Significance
Autoimmune diseases are characterized by overactivation of an organism’s immune system and subsequent tissue damage and pathogenesis. Although genome-wide association studies have identified genetic variations, such as single-nucleotide polymorphisms (SNPs), that are linked to autoimmunity, the exact function of the genes involved and molecular mechanisms underlying their contribution to autoimmunity remain not well understood. Here, we show that an evolutionarily conserved autoimmunity risk gene, TMEM39A, regulates lysosome dynamics through interacting and functioning with molecular motor dynein intermediate light chain. As lysosomes are implicated in various aspects of immune-system dysfunction, including abnormal cytokine secretion and antigen presentation, our study provides an important mechanistic insight suggesting how TMEM39A might contribute to autoimmunity like multiple sclerosis and lupus.
Keywords: TMEM39A, lysosome, dynein, autoimmunity, autoimmune disorders
Abstract
TMEM39A encodes an evolutionarily conserved transmembrane protein and carries single-nucleotide polymorphisms associated with increased risk of major human autoimmune diseases, including multiple sclerosis. The exact cellular function of TMEM39A remains not well understood. Here, we report that TMEM-39, the sole Caenorhabditis elegans (C. elegans) ortholog of TMEM39A, regulates lysosome distribution and accumulation. Elimination of tmem-39 leads to lysosome tubularization and reduced lysosome mobility, as well as accumulation of the lysosome-associated membrane protein LMP-1. In mammalian cells, loss of TMEM39A leads to redistribution of lysosomes from the perinuclear region to cell periphery. Mechanistically, TMEM39A interacts with the dynein intermediate light chain DYNC1I2 to maintain proper lysosome distribution. Deficiency of tmem-39 or the DYNC1I2 homolog in C. elegans impairs mTOR signaling and activates the downstream TFEB-like transcription factor HLH-30. We propose evolutionarily conserved roles of TMEM39 family proteins in regulating lysosome distribution and lysosome-associated signaling, dysfunction of which in humans may underlie aspects of autoimmune diseases.
Autoimmune diseases affect millions of people worldwide, while exact causes of the diseases remain not well understood, despite decades of research (1–3). Recently, genome-wide association studies have identified single-nucleotide polymorphisms (SNPs) in TMEM39A that are associated with enhanced risks of multiple sclerosis (MS), systemic lupus erythematosus (SLE), and autoimmune thyroid disease (4–8), indicating an important link between TMEM39A and autoimmune disease. TMEM39A is also overexpressed in glioma cell lines and glioma patient cells (9), implicating its possible role in glioma pathogenesis. TMEM39A encodes a predicted multitransmembrane protein, homologs of which are found in most multicellular organisms (Fig. 1 A and B and SI Appendix, Fig. S1A). A recent study reported that TMEM39A localizes to the endoplasmic reticulum (ER) and regulates autophagy through controlling trafficking of the PtdIns(4)P phosphatase SAC1 (10). To investigate TMEM39A’s physiological roles and whether its cellular functions might be evolutionarily conserved, we generated Caenorhabditis elegans (C. elegans) strains and HEK 293T human cell lines that lack respective TMEM39 homologs in each system. Phenotypic and protein-interactor analyses revealed conserved roles of TMEM39 in regulating lysosome dynamics and signaling, providing a plausible link of human TMEM39A to pathogenesis of autoimmune diseases.
Fig. 1.
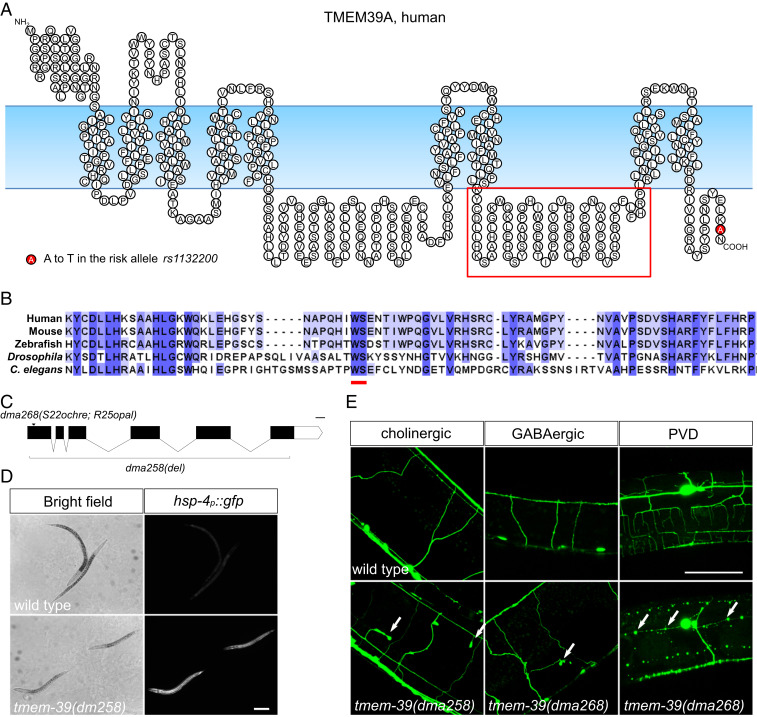
TMEM-39 belongs to a conserved family of transmembrane proteins in metazoans and its role in maintaining normal neuronal morphology in C. elegans. (A) Schematic diagram showing the predicted structure of human TMEM39A protein, which contains nine transmembrane domains and two large loops (by SOSUI structural prediction server: https://harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html). The autoimmune-risk allele rs1132200 confers an A-to-T substitution in the C terminus of the protein, which is represented by a red circle. The red box highlights the loop domain aligned in B. (B) Sequence alignment of the second loop of the TMEM39A protein from the worm to human. The conserved WS residues within the loop are highlighted by a red line. (C) Schematic diagram showing the genomic structure and loss-of-function alleles of C. elegans tmem-39. dma258 contains a deletion that removes most of the tmem-39 coding sequences, while dma268 contains two early stop codons in tmem-39 that likely render the allele null. (D) Bright-field and fluorescence images showing wild-type and tmem-39 mutant animals, with the ER stress reporter hsp-4p::gfp markedly up-regulated in tmem-39 mutants. (Scale bar, 100 μm.) (E) The tmem-39 mutants show broad morphological defects in the nervous system exemplified by ectopic branching (labeled by cholinergic reporter unc-17p::gfp and GABAergic reporter unc-47p::gfp; arrows) and neurodegeneration-like beading phenotypes (labeled by the PVD neuron reporter F49H12.4::gfp; arrows). (Scale bar, 40 μm.)
