alu cDNA (SI Appendix, Fig. S5 A–C). Using in situ hybridization, we detected an artificially synthesized single-stranded Alu DNA transfected into primary human RPE cells in a dose-dependent manner; this signal was abolished by single-strand–specific S1 nuclease (SI Appendix, Fig. S5B). We next developed a variation of nucleic acid blotting that we term “equator blotting”: a functional combination of northern and Southern blotting (SI Appendix, Supplementary Methods) to detect extrachromosomal DNAs and their size. An equator blot is similar to a Southern blot in that it probes for a target DNA sequence, yet unlike a typical Southern blot, does not involve restriction enzyme digestion of the DNA. Instead, the DNA is separated without enzyme digestion prior to hybridization, per the typical northern blot procedure. Hence, we refer to the procedure of hybridization of undigested DNA as an equator blot. Alu RNA and Alu cDNA were probed in nuclear and cytoplasmic fractions. A probe recognizing U6 RNA was hybridized to fractions before RNase A treatment to confirm successful nuclear fractionation. Equator blotting using the Alu-specific probe, we detected accumulation of Alu cDNA, ∼300 nt in length, after Alu RNA transfection in primary human RPE cells; this signal was resistant to RNase A and double-stranded DNase but sensitive to S1 nuclease (SI Appendix, Fig. S5C), consistent with its specificity for nongenomic Alu sequences.
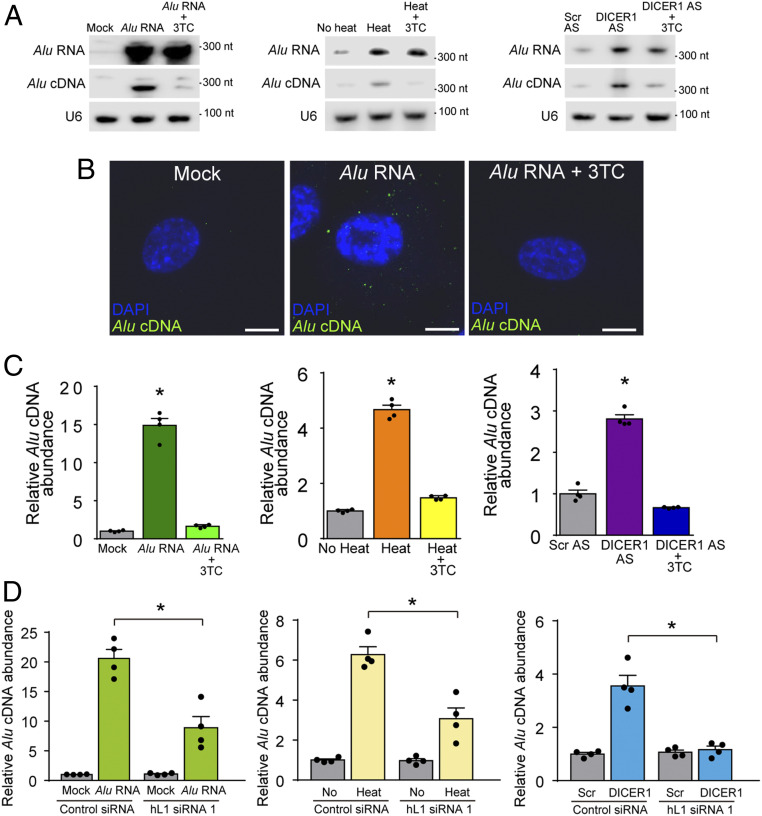
Using equator blotting and in situ hybridization, we assessed whether Alu cDNA synthesis is modulated by titrating Alu RNA levels. Increasing Alu RNA levels by any of three methods [transfection of in vitro transcribed synthetic RNA (4), heat shock (20), or DICER1 knockdown by antisense oligonucleotide (4)] induced Alu cDNA formation, which was abrogated by inhibition of endogenous RT with 3TC in primary human RPE cells (Fig. 2 A and B and SI Appendix, Figs. S6 and S7 A–D). This endogenous Alu cDNA was ∼300 nt long (Fig. 2A), suggesting it does not correspond to Alu sequences in fragmented genomic DNA. Little or no Alu cDNA was observed in untreated cells, suggesting it was newly synthesized and that the detection methodology was not identifying genomic DNA.
Fig. 2.
Endogenous L1 reverse-transcribed cytoplasmic Alu cDNA. (A) Northern blotting for Alu RNA (top bands), equator blotting for Alu cDNA (middle bands), and northern blotting for U6 RNA (bottom bands) of cytoplasmic fractions of primary human RPE cells transfected with Alu RNA, exposed to heat shock, or transfected with a DICER1-targeted antisense oligonucleotide (DICER1 AS) in the presence or absence of 3TC. Representative of n = 3 experiments. (B) Fluorescent micrographs of in situ hybridization of Alu cDNA in human RPE cells (green) colabeled with DAPI (blue) to identify nuclei. Cells were transfected with Alu RNA in the presence or absence of 3TC. Representative of n = 3 experiments. (Scale bar, 10 µm.) (C) Alu c-PCR of cytoplasmic fractions of primary human RPE cells transfected with Alu RNA, exposed to heat shock, or transfected with a DICER1 AS in the presence or absence of 3TC. Representative of n = 3 experiments. *P < 0.05, Mann–Whitney U test. (D) Direct amplification by real-time PCR (without reverse transcription) of Alu cDNA in primary human RPE cells treated with in vitro-transcribed Alu RNA, heat shock, or DICER1 AS after transfection with hL1 siRNA #1 compared with control siRNA. *P < 0.05 by Mann–Whitney U test. n = 4. Error bars show SEM.
Alu cDNA accumulation resulting from all three methods was predominantly cytoplasmic (Fig. 2B and SI Appendix, Fig. S6); however, following DICER1 knockdown, Alu cDNA was occasionally nuclear (SI Appendix, Fig. S7C). S1 nuclease eliminated the Alu cDNA signal, confirming it as single-stranded DNA (SI Appendix, Fig. S7D). These data confirm RT activity is required for generating Alu cDNA.
To quantify reverse transcription of Alu RNA into Alu cDNA, we subjected cytoplasmic fractions of primary human RPE cells to adaptor-based PCR quantification (Alu c-PCR) (SI Appendix, Fig. S8 A–F). This method avoids detecting circular forms of extrachromosomal Alu DNAs (SI Appendix, Fig. S8B) (21). 3TC down-regulated Alu cDNA in primary human RPE cells (SI Appendix, Fig. S8A), suggesting reverse transcription of endogenous Alu RNA generates Alu cDNA in human RPE cells. Increased levels of Alu cDNA in primary human RPE cells following transfection of in vitro transcribed Alu RNA, heat shock, or DICER1 knockdown were abolished by 3TC in primary human RPE cells (Fig. 2C), further confirming endogenous RT activity is required for generating Alu cDNA under conditions that elevate Alu RNA levels.
