Abstract
Anaplastic oligodendroglioma (AO), IDH‐mutant and 1p/19q codeleted (IDHmut+/1p19qcodel), is a high‐grade glioma with only limited prognostic markers. The primary objective of this study was to evaluate, by immunohistochemistry, the prognostic value of two proliferation markers, MCM6 and Ki‐67, in a large series of IDHmut+/1p19qcodel AO included in the POLA (“Prise en charge des Oligodendrogliomes Anaplasiques”) French national multicenter network. We additionally examined the transcriptome obtained from this series to understand the functional pathways dysregulated with the mRNA overexpression of these two markers. The labeling indices (LI) of MCM6 and Ki‐67 were obtained via computer‐assisted color image analyses on immunostained AO tissues of the cohort (n = 220). Furthermore, a subgroup of AO (n = 68/220) was used to perform transcriptomic analyses. A high LI of either MCM6 (≥50%) or Ki‐67 (≥15%) correlated with shorter overall survival, both in univariate (P = 0.013 and P = 0.004, respectively) and multivariate analyses (P = 0.027; multivariate Cox model including age, mitotic index, MCM6 and Ki‐67). MCM6 and Ki‐67 LI also correlated with overall survival in an additional retrospective cohort of 30 grade II IDHmut+/1p19qcodel oligodendrogliomas. The prognostic value of MCM6 mRNA level was confirmed in The Cancer Genome Atlas (TCGA) IDHmut+/1p19qcodel gliomas. The transcriptomic approach revealed that high transcriptional expressions of MCM6 and MKI67 were both linked positively with cell cycle progression, DNA replication, mitosis, pro‐neural phenotype as well as neurogenesis, and negatively with microglial cell activation, immune response, positive regulation of myelination, oligodendrocyte development, beta‐amyloid binding and postsynaptic specialization. In conclusion, the overexpression of MCM6 and/or Ki‐67 is independently associated to shorter overall survival in IDHmut+/1p19qcodel AO. These two easy‐to‐use and cost‐effective markers could thus be used concurrently in routine pathology practice. Additionally, the transcriptomic analyses showed that AO with high proliferation index have down‐regulated immune response and lower microglial cells activation, and bears pro‐neural phenotype.
Keywords: 1p/19q codeletion, anaplastic oligodendroglioma, glioma, immune response, immunohistochemistry, Ki‐67, MCM6, proliferation, pro‐neural, transcriptomics
Introduction
According to the World Health Organization (WHO), IDH‐mutant and 1p/19q‐codeleted (IDHmut+/1p19qcodel) anaplastic oligodendrogliomas (AOs) are high‐grade gliomas, corresponding histologically to WHO grade III primary brain tumors 35. AO are rare, accounting for 0.5% of all primary brain tumors and one‐third of oligodendroglial tumors 38. They are histologically defined as tumors of the oligodendroglial cells, with features of anaplasia, including increased cellularity and mitotic activity, microvascular proliferation and/or necrosis. Although Ki‐67 labeling index (LI) is usually used to help differentiate low‐grade from high‐grade oligodendroglioma, no clear cut‐off point has been established 12. By definition, and according to the 2016 update of the WHO Classification of Central Nervous System, AO carry both IDH1/2 mutations and 1p/19q codeletion 10, 43. The standard of care for these tumors includes surgical resection in conjunction with adjuvant therapies like combined adjuvant radiochemotherapy with procarbazine, CCNU (lomustine), and vincristine (RT‐PCV) to improve the overall survival (OS) 46.
The prognostic role of the WHO grading in IDH‐mutant gliomas remains debatable, and this is particularly true in the case of oligodendrogliomas 43. In AO patients, young age, high Karnofsky index/clinical performance status, complete surgical resection and adjuvant therapy such as RT‐PCV have been shown to associate with longer OS 45, 46. Histological features such as microvascular proliferation, mitotic index (MI) and necrosis have been found associated with three distinct prognostic subgroups of IDH‐mut+/1p19qcodel AO with other genomic alterations 19. These include mutations of CIC and TCF12, both linked to more aggressive AO 4, 22, 32, and allelic loss of 9p21.3, associated with shorter progression‐free survival (PFS) and shorter OS in IDH‐mut+/1p19qcodel AO 2. Omics approaches using transcriptome, genome and methylome have also been used to identify potential prognostic factors. One such study has successfully classified oligodendroglial tumors into three molecular subgroups with distinct clinical behaviors 29. Machine learning algorithms aiming to identify copy number variations have also been applied recently to AO samples; such technique seems promising in helping with prognostic stratification 41. Despite of these advances, however, it is worth noting that all the above‐mentioned markers of AO need expansive and time‐consuming molecular analyses, and thus cannot be widely applied to all samples in all laboratories.
In contrast, immunohistochemical proliferation markers such as Ki‐67, whose LI correlates strongly with prognosis of many tumor types, are simple, effective and economical means to assess tumor aggressivity. In the case of AO, past few evidence obtained using univariate—but not multivariate—analysis appeared to support the prognostic value of Ki‐67 LI 40. However, as less than 50% of the AO tissues in this latter study (performed prior to the 2016 revision of the WHO classification) actually harbored IDH mutations and 1p/19q codeletion, its validity shall be questioned. In a more recent study, Zeng et al evaluated the prognostic value of Ki‐67 upon classifying gliomas into subgroups with either the presence or the absence of IDH1/2; however, data concerning 1p/19q values codeletion were lacking in the studied gliomas 49.
Several proliferation markers other than Ki‐67 have also been studied in brain tumors. Notably, we previously reported that Minichromosome Maintenance Complex component 6 (MCM6) was overexpressed in meningioma, and was associated with higher histological grade and risk of recurrence 21. In adamantinomatous craniopharyngiomas, MCM6 correlated with a higher risk of long‐term recurrence 48. In gliomas, especially in glioblastomas of the Chinese Cancer Genome Atlas (CCGA), MCM6 mRNA overexpression was also reported to be correlated with poor overall survival 9.
Minichromosome Maintenance proteins (MCMs) play a key role in DNA synthesis and replication, forming a hexameric helicase complex around the DNA 31. All MCMs (MCM2‐7) are detectable during the different phases of the cell cycle, including G1, S, G2 and M, but are absent in G0 33. These proteins are also expressed earlier during G1, in comparison with Ki‐67. In other solid tumors, like non‐small cell lung carcinomas 47, hepatocellular carcinomas 34, endometrial carcinomas 26, low‐grade chondrosarcomas 24 and mantle cell lymphomas 42, a high MCM6 LI correlated with a worse prognosis. To our knowledge since the revision of the 4th WHO classification of 2016, MCMs, and notably MCM6, have never been specifically studied in IDHmut+/1p19qcodel AO.
The primary goal of this study was therefore to evaluate and compare, by immunohistochemistry, the prognostic value of MCM6 and Ki‐67 in a large series of IDHmut+/1p19qcodel AO obtained from the POLA (“Prise en charge des Oligodendrogliomes Anaplasiques”) French national multicenter network. We additionally examined the transcriptomes obtained from part of this series to understand the functional pathways dysregulated with the mRNA overexpression of these two markers.
Material and Methods
Population and clinicopathological data
Two hundred and thirty‐one cases of IDHmut+/1p19qcodel AO were retrieved from the French national multicenter POLA cohort. Clinical data, such as age, sex, extent of surgical removal, type of adjuvant treatment and OS are available in the database, as well as molecular data, including IDH mutation status and presence of 1p/19q codeletion. All cases included in this cohort have been centrally reviewed by the neuropathologists of the national board of French national POLA network and were classified according to the 2016 4th WHO classification update. The MI and Ki‐67 LI were evaluated in whole tissue sections, as previously described 18. In 220 out of 231 patients, tissue microarray (TMA) blocks were designed with representative samples of tumors, in order to perform immunohistochemical analyses. One to three spots were available for each case.
Additionally, in order to evaluate the prognostic value of MCM6 and Ki‐67 LI in low‐grade tumors, 30 cases of grade II IDHmut+/1p19qcodel oligodendrogliomas (IDHmut+/1p19qcodel OII) were retrieved from the files of the Department of Neuropathology at Pitié‐Salpétrière Hospital (AP‐HP, Paris; OncoNeuroTek database; Pitié‐Salpétrière) and the Department of Pathology at Nancy University Hospital (CHRU, Nancy; Centre de Ressources Biologiques, BB‐0033‐00035).
Ethics
Anonymity was strictly respected, according to the principles of the declaration of Helsinki and national ethical guidelines. Patients consent for clinical data collection and genetic analyses have been obtained prospectively, according to POLA network policies. The study was approved by the ethics committee of Hôpital Universitaire la Pitié‐Salpêtrière.
Immunohistochemistry
Paraffin sections of 5 μm thickness were immersed in a 10mM sodium citrate buffer (pH 6) for 20 minutes at 97°C for dewaxing and antigen retrieval. The following primary antibodies were used: MCM6 (1/400; goat polyclonal, Santa Cruz Biotechnology, Heidelberg, Germany), Ki‐67 (1/200; mouse monoclonal, MIB‐1, Dako Cytomation, Glostrup, Denmark).
Immunohistochemistry was performed with Dako Autostainer Plus (Dako) and the Flex + Envision revelation system (Dako).