Results
We identified the uncharacterized C. elegans gene D1007.5 (named as tmem-39 hereafter) in a previous genome-wide RNA interference (RNAi) screen for cellular-stress regulators (11). tmem-39 encodes the only homolog of mammalian TMEM39A in the C. elegans genome. Using CRISPR-Cas9, we either removed the entire coding region of tmem-39 or inserted stop codons in the first exon of the gene (Fig. 1C). Mutant animals carrying tmem-39 null alleles showed multiple developmental abnormalities, including slow growth, reduced body length, and a tendency to burst, indicating defects in cuticle development (Fig. 1D and SI Appendix, Fig. S1B). In addition, tmem-39 mutant animals also have markedly higher expression of ER stress reporter hsp-4p::gfp in the hypoderm, supporting an important role of TMEM-39 in normal protein homeostasis (Fig. 1D). To investigate if disruption of TMEM-39 impairs the nervous system, the primary target of MS, we examined neuronal morphology of the mutant animals using transgenic strains that express cholinergic (unc-17p::gfp), GABAergic (unc-47p::gfp), or somatosensory PVD interneuron (F49H12.4::gfp) reporters. About 10 to 20% of the wild-type 8animals showed mild morphological abnormalities (e.g., short filopodia-like structures) when they aged (Table 1 and SI Appendix, Fig. S1G). Strikingly, disruption of tmem-39 led to severe morphological defects of the nervous system in 50 to 90% of the mutant animals examined, including long ectopic branches, incorrect neurite projections, and beading of neuronal processes (Fig. 1E and Table 1). We also found that the dendritic structures of the PVD neurons were missing in L2 and L3 larva, despite the presence of the PVD soma, suggesting developmental defects of the neuronal processes (SI Appendix, Fig. S1H). Together, our observations indicate that TMEM-39 is important for maintaining normal ER homeostasis and neuronal morphology in the nervous system.
Table 1.
Neuronal defects of adult (2 d after L4 stage) tmem-39 mutants
| Neuronal reporter and genotype | Abnormal, % | n | Morphological defects |
| Cholinergic* | |||
| Wild type | 26 | 54 | Short filopodia-like structures |
| tmem-39(dma258) | 96 | 50 | Long ectopic branches; incorrect projections |
| tmem-39(dma268) | 80 | 50 | Long ectopic branches; incorrect projections |
| GABAergic† | |||
| Wild type | 16 | 32 | Short filopodia-like structures |
| tmem-39(dma258) | 56 | 50 | Beading on neurites |
| tmem-39(dma268) | 36 | 50 | Long ectopic branches; incorrect projections |
| PVD‡ | |||
| Wild type | 10 | 10 | Mild beading on neurites |
| tmem-39(dma258) | 73 | 11 | Extensive degeneration-like beading on neurites |
| tmem-39(dma268) | 77 | 13 | Extensive degeneration-like beading on neurites |
Contains vsIs48[unc-17p::gfp].
Contains oxIs12[unc-47p::gfp].
Contains wdIs52[F49H12.4::gfp].
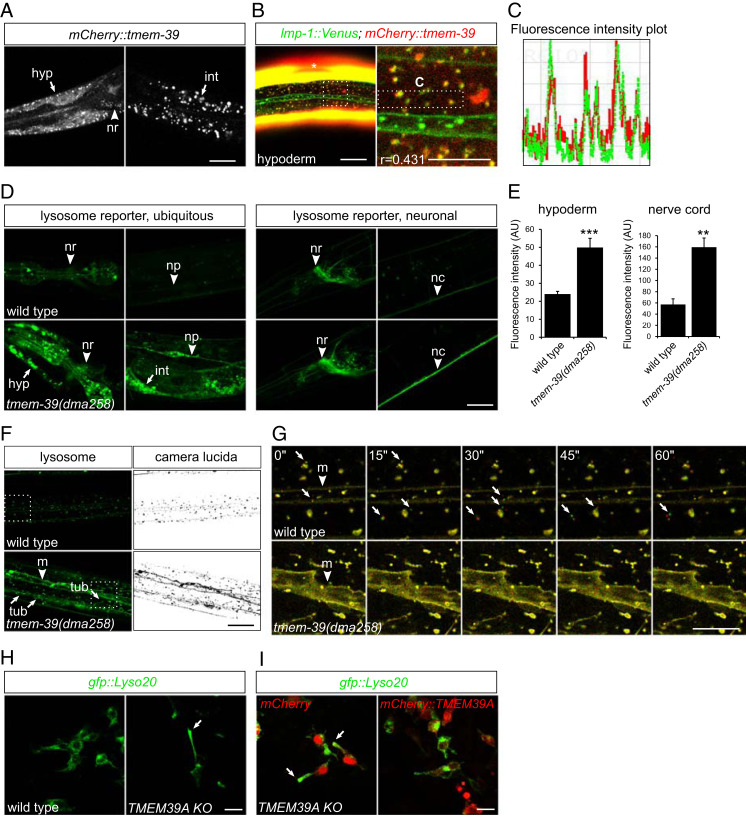
To determine the subcellular localization of TMEM-39 protein, we generated transgenic strains that express either a rescuing tmem-39 genomic transgene with mCherry tagged to the N terminus or N-mCherry–tagged tmem-39 complementary DNA (cDNA) under an hsp-16.48 heat-shock promoter. The tmem-39 genomic construct rescued developmental defects of tmem-39 mutant animals, including dumpiness and elevated hsp-4p::gfp ER stress reporter expression (SI Appendix, Fig. S1 D–F). We observed broad mCherry::TMEM-39 expression in multiple tissues, particularly the nervous system, hypoderm, and intestine (Fig. 2A). Both transgenic strains showed TMEM-39 localization to cytosolic puncta, largely colocalizing with GFP-tagged lysosome-associated membrane protein LMP-1 (Fig. 2 B and C). By contrast, TMEM-39 did not colocalize with a Golgi-targeted reporter mans::gfp (SI Appendix, Fig. S2 A and B), suggesting that TMEM-39 protein primarily localizes to compartments closely associated with lysosomes, at least in hypoderm. To investigate if disruption of TMEM-39 impairs lysosome morphology