Using in situ hybridization of RPE whole mounts, we tested whether Alu cDNA was generated in mice after subretinal Alu RNA transfection. We performed this study using mice functionally deficient in the inflammasome components caspase-1 and caspase-4 (termed Casp1/4 dko mice), which are protected from Alu RNA toxicity (6, 22), to visualize Alu cDNA signals free of distortions arising from degenerating cells. In these mice, Alu cDNA accumulation was detected after Alu RNA transfection and blocked by 3TC, suggesting Alu cDNA production in vivo required reverse transcription (SI Appendix, Fig. S9).
Alu cDNA Formation Requires Human L1 RT Activity.
Two human L1 (hL1) siRNAs that down-regulated endogenous L1 ORF2 in primary human RPE cells (SI Appendix, Fig. S10 A and B) prevented Alu cDNA production in primary human RPE cells after Alu RNA transfection, heat shock, or DICER1 antisense treatment, as monitored by in situ hybridization (SI Appendix, Fig. S11) and real-time PCR (Fig. 2D). In situ hybridization revealed hL1 siRNA down-regulated Alu cDNA in ARPE-19 cells after heat shock and DICER1 antisense treatments (SI Appendix, Fig. S12). Conversely, L1 overexpression enhanced Alu cDNA production in Alu RNA-transfected ARPE-19 cells (SI Appendix, Fig. S13).
Supportive of the idea that reverse transcription, but not retrotransposition, is essential for Alu RNA toxicity, the retrotransposition-deficient Alu G25C/G159C RNA (Fig. 1 B and C), which induced RPE degeneration in WT mice (Fig. 1D and SI Appendix, Fig. S2B), also induced Alu cDNA formation in mouse embryonic carcinoma F9 cells and primary human RPE cells (SI Appendix, Fig. S14 A and B) and in vivo in Casp1/4 dko mice (SI Appendix, Fig. S14C). These data demonstrate Alu cDNA is produced and exerts toxicity in the absence of L1-mediated Alu retrotransposition.
We investigated Alu cDNA formation in multiple human cell types using direct amplification by real-time PCR. Basal levels of endogenous Alu cDNA varied more than 100-fold among 11 different primary cells and cell lines (SI Appendix, Fig. S15). Among the cells tested, those with the highest expression of endogenous Alu cDNA were, in order of abundance, NTera2D cells, primary human peripheral mononuclear cells, ARPE-19 cells, primary human RPE cells, and human embryonic kidney 293-T cells.
Since Alus exhibit sequence heterogeneity, we investigated from which Alu subfamilies the identified Alu cDNA sequences were derived. Alu sequences are broadly grouped into J, S, and Y families based on sequence divergence throughout millions of years of genomic Alu element propagation (23–25). We performed next-generation sequencing of cytoplasmic fractions of primary human RPE cells, restricted to 200- to 800-nt-long species to exclude genomic DNA contamination and embedded Alus. AluS and AluJ sequences were overrepresented in the cytoplasmic fractions covering ∼92% of all obtained reads, whereas AluY reads comprised ∼8% (SI Appendix, Fig. S16A). These fractions are comparable to the distribution of subfamilies of expressed Pol III-derived Alu RNAs in multiple cell types (26–28) (SI Appendix, Fig. S16B).
Cytoplasmic Synthesis of Alu cDNA.
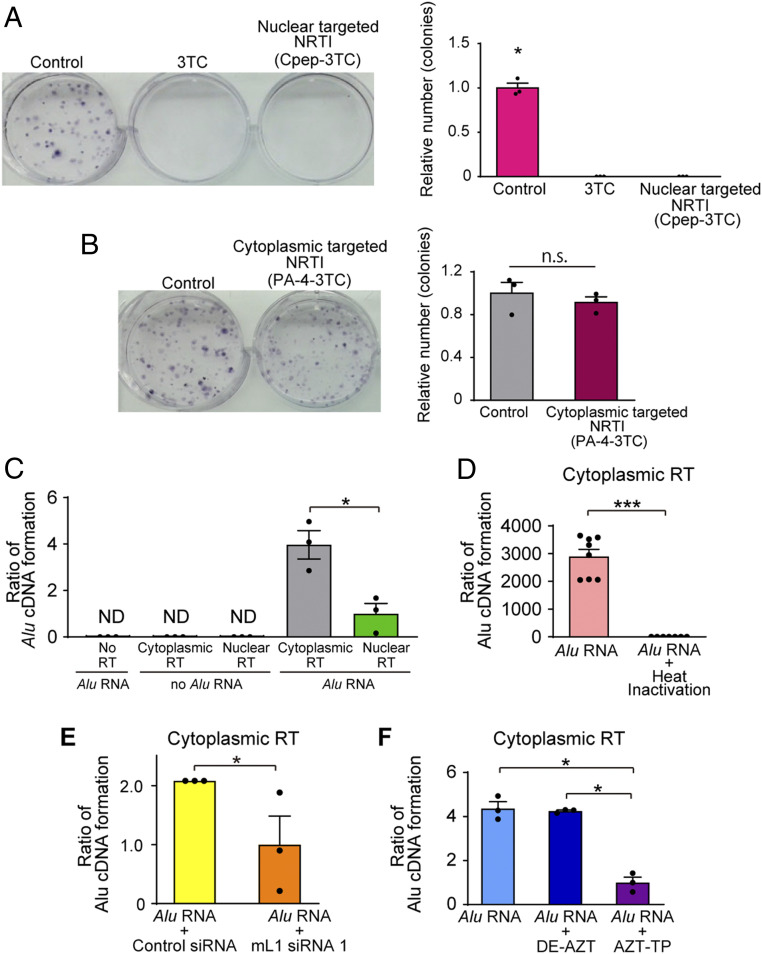
Canonically, reverse transcription of Alu RNA by L1 is thought to occur in the nucleus, coupled with genomic integration of Alu DNA. However, whether Alu cDNA is also synthesized in the cytoplasm is unknown. To determine the locus of synthesis of Alu cDNA that is not genomically integrated via TPRT, we used 3TC formulations that restrict it to either nuclear or cytoplasmic compartments. Conjugation of a cyclic peptide (Cpep) targets 3TC for nuclear localization (29), whereas a mixture of an amino acid/fatty acyl moiety (PA-4) restricts 3TC to the cytoplasm (30). Consistent with the model that TPRT of Alu RNA occurs in the nucleus and results in Alu retrotransposition, we confirmed that Cpep-3TC, but not PA-4–3TC, blocked Alu retrotransposition (Fig. 3 A and B). Conversely, cytoplasmic Alu cDNA formation in primary human RPE cells and Alu RNA-induced RPE degeneration in WT mice were blocked by PA-4–3TC but not Cpep-3TC (SI Appendix, Fig. S17 A and B), indicating that inhibition of cytoplasmic RT activity is critical for preventing Alu RNA toxicity and that toxic Alu cDNA did not leak from nucleus to cytoplasm following aborted Alu retrotransposition.
