Fanconi anemia (FA) is a rare multi-system disorder characterized by bone marrow failure, congenital abnormalities, and cancer predisposition.1 Pathogenic variants have been described in 22 known FA genes (FANCAFANCW) that are required for the proper repair of DNA interstrand crosslinks.2,3 A key step in the repair of interstrand crosslinks is FA pathway activation via monoubiquitination of FANCD2 and FANCI by FANCL, an E3 ubiquitin- ligase working with UBE2T/FANCT, an E2 ubiquitin- conjugating enzyme.4-7 Pathogenic germline variants in UBE2T have been described in three individuals with FA;6-8 thus, the knowledge of the phenotypic spectrum is limited for the FA-T complementation group. Here we describe a mild presentation of FA resulting from a hypomorphic missense variant in UBE2T that partially disrupts the function of the encoded protein. This report highlights the importance of an algorithmic approach to bone marrow failure that combines genetic testing and functional cellular assays.9
Three patients have previously been reported with biallelic pathogenic variants in UBE2T consistent with an autosomal recessive disease. All three patients presented with the classic features of FA (Online Supplementary Table S1).6-8 Hira et al. reported two unrelated patients both harboring a c.4C>G, p.Gln2Glu missense variant in trans with either a 23 kb whole gene deletion (patient 1) or a c.180+5G>A, p.Gln37Argfs*47 frameshift variant (patient 2). Both patients developed hematologic abnormalities and bone marrow failure. Patient 2 developed myelodysplastic syndrome which evolved to acute myeloid leukemia. Both patients required hematopoietic stem cell transplantation.7 Rickman et al. and Virts et al. reported the findings of a maternally inherited Alu-mediated duplication, c.-64_468dup, producing an unstable transcript and a paternally inherited Alu-mediated deletion, c.-64_468del, leading to loss of the majority of the gene. However, this patient did not develop bone marrow failure as a result of somatic mosaicism identified in his peripheral blood.6,8
The patient reported here is a 22-year-old Hispanic female who was unaffected at birth, had a normal developmental history, and a negative family history with no reported consanguinity. She originally presented to an another institution at 8 years of age and was reported to have mild neutropenia and thrombocytopenia; a bone marrow biopsy at that time was non-diagnostic. At 21 years, the patient presented with persistent neutropenia and macrocytosis, intermittent thrombocytopenia, episodic fevers, an urticarial erythematous rash, and metromenorrhagia (Online Supplementary Tables S1 and S2). No developmental anomalies or cutaneous hypo/hyperpigmentation were noted. Chromosomal breakage assays performed on peripheral blood lymphocytes showed increased breakage (Online Supplementary Table S1). A repeat bone marrow biopsy revealed moderate hypocellularity (40-50%) with no evidence of dysplasia or a lymphoproliferative process and a normal karyotype. A gene panel to investigate periodic fever was negative (Online Supplementary Table S3). Due to the patient’s undiagnosed neutropenia, panel-based nextgeneration sequencing was performed on whole blood (Online Supplementary Table S4) and revealed a homozygous c.196C>A, p.P66T (NM_014176.3, Chr1(GRCh37): 202302667G>T) missense variant of uncertain significance in UBE2T. This variant is absent from the gnomAD database. GeneDx exon level deletion/duplication calling from sequencing data (with manual verification) did not detect any evidence of a multi-exon copy number variant in UBE2T, suggesting the patient is not hemizygous. Parental samples were not available for testing.