and/or function, we generated a compound mutant tmem-39(null);dmaIs58[rpl-28p::lmp-1::gfp] and examined lysosome reporter expression (under the ubiquitous promoter rpl-28) in tmem-39 null mutant animals. We found that tmem-39 mutant animals showed markedly higher accumulation of LMP-1::GFP in intestine, hypoderm, pharynx, and neuronal processes along the body, but to a lesser extent in the nerve ring, indicating that the level of lysosome reporter up-regulation is likely tissue-type dependent (Fig. 2D). Consistent with the observations, a second transgenic strain that expresses lmp-1::gfp specifically in neurons (under a pan-neuronal promoter rab-3) showed a similar pattern of lysosome reporter accumulation in nerve ring and nerve cords upon tmem-39 disruption (Fig. 2 D and E). By contrast, the tmem-39 null mutants showed slightly decreased expression of a different reporter mCherry::pdr-1 (which encodes the ortholog of human PRKN protein) driven by the ubiquitous promoter rpl-28, indicating that the lysosome reporter up-regulation is likely caused by changes in protein dynamics rather than transcriptional up-regulation of the rpl-28 promoter (SI Appendix, Fig. S2 C and D). Unlike the punctal morphology of LMP-1::GFP-labeled lysosomes in the hypoderm of wild-type animals, lysosomes in tmem-39 mutants formed more tubular structures (Fig. 2F). Time-lapse confocal imaging revealed that the tubular lysosomes were more static compared with wild-type lysosomes (Fig. 2G and Movies S1 and S2). We conclude that TMEM-39 regulates lysosomal LAMP1 abundance and is important for maintaining normal morphology and motility of lysosomes.
Fig. 2.
TMEM-39 is associated with lysosomes and regulates lysosome positioning and morphology. (A) Confocal images showing expression and punctate distribution of the C. elegans mCherry::TMEM-39 fusion protein, detected in hypoderm (hyp), nerve ring (nr), and intestine (int). (Scale bar, 20 μm.) (B) Confocal images showing partial colocalization of mCherry::TMEM-39 with lysosomal marker LMP-1::Venus in C. elegans hypoderm. Asterisks denote the expression of muscle GFP or mCherry from coinjection plasmids. The Pearson’s correlation coefficient (r = 0.431) indicates moderate association of TMEM-39 with lysosomes. (Scale bar, 20 μm [B, Left] and 10 μm [B, Right].) (C) Intensity plot of mCherry::TMEM-39 and LMP-1::Venus fluorescence in the boxed region in B. (D) Ubiquitous lysosome reporter rpl-28p::lmp-1::Venus is strongly up-regulated in tmem-39 mutant animals, evident in hypoderm, intestine, nerve ring, and nerve cord (nc). The lysosome reporter induction in nerve ring is more modest compared with that in hypoderm or intestine, based on a neuronal lysosomal reporter rab-3p::lmp-1::gfp. (Scale bar, 20 μm.) (E) Quantification of lysosome reporter up-regulation in tmem-39 mutants. Data represent mean ± SEM. **P < 0.01; ***P < 0.001. AU, arbitrary units. (F) The lysosome LMP-1::Venus reporter labels cytoplasmic or endosomal membrane (m; arrowhead) more strongly in tmem-39 mutant hypoderm than does in the wild type and tends to form more tubular structures (tub; arrows). Dashed boxes represent areas enlarged in G. (Scale bar, 20 μm.) (G) Time-lapse confocal images showing lysosome dynamics. Two consecutive time-lapse images are pseudocolored with red and green and are superposed to reveal mobile (nonyellow) lysosomes (arrows). Compared with lysosomes in wild type that are mobile, tubular lysosomes in tmem-39 mutants are relatively static. The putative cytoplasmic or endosomal membrane (m) is labeled by arrowheads. (Scale bar, 10 μm.) (H) Confocal images showing redistribution of lysosomes from perinuclear regions to cell periphery in TMEM39A KO HEK 293T cells (arrow). (Scale bar, 20 μm.) (I) Confocal images showing the restoration of perinuclear localization of lysosomes in TMEM39A KO cells transfected with mCherry::TMEM39A, but not with mCherry plasmid (arrows). (Scale bar, 20 μm.)
