Abstract
Mice possess two types of teeth that differ in their cusp patterns; incisors have one cusp and molars have multiple cusps. The patterning of these two types of teeth relies on fine-tuning of the reciprocal molecular signaling between dental epithelial and mesenchymal tissues during embryonic development. The AP-2 transcription factors, particularly Tfap2a and Tfap2b, are essential components of such epithelial-mesenchymal signaling interactions that coordinate craniofacial development in mice and other vertebrates, but little is known about their roles in the regulation of tooth development and shape. Here we demonstrate that incisors and molars differ in their temporal and spatial expression of Tfap2a and Tfap2b. At the bud stage, Tfap2a is expressed in both the epithelium and mesenchyme of the incisors and molars, but Tfap2b expression is restricted to the molar mesenchyme, only later appearing in the incisor epithelium. Tissue-specific deletions show that loss of the epithelial domain of Tfap2a and Tfap2b affects the number and spatial arrangement of the incisors, notably resulting in duplicated lower incisors. In contrast, deletion of these two genes in the mesenchymal domain has little effect on tooth development. Collectively these results implicate epithelial expression of Tfap2a and Tfap2b in regulating the extent of the dental lamina associated with patterning the incisors and suggest that these genes contribute to morphological differences between anterior (incisor) and posterior (molar) teeth within the mammalian dentition.
Keywords: AP-2, Tfap2, odontogenesis, incisor, molar
1. Introduction
Teeth arise from a series of molecular and physical interactions between epithelial and mesenchymal tissues in the embryonic oral cavity (Kollar and Baird, 1969; Lumsden, 1988; Mina and Kollar, 1987). Genetic mutations that affect these tissue interactions (e.g., PITX2, MSX1, and PAX9) can cause profound disruptions to the development of the human dentition, including loss, gain, or mis-patterning of teeth (Alappat et al., 2003; Chen et al., 1996; Dressler et al., 2010; Mostowska et al., 2003; Peters et al., 1998; Satokata and Maas, 1994). The genetic basis for tooth development has been well-studied in mice (Ahn et al., 2010; Harada et al., 2002; Harjunmaa et al., 2012; Jernvall et al., 1994; Klein et al., 2008; Pispa et al., 1999; Thesleff et al., 2001; Tummers and Thesleff, 2003), however, few studies have explicitly compared gene expression between developing incisors and molars (Hu et al., 2013; Huang et al., 2014; Laugel-Haushalter et al., 2013; Tucker et al., 1998).
Activator protein-2 (AP-2) transcription factors are known to play an essential role in craniofacial development in numerous vertebrate species, including mice, zebrafish, and chickens (Brewer et al., 2004; Brewer and Williams, 2004; de Croze et al., 2011; Hoffman et al., 2007; Knight et al., 2005; Li and Cornell, 2007; Nottoli et al., 1998; Schorle et al., 1996; Van Otterloo et al., 2018; Zhang et al., 1996). Several human genetic studies have identified dental anomalies in patients with TFAP2A and TFAP2B mutations, which cause the human syndromic disorders branchio-oculo-facial syndrome and Char syndrome, respectively (Milunsky et al., 2008; Satoda et al., 2000; Tanasubsinn et al., 2017). Although several studies have examined aspects of Tfap2 expression in vertebrate tooth development (Laugel-Haushalter et al., 2013; Moser et al., 1997; Tanasubsinn et al., 2017; Uchibe et al., 2012; Wang et al., 2014), there has not been a comprehensive spatiotemporal analysis of their expression and little is known about the tissue-specific functions of AP-2 genes during dental development. To understand the roles of AP-2 genes in establishing the molecular and morphological identities of incisors and molars, here we investigate the expression dynamics and tissue-specific functions of two AP-2 paralogs, Tfap2a and Tfap2b.
We compared spatiotemporal differences in the expression of Tfap2a and Tfap2b between incisors and molars and used mouse conditional genetics to determine the tissue-specific roles of these genes in dental epithelium and mesenchyme of each tooth class. Though Tfap2a and Tfap2b are expressed in epithelial and mesenchymal tissues during tooth development, we found that epithelial-specific loss of Tfap2a and Tfap2b results in a loss or reduction of upper incisors along with a duplication of lower incisors, but deletion of these genes in the neural crest (NC)-derived mesenchyme does not perturb dental development. Despite major impacts on incisor development, molar development is essentially unaffected by epithelial loss of Tfap2a and Tfap2b. Collectively, our results identify a novel role for Tfap2 family members in dental development.
2. Materials and Methods
2.1. Mice
Animal experiments were conducted following the ‘Guide for the Care and Use of Laboratory Animals of the National Institutes of Health’ and approved by the Institutional Animal Care and Use Committees of the University of Florida or the University of Colorado – Denver. ICR (CD-1) “wild-type” laboratory mice (Envigo) were housed at the University of Florida, while Tfap2a and Tfap2b mutant lines were housed at the University of Colorado – Denver. See Supplementary Methods 5.1 for details on embryo collection and histology.