Ki‐67 and MCM6 evaluation
Ki‐67 and MCM6 labeling indices (LI) were defined as the percentage of cells with positive nuclear stain of the two individual markers, independently from the signal intensity. For each analyzed TMA spot, the field with the strongest immunostaining was selected by the observer, blinded to the clinical data and outcome of the respective patients. To limit inter‐observer variability, cell counting was performed with a computerized color image analyzer (Olympus Cellsens Dimension, Olympus Medical System and Micro‐Imaging Group, Hamburg, Germany). A x20 objective was used to take one microphotograph of the area of interest. A minimal object size of 50 pixels was required to count positive nuclei. Vessels and microcalcifications were excluded from the analysis. The percentage of positive and negative cells were automatically computed by the color image analyzer. A mean LI was calculated for each case. Additionally, in order to validate this computer‐based LI, manual Ki‐67 and MCM6 LI were evaluated in 50 randomly selected cases, by counting 1000 cells in TMA spots.
Statistical analysis
Statistical analyses were performed using IBM SPSS Statistics for Windows, Version 23.0 (IBM Corp., Armonk, NY, USA).
Because the quantitative variables did not pass the Kolmogorov–Smirnov normality test, non‐parametric tests were used. Spearman correlation test was used to explore the correlation between Ki‐67 LI, MCM6 LI and the MI. The concordance between image analysis and manual LI was evaluated with the intraclass correlation coefficient. The correlation between MCM6 and Ki‐67 LI and WHO grade was evaluated with the Mann–Whitney Wilcoxon test. Overall survival (OS) analyses were performed with Kaplan–Meier estimation (log‐rank test) and the Cox model using univariate and multivariate analyses. For Ki‐67 and MCM6 LI, the optimal threshold was computed using the Cutoff Finder online tool 8, in order to separate patients in two groups. For Cox multivariate analyses, only variables which were significant on univariate analyses and WHO grade were integrated into the model. Survival analyses were performed in the POLA cohort of IDHmut+/1p19qcodel AO, then in the IDHmut+/1p19qcodel OII cohort. A P‐value (P) of less than 0.05 was considered statistically significant.
Transcriptomic analyses
POLA cohort: Gene expression data from POLA 29 were downloaded from http://gliovis.bioinfo.cnio.es/ under the “Kamoun” and “POLA Network” tags. From these publicly available data, we identified 68 samples satisfying the following phenotypic criteria: histology = oligodendroglioma, grade = III (anaplastic), co‐deletion 1p‐19q status = yes, IDH1 or IDH2 status = mutant. These data were normalized (lowess algorithm) against a median profile of all 68 samples and further underwent descriptive statistics, which helped define groups of samples before proceeding with downstream analyses. Hence, all samples were individually labeled MCM6‐ and MKI67‐up or ‐down based on their relative MCM6 and MKI67 expression (Supplementary Figure S1).
K‐means clustering, functional annotations, enrichments computations, differential expression statistics and renderings were achieved using protocols and tools as described before 20. Further hierarchical clustering was applied for each significant k‐means cluster to evaluate the level of correlation and quickly delineate gene or sample outliers in each gene signature.
Differential expression P‐values were computed on normalized data using a two‐way t‐test between both MCM6 and MKI67 up and down groups. These were adjusted to allow for false discovery rate (FDR) using the Benjamini–Hochberg procedure and FDR < 0.01 was considered to indicate statistical significance.
Linear correlations between MCM6 and MKI67 expression levels were performed with Pearson's method, while Spearman's rank correlation coefficients were used to explore the dependence between both MCM6 and MKI67 gene expressions with their respective MCM6 and Ki‐67 immunohistochemical LI.
TCGA cohort: Additionally, in order to evaluate the results obtained on the POLA cohort, further analyses were run on a subset of The Cancer Genome Atlas Lower Grade Glioma cohort (TCGA‐LGG). From the available RNA‐Seq processed data (FPKM‐UQ normalized counts), 98 samples corresponded to unique cases with the following criteria: IDH1 and/or IDH2 mutated, presence of the 1p‐19q codeletion, and clearly clinically labeled as grade III and/or Anaplastic OD (35 samples) or grade II OD (63 samples). The original 60483 gene Ensembl ID were re‐annotated (using https://biotools.fr) and their expression values were aggregated to obtain a set of unique official gene symbols. Data were then filtered to remove genes with very low expression values across samples (FPKM‐UQ < 10 in more than 9/10th of the samples), which left 16345 genes. Values were re‐normalized according to library size and log2 transformed. Two datasets were then created, one set containing the 35 grade III samples, with the aim to conduct validation analyses of the POLA high‐grade OD transcriptomics, and another set with the entire selected TCGA cohort of 98 grade II + grade III samples for extending the results to lower grade OD. These two data sets finally underwent the same unsupervised workflow as described above (sample ordering according to MCM6 mRNA expression levels, k‐means clustering, functional annotations, enrichment analyses).
Results
Clinicopathological data
The mean age of the patients of the POLA cohort was 49 years (range 19–80) (Table 1), with a male‐to‐female ratio of 1.22:1. A large majority of patients had surgery first, with 33% (72/220) of them showing macroscopic total resection, 31% (68/220) subtotal resection and 21% (46/220) partial resection. As adjuvant therapy, 32% (71/220) received RT‐PCV therapy and 21% (46/220) combined radiotherapy with Temozolomide (Stupp Protocol). For 29% (64/220) of them, adjuvant radiotherapy was chosen. Fourteen patients did not receive any treatment (6%; 14/220). Follow‐up data were available for 220 patients and were collected over a median follow‐up of 40.9 (0.3–92) months. Disease‐related death occurred in 9% (19/220) of patients. Using univariate analyses, age was the only variable significantly associated with survival (P = 0.001; HR: 1.055 [95%CI: 1.021–1.091]) (Table 2). A high MI (at least eight mitoses per 1.6 mm2) was significantly associated with a shorter OS (P = 0.018; HR: 2.587 [95%CI: 1.177–5.686]). No significant correlation between OS and microvascular proliferation (P = 0.511; HR: 1.493 [95%CI: 0.451–4.939] or necrosis (P = 0.192; HR: 1.648 [95%CI: 0.778–3.492] was found.
Table 1.
Clinico‐pathological characteristics and molecular data.
| Variable | Results |
|---|---|
| Age (mean; min‐max) | 49; 19–80 years |
| Sex | Male‐to‐female ratio: 1.22:1 (121/99) |
| Surgery | Biopsy: 4.1% (9/220) |
| Total resection: 32.7% (72/220) | |
| Subtotal resection 30.9% (68/220) | |
| Partial resection 20.9% (46/220) | |
| Missing data 11.4% (25/220) | |
| Type of treatment | RT‐PCV: 32.3% (71/220) |
| Radiotherapy: 29.1% (64/220) | |
| PCV: 2.3% (5/220) | |
| Stupp protocol: 20.9% (46/220) | |
| Temozolomide: 4.1% (9/220) | |
| Other: 0.5% (1/220) | |
| No treatment: 6.4% (14/220) | |
| Missing data: 3.6% (8/220) | |
| Survival | Progression: 30.7% (71/220) |
| Death: 8.2% (19/220) | |
| Molecular data | TERT promoter mutation: 98.3% (216/220) |
| CIC loss: 61% (141/220) |
RT‐PCV = radiation therapy – procarbazine, CCNU (lomustine), and vincristine.
Table 2.
Univariate and multivariate Cox analyses for overall survival.
| Variable | Cox univariate (OS) | Cox multivariate (OS) Model 1 | Cox multivariate (OS) Model 2 | |||
|---|---|---|---|---|---|---|
| HR [95%CI] | P‐value | HR [95%CI] | P‐value | HR [95%CI] | P‐value | |
| Age | 1.055 [1.021–1.091] | 0.001* | 1.060 [1.020–1.103] | 0.003* | 1.051 [1.011–1.092] | 0.012 * |
| Mitotic index ≥ 8/1.6 mm2 | 2.587 [1.177–5.686] | 0.018* | 1.439 [0.538–3.851] | 0.469 | 1.588 [0.606–4.162] | 0.347 |
| MCM6 LI ≥ 50% | 3.283 [1.221–8.826] | 0.018* | 2.896 [0.964–8.702] | 0.058 | ||
| Ki‐67 LI ≥ 15% | 3.948 [1.442–10.41] | 0.008* | 2.713 [0.935–7.875] | 0.066 | ||
| MCM6 LI ≥ 50% and/or Ki‐67 LI ≥ 15% | 3.875 [1.603–9.370] | 0.003* | 2.872 [1.125–7.328] | 0.027 * | ||
HR = hazard ratio; LI = labeling index; OS = overall survival.
Statistically significant (P < 0.05).
In the grade II series, mean age was 44 years (range 18–81), with a male‐to‐female ratio of 1.21:1. Disease‐related death occurred in 13% (4/30) of patients, with a median follow‐up of 38.7 (2.4–79.9) months. When combining the two cohorts, WHO grade was not correlated to OS (P = 0.642).
MCM6 labeling index and correlation with survival
MCM6 staining was interpretable in 94% (206/220) of the cases with an average count of 1383 cells per case. Image analysis‐based LI was strongly correlated to manual counting (ICC: 0.864; 95%CI [0.772–0.920]). Mean MCM6 LI was 24% (range 0.1–87%; median 21.4; standard deviation 18.8) (Figure 1A,B). MCM6 LI was significantly correlated to MI (ρ = 0.253; P < 0.0001).
Figure 1.