Given the evolutionary conservation and homology of TMEM39 proteins between worms and humans, we next examined human TMEM39A protein localization in HEK 293T cells. Confocal imaging of cells transiently transfected with mCherry::TMEM39A and GFP-tagged reporters for various subcellular compartments revealed that TMEM39A partially colocalized with ER, Golgi apparatus, and Rab7-labeled late-endosomes/lysosomes (SI Appendix, Fig. S2 E–G). To determine if knocking out TMEM39A disrupts any of the subcellular organelles, we generated TMEM39A knockout (KO) HEK 293T cells by CRISPR-Cas9 and examined subcellular organelle morphology and localization by confocal microscopy. We found that disruption of TMEM39A led to redistribution of lysosomes from perinuclear regions to cell periphery, while other subcellular organelles, including ER, Golgi apparatus, and mitochondria, remained largely unaffected (Fig. 2H and SI Appendix, Fig. S2H). The peripheral distribution of lysosomes in TMEM39A KO cells was rescued by a wild-type mCherry::TMEM39A transgene (Fig. 2I). Together, our findings indicate that mammalian TMEM39A has an evolutionarily conserved role in regulating lysosome distribution.
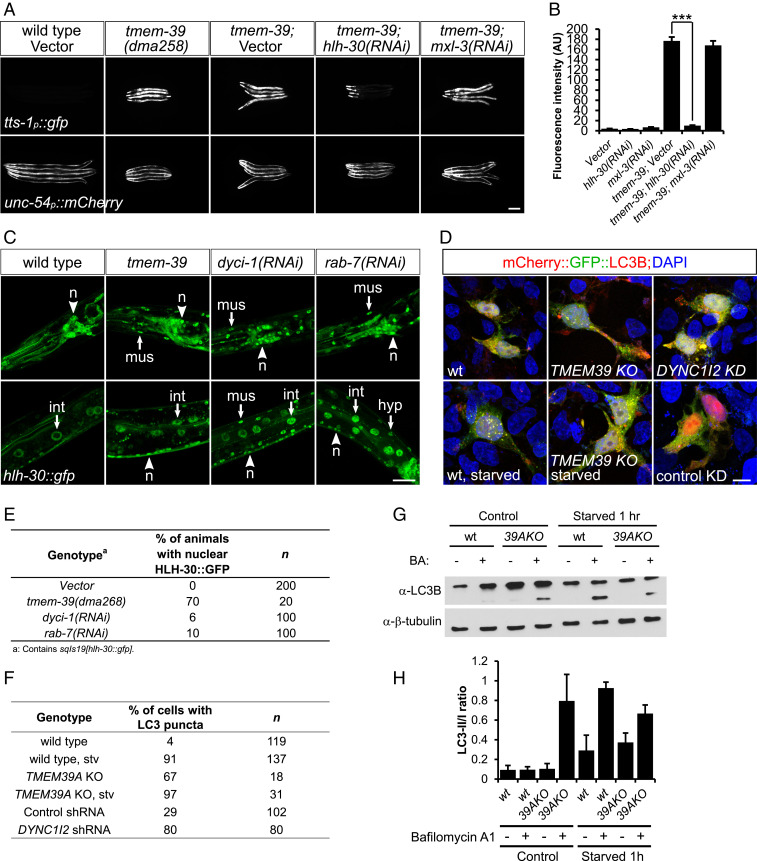
Lysosome distribution is highly regulated, and its positioning affects lysosomal functions in cellular material degradation, nutrient sensing, and autophagy signaling (12, 13). To determine if redistribution of lysosomes in TMEM39A KO cells and animals is associated with changes in lysosomal function, we examined lysosomal luminal enzyme activity in wild-type and mutant cells or worms using the Magic Red Cathepsin B assay (14). We did not detect a noticeable difference in Magic Red staining between wild-type and tmem-39 mutant worms or wild type and TMEM39A KO cells, suggesting that the degradative activity of lysosomal enzymes remains largely unimpaired in the absence of TMEM39A (SI Appendix, Fig. S2 I–L). In addition to its degradative function, the cytosolic surface of lysosomes also serves as an important hub for nutrient and mTOR signaling (15–19). Using a GFP reporter under the promoter of tts-1, which is activated by mTOR inhibition or starvation through the C. elegans transcription factor HLH-30 (ref. 20), we found that disruption of TMEM-39 in C. elegans strongly induced tts-1 reporter expression (Fig. 3 A and B). This indicates that mTOR signaling in tmem-39 mutant animals is reduced under nonstarved conditions in C. elegans.
Fig. 3.
TMEM-39 regulates mTOR signaling through the master regulator of lysosome biogenesis HLH-30/TFEB. (A) Expression of an mTOR reporter tts-1p::gfp is strongly up-regulated in tmem-39 mutant animals, while disruption of hlh-30/TFEB by RNAi abolishes tts-1p::gfp up-regulation. unc-54p::mCherry, coinjection reporter; mxl-3, bHLH transcription factor that antagonizes HLH-30 function in lipolysis upon starvation. (Scale bar, 100 μm.) (B) Quantification of tts-1p::gfp fluorescence intensity in A. Data represent mean ± SEM. ***P < 0.001. AU, arbitrary units. (C) Confocal images showing nuclear localization of HLH-30 in neurons (n), hypoderm (hyp), intestine (int), and muscle (mus) cells of animals deficient in tmem-39, dyci-1, or rab-7, but not in wild type. (Scale bar, 20 μm.) (D) Representative confocal images showing puncta formation of mCherry::gfp::LC3 in cells deficient in TMEM39A, DYNC1I2 or following 1 h of starvation. The presence of more red, but not yellow, LC3 puncta in TMEM39A KO cells indicates lysosome-mediated quenching of the GFP signal. (Scale bar, 10 μm.) (E) Quantification of HLH-30::GFP nuclear localization in C. (F) Quantification of mCherry::gfp::LC3 puncta formation in D. (G) Western blot analysis showing up-regulation of LC3-II/I ratio in nonstarved TMEM39A KO compared with wild-type HEK 293T cells, indicating enhanced autophagosome formation. BA, bafilomycin A1. (H) Quantification of the Western blot results. Data represent mean ± SEM. wt, wild type.