2.2. Conditional Deletion of Tfap2a and Tfap2b
To generate Tfap2 mutant embryos, we used either conditional floxed alleles or null alleles of Tfap2a and Tfap2b and two strains in which Cre recombinase was expressed in either the epithelium, Crect (Schock et al., 2017), or the neural crest (NC), Wnt1-Cre (Danielian et al., 1998) (Supplementary Figure 1). Females were homozygous for the Tfap2a floxed conditional allele, Tfap2atm2Will/J (Brewer et al., 2004) and the Tfap2b floxed conditional allele, Tfap2btm2Will (Martino et al., 2016; Seberg et al., 2017; Van Otterloo et al., 2018). Males were heterozygous for either the epithelial or NC Cre allele, and the Tfap2a and Tfap2b conditional alleles (i.e., Tfap2aflox/wt;Tfap2bflox/wt;Wnt1-Cre or Tfap2aflox/wt;Tfap2bflox/wt;Crect). In the second cross, males were heterozygous for conditional null alleles of Tfap2a (Zhang et al., 1996) and Tfap2b (Martino et al., 2016; Seberg et al., 2017; Van Otterloo et al., 2018) (i.e., Tfap2anull/wt;Tfap2bnull/wt;Wnt1-Cre or Tfap2anull/wt;Tfap2bnull/wt;Crect) and females were homozygous for the conditional alleles. See Supplementary Methods 5.2 for additional details on tissue-specific deletions and embryo genotypes. For all embryos examined, genotypes, phenotypes, and sample sizes are provided in Supplementary Table 1.
2.3. RNA in situ hybridization on cryosections
RNA probes for in situ hybridization (ISH) were generated for Tfap2a, Tfap2b, Yeats4, Kctd1, and Ets1 (see Supplementary Methods 5.3). ISH was performed as described previously (Acloque et al., 2008) with some modifications (see Supplementary Methods 5.3). Expression patterns reported here were detected in a minimum of 3 wild-type CD-1 embryos per stage. Because the expression patterns of these genes had been documented in the head, these regions were used as positive controls (Supplementary Figure 2 A–G). Negative (sense) controls were also conducted for each gene and produced no detectable signal (Supplementary Figure 2 H–L).
2.4. Micro-CT scanning and 3-D reconstruction of Tfap2 mutant and control embryos
Mouse embryos were prepared for micro-CT (μCT) (see Supplementary Methods 5.4). and scanned in a GE V|TOME|X M 240 Nano CT scanner (General Electric) at the University of Florida Nanoscale Research Facility. Tiff stacks were generated using Phoenix Datos2 software (General Electric) and VG Studio Max (Volume Graphics v3.3.4) was used for 3-D reconstructions (See Supplementary Methods 5.4 for details). Length and width ratios from 3-D reconstructed first upper and lower molar crowns (M1/1) of control and mutant embryos were calculated (see Supplementary Methods 5.5 for details).
3. Results and Discussion
3.1. Incisors and molars differ in temporal and spatial expression of Tfap2a and Tfap2b
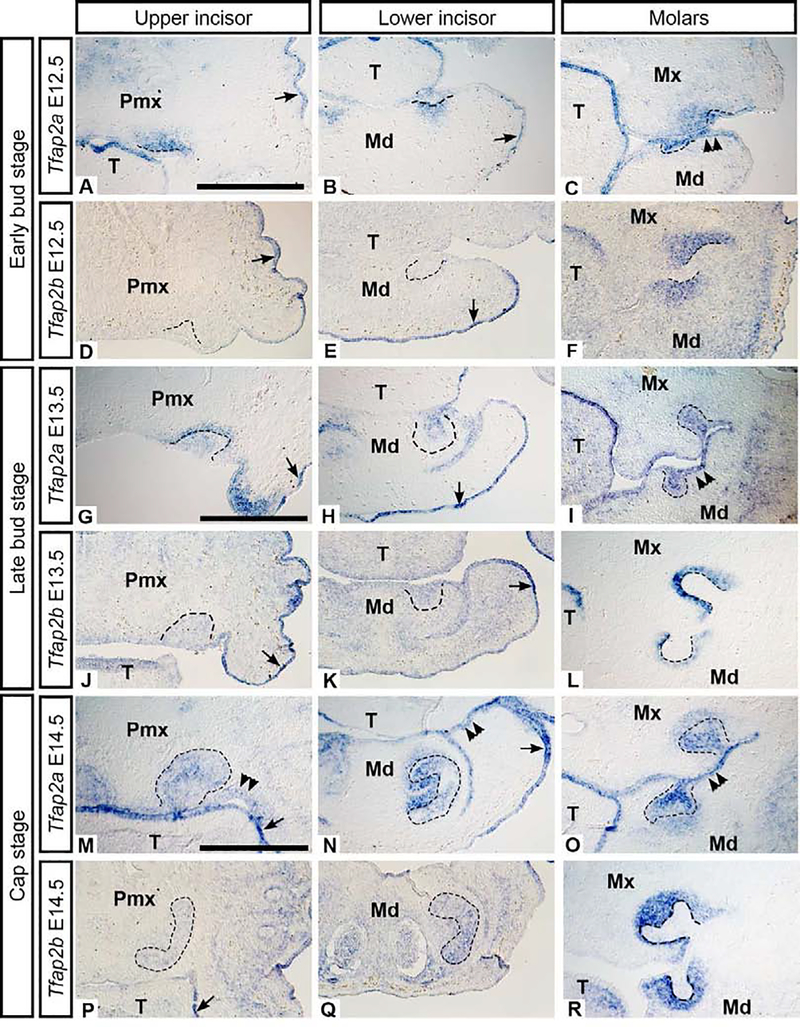