Fig. 3.
Alu cDNA is synthesized in the cytoplasm. (A and B) Alu retrotransposition assays in the presence of (A) 3TC or nuclear targeted-3TC (Cpep-3TC), or (B) cytoplasmic-targeted 3TC (PA-4–3TC). n = 3. *P < 0.05 by Mann–Whitney U test. Error bars show SEM. (C) Quantification of reverse transcription of Alu RNA by Alu-specific qPCR in cytoplasmic and nuclear fractions of mouse embryonal carcinoma F9 cells. (D) Quantification of Alu cDNA production in heat-inactivated cytoplasmic fractions. n = 7. (E) Quantification of Alu cDNA production in cytoplasmic fractions isolated from mouse L1 siRNA-treated cells. (F) Quantification of Alu cDNA production in cytoplasmic fractions of WT mouse RPE cells incubated with AZT-triphosphate (AZT-TP) or diethyl-AZT (DE-AZT). n = 3. *P < 0.05; ***P < 0.001 by Mann–Whitney U test. Error bars show SEM.
We then performed an ex vivo reverse transcription assay of Alu cDNA synthesis by incubating Alu RNA with protein extracts of nuclear or cytoplasmic fractions of WT mouse RPE cells or F9 cells, which have robust L1 expression (31). We observed higher amounts of synthesized Alu cDNA with cytoplasmic fractions than with nuclear fractions in both cells (Fig. 3C). Heat denaturation of cytoplasmic fractions eliminated Alu cDNA synthesis (Fig. 3D), compatible with its formation by a heat-labile enzyme. Cytoplasmic fractions isolated from mL1 siRNA-treated cells produced less Alu cDNA than from control siRNA-treated cells (Fig. 3E). Treatment of cytoplasmic extracts with AZT-triphosphate (AZT-TP), the active form of the NRTI zidovudine (AZT) that inhibits reverse transcription, reduced Alu cDNA synthesis, whereas diethyl-AZT (DE-AZT), an alkyl-modified NRTI derivative that does not block RT (11, 32), did not inhibit Alu cDNA formation (Fig. 3F). Furthermore, mouse platelets, which lack a nucleus and contain L1 ribonucleoprotein particles harboring endogenous L1 RT activity (33), also synthesize Alu cDNA following Alu RNA transfection (SI Appendix, Fig. S18 A–C). These data support the conclusion that Alu cDNA can be synthesized via L1-mediated reverse transcription in the cytoplasm and that this cytoplasmic Alu cDNA is responsible for its retinal cytotoxicity.
To monitor cytoplasmic Alu cDNA formation, we probed the association between L1 ORF2p, Alu RNA, and Alu cDNA in RNase H2-deficient biotinylated-Alu RNA-transfected HeLa cells expressing V5-tagged rat L1 ORF2p. Using pull-down assays, we captured the association of both Alu RNA and Alu cDNA with L1 ORF2p in cytoplasmic extracts (SI Appendix, Fig. S19 A–C). These data support a model in which Alu RNA associates with L1 ORF2p in the cytoplasm and is reverse transcribed into Alu cDNA.
Consistent with this concept, we detected RNA–DNA hybrids following Alu RNA transfection in mouse embryonic fibroblasts (MEFs) (SI Appendix, Fig. S20). Following biotin-labeled Alu RNA transfection and streptavidin-affinity pulldown, we detected Alu cDNA in the biotin-bound fraction, but a ∼13-fold greater amount of Alu cDNA in the biotin-unbound fraction (SI Appendix, Fig. S20A), suggesting Alu RNA–Alu cDNA hybrids are transient, as the majority of Alu cDNA is not bound to Alu RNA. Evidence supportive of Alu RNA–Alu cDNA hybrid formation was also found by immunostaining using an antibody that recognizes RNA–DNA hybrids (34) (SI Appendix, Fig. S20B). Biotin-labeled Alu RNA stability was greater in Rnaseh2−/− MEFs compared to WT MEFs, suggesting RNaseH2 is involved in degrading Alu RNA in these hybrids (SI Appendix, Fig. S20C).
Alu cDNA Formation via Self-Priming.
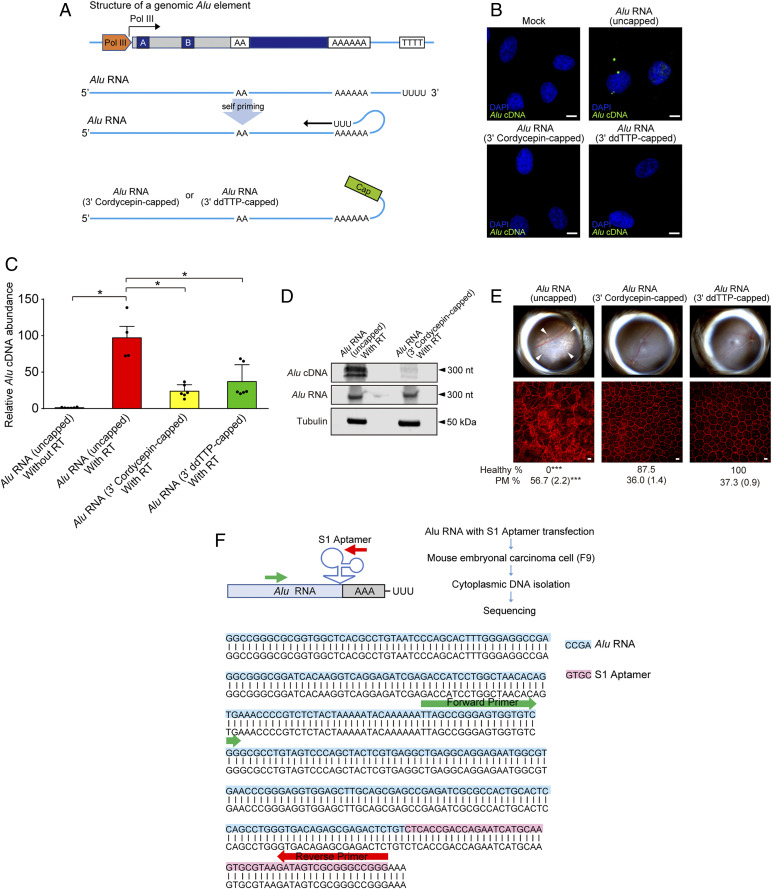
We sought to determine how priming of Alu reverse transcription might occur in the cytoplasm given that the canonical model of Alu retrotransposition holds that reverse transcription of Alu RNA by L1-ORF2p occurs in the nucleus and via TPRT, wherein the endonuclease activity of L1 exposes an oligo-T stretch of genomic DNA that serves to prime reverse transcription of the 3′ oligo-A stretch of Alu RNA (35). Alu RNA is capable of intramolecular base pairing (36); therefore, we hypothesized Alu could be capable of self-priming using its 3′ polyU-stretch (Fig. 4A). Indeed, the repetitive rodent BC1 RNA can prime its own reverse transcription (37).