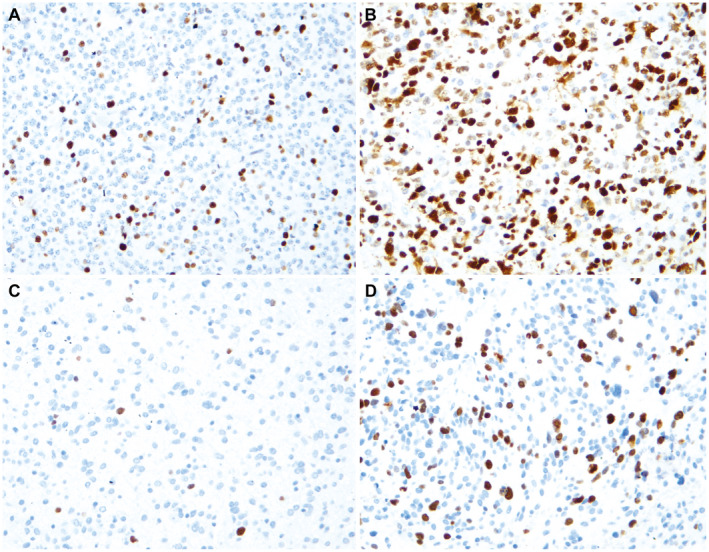
Immunolabeling for MCM6 and Ki‐67 (immunohistochemistry, x200). A. Low MCM6 labeling index (LI). B. High MCM6 LI. C. Low Ki‐67 LI. D. High Ki‐67 LI (same case as B).
In the POLA cohort, log‐rank survival analyses showed that patients with high MCM6 expression (upper than 50%) had significantly shorter overall survival (OS) (P = 0.013) (Figure 2A). Using univariate Cox model, MCM6 expression correlated inversely with OS (P = 0.018; HR: 3.28; 95%CI [1.22–8.83]) (Table 2). In the group of patients with grade II oligodendroglioma, a high MCM6 LI was also correlated to a shorter OS (P = 0.001) (Figure 3A). MCM6 LI was not significantly higher in grade III than in grade II oligodendrogliomas (23% vs. 18%; P = 0.232).
Figure 2.
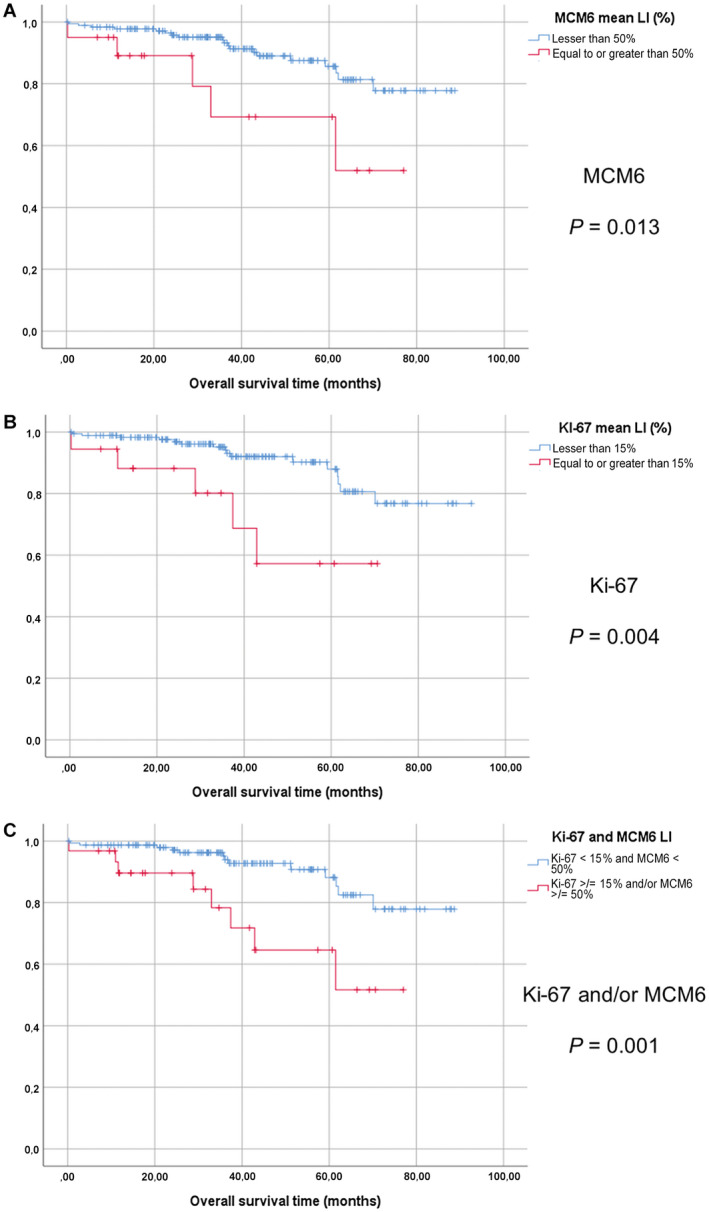
Survival analyses in the POLA cohort of anaplastic oligodendroglimas. Kaplan‐Meier curves with log‐rank tests (overall survival). A. MCM6 labeling index, 50% threshold (LI). B. Ki‐67 LI, 15% threshold. C. MCM6 LI ≥ 50% and/or Ki‐67 LI ≥ 15% vs. MCM6 < 50% and Ki‐67 LI < 15%.
Figure 3.
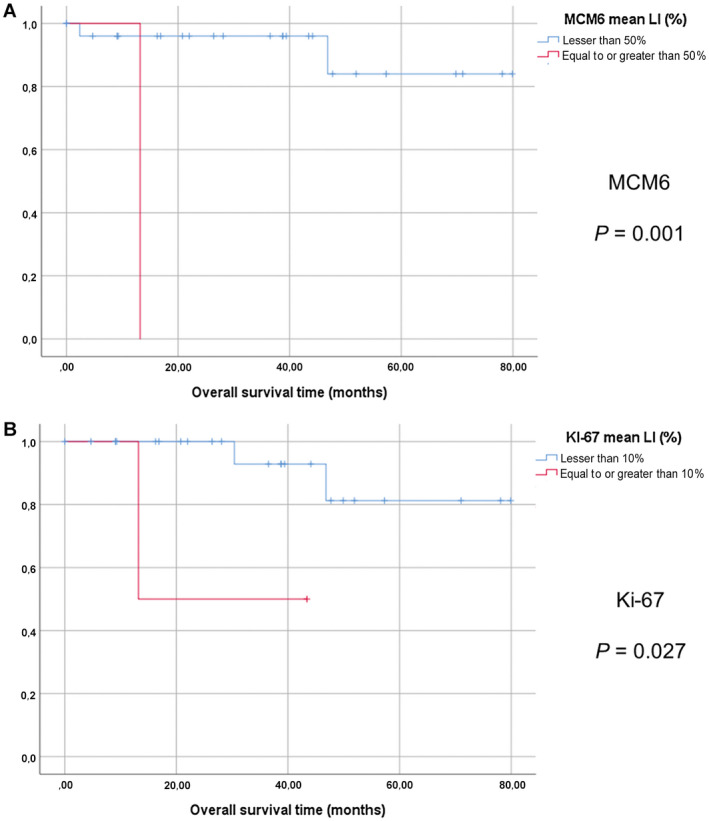
Survival analyses the cohort of grade II oligodendrogliomas. Kaplan‐Meier curves with log‐rank tests (overall survival). A. MCM6 labeling index, 50% threshold (LI). B. Ki‐67 LI, 10% threshold.
Ki‐67 labeling index and correlation with survival
For Ki‐67 LI, 91% (199/220) of the cases were interpretable. We found a high reproducibility between image analysis and manual counting (ICC: 0.900; 95%CI [0.827–0.943]). Ki‐67 LI evaluated by image analysis was also strongly correlated to the Ki‐67 LI previously obtained from whole slides (ρ = 0.553; P < 0.0001). The mean number of cells counted for each TMA was 1121. The mean Ki‐67 LI was 6.3% (range 0.1–36.9; median 3.7; standard deviation 6.7) (Figure 1C,D). Ki‐67 LI was significantly correlated to MI (ρ = 0.275; P < 0.0001) and MCM6 LI (ρ = 0.449; P < 0.0001).
The log‐rank test and the univariate regression Cox model revealed that a Ki‐67 LI equal to or greater than 15% was correlated with a shorter OS (log‐rank: P = 0.004; Cox: P = 0.008; HR = 3.948; 95%CI [1.442–10.41]) (Figure 2B; Table 2). Similarly, in the group of patients with grade II oligodendroglioma, a high Ki‐67 LI was correlated to shorter OS (P = 0.027; log‐rank test) (Figure 3B). Ki‐67 LI was significantly higher in grade III than in grade II tumors (6% vs. 3%; P = 0.001).
Multivariate survival analyses
The variables that were significant in univariate analyses (age, MI ≥ 8/1.6 m2, MCM6 LI ≥ 50%, Ki‐67 LI ≥ 15%) were included in the first multivariate Cox model. Only age remained significantly correlated to OS (P = 0.003), whereas MCM6 and Ki‐67 LI were close to significance (P = 0.058 and P = 0.066, respectively) (Table 2).
In order to evaluate if MCM6 could be an interesting marker to use in complement to Ki‐67, we designed a second Cox multivariate model, with the variable “MCM6 LI ≥ 50% and/or Ki‐67 LI ≥ 15%”, that was strongly significant in univariate analysis (log‐rank: P = 0.001; Cox: P = 0.003; HR: 3.875; 95%CI [1.603–9.370]). With this model, age and “MCM6 LI ≥ 50% and/or Ki‐67 LI ≥ 15%” were significantly correlated to OS (P = 0.012 and P = 0.027, respectively), but not MI (P = 0.347) (Figure 2C; Table 2).
Additionally, we evaluated the prognostic value of MCM6 and Ki‐67 LI in all cases of IDHmut+/1p19qcodel oligodendrogliomas, including grade II and III tumors. In a multivariate model including age, MI, WHO grade and the two proliferation markers, we found that age (P = 0.022) and MCM6 LI ≥ 50% and/or Ki‐67 LI ≥ 15% (P = 0.001; HR: 4.148; 95%CI [1.735–9.920] were significantly associated with OS, but not WHO grade (P = 0.108) or MI (P = 0.637).