The mTOR complex phosphorylates a variety of downstream substrates, including the transcription factor EB (TFEB), a master regulator of lysosome biogenesis (21). Starvation or pharmaceutical inhibition of mTOR induces dephosphorylation of TFEB, which then translocates to nucleus and activates lysosome biogenesis-related gene transcription (22). Similarly, the C. elegans TFEB ortholog HLH-30 responds to nutritional status and activates autophagic gene expression upon starvation (23, 24). We reasoned that HLH-30 might be activated in mutant animals lacking TMEM-39 and examined HLH-30 expression in wild-type and tmem-39 mutant animals using a stably integrated hlh-30::gfp transgene. The HLH-30::GFP was primarily detected in the cytosol of neurons, muscles, and intestinal and hypodermal cells of wild-type animals (Fig. 3C). By contrast, HLH-30::GFP accumulated in nuclei of neuronal and nonneuronal cells of tmem-39 mutants, indicating activation of the transcription factor (Fig. 3 C and E). We then asked if HLH-30 is required for the strong tts-1p::gfp mTOR reporter expression induced by tmem-39 disruption. Feeding tmem-39 mutant animals with hlh-30 small interfering RNA (siRNA)-expressing bacteria, but not with mxl-3 (another bHLH factor) siRNA-expressing or vector bacteria, nearly abolished the mTOR reporter expression (Fig. 3 A and B). Therefore, we conclude that tmem-39 disruption in C. elegans likely inhibits lysosome-associated mTOR signaling, which induces strong tts-1p::gfp mTOR reporter expression through nuclear translocation and activation of HLH-30/TFEB.
Another cellular program regulated by mTOR signaling is autophagy, which is enhanced upon starvation or pharmaceutical inhibition of mTOR complex (17, 18). To find out if cells lacking TMEM39A have altered autophagy, we examined expression patterns of mCherry::gfp::LC3B, a tandem-fluorescence-tagged autophagosome protein in wild-type and TMEM39A KO HEK 293T cells (25). LC3B was primarily diffused in the cytoplasm of wild-type cells, and 1 h of starvation of the cells in the serum-free Earle’s Balanced Salt Solution medium induced strong formation of LC3B puncta that was indicative of autophagosome formation (Fig. 3 D and F). Strikingly, in cells lacking TMEM39A, LC3B clustered in puncta even under nonstarved conditions, and starvation further enhanced puncta formation (Fig. 3 D and F). Moreover, Western blot analysis using an anti-LC3B antibody detected enhanced accumulation of LC3-II in nonstarved TMEM39A KO cells, indicating enhanced autophagosome formation in the absence of TMEM39A (Fig. 3 G and H). Consistent with the observations, phosphorylation of Ser15 on Beclin-1, a molecular event that is indicative of amino acid starvation and mTOR inhibition and required for autophagy initiation (26), was enhanced in nonstarved TMEM39A KO cells (SI Appendix, Fig. S3C). Together, these findings support an important role of TMEM39A in maintaining normal mTOR signaling and suppressing autophagy initiation, and disrupting TMEM39A enhances autophagy, at least in HEK 293T cells.
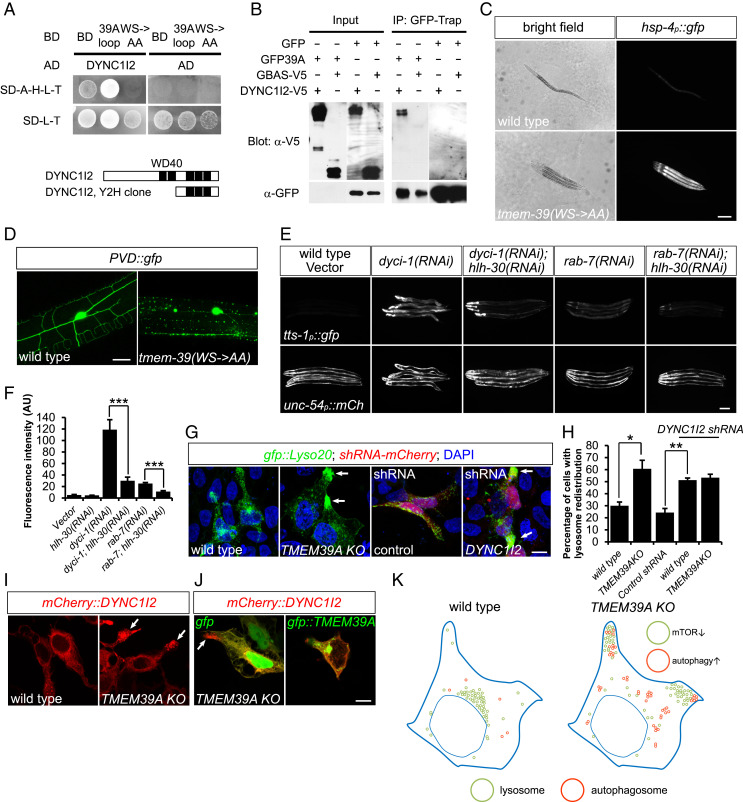
TMEM39A is predicted to contain nine transmembrane domains and two large loops (https://harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html), with the second loop highly conserved from C. elegans to human (Fig. 1A). To identify molecular mechanisms of how TMEM39A regulates lysosome dynamics, we performed yeast two-hybrid (Y2H) screens to identify proteins that interact with the second loop of TMEM39A. From a normalized universal human cDNA library, we identified 20 candidate interacting proteins, including the dynein intermediate light chain DYNC1I2 (Fig. 4A and SI Appendix, Table S1). The interaction appeared to require the conserved tryptophan-serine (WS) residues within the loop, as mutations of these amino acids to alanines abolished the interaction (Fig. 4A). We further confirmed the interaction between TMEM39A and DYNC1I2 in mammalian cells using coimmunoprecipitation (Fig. 4B). To test whether the interaction is functionally important in vivo, we generated CRISPR-knockin C. elegans strains, in which the conserved WS residues were replaced by alanines. Strains that carry such mutations in tmem-39 showed markedly stronger hsp-4p::gfp ER stress-reporter expression and PVD neuronal morphological defects, closely resembling the abnormalities seen in tmem-39 null mutant animals (Fig. 4 C and D).