Fig. 4.
Alu cDNA is synthesized via self-priming. (A) Schematic of putative Alu RNA transcription and self-priming by 3′ complementary hybridization. Alu RNA was synthetically capped on the 3′ end via dideoxy thymidine base (ddTTP) or cordycepin triphosphate to prevent extension via RT. (B) In situ hybridization of Alu cDNA after transfection of uncapped and or 3′-capped Alu RNAs into WT mouse RPE cells. Green, Alu cDNA; blue, DAPI. Representative of n = 3 experiments. (Scale bar, 10 µm.) (C) Alu cDNA abundance in cytoplasmic fractions of WT mouse RPE cells via an ex vivo RT activity assay (SI Appendix, Supplementary Methods), in the absence of external primers, followed by Alu-specific real-time PCR. PCR was performed with Alu RNA that was uncapped, or 3′-capped with ddTTP or cordycepin triphosphate. *P < 0.05 by Mann–Whitney U test. (D) Blotting analysis of cytoplasmic fractions of WT mouse RPE cells to detect Alu cDNA, Alu RNA, and tubulin after performing an ex vivo RT activity assay in the absence of external primers (see above). Fractions were incubated with uncapped or 3′-cordycepin triphosphate-capped Alu RNA. (E) Fundus photographs (Top) and corresponding representative RPE sheet micrographs (Bottom) of WT mice administered uncapped Alu RNA or Alu RNAs capped on the 3′ end with the chain ddTTP or cordycepin triphosphate. (Scale bars, 10 μm.) The arrowheads in fundus image denote the boundaries of RPE hypopigmentation. Binary and morphometric quantification of RPE degeneration are shown. *P < 0.05; **P < 0.01; ***P < 0.001, Fisher’s exact test for binary; two-tailed t test for morphometry. PM, polymegethism [mean (SEM)]. n = 6–8. (F) Alu RNA fused with S1 RNA aptamer at the 3′ end (Alu-S1) was transfected into mouse embryonic carcinoma cells (mF9) cells. Alu cDNA was detected in the RNase-treated cytoplasmic fraction of these cells, and sequencing using an Alu specific-forward and S1 aptamer-specific reverse primer confirmed the presence of the complementary S1 aptamer sequence in this Alu cDNA.
We disabled the potential self-priming capability of an in vitro-synthesized Alu RNA via 3′ capping with the chain terminators 2′,3′-dideoxythymidine-5′-triphosphate (ddTTP) or cordycepin (3′-deoxyadenosine) (Fig. 4A). Consistent with our hypothesis, transfection of uncapped Alu RNA into WT mouse RPE cells supported Alu cDNA formation, whereas transfection of 3′-capped Alu RNA species did not (Fig. 4B). Also supportive, incubating WT mouse RPE cell cytoplasmic protein extracts with uncapped Alu RNA in an in-tube RT assay performed in the absence of external DNA or RNA primers resulted in far more Alu cDNA synthesis than with 3′-capped Alu RNA (Fig. 4 C and D). As in vivo corroboration, subretinal administration of 3′-capped Alu RNA species did not induce RPE degeneration in WT mice (Fig. 4E).
Next, we tagged an Alu RNA by inserting an S1 aptamer sequence into its 3′ end (Fig. 4F) and transfected it into mouse F9 cells. We detected Alu cDNA formation in the cytoplasm of these cells and confirmed that the complementary S1 aptamer sequence was present in this Alu cDNA by using an Alu-specific forward primer and an S1 aptamer-specific reverse primer (Fig. 4F). These data demonstrate that Alu RNA can undergo self-priming to form Alu cDNA and suggest self-priming is one mechanism by which Alu reverse transcription is initiated in the cytoplasm.
NRTIs Associated with Lower Risk of Atrophic Macular Degeneration in Humans.
We analyzed whether NRTI use was associated with altered risk of atrophic AMD in humans by analyzing four longitudinal health insurance databases in the United States: Truven MarketScan Commercial Claims; US Veterans Health Administration; PearlDiver Humana; and PearlDiver Mariner (SI Appendix, Tables S1–S5). In all four databases, fewer HIV-negative persons taking NRTIs for preexposure prophylaxis developed atrophic AMD compared to HIV-negative persons not taking NRTIs (SI Appendix, Table S1).