Transcriptomics in the POLA cohort
For both MCM6 and MKI67, we expected positive correlations between their LI and mRNA level in AO cases for which both transcriptomic data and LI were available. The Spearman tests indeed confirmed such correlations (ρ = 0.43, P = 0.003, n = 46; and ρ = 0.63, P = 4.7 × 10−5, n = 38, respectively) (Supplementary Figure S2). A highly positive correlation was also found between the mRNA level of MCM6 and that of the MKI67 in the AO transcriptomes of the POLA network (n = 68; Pearson's test: 0.69; P = 5.6 × 10−11) (Supplementary Figure S3).
Molecular events associated with differential expressions of either MCM6 or MKI67 in these AO samples were subsequently analyzed (n = 68). This was achieved using the median expression levels for MCM6 and MKI67 as “up” and "down" subgroup delimiters (Supplementary Figure S1) in two separate analyses based on the relative expressions of MCM6 and MKI67. When the median MCM6 mRNA level was used to organize the 68 AO samples, 34 were classified and labeled as MCM6‐down (sample expression < median expression), and the remaining 34 as MCM6‐up (expression > median). The same was applied for MKI67. In these two analyses, k‐means clustering coupled with a distance computation based on the weighted correlation method, permitted the identification of clusters with distinct molecular functions differentiating the “up” from the “down” MCM6 or MKI67‐expresser AO (Figure 4). Remarkably, these MCM6‐ and MKI67 gradient‐driven clusterings resulted in individual clusters sharing high level of identity, as the clusters produced between the MCM6 and MKI67 series of experiments overlapped with each other (ranging from 85% to 99%, the most significant clusters presenting with the greatest identity) (Figure 4). Moreover, among the 10 clusters identified in each series (from c0 to c9), 2 clusters of upregulated genes (c3 and c7) and 3 clusters of downregulated genes (c1, c2 and c6) were highly conserved between the two series (98%, 99%, 97%, 97%, 96%, respectively; Figure 4). Further functional annotation of the up‐ and downregulated clusters revealed that tumors expressing higher levels of MCM6 and MKI67 were also enriched with genes controlling replication and cell cycle (c3 and c7), but downregulated with genes associated with synaptic activity (c1), neuron ensheathment, glial cell development, immune and inflammatory response, vitamin biosynthetic process (c2), and alpha amino acid metabolic process (c6) (FDR < 0.0001) (Supplementary Table S1).
Figure 4.
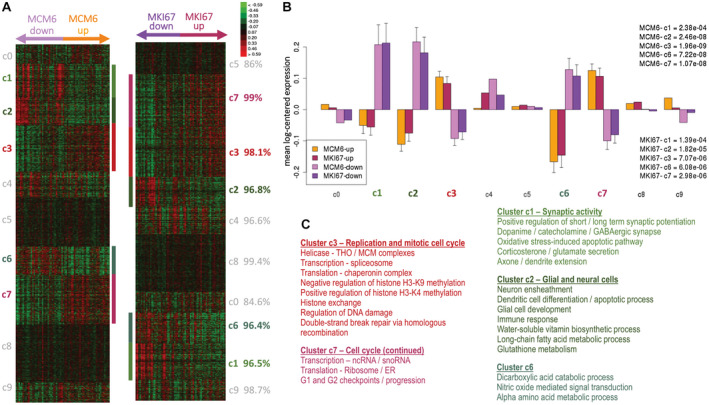
K‐means clustering from MCM6 and MKI67 expression gradients (from lowest to highest) of 68 transcriptomes of anaplastic oligodendrogliomas from the POLA network. The two independent K‐means clustering each delineated five significant groups of co‐expressed genes (P < 0.01) showing highly correlated expression profiles. A. Heatmap of the ten identified k‐means clusters in the two clustering results, and lowest identity (in %) found for each cluster (green, red and black indicate downregulated, upregulated, and median genes, respectively). B. Mean gene expression (overall with standard error at the mean) for each cluster in MCM6 and MKI67 analyses where both upregulated samples for MCM6 or MKI67 are compared to their downregulated counterparts (AU – arbitrary units). Upregulated and downregulated samples are defined relatively to the median gene expression for either MCM6 or MKI67. C. Overview of the most relevant functional annotations for the significantly over‐ and under‐expressed clusters (in red and green hues, respectively, all p(FDR) ≤ 0.01).
Additionally, we explored differential expression statistics (two‐sided t‐test) between MCM6‐up and MCM6‐down, and between MKI67‐up and MKI67‐down samples. Four significant gene lists were produced (all FDR < 0.01), one list for over‐ and one for under‐expressed genes in MCM6‐up samples (871 and 461 genes; fold changes > 4/3 and < 3/4, respectively), and likewise for MKI67‐up samples (352 and 237 genes, respectively) (Supplementary Table S2). MCM6 overexpressed samples presented with a significant enrichment in genes involved in cell cycle functions (Table 3), including most notably DNA replication, mitotic centrosome separation and mitotic chromosome condensation and genes involved in internal ribosome entry site (IRES) dependent translational initiation. Tumors harboring lower MCM6 expression showed significant upregulation of pathways involved in glial differentiation and immune response, such as microglial cell activation, myelination, oligodendrocyte development and oligodendrocyte differentiation. Similarly, tumors with higher expression of MKI67 (Table 4) were enriched in genes involved with cell cycle functions such as DNA strand elongation in DNA replication during cell cycle, pre‐replicative complex assembly, mitotic chromosome condensation and MCM complex. Tumors with lower MKI67 expression were significantly enriched in genes involved in beta‐amyloid binding, synaptic vesicle, axon terminus, trans‐synaptic signaling, neuronal cell body and cell projection.
Table 3.
Top 12 Gene Ontology functional annotations associated with the expression of MCM6 (sorted by fold enrichment).
| GO biological process | Fold enrichment | FDR |
|---|---|---|
| Enrichment in tumors showing MCM6 higher expression | ||
| Cell cycle DNA replication initiation (GO:1902292) | 22.1 | <0.0001 |
| DNA strand elongation involved in cell cycle DNA replication (GO:1902296) | 21.7 | 0.0001 |
| Ndc80 complex (GO:31262) | 21.7 | 0.0001 |
| Pre‐replicative complex assembly (GO:36388) | 21.3 | <0.0001 |
| Condensed nuclear chromosome kinetochore (GO:778) | 15.2 | <0.0001 |
| Mitotic centrosome separation (GO:7100) | 15.2 | 0.001 |
| Mitotic chromosome condensation (GO:7076) | 14.8 | <0.0001 |
| IRES‐dependent translational initiation (GO:2192) | 14.6 | 0.0001 |
| Aster (GO:5818) | 13.8 | 0.0001 |
| Exodeoxyribonuclease activity (GO:4529) | 12.6 | 0.003 |
| Mitotic prophase (GO:88) | 12.1 | <0.0001 |
| Regulation of transcription involved in G1/S transition (GO:83) | 11.9 | <0.0001 |
| Enrichment in tumors showing MCM6 lower expression | ||
| Myelin sheath adaxonal region (GO:35749) | 11.2 | 0.009 |
| Negative regulation of inclusion body assembly (GO:90084) | 8.7 | 0.02 |
| Sodium ion export from cell (GO:36376) | 8.7 | 0.02 |
| Positive regulation of organic acid transport (GO:32892) | 8.2 | 0.02 |
| Microglial cell activation (GO:1774) | 8.4 | <0.0001 |
| Macrophage activation involved in immune response (GO:2281) | 8.2 | 0.009 |
| Central nervous system myelination (GO:22010) | 7.6 | 0.002 |
| Positive regulation of myelination (GO:31643) | 6.5 | 0.02 |
| Main axon (GO:44304) | 4.6 | <0.0001 |
| Proteoglycan binding (GO:43394) | 4.5 | 0.01 |
| Oligodendrocyte development (GO:14003) | 4.2 | 0.005 |
| Positive regulation of glial cell differentiation (GO:45687) | 4.0 | 0.02 |
FDR = false discovery rate; GO = gene ontology.
Table 4.
Top 12 Gene Ontology functional annotations associated with the expression of MKI67 (sorted by fold enrichment).
| GO biological process | Fold enrichment | FDR |
|---|---|---|
| Enrichment in tumors showing MKI67 higher expression | ||
| Condensin complex (GO:796) | 50.2 | <0.0001 |
| Replication fork protection complex (GO:31298) | 42.1 | <0.0001 |
| DNA strand elongation involved in cell cycle DNA replication (GO:1902296) | 40.1 | <0.0001 |
| Ndc80 complex (GO:31262) | 40.1 | <0.0001 |
| Pre‐replicative complex assembly (GO:36388) | 35.1 | <0.0001 |
| Leading strand elongation (GO:6272) | 35.1 | <0.0001 |
| Condensed nuclear chromosome kinetochore (GO:778) | 31.6 | <0.0001 |
| MCM complex (GO:42555) | 28.1 | 0.0001 |
| Mitotic centrosome separation (GO:7100) | 28.1 | 0.0001 |
| Mitotic chromosome condensation (GO:7076) | 27.5 | <0.0001 |
| Kinetochore microtubule (GO:5828) | 25.5 | 0.0001 |
| Aster (GO:5818) | 25.5 | 0.0001 |
| Enrichment in tumors showing MKI67 lower expression | ||
| Regulation of inclusion body assembly (GO:90083) | 12.3 | 0.03 |
| Beta‐amyloid binding (GO:1540) | 10.5 | 0.005 |
| Neurotrophin TRK receptor signaling pathway (GO:48011) | 7.9 | 0.04 |
| Import into cell (GO:98657) | 6.6 | 0.01 |
| Sodium ion transmembrane transport (GO:35725) | 4.7 | 0.03 |
| Synaptic vesicle (GO:8021) | 4.1 | 0.007 |
| Postsynaptic specialization (GO:99572) | 3.7 | 0.001 |
| Axon part (GO:33267) | 3.7 | <0.0001 |
| Excitatory synapse (GO:60076) | 3.7 | 0.001 |
| Myelin sheath (GO:43209) | 3.4 | 0.01 |
| Dendrite (GO:30425) | 2.6 | 0.001 |
| Trans‐synaptic signaling (GO:99537) | 2.5 | 0.001 |
FDR = false discovery rate; GO = gene ontology.