Fig. 4.
TMEM39A regulates lysosome distribution and function through dynein intermediate light chain DYNC1I2. (A) Y2H assays showing the interaction between TMEM39A loop with DYNC1I2, which is dependent on the conserved WS residues within the TMEM39A loop. Schematic diagrams of the WD domain containing DYNC1I2 fragment recovered from the Y2H screen are shown. The 39A loop is the second cytosolic loop of TMEM39A. (B) Full-length DYNC1I2 was coimmunoprecipitated (IP) with GFP::TMEM39A, but not with GFP from transiently transfected HEK 293T cells. GBAS, another candidate protein identified from the Y2H screen, did not coimmunoprecipitate with GFP::TMEM39A. GFP::TMEM39A was detected in GFP-trap fraction, but not in total lysate, likely because of low expression levels. (C) Bright-field and compound fluorescence images showing the dumpy phenotype and elevated hsp-4p::gfp ER stress-reporter expression in tmem-39(WS->AA) CRISPR-knockin animals. (Scale bar, 100 μm.) (D) Confocal images showing PVD degeneration-like beading phenotype in tmem-39(WS->AA) CRISPR-knockin animals. (Scale bar, 20 μm.) (E) Disruption of dyci-1 or rab-7 by RNAi induces tts-1p::gfp mTOR reporter expression, which is abolished by hlh-30/TFEB deficiency. (Scale bar, 100 μm.) (F) Quantification of tts-1p::gfp intensity in E. Data represent mean ± SEM. ***P < 0.001. AU, arbitrary units. (G) Expression of DYNC1I2-targeting shRNA in HEK 293T cells induces redistribution of lysosomes to cell periphery, similar to that observed in TMEM39A KO cells (arrows). (Scale bar, 10 μm.) (H) Quantification of percentage of cells with lysosome redistribution in G. Data represent mean ± SEM. *P < 0.05; **P < 0.01. (I) Confocal images showing abnormal accumulation of mCherry::DYNC1I2 in cell periphery of TMEM39A KO cells (arrows). (J) Expression of gfp::TMEM39A, but not gfp, rescued mCherry::DYNC1I2 localization in TMEM39A KO cells. (Scale bar, 10 μm.) (K) Schematic diagram showing a model in which disruption of TMEM39A leads to lysosome redistribution to cell periphery, which affects lysosome-associated mTOR signaling and autophagy initiation.
Dynein is a family of cytoskeletal motor proteins responsible for cargo trafficking toward the minus end of microtubules and can be recruited to lysosomes via dynein light intermediate chain DYNC1LI1 and Rab-interacting lysosomal protein RILP (27, 28). To find out if dynein functions downstream of TMEM-39 to regulate lysosome distribution and biogenesis, we performed feeding RNAi to disrupt the expression of the sole homolog of dynein intermediate light chain dyci-1 in C. elegans. Knockdown of dyci-1 led to partial embryonic lethality and growth arrest, consistent with an essential role of the dynein subunit in early embryonic development (29). Strikingly, dyci-1 deficiency induced a strong increase in mTOR reporter tts-1p::GFP expression, as well as a moderate, but significant, increase in LMP-1::GFP, suggesting a starvation-like status in the RNAi-treated animals (Fig. 4 E and F and SI Appendix, Fig. S3 A and B). Disruption of hlh-30 by RNAi in the dyci-1–deficient animals abolished mTOR reporter tts-1p::GFP up-regulation, indicating that HLH-30 functions downstream of both TMEM-39 and DYCI-1 to activate mTOR reporter expression (Fig. 4 E and F). We further tested if RAB-7, RILP-1, or dynactin DNC-1 are required for regulating mTOR reporter expression and/or lysosome biogenesis, given their involvement in dynein–dynactin complex formation and recruitment of the complex to lysosomes (30). We found that disruption of the expression of rab-7 by RNAi induced a marked increase in the mTOR reporter tts-1p::GFP and lysosomal LMP-1::GFP expression, while deficiency in rilp-1 or dnc-1 caused only a moderate effect (Fig. 4 E and F and SI Appendix, Fig. S3 A and B).