To determine the spatial and temporal expression of Tfap2a and Tfap2b, we compared mRNA localization in mouse incisors and molars at the bud stage (E12.5-E13.5), the cap stage (E14.5), and the bell stage (E16.5–17). Tfap2a expression was detected throughout the bud and cap stages in the incisor epithelium and mesenchyme (Figure 1 A–B, G–H, M–N). Tfap2b, however, was first detected in the incisor epithelium at the late bud stage (Figure 1 D–E, J–K) in some (but not all) embryos, suggesting that Tfap2b transcripts were just beginning to accumulate at E13.5. In the molars, by contrast, both Tfap2a and Tfap2b were expressed prominently in the mesenchyme at the bud and cap stages (Figure 1 C, F, I, L, O, R), in agreement with previous reports (Tanasubsinn et al., 2017; Uchibe et al., 2012). At the cap stage, Tfap2b was faintly detected in the incisor epithelium (Figure 1 P, Q). At the bud and cap stages, both genes were expressed in the oral and/or surface epithelium (Figure 1 A, B, D, E, G, H, J, K), consistent with previous studies showing that at E10.5 Tfap2a is expressed in the NC-derived mesenchyme of the first branchial arch, and both Tfap2a and Tfap2b are expressed in the surrounding surface epithelium (Zhang and Williams, 2003; Zhao et al., 2011).
Figure 1. Bud stage (E12.5 and 13.5) and cap stage (E14.5) mRNA expression of Tfap2a and Tfap2b in wild-type mouse embryos.
Images showing mRNA transcripts detected by in situ hybridization on frontal cryosections through the upper incisor (left column), lower incisor (middle column), and molars (right column). The dental epithelium is outlined. There is minimal expression of Tfap2b in the bud stage upper and lower incisors (D-E, J-K) compared to the molar buds (F, L). Both Tfap2a and Tfap2b were detected in the surface epithelium (arrows) but only Tfap2a was present in the oral epithelium (M, N double arrowheads). Scale bars in A, G, M: 500um, all images are at the same scale. Pmx: premaxilla, Mx: maxilla, Md: mandible, T: tongue.
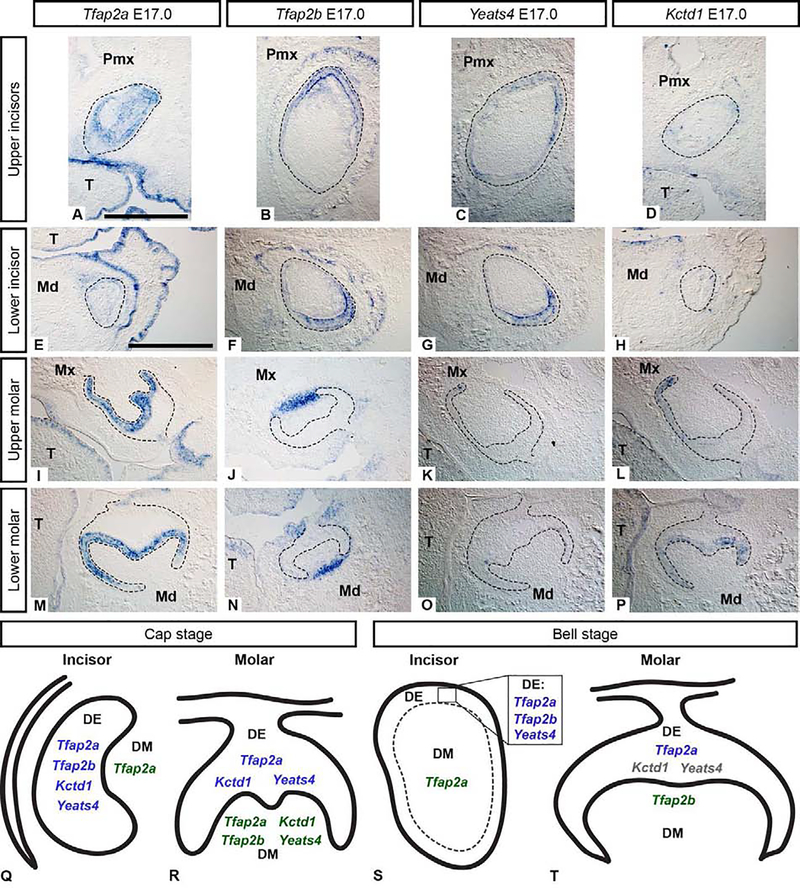
At the bell stage, in the upper incisor Tfap2a was expressed in the epithelium and the mesenchyme (Figure 2 A), whereas in the lower incisor, Tfap2a was only weakly expressed in the incisor epithelium, unlike at earlier stages when it was more prominently expressed (Figure 1 H, N). In the molars, Tfap2a expression was restricted to the inner enamel epithelium directly adjacent to the dental mesenchyme (Figure 2 I, M). Tfap2b transcripts were detected prominently in bell stage incisors within epithelial-derived ameloblasts and faintly in the mesenchyme (Figure 2 B, F); however, in the molars, Tfap2b expression became limited to mesenchymal cells closer to the outer regions of the tooth germ (Figure 2 J, N). Restricted expression of Tfap2a in the molar inner enamel epithelium suggests it may be associated with cusp formation which is facilitated by enamel knots (Cho et al., 2007; Matalova et al., 2005; Thesleff et al., 2001).
Figure 2. Bell stage (E17.0) mRNA expression of Tfap2a, Tfap2b, Yeats4, and Kctd1 (A-P) in wild-type mouse embryos.