When focusing on genes associated with a pro‐neural signature, such as SOX2, SOX4, SOX11, OLIG1, OLIG2, INSM1, FERMT1, DCX, CTTNBP2, ATOH8 and ASCL1, we found a significant overexpression of these genes in tumors showing higher levels of MCM6 and MKI67 (FDR < 0.01) (Supplementary Figure S4A). Surprisingly, a number of genes involved in immunological responses were significantly downregulated in these tumors (CX3CR1, TLR4, SYK, FAS, HSPA2, CEBPB, TRIM59, ITGB5, PLP1, C3AR1, PREX1, MNDA, GAB2, VAMP3; FDR < 0.01) (Supplementary Figure S4B). Genes controlling the production of molecular mediators involved in inflammatory response were more significantly downregulated with MCM6‐up than MKI67‐up (EPHX2, APOD, VAMP3, SYK, ALOX5, DUSP10, SNX6, ADORA3, ALOX5AP, TNF, LYN, BTK, FCER1G, IL17RA and the pro‐inflammatory mediator IL17D; FDR < 0.01) (Supplementary Table S2).
Other gene signatures were also found to overexpress with MCM6 and MKI67 high transcription, such as epigenetic markers (DNMT1A, EZH2, TYMS, DHFR; FDR < 0.01) and cell cycle progression key protagonists (all MCMs, PCNA, CCNB1, CCND1, CDK1, CDK4; FDR < 0.01), whereas myelination and glial cell differentiation genes were under‐expressed with MCM6 (FDR < 0.01) but less clearly with MKI67 high levels, which is in accordance with the functional annotations reported above (Supplementary Figures S4B,C,E,F). Strongly upregulated KEGG pathways were all associated with an MCM6 or MKI67 overexpression (ribosome, RNA transport, splicing and degradation, DNA replication and repair, P53 signaling; q‐value < 0.05). Both MCM6‐up and MKI67‐up sets of significantly disturbed pathways were nearly identical, with cell cycle controlling genes showing the same overexpression patterns (Supplementary Figure S5).
Confirmation analyses based on TCGA data
To validate the observations made on the POLA cohort, we ran a validation analysis with 98 similar cases identified in TCGA (The Cancer Genome Atlas) database. These were all identified as IDH1/IDH2 codeleted OD cases and included 35 fully annotated grade III and 63 grade II samples. Our survival analyses of the 98 TCGA cases showed that similar to that observed in our own grade II/III cohort, high levels of MCM6 mRNA (equal to or greater than the median expression; arbitrary unit) indeed correlated to a shorter OS (P = 0.027) (Supplementary Figure S6A). However, the mRNA level of MKI67 in the TCGA samples did not correlate with OS (P = 0.398) (Supplementary Figure S6B). WHO grade did not correlate to survival (P = 0.617) (Supplementary Figure S6C). Only age was independently correlated to survival (Cox multivariate analysis, P = 0.007). MCM6 and MKI67 mRNA levels were strongly correlated (ρ = 0.623; P < 0.0001). No significant correlation existed between the WHO grade and the mRNA level of either MCM6 or MKI67 (P > 0.50).
We also compared the transcriptomic signatures obtained with anaplastic POLA samples with the 35 grade III independent cases retrieved from TCGA (Supplementary Figure S7). We did so by conducting the analyses with the same unsupervised protocols on the TCGA data set as were applied to the POLA study. As sample grouping according to MCM6 and MKI67 median expression levels led to the same configuration, results for both comparisons were fused as one. Similar molecular signatures were indeed uncovered in TCGA samples, each signature presenting with an enrichment with its POLA counterpart ranging from 3.3 to 7.6 times higher than expected (all P < 1 × 10−100, Fisher's exact test). These showed a downregulation of inflammation, glial differentiation, myelin sheath and synaptic activity in tumors that overexpressed MCM6 and/or MKI67, and an upregulation of mitotic cell cycle and DNA replication (all FDR < 0.01).
We further asked if these results extended to grade II cases and applied the same experimental protocol on TCGA combined grades II/III cohort (n = 98; Supplementary Figure S8). Clusters of genes involved in mitotic cell cycle/replication, myelin sheath/glial cell differentiation and axoneme/oxacid metabolic process were indeed strongly differential between MCM6‐down and MCM6‐up groups (all FDR < 0.01), and presented with high identities with their POLA counterparts (73.6%, 60.6% and 48.2%, respectively). However, we did not find well‐defined signatures such as those reported for grade III alone of both POLA and TCGA cohorts for the other clusters. The immune response/inflammation signature, in particular, was lost when including grade II samples to the mixture.
Discussion
The aim of our study was to evaluate the prognostic value of MCM6 and Ki‐67 LI in AOs. Previously, other members of MCM family, such as MCM2, MCM3 and MCM7, have indeed been studied in gliomas, and their overexpression was associated with a poor overall prognosis; however, their applicability in clinical practice seems to be unclear 14, 16, 27. The prognostic value of MCM6 has been confirmed in various types of brain tumors like meningiomas, adamantinomatous craniopharyngiomas and gliomas 9, 21, 48. In the present study, we specifically focused on AOs with the presence of defined mutations of IDH1/2 mutations and 1p/19q codeletion to avoid confusing factors due to molecular alterations. Indeed, IDH1/2 mutations and 1p/19q codeletion are known to correlate with better outcome, due notably to a better response to adjuvant chemotherapy. Only few prognostic markers have been reported in the specific group of IDHmut+/1p19qcodel AO. In this work, PFS was not assessed, because a consensual definition of progression in glioma is still lacking 1, 7, 28.
In this study, we found a good correlation between the mean LIs of MCM6 and Ki‐67. MCM6 LI was always higher than Ki‐67 LI in a given tumor (Figure 1). This is consistent with data of the literature, and is due to the fact that MCMs are expressed earlier in the cell cycle 15, 30. Furthermore, we have identified a cutoff point at 50% MCM6 LI as the optimal criteria for survival prediction: at greater than a LI of 50%, MCM6 overexpression correlated with a shorter overall survival. Based on the results we obtained earlier in meningioma, in which high MCM6 LI were found correlated with survival 21, this marker may even be applicable to other brain tumor types. Consequently, this high labeling index of MCM6 at 50% threshold can be considered as an easy‐to‐use tool for routing practice of the pathology laboratories, in complement to Ki‐67.
We also found that high mean LIs of Ki‐67 were associated with poor outcome, with a threshold of 15%. This result is consistent with previous studies using the EORTC (European Organization for Research and Treatment of Cancer) Brain Tumor Group 40.
Given the results reported above, it shall be noted that although Ki‐67 LI had good inter‐observer agreement in previous studies 11, 39, there is no consensual method for counting Ki‐67 LI in AO. Usually, most of the neuropathologists select areas showing the highest proliferation. Here, in order to have a more reproducible counting method for both Ki‐67 and MCM6, we adapted computerized color image analyses based on previous results for Ki‐67 cell scoring 6, 36. Consequently, a wide area was analyzed for each TMA spot. However, as we were not sure whether the selected spots corresponded to areas with the highest cell proliferation (despite the fact that individual spots were made with histologically representative samples of the tumors), we determined in our series the correlation between the Ki‐67 LI obtained from TMA and the one obtained from whole slides; we found that these values correlated significantly (ρ = 0.553; P < 0.0001; data not shown), confirming the adequacy of our prepared TMA spots.
Multivariate analyses showed that MCM6 may be an interesting marker to use in association to Ki‐67. Indeed, tumors harboring MCM6 LI ≥ 50% and/or Ki‐67 LI ≥ 15% were significantly correlated to shorter survival, both in univariate and multivariate analyses. High MCM6 and Ki‐67 LI were also correlated to shorter survival in grade II tumors, while in our study WHO grade was not significantly correlated to survival. However, these results are limited by a relatively short median follow‐up time and a low number of OS events, thus the observed survival differences were based on few patients. Another limitation is that we used a statistical method that specifically seeks for the best cutoff in a given data set, which may not be applicable in another cohort. Further studies are needed in order to confirm the validity of these thresholds.
Analyses of the transcriptome data of the POLA grade III cohort showed that MCM6 and/or MKI67 overexpressing tumors share near identical whole transcriptional profiles, in spite of a correlation between MCM6 and MKI67 mRNA levels not being perfect. These tumors upregulated genes linked to DNA replication during the cell cycle (including all MCMs), DNA strand elongation, pre‐replicative complex assembly, mitotic centrosome condensation and separation or regulation of helicase activity. This is consistent with the highly proliferative activity of these aggressive cancers. In addition, the MCM6‐up and MKI67‐up tumors of the POLA AO cohort overexpressed pro‐neuronal differentiation genes such as SOX4, SOX11, INSM1 and ASCL1. These findings are consistent with the pro‐neural signature of the AO previously highlighted by Bielle et al These authors identified a subgroup of AO overexpressing neuronal intermediate progenitor genes, associated with immunohistochemical similarities to embryonic subventricular zone, expression of INSM1 and no expression of SOX9 5.