We next asked if the role of dynein is also conserved in mammalian cells. The expression of DYNC1I2 in HEK 293T cells could be efficiently knocked down by using small-hairpin RNA (shRNA)-expressing plasmids (SI Appendix, Fig. S3 D and E). We found that disruption of DYNC1I2 induced lysosome redistribution to cell periphery in 51.2 ± 1.8% of the transfected cells, similar to that observed in TMEM39A KO cells (60.7 ± 7.1%) (Fig. 4 G and H), while knocking down DYNC1I2 in TMEM39A KO cells did not further enhance lysosome redistribution (53.3 ± 2.8%) (Fig. 4H). In addition, expression of shRNA against DYNC1I2, but not GFP control shRNA plasmid, induced strong puncta formation of autophagosome protein LC3B, indicating that DYNC1I2 and TMEM39A function similarly to regulate lysosome positioning and autophagy initiation (Fig. 3 D and F). Interestingly, mCherry::DYNC1I2 was redistributed to distal cell periphery, similar to that seen with lysosome reporter in TMEM39A KO cells, suggesting that TMEM39A promotes centripedal locomotion of both dynein and lysosomes (Fig. 4 I and J). Taken together, our findings support a model in which TMEM39A regulates lysosome positioning and associated signaling, at least in part through its interaction with the dynein intermediate light chain DYNC1I2 (Fig. 4K).
Discussion
TMEM39A is an evolutionarily conserved protein in which SNPs were previously identified in several human genetic studies with enhanced risks to MS and lupus. A recent study revealed a critical role of TMEM39A in regulating autophagy initiation (10). Here, we describe a key role of TMEM39A in regulating lysosome dynamics and mTOR signaling, in addition to autophagy initiation. We have also identified the dynein intermediate light chain DYNC1I2 as a mechanistic link between TMEM39A and downstream cellular processes. As MS is characterized by demyelination of neurons, owing to the immune-system attack of oligodendrocytes, the broad expression of TMEM39A and function in lysosomes indicate that it may contribute to MS pathology via lysosomal regulation in both a cell-autonomous (neurons) and/or nonautonomous manner (immune cells).
As an essential organelle in which antigens undergo processing and presentation through the major histocompatibility complex, lysosomes are essential for normal immune response (31). Dysregulation of lysosomes is also connected to pathological cytokine release, linking lysosome dysfunction to tissue destruction and autoimmunity (32, 33). In both SLE and MS, aberrant lysosomal activities have been postulated to underlie accumulation of autoantigens in lysosomes, which leads to aberrant immune responses and pathogenesis of the diseases (34, 35). In addition, dysregulated lysosomes also contribute to impaired cellular material and organelle degradation under conditions of aging, fertilization, and neurodegenerative diseases (18, 28, 36–40). Interestingly, TMEM39A RNAi has been shown to affect mitophagy (to degrade mitochondria by autophagy) mediated by the Parkinson’s disease protein Parkin (41, 42), and inhibition of mitochondrial ribosomes together with impaired mitochondrial fission or fusion activates HLH-30 and increases C. elegans lifespan (43).
TMEM39A has been shown to promote ER-to-Golgi transport by interacting with the COPII complex subunits SEC23/SEC24 (10). From our Y2H screen, we also identified SEC23A as an interacting partner of TMEM39A (SI Appendix, Table S1), supporting an important role of TMEM39A in regulating ER-to-Golgi trafficking. This is also consistent with our observation that disruption of TMEM-39 in C. elegans induces strong activation of the ER stress reporter hsp-4p::gfp (Figs. 1D and 4C). Interestingly, the C terminus of mammalian TMEM39A contains a putative COPI (which regulates Golgi-to-ER trafficking)-interacting lysine motif KAN, and the autoimmune risk allele rs1132200 confers an A-to-T mutation in the lysine motif that is predicted to impair interaction with COPI and possibly causes a loss-of-function of the protein (44). We speculate that TMEM39A is involved not only in anterograde (from ER to Golgi), but also in retrograde (from Golgi to ER), trafficking. These observations, together with our findings, support important roles of TMEM39A in regulating proper functions of multiple subcellular organelles, including the ER, lysosome, and autophagosome. Detailed mechanisms of how such multifaceted roles of TMEM39A are executed, coordinated, and regulated warrant further investigation.
To summarize, our results identify a previously uncharacterized function of TMEM39A in lysosome regulation. We propose a broadly evolutionarily conserved role of TMEM39A in promoting proper lysosome positioning and accumulation, which are important for regulation of mTOR signaling, autophagy initiation, and immune-cell responses under a variety of physiological and pathological conditions (28, 32, 36, 45–47).