mRNA transcripts detected by in situ hybridization on cryosections in the frontal plane are shown in the upper incisor (A-D), lower incisor (E-H), upper molar (I-L), and lower molar (M-P). The dental epithelium is outlined. Note the spatially restricted expression of Tfap2a in the molars (I, M) and the overlapping expression domains of Tfap2b and Yeats4 in the incisors (B, C, F, G). Scale bar in A, G: 500um, all images are at the same scale. Pmx: premaxilla, Mx: maxilla, Md: mandible, T: tongue. Summary of gene expression results highlighting differences between incisors and molars at the cap (Q, R) and bell stages (S, T). In cap stage incisors (Q), Tfap2a, Tfap2b, Kctd1, and Yeats4 are expressed in the epithelium but only Tfap2a is expressed throughout the mesenchyme. In the molars (R), Tfap2b is expressed in the mesenchyme, while Tfap2a, Kctd1, and Yeats4 transcripts are present in both the epithelium and mesenchyme. In bell stage incisors (S), the expression of each gene is similar to that of the cap stage incisors, however, Kctd1 was not detected. In molars at the bell stage (T), Tfap2a was expressed in the epithelium only, Tfap2b expression was restricted to the mesenchyme, and minimal to no expression of Kctd1 and Yeats4 was detected, indicated by gray text (T). DE: dental epithelium, DM: dental mesenchyme.
As a first step towards connecting the dynamic spatiotemporal expression of Tfap2a and Tfap2b to their potential function(s) in tooth development, we next asked whether Tfap2a/Tfap2b expression is associated with two modulators of AP-2 activity, Yeats4, which encodes an AP-2 activator protein (Ding et al., 2006), and Kctd1 which encodes a transcriptional target of AP-2 (Marneros, 2020) or inhibitor (Ding et al., 2009). Tfap2a, Kctd1, and Yeats4 exhibit similar expression patterns in the dental epithelium of the incisors and molars at bud-cap stages (E13.5 and E14.5) (Figure 1, Supplementary Figure 3, Supplementary Results 6.1); however, expression of Tfap2b is co-expressed with Kctd1, and Yeats4 only at E14.5 in the incisor epithelium (Figure 1 P, Q; Supplementary Figure 3 G–H, J–K). There was limited expression of Kctd1 and Yeats4 in the dental mesenchyme at E14.5, where their expression domains were more similar to that of Tfap2a rather than Tfap2b (Supplementary Figure 3 I, L). At the bell stage, the incisors did not express Kctd1, whereas Yeats4 was expressed in the ameloblast layer, but not the mesenchyme, similar to Tfap2b (Figure 2 B, C). In contrast to the incisors, both Kctd1 and Yeats4 expression were detected to varying extents in bell stage molar epithelium in patterns more similar to Tfap2a (Figure 2 K, L, O, P; Supplementary Figure 4 I, J, K, L). The overlapping expression domains between Tfap2a, Kctd1, and Yeats4 in the dental epithelium, and to a lesser extent in the molar mesenchyme, suggest that Kctd1 and Yeats4 may interact directly with Tfap2a to modulate the transcriptional activity of the latter in developing teeth, though this prediction remains to be tested (Figure 2 Q–T).