On the contrary, in AO cases showing an overall lower expression of MCM6 and/or MKI67, transcriptomic analyses revealed enrichments of genes involved in myelin sheath production, myelinization or oligodendroglial differentiation. These results suggest that both MCM6‐down and MKI67‐down tumors lack a pro‐neural signature, and unlike their MCM6‐MKI67‐up counterparts, are well‐differentiated oligodendroglial‐like tumors.
In the MCM6‐down subgroup, we also found genes like CX3CR1, TLR4 and SYK, involved in immune response and microglial cells activation. Such downregulations in immune responsive genes have been observed in other types of glioma. For example, in murine models, loss of CX3CR1 expression promoted gliomagenesis in glioblastoma with pro‐neural signature through larger inflammatory monocytes accumulation 17. Lower levels of TLR4, a member of the toll‐like receptors involved in macrophagic immune response, have been implicated in the immune evasion of glioblastoma stem cells 3. The immune evasion in high‐grade gliomas is well documented. The non‐neoplastic cells of the glioma microenvironment, such as fibroblasts, endothelial cells or microglial cells, produce survival and growth factors, helping tumors cells to grow and infiltrate the brain parenchyma 23. The stimulatory role of spleen tyrosine kinase receptors (SYK) in gliomagenesis, brain invasion, and recruitment of immune cells has recently been studied in glioblastoma stem cells by Moncayo et al using SYK‐inhibitors, but their involvement has never been specifically studied in AO 37. In another study, SYK appeared as a tumor suppressor gene in breast carcinomas 13. First‐generation SYK‐inhibitors have been successfully tested in some hematologic malignancies, as B‐acute lymphoid leukemia and chronic lymphocytic lymphoma 25, 44. The assessment of their efficiency in high‐grade gliomas needs further investigations.
Finally, the predictive prognostic power of MCM6 and/or MKI67 was tested using a TCGA cohort of IDH1‐IDH2‐codel glioma composing of both grade II (n = 68) and III (n = 35) tumors. We found that high MCM6 expression indeed correlated with OS, but not MKI67.
Conclusion
In this study, we assessed the prognostic value of MCM6 and Ki‐67 LI in IDH‐mutant and 1p/19q codeleted AOs of the POLA cohort. Our multivariate analyses showed that the overexpression of MCM6 and/or Ki‐67 was independently correlated to shorter survival. The prognostic value of MCM6 was confirmed in TCGA grade II–III IDH‐mutant and 1p/19q codeleted gliomas. These two easy‐to‐use and cost‐effective markers could thus be used concurrently in routine pathology practice. Potentially, these markers could also be integrated into therapeutic as well as clinicoradiological monitoring strategies to improve disease outcomes. We also would like to underline the important new insight gained from the transcriptomic analyses of the AO, which is that AO with high proliferation have downregulated immune response and lower microglial cells activation.
Conflict of Interest
The authors have no conflict of interest to declare.
Supporting information
Figure S1 . MCM6 and MKI67 mRNA level distributions into two subgroups of higher (“‐up”) and lower (“‐down”) expressions. Transcriptomic samples (n = 68) are separated at the median expression for each gene and ordered according to their expression gradient for either MCM6 or MKI67.
Figure S2 . Correlations between immunohistochemistry (IHC) and mRNA level for Ki‐67 (MKI67) and MCM6.
Figure S3 . Correlation between mRNA expression levels of MKI67 and MCM6.
Figure S4 . Log2‐normalized gene expression levels for the upregulated pro‐neural (A), epigenetic (B), and cell cycle (C) signatures, as well as the downregulated immune response (D), myelination (E) and glial cells differentiation (F) signatures for both MCM6 and MKI67 “‐down” and “‐up” samples (transcriptomics).
Figure S5 . Significantly dysregulated KEGG pathways with an overall upregulation (Table as obtained with Gage) and map of the cell cycle (GraphViz rendered with Pathview) in both MCM6 (A) and MKI67 (B) upregulated samples. Significantly (FDR < 0.01) down‐ and upregulated genes are colored from green to red, with respect to their fold change of expression (log2).
Figure S6 . Survival analyses in grades II/III IDH‐mutant/1p19q codeleted oligodendrogliomas selected from TCGA database. Kaplan–Meier curves with log‐rank tests (overall survival). A. MCM6 mRNA level, median threshold (arbitrary units). B. MKI67 mRNA level, median threshold (arbitrary units). C. WHO 2016 grade.
Figure S7 . K‐means clustering based on MCM6 and/or MKI67 mRNA expression gradient (from lowest to highest) of the 35 transcriptomes of anaplastic IDH‐mutant/1p19q codeleted oligodendrogliomas selected from TCGA bank. The K‐means clustering delineated five significant clusters of co‐expressed genes (P < 0.05) with highly correlated molecular functions. A. Heatmap of the 10 k‐means clusters organized based on the extent of MCM6 and MKI67 expression (green, red and black indicate downregulated, upregulated and median genes, respectively). B. Overview of the most relevant functional annotations for the significantly over‐ and under‐expressed clusters (in red and green hues, respectively, all P(FDR) ≤ 0.01). Each of these clusters was compared to the clusters found of the POLA cohort, and their identities calculated based on the corresponding POLA clusters. C. Mean gene expression (overall with standard error at the mean) for each cluster where both upregulated samples for MCM6 and/or MKI67 are compared to their downregulated counterparts (AU—arbitrary units). Upregulated and downregulated samples are defined relatively to MCM6 median gene expression.
Figure S8 . K‐means clustering based on MCM6 and/or MKI67 mRNA expression gradient (from lowest to highest) of the 98 transcriptomes of grades II and III IDH‐mutant/1p19q codeleted oligodendrogliomas selected from TCGA bank. The K‐means clustering delineated five significant clusters of co‐expressed genes (P < 0.05) with highly correlated molecular functions. A. Heatmap of the ten k‐means clusters organized based on the extent of MCM6 and MKI67 expression (green, red and black indicate downregulated, upregulated and median genes, respectively). B. Overview of the most relevant functional annotations for the significantly over‐ and under‐expressed clusters (in red and green hues, respectively, all P(FDR) ≤ 0.01). Each of these clusters was compared to the clusters found of the POLA cohort, and their identities calculated based on the corresponding POLA clusters. C. Mean gene expression (overall with standard error at the mean) for each cluster where both upregulated samples for MCM6 and/or MKI67 are compared to their downregulated counterparts (AU—arbitrary units). Upregulated and downregulated samples are defined relatively to MCM6 median gene expression.
Table S1 . K‐means clustering from MCM6 and MKI67 expression gradients of 68 transcriptomes of anaplastic oligodendrogliomas from the POLA network. Functional annotations of the genes present in the five significant groups of co‐expressed genes (P < 0.01), sorted by gene ontology enrichment (FDR, false discovery rate; GO, gene ontology).
Table S2 . Differential gene expression statistics (t‐test) between MCM6‐up and MCM6‐down, and between MKI67‐up and MKI67‐down samples, sorted by P‐value (BH, Benjamini–Hochberg; FDR, false discovery rate; GO, gene ontology; SD, standard deviation).
Table S3 . K‐means clustering from MCM6 (or MKI67) expression gradients of 35 transcriptomes of grade III oligodendrogliomas from the TCGA bank. Functional annotations of the genes present in the five significant groups of co‐expressed genes (P < 0.01), sorted by gene ontology enrichment (FDR, false discovery rate; GO, gene ontology).
Table S4 . K‐means clustering from MCM6 expression gradients of 98 transcriptomes of grade II and grade III oligodendrogliomas from the TCGA bank. Functional annotations of the genes present in the six significant groups of co‐expressed genes (P < 0.01), sorted by gene ontology enrichment (FDR, false discovery rate; GO, gene ontology).
Acknowledgments
The authors wish to thank all the team of the CHRU Nancy Pathology Department for technical support, the members of the POLA network for contributing case material and Marion Divoux for her help in manuscript preparation. The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
List of investigators
POLA Network: Amiens (Christine Desenclos, H. Sevestre), Angers (Philippe Menei, A. Rousseau), Annecy (T. Cruel, S. Lopez), Besançon (M.I. Mihai, A. Petit), Brest (R. Seizeur, I. Quintin‐Roué), Bicêtre (C. Adam, F. Parker), Bordeaux (S. Eimer, H. Loiseau), Caen (L. Bekaert, F. Chapon), Clamart (D. Ricard), Clermont‐Ferrand (C. Godfraind, T. Khallil), Clichy (D. Cazals‐Hatem, T. Faillot), Colmar (C. Gaultier, M. C. Tortel), Cornebarrieu (I. Carpiuc, P. Richard), Créteil (W. Lahiani), Dijon (H. Aubriot‐Lorton, F. Ghiringhelli), Lille (C‐A. Maurage, E. Le Rhun), Limoges (E. M. Gueye, F. Labrousse), Lyon (F. Ducray, D. Meyronet), Marseille (O. Chinot), Montpellier (L. Bauchet, V. Rigau), Nancy (P. Beauchesne), Nantes (M. Campone, D. Loussouarn), Nice (D. Fontaine, F. Vandenbos‐Burel), Nîmes (A. Le. Floch, P. Roger), Orléans (C. Blechet, M. Fesneau), Paris (A. Carpentier, A. Idbaih, J. Y. Delattre [POLA Network National coordinator], K. Mokhtari, F. Bielle, S. Hamdi, M. Polivka), Poitiers (S. Milin), Reims (P. Colin, M. D. Diebold), Rennes (D. Chiforeanu, E. Vauleon), Rouen (O. Langlois, A. Laquerriere), Saint‐Etienne (F. Forest, M. J. Motso‐Fotso), Saint‐Pierre de la Réunion (M. Andraud, G. Runavot), Strasbourg (B. Lhermitte, G. Noel), Suresnes (S. Gaillard, C. Villa), Toulon (N. Desse), Tours (C. Rousselot‐Denis, I. Zemmoura), Toulouse (E. Cohen‐Moyal, E. Uro‐Coste), Villejuif (F. Dhermain).