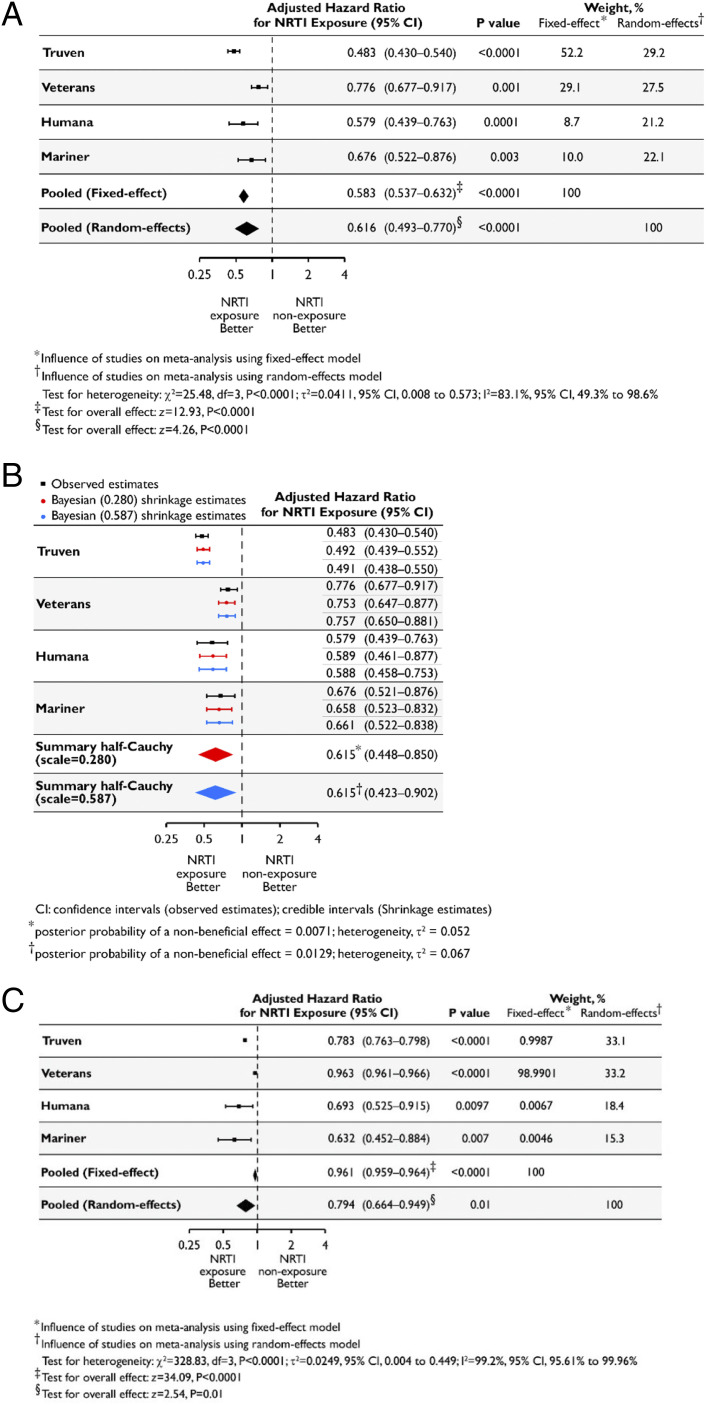
We analyzed the risk of developing atrophic AMD using Cox proportional hazards models, adjusting for age, gender, race, body mass index, smoking, and Charlson Comorbidity Index score. NRTI exposure was associated with a reduced hazard of developing atrophic AMD (Fig. 5A) in Humana [adjusted hazard ratio (aHR) = 0.579; 95% confidence interval (CI), 0.439, 0.763] and Mariner (aHR = 0.676, 95% CI, 0.522, 0.876). Given the low event rate (38) among NRTI users in Truven and Veterans databases, we employed Bayesian proportional hazards Cox regression models: NRTI exposure was associated with a reduced hazard of atrophic AMD in Truven (aHR = 0.483; 95% CI, 0.430, 0.540) and Veterans (aHR = 0.776; 95% CI, 0.677, 0.917). Sensitivity analyses varying the weight allotted to the exponential distribution underlying the baseline hazard function in the Bayesian Cox models also revealed protective associations of NRTI use (SI Appendix, Table S6). We estimate the combined risk across databases based on an inverse-variance–weighted meta-analysis using random effects for two reasons. First, a large amount of the variance across studies is attributable to heterogeneity (I2 = 83.1%; 95% CI, 49.3–98.6%; P < 0.001, test of heterogeneity). Second, the random-effects model is more appropriate as the databases represent populations with different underlying true effects (39–41). The random-effects meta-analysis revealed a protective effect of NRTIs (pooled aHR = 0.616; 95% CI, 0.493–0.770; P < 0.0001) (Fig. 5A). For completeness, we also estimate a fixed-effect model, which also revealed a protective NRTI effect.
Fig. 5.
NRTI use is associated with reduced hazard of incident atrophic AMD. (A–C) Using methodology employed in ref. 58, aHRs estimated separately for each database shown in black along with their 95% CIs, along with fixed-effect and random-effects meta-analyses shown in diamonds. The dashed vertical line denotes a HR of 1.0, which represents no difference in risk between nucleoside reverse-transcriptase inhibitor (NRTI) exposure and nonexposure. The horizontal bars represent 95% CIs. P values derived from z statistics for individual databases are reported. The estimates of heterogeneity (τ2), results of the statistical test of heterogeneity using the χ2 test statistic and its degrees of freedom (df), and posterior probabilities of a nonbeneficial effect for each model are shown below the plot. The Higgins I2 statistic and its 95% CI are presented. The results of the statistical tests of overall effect, the z-test statistics, and corresponding P values are presented. (A) HRs based on a Cox proportional-hazards model and adjusted for the confounding variables listed in SI Appendix were estimated separately for each database. Inverse-variance–weighted random-effects and fixed-effect meta-analyses were performed to obtain a pooled estimate of the aHR of incident atrophic AMD for NRTI exposure (ever vs. never). (B) HRs based on a Cox proportional-hazards model and adjusted for the confounding variables listed in SI Appendix, Supplementary Methods, were estimated separately for each database and are shown in black along with their 95% CIs. A Bayesian meta-analysis was performed using a random-effects model and a weakly informative hierarchical half-Cauchy prior distribution for between-study variance with the assumption that it was unlikely for the between-study HRs to vary by more than threefold (scale = 0.280). A sensitivity analysis to the choice of the prior by assuming that it was unlikely for the between-study HRs to vary by more than 10-fold was also performed (scale = 0.587). The Bayesian shrinkage estimates and the summary estimates of the aHR of incident atrophic AMD for NRTI exposure (ever vs. never), along with the 95% credible intervals, are shown in red (scale = 0.280) and blue (scale = 0.587). (C) HR estimates derived from propensity score-matched models adjusted for the confounding variables listed in SI Appendix, Supplementary Methods, were estimated separately for each database. Inverse-variance–weighted random-effects and fixed-effect meta-analyses were performed to obtain a pooled estimate of the aHR of incident atrophic AMD for NRTI exposure (ever vs. never).