We also compared these patterns with Ets1, which, like Tfap2a and Tfap2b, is expressed in migrating NC cells, where it acts downstream of Tfap2a in the chick (Barembaum and Bronner, 2013). In incisors and molars, Ets1 was not strongly expressed in mesenchyme associated with the dentition, but instead appeared to be associated with erythrocytes (Supplementary Results 6.1; Supplementary Figure 4 A–H). Altogether, these findings highlight regions of both co-expression and divergent expression domains of Tfap2a/Tfap2b and two regulators of AP-2 function during development of incisors and molars.
3.2. Epithelial deletion of Tfap2a and Tfap2b leads to extra incisors and misshapen teeth
The dynamic expression patterns of both Tfap2a and Tfap2b in the dental epithelium and mesenchyme suggested that these factors could play tissue-specific roles in tooth development. To test whether epithelial-specific expression of Tfap2a and Tfap2b is required for the development of properly shaped teeth, we used an epithelial-specific Cre recombinase allele, Crect (Schock et al., 2017). Mutant embryos were homozygous for Tfap2a and Tfap2b conditional alleles and heterozygous for the Crect transgene (Tfap2aflox/flox;Tfap2bflox/flox;Crect), resulting in deletion of Tfap2a and Tfap2b exclusively from the ectoderm, including the presumptive dental epithelium (Supplementary Figure 1 A).
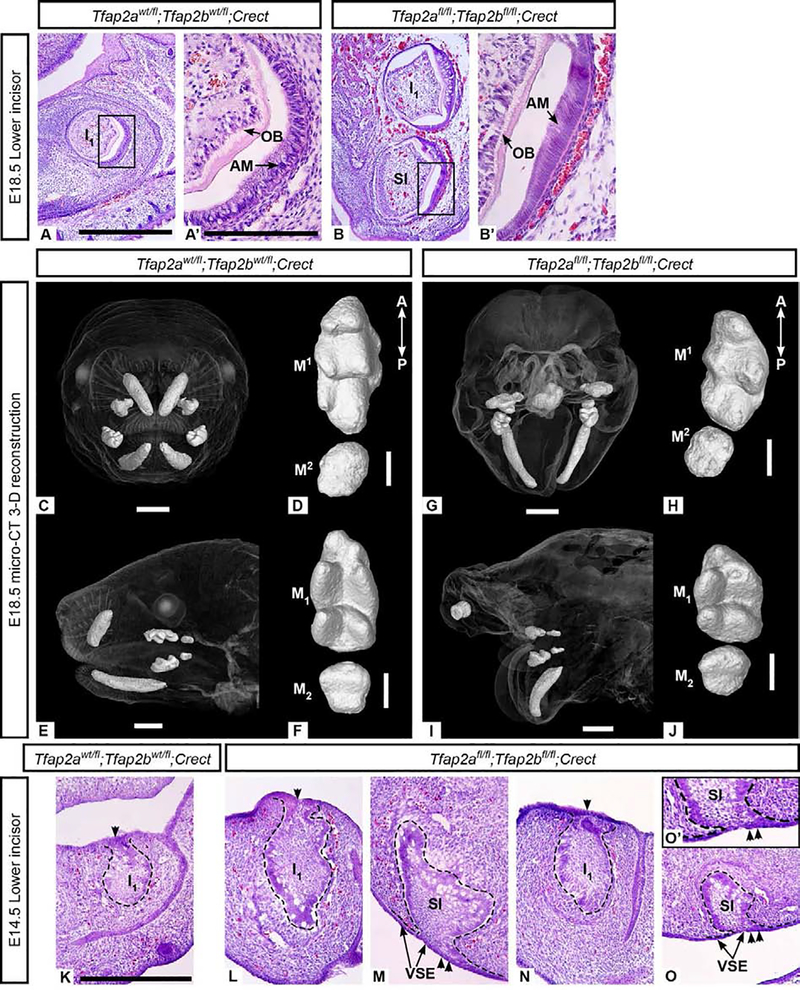
The most striking difference in E18.5 embryos lacking Tfap2a and Tfap2b in the epithelium were changes in the number and/or morphology of the lower incisors (Supplementary Table 1). Control embryos at E18.5 had a single upper and single lower incisor (I1/1) on each side of the jaw (Figure 3 A, C, E) whereas mutant embryos often displayed an additional bilateral lower incisor ventral to I1 (N=2/4 embryos) (Figure 3 B). The additional teeth appeared similar to I1 and a complete repertoire of differentiated cell types were found in the supernumerary incisors, including enamel-forming ameloblasts and dentin-forming odontoblasts (Figure 3 B, B’). Mutants that lacked duplicated incisors had aberrantly shaped lower incisors (I1) that exhibited ventral curvature (N=2/4 embryos; bilaterally symmetrical) (Figure 3 G, I; Supplementary Figure 5 D, D‘).