Materials and Methods
All C. elegans strains were handled and maintained at 22 °C, and the N2 Bristol strain was used as the wild type. The dma258 and dma268 loss-of-function alleles and dma308 (WS->AA) knockin allele of tmem-39 were generated using the CRISPR-Cas9 system via germline transformation. The Ahringer and ORFeome RNAi libraries were used to knock down target gene expression in C. elegans. For mammalian cell experiments, HEK 293T cells were maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum and were grown at 37 °C with 5% CO2. The TMEM39A KO HEK 293T cells were generated using the CRISPR-Cas9 system via transfection and screening for deletion clones. The Sigma-Aldrich MISSION shRNA was used to knock down target gene expression in HEK 293T cells. Compound images were obtained using either an EVOS inverted microscope (Life Technologies) or a Leica CTR5000 compound microscope (Leica), and confocal images were obtained using a Zeiss LSM 700 or Leica SPE microscope. The processing, quantification and colocalization analysis of images were performed in Fiji software (NIH). Detailed materials and methods are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center and National BioResource Project in Japan for C. elegans strains, and Drs. Hong Zhang, Zimu Li, Anthony Shum, Scott Zamvil, and Jorge Oksenberg for helpful discussions. The work was supported by NIH Grant R01GM117461, a Pew Scholar Award, and a Packard Fellowship in Science and Engineering (to D.K.M).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011379118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Cooper G. S., Bynum M. L. K., Somers E. C., Recent insights in the epidemiology of autoimmune diseases: Improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun. 33, 197–207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng M. H., Anderson M. S., Monogenic autoimmunity. Annu. Rev. Immunol. 30, 393–427 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutierrez-Arcelus M., Rich S. S., Raychaudhuri S., Autoimmune diseases—connecting risk alleles with molecular traits of the immune system. Nat. Rev. Genet. 17, 160–174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao Q., et al., Genetic variants in TMEM39A gene are associated with autoimmune thyroid diseases. DNA Cell Biol. 38, 1249–1256 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Varadé J., et al., Replication study of 10 genes showing evidence for association with multiple sclerosis: Validation of TMEM39A, IL12B and CBLB [correction of CLBL] genes. Mult. Scler. 18, 959–965 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Cai X., Huang W., Liu X., Wang L., Jiang Y., Association of novel polymorphisms in TMEM39A gene with systemic lupus erythematosus in a Chinese Han population. BMC Med. Genet. 18, 43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCauley J. L., Hussman J. P.; International Multiple Sclerosis Genetics Consortium (IMSGC) , Comprehensive follow-up of the first genome-wide association study of multiple sclerosis identifies KIF21B and TMEM39A as susceptibility loci. Hum. Mol. Genet. 19, 953–962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessard C. J.et al.; BIOLUPUS Network; GENLES Network , Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am. J. Hum. Genet. 90, 648–660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J., et al., Recognition of transmembrane protein 39A as a tumor-specific marker in brain tumor. Toxicol. Res. 33, 63–69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miao G., Zhang Y., Chen D., Zhang H., The ER-localized transmembrane protein TMEM39A/SUSR2 regulates autophagy by controlling the trafficking of the PtdIns(4)P phosphatase SAC1. Mol. Cell 77, 618–632.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., et al., Broadly conserved roles of TMEM131 family proteins in intracellular collagen assembly and secretory cargo trafficking. Sci. Adv. 6, eaay7667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korolchuk V. I., et al., Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., et al., A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 18, 404–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J., et al., Metformin extends C. elegans lifespan through lysosomal pathway. eLife 6, e31268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J., et al., KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature 557, 585–589 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Wolfson R. L., et al., KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence R. E., Zoncu R., The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133–142 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Efeyan A., Zoncu R., Sabatini D. M., Amino acids and mTORC1: From lysosomes to disease. Trends Mol. Med. 18, 524–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mak K. H., et al., Lysosomal nucleotide metabolism regulates ER proteostasis through mTOR signaling. bioRxiv [Preprint] (2020) 10.1101/2020.04.18.048561 (Accessed 10 June 2020). [DOI] [Google Scholar]
- 20.Nakamura S., et al., Mondo complexes regulate TFEB via TOR inhibition to promote longevity in response to gonadal signals. Nat. Commun. 7, 10944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Settembre C., et al., TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Settembre C., et al., A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapierre L. R., et al., The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Rourke E. J., Ruvkun G., MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 15, 668–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura S., Noda T., Yoshimori T., Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Russell R. C., et al., ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan S. C., Scherer J., Vallee R. B., Recruitment of dynein to late endosomes and lysosomes through light intermediate chains. Mol. Biol. Cell 22, 467–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lie P. P. Y., Nixon R. A., Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol. Dis. 122, 94–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Rourke S. M., Dorfman M. D., Carter J. C., Bowerman B., Dynein modifiers in C. elegans: Light chains suppress conditional heavy chain mutants. PLoS Genet. 3, e128 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pu J., Guardia C. M., Keren-Kaplan T., Bonifacino J. S., Mechanisms and functions of lysosome positioning. J. Cell Sci. 129, 4329–4339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trombetta E. S., Mellman I., Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 23, 975–1028 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Ge W., Li D., Gao Y., Cao X., The roles of lysosomes in inflammation and autoimmune diseases. Int. Rev. Immunol. 34, 415–431 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Rigante D., Cipolla C., Basile U., Gulli F., Savastano M. C., Overview of immune abnormalities in lysosomal storage disorders. Immunol. Lett. 188, 79–85 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Nagata S., Hanayama R., Kawane K., Autoimmunity and the clearance of dead cells. Cell 140, 619–630 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Haves-Zburof D., et al., Cathepsins and their endogenous inhibitors cystatins: Expression and modulation in multiple sclerosis. J. Cell. Mol. Med. 15, 2421–2429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraldi A., Klein A. D., Medina D. L., Settembre C., Brain disorders due to lysosomal dysfunction. Annu. Rev. Neurosci. 39, 277–295 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Zhou Q., Li H., Xue D., Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res. 21, 1662–1669 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohnert K. A., Kenyon C., A lysosomal switch triggers proteostasis renewal in the immortal C. elegans germ lineage. Nature 551, 629–633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savini M., Zhao Q., Wang M. C., Lysosomes: Signaling hubs for metabolic sensing and longevity. Trends Cell Biol. 29, 876–887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y., et al., Lysosome activity is modulated by multiple longevity pathways and is important for lifespan extension in C. elegans. eLife 9, e55745 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orvedahl A., et al., Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480, 113–117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran Q., et al., TMEM39A and human diseases: A brief review. Toxicol. Res. 33, 205–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali L., Haynes C. M., Mitochondrial translation, dynamics, and lysosomes combine to extend lifespan. J. Cell Biol. 219, 1–2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma W., Goldberg J., Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J. 32, 926–937 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonam S. R., Wang F., Muller S., Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 18, 923–948 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korolchuk V. I., Rubinsztein D. C., Regulation of autophagy by lysosomal positioning. Autophagy 7, 927–928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y. G., Zhang H., Core autophagy genes and human diseases. Curr. Opin. Cell Biol. 61, 117–125 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.