A Bayesian meta-analysis employing a random-effects normal–normal hierarchical model (42, 43) was performed. A half-Cauchy prior distribution for between-study variability assuming hazard ratios (HRs) between studies were unlikely to vary greater than threefold (scale = 0.280) was used. Collectively in the four databases, NRTI users had a reduced hazard of atrophic AMD [aHR, 0.615; 95% credible interval, 0.448, 0.850; P(HR > 1) = 0.007] (Fig. 5B). A sensitivity analysis assuming HRs between studies were unlikely to vary greater than 10-fold (scale = 0.587) confirmed the directionality of the summary effect: NRTI use was protective [aHR, 0.615; 95% credible interval, 0.423, 0.902; P(HR > 1) = 0.013] (Fig. 5B).
We used falsification outcomes to detect residual confounding (44–46). Appendicitis and hernia outcomes were selected as they are causally unrelated to NRTI exposure. Among HIV-negative patients not previously diagnosed with these outcomes, NRTI exposure was not associated with reduced risk of appendicitis or hernia (SI Appendix, Table S7).
Next, we used propensity-score matching, a causal inference approach used in observational studies (47–50), to create cohorts with similar baseline characteristics, reducing potential bias in estimating treatment effects (SI Appendix, Tables S8–S11 and Figs. S21–S24). To control further for residual covariate imbalance, we adjusted for all factors employed in the unmatched analyses. In all four databases, NRTI users had a reduced hazard of atrophic AMD (Fig. 5C and SI Appendix, Table S12). The combined risk, based on random-effects meta-analysis, revealed a protective effect of NRTIs (pooled aHR = 0.794; 95% CI, 0.664, 0.949; P = 0.01), as did a fixed-effect model (Fig. 5C). The difference in the summary HR estimates between unmatched and propensity-score–matched analyses (0.616 vs. 0.794) suggests there could be residual bias in the unmatched analyses that was captured by propensity-score matching. However, the overlapping CIs of these summary HRs (0.493–0.770 vs. 0.664–0.949) suggests such residual bias is not significant.
Discussion
The principal hazard of L1 to the human genome is perceived as mutagenic retrotransposition and enzymatic activity of the L1 ORF2-encoded endonuclease (EN) domain. Our findings suggest L1 RT activity itself may contribute to the pathological process of RPE degeneration independent of retrotransposition, thus potentially revealing a mechanism of human disease driven by reverse transcription of host genetic material. Previously, we demonstrated NRTIs could, by virtue of inhibiting inflammasome activation, block Alu RNA-induced RPE degeneration even when robbed of their RT-inhibitory activity (11). Our findings suggest NRTIs also protect against RPE degeneration by intercepting RT-dependent Alu cDNA synthesis upstream of inflammasome activation.
The evolutionarily recent AluY elements are the most retrotranspositionally active (18, 51). In contrast, the majority of Pol III-transcribed Alu RNAs is expressed from the more ancient AluJ and AluS elements (27–29). The similarity in the distribution of cytoplasmic Alu cDNA and Alu RNA subfamilies, i.e., the finding that the majority of both pools comprises elements of the more ancient J and S subfamilies, suggests that although the majority of Pol III-transcribed Alu elements lack sequence features important for retrotransposition capability (29) they remain reverse transcription competent. It also suggests that, in the cytoplasm, L1 RT has no preference for any Alu subfamily as a substrate and that only the concentration of Alu transcripts from the respective Alu subfamily determines the fraction of Alu cDNAs resulting from reverse transcription of Alu RNAs. The fact that, contrarily, RNAs from young AluY elements that are underrepresented in the cytoplasmic pool of Alu RNAs and cDNAs are transmobilized by the L1 protein machinery more efficiently than the remaining Alu subfamilies suggests that there are either host-encoded nuclear factors necessary for the retrotransposition steps or cytoplasmic factors relevant for the transfer of the Alu ribonucleoprotein into the nucleus but not relevant for reverse transcription that have a bias for AluY sequences.
Our finding that Alu cDNA can be synthesized in the cytoplasm expands our understanding of the Alu replication cycle beyond the canonical model of TPRT-based Alu retrotransposition (35). More broadly, it will be interesting to address the possibility that various cytoplasmic RNAs (host or foreign) could be templates for cDNA formation. mRNAs can serve as substrates for L1-mediated retrotransposition, albeit at lower efficiency than Alu or L1 substrates (12, 52); future studies can determine the relative efficiency of cDNA formation from retroelements vs. other RNAs (53, 54). Although we demonstrate a pathogenic role for Alu cDNA, it might have functional roles in other settings. In high abundance, Alu cDNA could induce cell death to counteract excess L1 activity in cancers (55–57). Conversely, at low abundance, Alu cDNA might promote immunological self-tolerance to endogenous DNAs or prime immune factors for more rapid pathogen responses.
A strength of our health insurance database analyses is that findings were replicated in four independent cohorts diverse in age, gender, race, and time period, collectively representing a sizeable fraction of American adults with health insurance. The results of propensity score matching and falsification testing (which detects confounding, measurement error, selection bias) (44–46) increase the internal validity of this conclusion. Limitations include those inherent to health insurance database analyses, particularly accurate documentation, coding, and granularity of clinical phenotyping. Furthermore, despite confounder adjustment and robust propensity score matching, we cannot exclude the possibility of residual confounding or selection bias. Randomized controlled trials can yield better causal insights. Recently, we demonstrated NRTI use is associated with reduced development of type 2 diabetes (58). The current findings provide a rationale for prospective testing of NRTIs or alkylated NRTI derivatives, which block inflammasome activation but are less toxic than NRTIs (11), as potential therapies for geographic atrophy as well.
Materials and Methods
Subretinal injections (1 μL) and/or intravitreous injections (0.5 μL) were performed in mice using a 35-gauge needle. Seven days after subretinal injection, RPE health was assessed by fundus photography and immunofluorescence staining of zonula occludens-1 (ZO-1) on RPE flat mounts. Quantification of RPE degeneration was performed as described previously (59).
For in situ hybridization the RPE from mice was collected at 24 h after subretinal injection. Cells in culture were collected after 6 to 8 h after Alu RNA transfection. Visualization of fluorescein-labeled probe was performed with the TSA plus fluorescence system under a confocal microscope.
The reaction to evaluate self-priming activity of Alu RNA was carried out in the absence of priming oligos in a 20-µL reaction mix containing the following: Alu RNA with 3′-U tail; dNTP mix; cytoplasmic protein from mouse RPE cells; and Quantiscript RT Buffer (Qiagen). The resulting cDNA product was quantified by qPCR using Alu RNA template-specific primers.