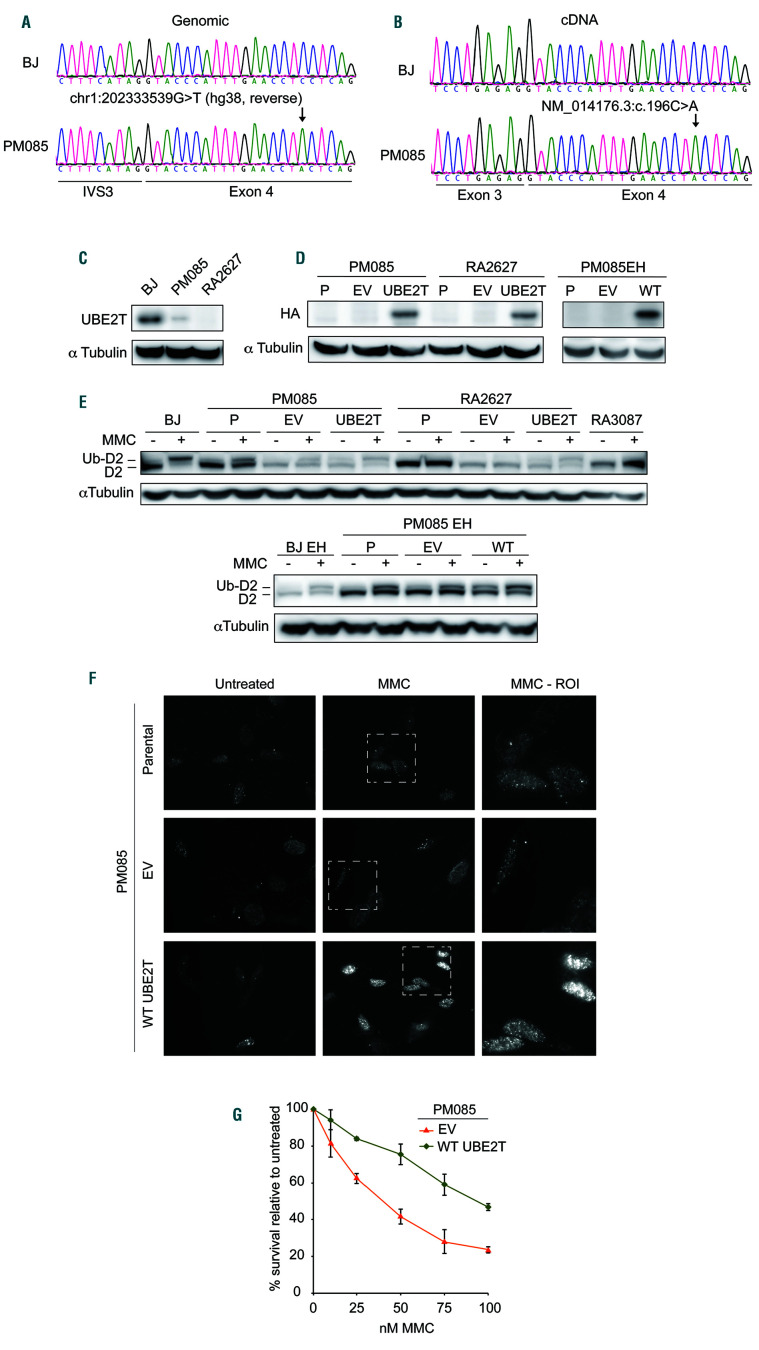
Figure 1.
The proband carries a likely pathogenic UBE2T variant expressed at low levels conferring defective interstrand crosslink repair. (A) Sequencing of genomic DNA extracted from primary fibroblasts (PM085) of the affected individual indicating a homozygous chr1:202333539G>T variant (hg38, reverse). (B) Sequencing of complementary DNA (cDNA) from the proband’s fibroblasts indicating the presence of a variant NM_014176.3:c.196C>A and no evidence of aberrant splicing. Exon numbering reflects ref seq NM_014176.3 since the primers were designed against this transcript.6 (C) Immunoblot with anti-UBE2T antibody in whole cell extract from the proband’s primary fibroblasts (PM085), wildtype BJ fibroblasts (from the American Type Culture Collection) and fibroblasts from an UBE2T/FANCT-null Fanconi anemia patient (RA2627).6 (D) Immunoblot with anti-HA antibody in PM085 (proband) and RA2627 (UBE2T-/-) primary fibroblasts and PM085 EH (immortalized fibroblasts) expressing C-HAFLAG empty vector (EV) or wild-type (WT) UBE2T. HA expression in parental (P) (nontransduced) cells and cells expressing EV or WT UBE2T. (E) Immunoblot with anti-FANCD2 antibody on whole cell extracts of cells treated or not with mitomycin C (MMC). Ub-D2 indicates the monoubiquitinated band. (F) Formation of foci of FANCD2 after MMC treatment in patient-derived PM085 cells (nontransduced parental cells) or cells expressing EV, or WT UBE2T. (G) Cell survival of the proband’s PM085 fibroblasts expressing EV or WT UBE2T after treatment with MMC.