Figure 3. Tfap2afl/fl;Tfap2bfl/fl ;Crect mutant embryos have duplicated or ventrally curved lower incisors.
Hematoxylin and eosin (H&E) staining of bell stage lower incisors from control (A, A’) and epithelial mutant (B, B’) embryos revealed a supernumerary incisor ventral to I1 in the mutant mandible. This additional incisor undergoes cytodifferentiation (B’) similar to I1 in mutant (B) and control (A, A’) embryos. All mutant mandibles examined exhibited ventral curvature as seen in G and I. In the 3-D reconstructed embryo (G, I), instead of duplicated lower incisors, a single ventrally curved lower incisor was present on the right and left sides and two small right and left upper incisors were also observed. 3-D reconstructions of upper (D, H) and lower (F, J) molars show that first (M1/1) and second (M2/2) molars develop in mutants lacking epithelial Tfap2a and Tfap2b (H, J) and that the main cusps are present, though less distinct, in mutants compared with the controls (D, F). The mutant molars appear shorter along the anterior-posterior (A-P) axis than the control molars. H&E staining of frontal cryosections through cap stage lower incisors from control (K) and epithelial mutants, where I1 and a supernumerary incisor are shown in two different individuals (L-M and N-O, respectively). I1 is attached to the dorsal dental lamina (single arrowhead) in the control (K) and mutants (L, N). The supernumerary incisors are tethered to the ventral surface epithelium (double arrowheads, see the region of attachment shown in M, O, and enlarged in O’) and are positioned ventral and slightly posterior to I1. Scale bars: 500um (A), 150um (A’), 1mm (C, E, G, I), 300um (D, F, H, J, K). A and B, are to scale; A’ and B’ are to scale; K-O are to scale. AM: ameloblasts, OB: odontoblasts, SI: supernumerary incisor, VSE: ventral surface epithelium.
The faces of the epithelium-specific mutants were highly dysmorphic (Van Otterloo et al., unpublished observations) which made it difficult to assess the upper incisors. In some mutants, upper incisors were not observed at E18.5 (N=3/4 embryos) (Supplementary Figure 6 G–I), but in one embryo we observed two diminutive upper incisors (Figure 3 G, I; Supplementary Figure 5 B, B‘; Supplementary Table 1). In contrast to the incisors, the first and second upper and lower molars of the mutants (Figure 3 H, J; Supplementary Figure 7 H, I, K, L) were structurally similar to those of the controls (Figure 3 D, F; Supplementary Figure 7 G, J).
In mutant embryos from a similar genetic cross in which one of the conditional alleles was null (Tfap2aflox/null;Tfap2bflox/null;Crect), we observed the same duplicated lower incisor phenotype as in the first cross (Supplementary Figure 8 D, E; Supplementary Table 1) and a single upper incisor was present (Supplementary Figure 8 B), whereas the molars appeared similar to the controls (N=2/2) (Supplementary Figure 8 F–K).
To determine how this supernumerary lower incisor develops, we examined the histological structure of cap stage (E14.5) teeth in Tfap2afl/fl;Tfap2bfl/fl;Crect embryos. In E14.5 control embryos, we observed bilateral I1 (Figure 3 K), but in mutants, bilateral duplicated incisors were observed at the cap (N=2/3; Figure 3 L–O) or bud (N=1/3; data not shown) stage. In these mutants, I1 appeared tethered to the dorsal dental lamina (Figure 3 L, N) as in the controls, but the duplicated (ventral) incisor was tethered to the ventral surface epithelium, a region that does not normally have the characteristics of the dental lamina (Figure 3 M, O). Upper incisors were not observed in E14.5 mutant embryos (Supplementary Figure 6 B–E) indicating that they failed to form prior to E14.5. The molars at E14.5 appeared similar to those of the controls (Supplementary Figure 7 A–F). In E14.5 “single” mutants that still contained one wild-type Tfap2a or Tfap2b allele (Tfap2afl/fl;Tfap2bfl/wt;Crect or Tfap2afl/wt;Tfap2bfl/fl;Crect), the dentition was similar to that of the control embryos (N=3/3) (Supplementary Figure 9; Supplementary Results 6.2).