This study used claims from the 1) Truven MarketScan Commercial Claims Database (IBM), which contains health care claims and medication usage obtained from analysis of commercial insurance claims from employer-based health insurance beneficiaries over the time period 2006 to 2018; 2) Veterans Health Administration system from 2000 to 2019, which contains data extracted from the Veterans Affairs Informatics and Computing Infrastructure (VINCI); 3) PearlDiver Patient Records Database, which captures health care claims and medication usage for persons in the Humana network between 2007 and the first quarter of 2017; and 4) PearlDiver Mariner database, which captures health care claims and medication usage for persons in provider networks from 2010 to the second quarter of 2018. Study approval and waiver of Health Insurance Portability and Accountability Act authorization for the Veterans dataset were provided by the Dorn Veterans Affairs Medical Center Institutional Review Board. All data within the Truven and PearlDiver databases are Health Insurance Portability and Accountability Act-compliant and were thus deemed exempt from institutional review board’s approval by the University of Virginia and University of South Carolina Institutional Review Boards. The completeness, utility, accuracy, validity, and access methods are described on these websites: https://www.virec.research.va.gov; https://www.ibm.com/products/marketscan-research-databases; and http://www.pearldiverinc.com/researchinfo.html.
The full materials and methods are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank J. L. Goodier and H. H. Kazazian for valuable discussions that improved the manuscript; A. P. Jackson, H. H. Kazazian, J. V. Moran, and M. A. Reijns for reagents and mice; and D. Robertson, K. Langberg, X. Zhou, R. Hankins, G. Pattison, Q. Zhong, K. A. Fox, and C. Spee for technical assistance. J.A. received support from NIH grants (DP1GM114862, R01EY022238, R01EY024068, R01EY028027, R01EY29799, and R01EY031039), the DuPont Guerry III Professorship, the University of Virginia Strategic Investment Fund, John Templeton Foundation Grant 60763, Doris Duke Distinguished Clinical Scientist Award, Ellison Medical Foundation Senior Scholar in Aging Award, the DuPont Guerry III Professorship, Dr. E. Vernon Smith and Eloise C. Smith Macular Degeneration Endowed Chair, and a gift from Mr. and Mrs. Eli W. Tullis; S.F., from Japan Society for the Promotion of Science Fund for the Promotion of Joint International Research (Home-Returning Researcher Development Research) and Research Grant of Japan Eye Bank Association; N.K. received support from NIH Grants K99EY024336, R00EY024336, and R21EY030651, and the Beckman Initiative for Macular Research; B.J.F. received support from NIH Grants T32HL091812 and UL1RR033173; R.Y. received support from Association for Research in Vision and Ophthalmology/Alcon Early Career Clinician-Scientist Research Award; T.Y. received support from Fight for Sight postdoctoral award; R.K.S. received support from the Fulbright Visiting Scholar Program; D.R.H. received support from NIH Grant R01EY001545 and an unrestricted departmental grant from Research to Prevent Blindness; S.S.S. received support from resources and the use of facilities at the W. J. B. Dorn Veterans Affairs Medical Center, Dorn Research Institute; B.D.G. received support from NIH Grants R01EY028027 and R01EY031039, BrightFocus Foundation, the Owens Family Foundation, the American Heart Association, and the National Center for Research Resources and the National Center for Advancing Translational Sciences, NIH, through Grant UL1TR000117; and G.G.S. is supported by a grant from the Ministry of Health of the Federal Republic of Germany (FKZ2518FSB403). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the US Department of Veterans Affairs, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government. This paper presents, in part, original research conducted using data from the Department of Veterans Affairs and is, in part, the result of work supported with resources and the use of facilities at the Dorn Research Institute, Columbia Veterans Affairs Health Care System (Columbia, SC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interest statement: J.A. is a co-founder of iVeena Holdings, iVeena Delivery Systems, and Inflammasome Therapeutics, and has been a consultant for Allergan, Biogen, Boehringer-Ingelheim, Immunovant, Janssen, Olix Pharmaceuticals, Retinal Solutions, and Saksin LifeSciences unrelated to this work. J.A., B.D.G., B.J.F., S.N., K.A., S.-b.W., I.A., M.A., F.P., N.K., and S.F. are named as inventors on patent applications on macular degeneration filed by the University of Virginia or the University of Kentucky. J.W.H. has received consulting fees from Celgene Corporation unrelated to this work. S.S.S. has received research grants from Boehringer Ingelheim, Gilead Sciences, Portola Pharmaceuticals, and United Therapeutics unrelated to this work. J.A. and B.D.G. are co-founders of DiceRx.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022751118/-/DCSupplemental.
Data Availability.
All data needed to evaluate the conclusions in this paper are available in the main text and SI Appendix.