Contributor Information
Guillaume Gauchotte, Email: g.gauchotte@chru-nancy.fr.
Investigators: The POLA Network:
Christine Desenclos, H. Sevestre, Philippe Menei, A. Rousseau, T. Cruel, S. Lopez, M.I. Mihai, A. Petit, R. Seizeur, I. Quintin‐Roué, C. Adam, F. Parker, S. Eimer, H. Loiseau, L. Bekaert, F. Chapon, D. Ricard, C. Godfraind, T. Khallil, D. Cazals‐Hatem, T. Faillot, C. Gaultier, M. C Tortel, I. Carpiuc, P. Richard, W. Lahiani, H. Aubriot‐Lorton, F. Ghiringhelli, C‐A Maurage, E. Le Rhun, E. M. Gueye, F. Labrousse, F. Ducray, D. Meyronet, O. Chinot, L. Bauchet, V. Rigau, P. Beauchesne, M. Campone, D. Loussouarn, D. Fontaine, F. Vandenbos‐Burel, A. Le. Floch, P. Roger, C. Blechet, M. Fesneau, A. Carpentier, A. Idbaih, J. Y. Delattre, K. Mokhtari, F. Bielle, S. Hamdi, M. Polivka, S. Milin, P. Colin, M. D. Diebold, D. Chiforeanu, E. Vauleon, O. Langlois, A. Laquerriere, F. Forest, M. J. Motso‐Fotso, M. Andraud, G. Runavot, B. Lhermitte, G. Noel, S. Gaillard, C. Villa, N. Desse, C. Rousselot‐Denis, I. Zemmoura, E. Cohen‐Moyal, E. Uro‐Coste, and F. Dhermain
References
- 1. Abdulla S, Saada J, Johnson G, Jefferies S, Ajithkumar T (2015) Tumour progression or pseudoprogression? A review of post‐treatment radiological appearances of glioblastoma. Clin Radiol 70:1299–1312. [DOI] [PubMed] [Google Scholar]
- 2. Alentorn A, Dehais C, Ducray F, Carpentier C, Mokhtari K, Figarella‐Branger D et al (2015) Allelic loss of 9p21.3 is a prognostic factor in 1p/19q codeleted anaplastic gliomas. Neurology 85:1325–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alvarado AG, Thiagarajan PS, Mulkearns‐Hubert EE, Silver DJ, Hale JS, Alban TJ et al (2017) Glioblastoma cancer stem cells evade innate immune suppression of self‐renewal through reduced TLR4 expression. Cell Stem Cell 20:450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH et al (2011) Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 333:1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bielle F, Ducray F, Mokhtari K, Dehais C, Adle‐Biassette H, Carpentier C et al (2017) Tumor cells with neuronal intermediate progenitor features define a subgroup of 1p/19q co‐deleted anaplastic gliomas. Brain Pathol 27:567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blaker YN, Brodtkorb M, Maddison J, Hveem TS, Nesheim JA, Mohn HM et al (2015) Computerized image analysis of the Ki‐67 proliferation index in mantle cell lymphoma. Histopathology 67:62–69. [DOI] [PubMed] [Google Scholar]
- 7. Brandsma D, Stalpers L, Taal W, Sminia P, Van den Bent MJ (2008) Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol 9:453–461. [DOI] [PubMed] [Google Scholar]
- 8. Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb‐Esfahani S, Denkert C (2012) Cutoff finder: A comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS ONE 7:e51862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai HQ, Chen ZJ, Zhang HP, Wang PF, Zhang Y, Hao JJ et al (2018) Overexpression of MCM6 predicts poor survival in patients with glioma. Hum Pathol 78:182–187. [DOI] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research Network , Brat DJ, Verhaak RGW, Aldape KD, Yung WKA, Salama SR et al (2015). Comprehensive, integrative genomic analysis of diffuse lower‐grade gliomas. N Engl J Med 372:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coleman KE, Brat DJ, Cotsonis GA, Lawson D, Cohen C (2006) Proliferation (MIB‐1 expression) in oligodendrogliomas: assessment of quantitative methods and prognostic significance. Appl Immunohistochem Mol Morphol 14:109–114. [DOI] [PubMed] [Google Scholar]
- 12. Coons SW, Johnson PC, Pearl DK (1997) The prognostic significance of Ki‐67 labeling indices for oligodendrogliomas. Neurosurgery 41:878–884; discussion 884–885. [DOI] [PubMed] [Google Scholar]
- 13. Coopman PJ, Do MT, Barth M, Bowden ET, Hayes AJ, Basyuk E et al (2000) The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature 406:742–747. [DOI] [PubMed] [Google Scholar]
- 14. Erkan EP, Ströbel T, Lewandrowski G, Tannous B, Madlener S, Czech T et al (2014) Depletion of minichromosome maintenance protein 7 inhibits glioblastoma multiforme tumor growth in vivo. Oncogene 33:4778–4785. [DOI] [PubMed] [Google Scholar]
- 15. Eward KL, Obermann EC, Shreeram S, Loddo M, Fanshawe T, Williams C et al (2004) DNA replication licensing in somatic and germ cells. J Cell Sci 117:5875–5886. [DOI] [PubMed] [Google Scholar]
- 16. Facoetti A, Ranza E, Benericetti E, Ceroni M, Tedeschi F, Nano R (2006) Minichromosome maintenance protein 7: a reliable tool for glioblastoma proliferation index. Anticancer Res 26:1071–1075. [PubMed] [Google Scholar]
- 17. Feng X, Szulzewsky F, Yerevanian A, Chen Z, Heinzmann D, Rasmussen RD et al (2015) Loss of CX3CR1 increases accumulation of inflammatory monocytes and promotes gliomagenesis. Oncotarget 6:15077–15094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Figarella‐Branger D, Mokhtari K, Dehais C, Carpentier C, Colin C, Jouvet A et al (2016) Mitotic index, microvascular proliferation, and necrosis define 3 pathological subgroups of prognostic relevance among 1p/19q co‐deleted anaplastic oligodendrogliomas. Neuro Oncol 18:888–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Figarella‐Branger D, Mokhtari K, Dehais C, Jouvet A, Uro‐Coste E, Colin C et al (2014) Mitotic index, microvascular proliferation, and necrosis define 3 groups of 1p/19q codeleted anaplastic oligodendrogliomas associated with different genomic alterations. Neuro Oncol 16:1244–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gauchotte G, Hergalant S, Vigouroux C, Casse JM, Houlgatte R, Kaoma T et al (2017) Cytoplasmic overexpression of RNA‐binding protein HuR is a marker of poor prognosis in meningioma, and HuR knockdown decreases meningioma cell growth and resistance to hypoxia. J Pathol 242:421–434. [DOI] [PubMed] [Google Scholar]
- 21. Gauchotte G, Vigouroux C, Rech F, Battaglia‐Hsu SF, Soudant M, Pinelli C et al (2012) Expression of minichromosome maintenance MCM6 protein in meningiomas is strongly correlated with histologic grade and clinical outcome. Am J Surg Pathol 36:283–291. [DOI] [PubMed] [Google Scholar]
- 22. Gleize V, Alentorn A, Connen de Kérillis L, Labussière M, Nadaradjane AA, Mundwiller E et al (2015) CIC inactivating mutations identify aggressive subset of 1p19q codeleted gliomas. Ann Neurol 78:355–374. [DOI] [PubMed] [Google Scholar]
- 23. Hambardzumyan D, Gutmann DH, Kettenmann H (2016) The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci 19:20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Helfenstein A, Frahm SO, Krams M, Drescher W, Parwaresch R, Hassenpflug J (2004) Minichromosome maintenance protein (MCM6) in low‐grade chondrosarcoma: distinction from enchondroma and identification of progressive tumors. Am J Clin Pathol 122:912–918. [DOI] [PubMed] [Google Scholar]
- 25. Hoellenriegel J, Coffey GP, Sinha U, Pandey A, Sivina M, Ferrajoli A et al (2012) Selective, novel Spleen tyrosine kinase (Syk) inhibitors suppress chronic lymphocytic leukemia B cell activation and migration. Leukemia 26:1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hotton J, Agopiantz M, Leroux A, Charra‐Brunaud C, Marie B, Busby‐Venner H et al (2018) Minichromosome maintenance complex component 6 (MCM6) expression correlates with histological grade and survival in endometrioid endometrial adenocarcinoma. Virchows Arch 472:623–633. [DOI] [PubMed] [Google Scholar]
- 27. Hua C, Zhao G, Li Y, Bie L (2014) Minichromosome Maintenance (MCM) Family as potential diagnostic and prognostic tumor markers for human gliomas. BMC Cancer 14:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jain R, Narang J, Sundgren PM, Hearshen D, Saksena S, Rock JP et al (2010) Treatment induced necrosis versus recurrent/progressing brain tumor: going beyond the boundaries of conventional morphologic imaging. J Neurooncol 100:17–29. [DOI] [PubMed] [Google Scholar]
- 29. Kamoun A, Idbaih A, Dehais C, Elarouci N, Carpentier C, Letouzé E et al (2016) Integrated multi‐omics analysis of oligodendroglial tumours identifies three subgroups of 1p/19q co‐deleted gliomas. Nat Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kingsbury SR, Loddo M, Fanshawe T, Obermann EC, Prevost AT, Stoeber K, Williams GH (2005) Repression of DNA replication licensing in quiescence is independent of geminin and may define the cell cycle state of progenitor cells. Exp Cell Res 309:56–67. [DOI] [PubMed] [Google Scholar]