The p.P66T variant identified causes a substitution of a hydrophobic to polar uncharged amino acid at a highly conserved position in the UBC fold domain (Online Supplementary Figure S1). Multiple in silico tools predict that this variant is likely to be damaging (Online Supplementary Table S5). The proline 66 resides at the base of one of multiple loops comprising the FANCL binding region10 (Online Supplementary Figure S2A and B). When modeled, P66T is predicted to change the position of the loop because of changes in the backbone phi/psi (ϕ/ψ) angles. The loop is moved out, as compared to the wild-type (WT) structure, and the interacting residues are moved away from the UBE2T and FANCL interface (Online Supplementary Figure S2C). As P66T changes the range of peptide backbone flexibility, making the base of the loop much more flexible, the binding with FANCL is expected to be dysregulated from a stricter cis/trans switch.
In order to confirm the pathogenicity of the c.196C>A (p.P66T) variant in UBE2T, functional in vitro studies were performed. Sanger sequencing of genomic DNA and complementary DNA from patient-derived fibroblasts (PM085) confirmed the presence of this variant and absence of splicing defects (Figure 1A and B). Immunoblotting of whole cell extract from these cells demonstrated decreased, but not absent, UBE2T protein expression (Figure 1C). This is consistent with the p.P66T missense variant causing instability in the UBE2T protein, resulting in the observed decrease in protein level.
To determine whether the c.196C>A (p.P66T) variant affects the E2 function of UBE2T, FANCD2 monoubiquitination was assessed after treatment with the DNA interstrand crosslinking agent mitomycin C. Normal FANCD2 monoubiquitination was observed in the WT control cell line (BJ fibroblasts), was absent in UBE2T-/- (RA2627) and FANCA-/- (RA3087) fibroblasts and was reduced in the proband’s fibroblasts (Figure 1E). Expression of WT UBE2T in the patient’s fibroblasts fully rescued FANCD2 monoubiquitination (Figure 1D and E), recruitment of FANCD2 to chromatin after mitomycin C treatment, and sensitivity of the proband’s fibroblasts to mitomycin C (Figure 1F-H). These results indicate that the proband belongs to FA-T complementation group and suggest that the patient’s missense variant is hypomorphic, resulting in reduced function.
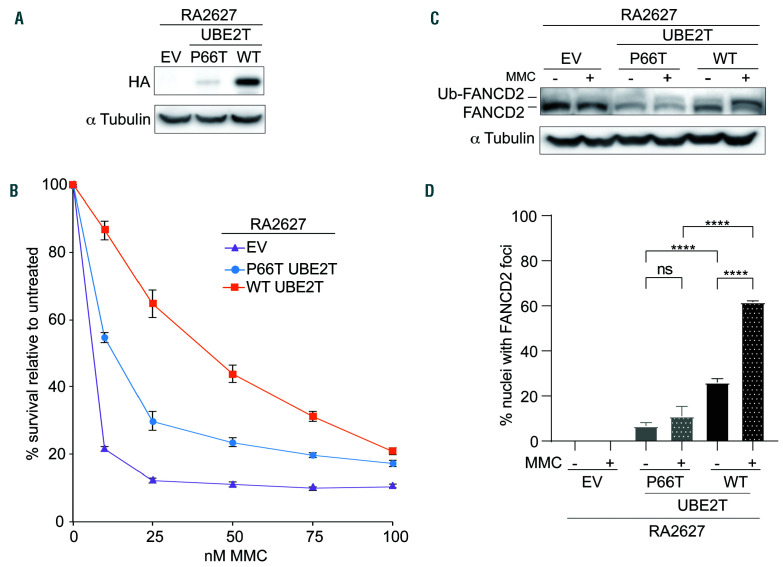
Figure 2.
P66T UBE2T is a partial loss-of-function variant. (A) Immunoblot with anti-HA antibody of RA2627 (UBE2T-/-) primary fibroblasts expressing empty vector (EV) or C-HA-FLAG P66T UBE2T or wild-type (WT) UBE2T. (B) Cell survival of RA2627 (UBE2T-/-) fibroblasts expressing EV, P66T UBE2T, or WT UBE2T after treatment with mitomycin C (MMC). (C) FANCD2 ubiquitination with and without MMC treatment in RA2627 (UBE2T-/-) fibroblasts expressing EV, P66T UBE2T, or W T UBE2T. (D) Quantification of FANCD2 foci formation af ter MMC treatment in RA2627 (UBE2T-/-) fibroblasts expressing EV, P66T UBE2T, or W T UBE2T. Approximately 300 HA-expressing cells were analyzed for the presence of FANCD2 foci in three separate coverslips. The mean percent nuclei with FANCD2 foci was plotted and tested for signif icance using one-way analysis of variance with multiple comparisons. ns: not significant, ****P=≤0.0001.