References
- 1.H. H. Kazazian, Jr, Moran J. V., Mobile DNA in health and disease. N. Engl. J. Med. 377, 361–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong W. L., et al., Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2, e106–e116 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Ambati J., Ambati B. K., Yoo S. H., Ianchulev S., Adamis A. P., Age-related macular degeneration: Etiology, pathogenesis, and therapeutic strategies. Surv. Ophthalmol. 48, 257–293 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Kaneko H., et al., DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471, 325–330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dridi S., et al., ERK1/2 activation is a therapeutic target in age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 109, 13781–13786 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarallo V., et al., DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149, 847–859 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerur N., et al., TLR-independent and P2X7-dependent signaling mediate Alu RNA-induced NLRP3 inflammasome activation in geographic atrophy. Invest. Ophthalmol. Vis. Sci. 54, 7395–7401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke J. D., LINEs and Alus—the polyA connection. Nat. Genet. 16, 6–7 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Deininger P., Alu elements: Know the SINEs. Genome Biol. 12, 236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.H. H. Kazazian, Jr, Mobile elements: Drivers of genome evolution. Science 303, 1626–1632 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Fowler B. J., et al., Nucleoside reverse transcriptase inhibitors possess intrinsic anti-inflammatory activity. Science 346, 1000–1003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewannieux M., Esnault C., Heidmann T., LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Hagan C. R., Sheffield R. F., Rudin C. M., Human Alu element retrotransposition induced by genotoxic stress. Nat. Genet. 35, 219–220 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Kleinman M. E., et al., Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature 452, 591–597 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinman M. E., et al., Short-interfering RNAs induce retinal degeneration via TLR3 and IRF3. Mol. Ther. 20, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An W., et al., Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mob. DNA 2, 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirilyuk A., et al., Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res. 36, 648–665 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett E. A., et al., Active Alu retrotransposons in the human genome. Genome Res. 18, 1875–1883 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyoshi T., Makino T., Moran J. V., Poly(ADP-ribose) polymerase 2 recruits replication protein A to sites of LINE-1 integration to facilitate retrotransposition. Mol. Cell 75, 1286–1298.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W. M., Chu W. M., Choudary P. V., Schmid C. W., Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 23, 1758–1765 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krolewski J. J., Schindler C. W., Rush M. G., Structure of extrachromosomal circular DNAs containing both the Alu family of dispersed repetitive sequences and other regions of chromosomal DNA. J. Mol. Biol. 174, 41–54 (1984). [DOI] [PubMed] [Google Scholar]
- 22.Kerur N., et al., cGAS drives noncanonical-inflammasome activation in age-related macular degeneration. Nat. Med. 24, 50–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin C. M., Houck C. M., Deininger P. L., Friedmann T., Schmid C. W., Partial nucleotide sequence of the 300-nucleotide interspersed repeated human DNA sequences. Nature 284, 372–374 (1980). [DOI] [PubMed] [Google Scholar]
- 24.Batzer M. A., et al., Standardized nomenclature for Alu repeats. J. Mol. Evol. 42, 3–6 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Batzer M. A., Deininger P. L., Alu repeats and human genomic diversity. Nat. Rev. Genet. 3, 370–379 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Zhang X. O., Gingeras T. R., Weng Z., Genome-wide analysis of polymerase III-transcribed Alu elements suggests cell-type-specific enhancer function. Genome Res. 29, 1402–1414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conti A., et al., Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-seq data. Nucleic Acids Res. 43, 817–835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oler A. J., et al., Alu expression in human cell lines and their retrotranspositional potential. Mob. DNA 3, 11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandal D., Nasrolahi Shirazi A., Parang K., Cell-penetrating homochiral cyclic peptides as nuclear-targeting molecular transporters. Angew. Chem. Int. Ed. Engl. 50, 9633–9637 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Nasrolahi Shirazi A., et al., Peptide amphiphile containing arginine and fatty acyl chains as molecular transporters. Mol. Pharm. 10, 4717–4727 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin S. L., Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 11, 4804–4807 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ambati J., Fowler B. J., Ambati K., “Compositions and methods for treating retinal degradation” (University of Kentucky Research Foundation). US Patent 10,294,220 (2019).
- 33.Schwertz H., et al., Endogenous LINE-1 (long interspersed nuclear element-1) reverse transcriptase activity in platelets controls translational events through RNA-DNA hybrids. Arterioscler. Thromb. Vasc. Biol. 38, 801–815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boguslawski S. J., et al., Characterization of monoclonal antibody to DNA⋅RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89, 123–130 (1986). [DOI] [PubMed] [Google Scholar]
- 35.Feng Q., Moran J. V., H. H. Kazazian, Jr, Boeke J. D., Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Ahl V., Keller H., Schmidt S., Weichenrieder O., Retrotransposition and crystal structure of an Alu RNP in the ribosome-stalling conformation. Mol. Cell 60, 715–727 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Shen M. R., Brosius J., Deininger P. L., BC1 RNA, the transcript from a master gene for ID element amplification, is able to prime its own reverse transcription. Nucleic Acids Res. 25, 1641–1648 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vittinghoff E., McCulloch C. E., Relaxing the rule of ten events per variable in logistic and Cox regression. Am. J. Epidemiol. 165, 710–718 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Anello C., Fleiss J. L., Exploratory or analytic meta-analysis: Should we distinguish between them? J. Clin. Epidemiol. 48, 109–116, discussion 117–118 (1995). [DOI] [PubMed] [Google Scholar]
- 40.DerSimonian R., Laird N., Meta-analysis in clinical trials. Control. Clin. Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- 41.Lau J., Ioannidis J. P., Schmid C. H., Summing up evidence: One answer is not always enough. Lancet 351, 123–127 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Sutton A. J., Abrams K. R., Bayesian methods in meta-analysis and evidence synthesis. Stat. Methods Med. Res. 10, 277–303 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Dawid A. P., The well-calibrated Bayesian. J. Am. Stat. Assoc. 77, 605–610 (1982). [Google Scholar]
- 44.Prasad V., Jena A. B., Prespecified falsification end points: Can they validate true observational associations? JAMA 309, 241–242 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Lipsitch M., Tchetgen Tchetgen E., Cohen T., Negative controls: A tool for detecting confounding and bias in observational studies. Epidemiology 21, 383–388 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnold B. F., Ercumen A., Benjamin-Chung J., J. M. Colford, Jr, Brief report: Negative controls to detect selection bias and measurement bias in epidemiologic studies. Epidemiology 27, 637–641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenbaum P. R., Rubin D. B., The central role of the propensity score in observational studies for causal effects. Biometrika 70, 41–55 (1983). [Google Scholar]
- 48.Haukoos J. S., Lewis R. J., The propensity score. JAMA 314, 1637–1638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohlsson H., Kendler K. S., Applying causal inference methods in psychiatric epidemiology: A review. JAMA Psychiatry 77, 637–644 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imai K., van Dyk D. A., Causal inference with general treatment regimes. J. Am. Stat. Assoc. 99, 854–866 (2004). [Google Scholar]
- 51.Konkel M. K.et al.; 1000 Genomes Consortium , Sequence analysis and characterization of active human Alu subfamilies based on the 1000 Genomes pilot project. Genome Biol. Evol. 7, 2608–2622 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei W., et al., Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21, 1429–1439 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dhellin O., Maestre J., Heidmann T., Functional differences between the human LINE retrotransposon and retroviral reverse transcriptases for in vivo mRNA reverse transcription. EMBO J. 16, 6590–6602 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esnault C., Maestre J., Heidmann T., Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Carreira P. E., Richardson S. R., Faulkner G. J., L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J. 281, 63–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Di Ruocco F., et al., Alu RNA accumulation induces epithelial-to-mesenchymal transition by modulating miR-566 and is associated with cancer progression. Oncogene 37, 627–637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez-Martin B.et al.; PCAWG Structural Variation Working Group; PCAWG Consortium , Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 52, 306–319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ambati J., et al., Repurposing anti-inflammasome NRTIs for improving insulin sensitivity and reducing type 2 diabetes development. Nat. Commun. 11, 4737 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Narendran S., et al., A clinical metabolite of azidothymidine inhibits experimental choroidal neovascularization and retinal pigmented epithelium degeneration. Invest. Ophthalmol. Vis. Sci. 61, 4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in this paper are available in the main text and SI Appendix.