- 31. Labib K, Kearsey SE, Diffley JF (2001) MCM2‐7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1‐phase and are required to establish, but not maintain, the S‐phase checkpoint. Mol Biol Cell 12:3658–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Labreche K, Simeonova I, Kamoun A, Gleize V, Chubb D, Letouzé E et al (2015) TCF12 is mutated in anaplastic oligodendroglioma. Nat Commun 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lindner K, Gregán J, Montgomery S, Kearsey SE (2002) Essential role of MCM proteins in premeiotic DNA replication. Mol Biol Cell 13:435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Z, Li J, Chen J, Shan Q, Dai H, Xie H et al (2018) MCM family in HCC: MCM6 indicates adverse tumor features and poor outcomes and promotes S/G2 cell cycle progression. BMC Cancer 18:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella‐Branger D, Cavenee WK et al (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131:803–820. [DOI] [PubMed] [Google Scholar]
- 36. Markiewicz T, Grala B, Kozlowski W, Osowski S (2010) Computer system for cell counting in selected brain tumors at Ki‐67 immunohistochemical staining. Anal Quant Cytol Histol 32:323–332. [PubMed] [Google Scholar]
- 37. Moncayo G, Grzmil M, Smirnova T, Zmarz P, Huber RM, Hynx D et al (2018) SYK inhibition blocks proliferation and migration of glioma cells and modifies the tumor microenvironment. Neuro‐Oncology 20:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ostrom QT, Gittleman H, Liao P, Vecchione‐Koval T, Wolinsky Y, Kruchko C, Barnholtz‐Sloan JS (2017) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro‐Oncology 19:v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prayson RA, Castilla EA, Hembury TA, Liu W, Noga CM, Prok AL (2003) Interobserver variability in determining MIB‐1 labeling indices in oligodendrogliomas. Ann Diagn Pathol 7:9–13. [DOI] [PubMed] [Google Scholar]
- 40. Preusser M, Hoeftberger R, Woehrer A, Gelpi E, Kouwenhoven M, Kros JM et al (2012) Prognostic value of Ki67 index in anaplastic oligodendroglial tumours–a translational study of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Histopathology 60:885–894. [DOI] [PubMed] [Google Scholar]
- 41. Rosenberg S, Ducray F, Alentorn A, Dehais C, Elarouci N, Kamoun A et al (2018) Machine learning for better prognostic stratification and driver gene identification using somatic copy number variations in anaplastic oligodendroglioma. Oncologist 23(12):1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schrader C, Janssen D, Klapper W, Siebmann JU, Meusers P, Brittinger G et al (2005) Minichromosome maintenance protein 6, a proliferation marker superior to Ki‐67 and independent predictor of survival in patients with mantle cell lymphoma. Br J Cancer 93:939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y et al (2015) Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 47:458–468. [DOI] [PubMed] [Google Scholar]
- 44. Uckun FM, Qazi S (2014) SYK as a new therapeutic target in B‐cell precursor acute lymphoblastic leukemia. J Cancer Ther 5:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van den Bent MJ, Brandes AA, Taphoorn MJB, Kros JM, Kouwenhoven MCM, Delattre JY et al (2013) Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long‐term follow‐up of EORTC brain tumor group study 26951. J Clin Oncol 31:344–350. [DOI] [PubMed] [Google Scholar]
- 46. Van den Bent MJ, Carpentier AF, Brandes AA, Sanson M, Taphoorn MJB, Bernsen HJJA et al (2006) Adjuvant procarbazine, lomustine, and vincristine improves progression‐free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol 24:2715–2722. [DOI] [PubMed] [Google Scholar]
- 47. Vigouroux C, Casse JM, Battaglia‐Hsu SF, Brochin L, Luc A, Paris C et al (2015) Methyl(R217)HuR and MCM6 are inversely correlated and are prognostic markers in non small cell lung carcinoma. Lung Cancer 89:189–196. [DOI] [PubMed] [Google Scholar]
- 48. Xu J, Zhang S, You C, Huang S, Cai B, Wang X (2007) Expression of human MCM6 and DNA Topo II alpha in craniopharyngiomas and its correlation with recurrence of the tumor. J Neurooncol 83:183–189. [DOI] [PubMed] [Google Scholar]
- 49. Zeng A, Hu Q, Liu Y, Wang Z, Cui X, Li R et al (2015) IDH1/2 mutation status combined with Ki‐67 labeling index defines distinct prognostic groups in glioma. Oncotarget 6:30232–30238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 . MCM6 and MKI67 mRNA level distributions into two subgroups of higher (“‐up”) and lower (“‐down”) expressions. Transcriptomic samples (n = 68) are separated at the median expression for each gene and ordered according to their expression gradient for either MCM6 or MKI67.
Figure S2 . Correlations between immunohistochemistry (IHC) and mRNA level for Ki‐67 (MKI67) and MCM6.
Figure S3 . Correlation between mRNA expression levels of MKI67 and MCM6.
Figure S4 . Log2‐normalized gene expression levels for the upregulated pro‐neural (A), epigenetic (B), and cell cycle (C) signatures, as well as the downregulated immune response (D), myelination (E) and glial cells differentiation (F) signatures for both MCM6 and MKI67 “‐down” and “‐up” samples (transcriptomics).
Figure S5 . Significantly dysregulated KEGG pathways with an overall upregulation (Table as obtained with Gage) and map of the cell cycle (GraphViz rendered with Pathview) in both MCM6 (A) and MKI67 (B) upregulated samples. Significantly (FDR < 0.01) down‐ and upregulated genes are colored from green to red, with respect to their fold change of expression (log2).
Figure S6 . Survival analyses in grades II/III IDH‐mutant/1p19q codeleted oligodendrogliomas selected from TCGA database. Kaplan–Meier curves with log‐rank tests (overall survival). A. MCM6 mRNA level, median threshold (arbitrary units). B. MKI67 mRNA level, median threshold (arbitrary units). C. WHO 2016 grade.
Figure S7 . K‐means clustering based on MCM6 and/or MKI67 mRNA expression gradient (from lowest to highest) of the 35 transcriptomes of anaplastic IDH‐mutant/1p19q codeleted oligodendrogliomas selected from TCGA bank. The K‐means clustering delineated five significant clusters of co‐expressed genes (P < 0.05) with highly correlated molecular functions. A. Heatmap of the 10 k‐means clusters organized based on the extent of MCM6 and MKI67 expression (green, red and black indicate downregulated, upregulated and median genes, respectively). B. Overview of the most relevant functional annotations for the significantly over‐ and under‐expressed clusters (in red and green hues, respectively, all P(FDR) ≤ 0.01). Each of these clusters was compared to the clusters found of the POLA cohort, and their identities calculated based on the corresponding POLA clusters. C. Mean gene expression (overall with standard error at the mean) for each cluster where both upregulated samples for MCM6 and/or MKI67 are compared to their downregulated counterparts (AU—arbitrary units). Upregulated and downregulated samples are defined relatively to MCM6 median gene expression.
Figure S8 . K‐means clustering based on MCM6 and/or MKI67 mRNA expression gradient (from lowest to highest) of the 98 transcriptomes of grades II and III IDH‐mutant/1p19q codeleted oligodendrogliomas selected from TCGA bank. The K‐means clustering delineated five significant clusters of co‐expressed genes (P < 0.05) with highly correlated molecular functions. A. Heatmap of the ten k‐means clusters organized based on the extent of MCM6 and MKI67 expression (green, red and black indicate downregulated, upregulated and median genes, respectively). B. Overview of the most relevant functional annotations for the significantly over‐ and under‐expressed clusters (in red and green hues, respectively, all P(FDR) ≤ 0.01). Each of these clusters was compared to the clusters found of the POLA cohort, and their identities calculated based on the corresponding POLA clusters. C. Mean gene expression (overall with standard error at the mean) for each cluster where both upregulated samples for MCM6 and/or MKI67 are compared to their downregulated counterparts (AU—arbitrary units). Upregulated and downregulated samples are defined relatively to MCM6 median gene expression.
Table S1 . K‐means clustering from MCM6 and MKI67 expression gradients of 68 transcriptomes of anaplastic oligodendrogliomas from the POLA network. Functional annotations of the genes present in the five significant groups of co‐expressed genes (P < 0.01), sorted by gene ontology enrichment (FDR, false discovery rate; GO, gene ontology).
Table S2 . Differential gene expression statistics (t‐test) between MCM6‐up and MCM6‐down, and between MKI67‐up and MKI67‐down samples, sorted by P‐value (BH, Benjamini–Hochberg; FDR, false discovery rate; GO, gene ontology; SD, standard deviation).
Table S3 . K‐means clustering from MCM6 (or MKI67) expression gradients of 35 transcriptomes of grade III oligodendrogliomas from the TCGA bank. Functional annotations of the genes present in the five significant groups of co‐expressed genes (P < 0.01), sorted by gene ontology enrichment (FDR, false discovery rate; GO, gene ontology).
Table S4 . K‐means clustering from MCM6 expression gradients of 98 transcriptomes of grade II and grade III oligodendrogliomas from the TCGA bank. Functional annotations of the genes present in the six significant groups of co‐expressed genes (P < 0.01), sorted by gene ontology enrichment (FDR, false discovery rate; GO, gene ontology).