TFAP2A and TFAP2B are able to form heterodimers (Ding et al., 2009; Williams and Tjian, 1991), can bind the same DNA consensus sequences (Williams and Tjian, 1991), and are capable of functioning redundantly in tissues in which they are co-expressed (Hoffman et al., 2007; Li and Cornell, 2007; Rothstein and Simoes-Costa, 2020; Seberg et al., 2017; Van Otterloo et al., 2018; Wang et al., 2008). Consistent with these observations, our analysis of Tfap2a and Tfap2b mutants shows that TFAP2A and TFAP2B cooperatively function within the craniodental ectoderm, including the dental epithelium, to regulate incisor development.
One possible explanation for the incisor duplication is that in the absence of Tfap2a and Tfap2b, the incisor odontogenic domain is expanded ventrally, permitting a second incisor to form in the mandible. A second possibility is that there is a duplication of the odontogenic domain so that a new and separate domain is formed on the aboral surface of the mandible, thereby allowing an ectopic incisor to form. Such a domain would need to be induced only at the distal end of the mandible, as the molars are not duplicated. Ventral curvature of I1 in the mutants lacking duplicated incisors is suggestive of dorsoventral mis-patterning, however, this may be secondary to changes in mandible shape, which also curves ventrally compared to the control (Figure 3 G, I) (Van Otterloo et al., unpublished observations). In mutants in which a supernumerary cap stage lower incisor was observed at E14.5, it appeared to be connected to the ventral surface epithelium (Figure 3 M, O), raising the possibility that the surface epithelium has properties of an ectopic dental lamina. This suggests that the ectopic tooth was initiated in the ventral epithelium, but further studies will be required to ascertain how odontogenic potential is altered by the loss of ectodermal expression of these two AP-2 genes.
3.3. Mesenchymal Tfap2a and Tfap2b is dispensable for tooth development
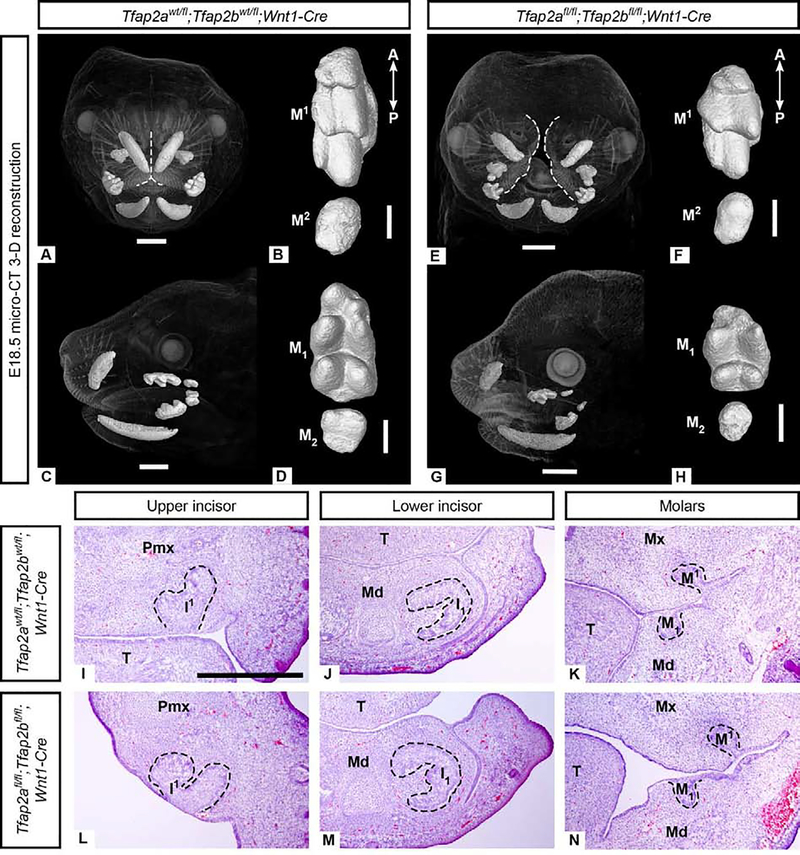
Given that robust Tfap2a and Tfap2b expression was detected in the dental mesenchyme (Figures 1–2), we predicted that AP-2 activity within the cranial NC-derived mesenchyme is also required for normal tooth development. To test this hypothesis, we generated mice with conditional deletions of Tfap2a and Tfap2b in the NC-derived mesenchyme using the Wnt1-Cre allele (Danielian et al., 1998) and floxed or null alleles of Tfap2a and Tfap2b (Van Otterloo et al., 2018) (Supplementary Figure 1 B, D). Despite an upper midfacial cleft (Figure 4 E, Supplementary Figure 10 E), mutant embryos did not possess an aberrant incisor phenotype at E18.5 (N=3/3) (Figure 4 E, G; Supplementary Figure 10 C, D; Supplementary Table 1) or at E14.5 (N=3/3) (Figure 4 I, J, L, M). Histological and μCT analysis showed that the molars also appeared unperturbed and were similar to controls at the cap stage (Figure 4 K, N; Supplementary Table 1) and the bell stage (Figure 4 A–H; Supplementary Figure 10 F–I). In E14.5 “single” mutants with one wild-type allele of either Tfap2a or Tfap2b in the NC-derived mesenchyme (e.g, Tfap2afl/fl;Tfap2bfl/wt;Wnt1-Cre), we also observed the proper number of cap stage incisors and molars (N=3/3) (Supplementary Figure 9; Supplementary Results 6.2). In contrast to the requirement for Tfap2a and Tfap2b in the dental epithelium, these results indicate that Tfap2a and Tfap2b in the mesenchyme are not necessary, either individually or collectively, for tooth development.