To further demonstrate that the missense variant reduces UBE2T function and is indeed likely pathogenic, UBE2T-/- cells were transduced with either WT or P66T HA-tagged UBE2T (Figure 2A). The P66T variant expressed at a lower level compared to WT UBE2T, consistent with decreased stability of UBE2T carrying that variant. Expression of P66T UBE2T also only partially rescued cell survival, FANCD2 ubiquitination, and FANCD2 foci formation upon treatment with mitomycin C compared to WT UBE2T expression (Figure 3B-D). This provides further evidence that the missense variant is a likely pathogenic hypomorph.
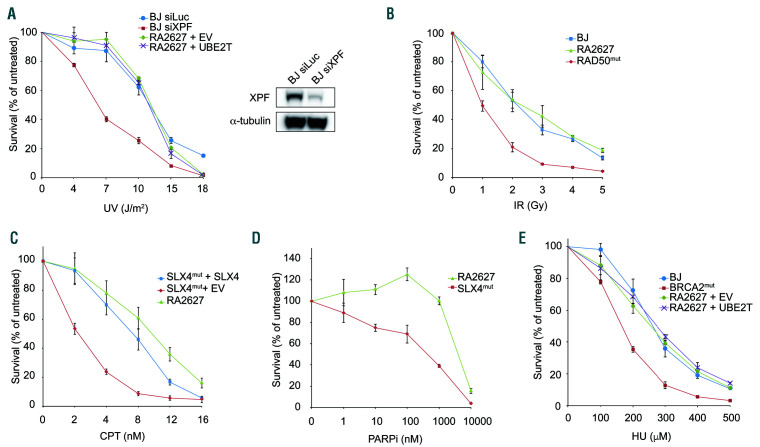
The cellular and patients’ phenotypes described for the FA-T complementation group are thus far consistent with defective FA pathway activation and a defect in interstrand crosslink repair. However, it was previously reported that UBE2T-deficient DT40 cells were sensitive to ultraviolet irradiation and the replication stress-inducing agent, hydroxyurea.11 To determine whether UBE2T is important for resistance to other types of DNA damage, RA2627 cells were tested for sensitivity to a number of other genotoxic agents. RA2627 cells were not found to be hypersensitive to ultraviolet irradiation, ionizing radiation, camptothecin, hydroxyurea, or the PARP inhibitor olaparib (Figure 3A-E). These data suggest that UBE2T does not have a major role in responding to DNA lesions or replication stress produced by these agents and its primary function is in interstrand crosslink repair and that the patients’ phenotypes reflect defects in the repair of interstrand crosslink lesions.
In conclusion, we report a novel presentation of FA-T complementation group resulting from a likely pathogenic missense variant (c.196C>A) in UBE2T. The patient presented with atypical, mild FA, characterized by persistent macrocytosis and neutropenia with intermittent thrombocytopenia but no severe bone marrow failure (without evidence of somatic reversion in blood) or congenital abnormalities common to FA. Clinical chromosomal breakage assays were consistent with a diagnosis of FA and subsequent functional analysis of patient-derived fibroblasts and the p.P66T UBE2T variant performed here demonstrate that the hypomorphic variant is the likely cause of disease in this patient and can be classified as likely pathogenic following the recommendations of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology.12
Figure 3.
UBE2T does not have a major role in the repair of non-interstrand crosslink DNA lesions. (A) Cell survival assay after ultraviolet treatment of complemented pairs of RA2627 fibroblasts compared to BJ wild-type (WT) fibroblasts depleted of XPF used as a positive control. The immunoblot shows decreased XPF levels after siRNA depletion. (B) Cell survival assay of RA2627 fibroblasts after treatment with irradiation. HA239F fibroblasts with RAD50 mutations are sensitive to irradiation and act as a positive control (RAD50mut). (C, D) Cell sensitivity assays comparing RA2627 fibroblasts to RA3331 Fanconi anemia patient-derived fibroblasts with SLX4 mutations (SLX4mut) expressing WT SLX4 or empty vector exposed to camptothecin (C) and the PARP inhibitor, olaparib (D). (E) Cell survival assay after hydroxyurea treatment of RA2627 cells compared to the RA3226 BRCA2 patient cell line (BRCA2mut). Error bars indicate the standard deviation. EV: empty vector; UV: ultraviolet irradiation; IR: irradiation; CPT: camptothecin; PARPi: PARP inhibitor; HU: hydroxyurea.
The c.196C>A (p.Pro66Thr) UBE2T variant is likely damaging to UBE2T function by conferring both reduced E2 activity and reduced stability as immunoblotting demonstrated decreased protein levels. The p.P66T variant affects a residue highly conserved across various E2 and likely affects the interaction with FANCL due to the amino acid residue substitution being at the hydrophobic E2-E3 interface.10 The patients previously reported by Hira et al. who also had a missense variant, p.Q2E, were likewise demonstrated to be hypomorphic in RA2627 cells,13 but heterozygous and in trans to loss of function variants suggesting the possibility of UBE2T dosage sensitivity, as the two patients presented with more severe disease. Disease severity may also be increased in those two patients because of the presence of the ALDH2* variant which is known to interact genetically with the FA pathway.14
We hypothesize that the hypomorphic variant and resulting residual function of the c.196C>A (p.P66T) variant in UBE2T explains our patient’s mild phenotype. This case adds to the limited knowledge regarding this rare FA-T complementation group. It is possible that there are other undiagnosed patients with mild phenotypes, emphasizing the utility of an algorithmic approach utilizing genomic sequencing and functional analysis for patients with non-specific hematologic phenotypes.
Supplementary Material
Acknowledgments
We thank the proband for participating in this study.
Funding Statement
Funding: this work was supported in part by the Mayo Clinic Center for Individualized Medicine and the “Henry Predolin Leukemia Foundation” and by a Starr Cancer Consortium grant (to AS), National Institutes of Health (NIH) RO1 HL120922 (to AS), and grant # UL1TR001866 from the National Center for Advancing Translational Sciences, NIH Clinical and Translational Science Award program. KAR was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. AS is a Howard Hughes Medical Institute.Faculty Scholar. The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Fiesco-Roa MO, Giri N, McReynolds LJ, Best AF, Alter BP. Genotype-phenotype associations in Fanconi anemia: a literature review. Blood Rev. 2019;37:100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493(7432):356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niraj J, Farkkila A, D'Andrea AD. The Fanconi anemia pathway in cancer. Annu Rev Cancer Biol. 2019;3:457-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meetei AR, de Winter JP, Medhurst AL, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35(2):165-170. [DOI] [PubMed] [Google Scholar]
- 5.Machida YJ, Machida Y, Chen Y, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23(4):589-596. [DOI] [PubMed] [Google Scholar]
- 6.Rickman KA, Lach FP, Abhyankar A, et al. Deficiency of UBE2T, the E2 ubiquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T subtype of Fanconi anemia. Cell Rep. 2015;12(1):35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hira A, Yoshida K, Sato K, et al. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am J Hum Genet. 2015;96(6):1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Virts EL, Jankowska A, Mackay C, et al. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum Mol Genet. 2015;24(18):5093-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mangaonkar AA, Ferrer A, Pinto EVF, et al. Clinical applications and utility of a precision medicine approach for patients with unexplained cytopenias. Mayo Clin Proc. 2019;94(9):1753-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodson C, Purkiss A, Miles JA, Walden H. Structure of the human FANCL RING-Ube2T complex reveals determinants of cognate E3- E2 selection. Structure. 2014;22(2):337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelsall IR, Langenick J, MacKay C, Patel KJ, Alpi AF. The Fanconi anaemia components UBE2T and FANCM are functionally linked to nucleotide excision repair. PLoS One. 2012;7(5):e36970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv Z, Rickman KA, Yuan L, et al. S. pombe Uba1-Ubc15 structure reveals a novel regulatory mechanism of ubiquitin E2 activity. Mol Cell. 2017;65(4):699-714.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hira A, Yabe H, Yoshida K, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122(18):3206-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.