Figure 4. Incisors and molars in Tfap2afl/fl;Tfap2bfl/fl;Wnt1-Cre embryos lacking Tfap2a and Tfap2b expression in the NC-derived mesenchyme lack major morphological defects based on μCT (A-H) and histological (I-N) analyses.
3-D reconstructions of μCT data comparing morphology of control upper (B: M1−2) and lower (D: M1–2) molars with mutant upper (F: M1−2) and lower (H: M1–2) molars. The correct number of cusps are present but the molars appear shorter along the anterior-posterior (A-P) axis than the controls. Note the midface cleft in the mutant (E), outlined in white, compared to the control (A). H&E stained cryosections in the frontal plane (I-N) showing that mesenchyme-specific mutant incisors (L, M) and molars (N) are similar to those of the control (I-K) at the cap stage (E14.5). Due to the cleft midface and palate, in the anterior-most frontal section shown here (L) the medial aspect of the premaxilla is at an angle. Scale bars are 1mm (A, C, E, G) and 300um (B, D, F, H), 500um (I-N). Pmx: premaxilla, Mx: maxilla, Md: mandible, T: tongue.
3.4. Molar development is largely unaffected following loss of Tfap2a and Tfap2b in either epithelium or mesenchyme
Loss of Tfap2a and Tfap2b in either the epithelium or the mesenchyme appeared to have little effect on the molar teeth; however, 3-D reconstructions of E18.5 molar crowns (M1/1) revealed that the mutant molar crowns were shorter along the anterior-posterior (mesiodistal) axis compared to the controls (Figure 3 D, F, H, J; Figure 4 B, D, F, H). Quantification of the upper and lower first molar crowns (ratio of molar crown length to width, see Supplementary Methods 5.5) suggested that for both crosses, anterior-posterior molar length was reduced in both the epithelium-specific and mesenchyme-specific mutants compared to the controls (Supplementary Table 4) (N=2 teeth/individual), although, given the sample size, it was not possible to determine if mutant molar ratios were statistically different from control molar ratios.
Though deletion of Tfap2a and Tfap2b may affect molar length, the mutations had the most profound effect on the incisors. Possible explanations for the relative lack of molar defects are that AP-2 function is not required for molar development, or alternatively, that additional Tfap2 family members, such as Tfap2c, which is expressed within the oral epithelium and dental mesenchyme (Chazaud et al., 1996), may compensate for the loss of Tfap2a and Tfap2b. Finally, given that Tfap2a homozygous null mice are so severely affected that they lack a mouth, among other ventral craniofacial structures (Zhang et al., 1996), we cannot exclude the possibility that Tfap2a and Tfap2b have an earlier role in patterning the molars, in addition to their effect on molar size.
Supplementary Material
Highlights.
During tooth development, transcription factors Tfap2a and Tfap2b are expressed in spatially and temporally dynamic patterns and differ between incisor and molar tooth germs.
Epithelial expression of Tfap2a and Tfap2b is necessary for incisor development, but mesenchymal expression of these genes is not required.
Acknowledgements
We thank Dr. Brooke Armfield for her assistance with experimental design, mouse breeding, and for teaching EDW to perform various assays. We thank Alyssa Mangino for assistance with sectioning and in situ hybridization, and Emily Merton for technical support. We also acknowledge Dr. Gary Scheiffle and Dr. Edward Stanley at the University of Florida Nanoscale Research Facility for their assistance with μCT scanning. Finally, we thank all members of the Cohn laboratory for valuable insights and critical discussion of this work.
Funding
This work was supported by an NSF DDRI 1455572 to EDW and MJC, an American Society of Mammalogists grant to EDW, an NIDCR K99/R00 DE026823 to EVO, and NIH 2R01 DE12728 to TW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
The μCT data from this study will be freely available to the public on FaceBase3 (https://www.facebase.org/).
References
- Acloque H, Wilkinson DG, Nieto MA, 2008. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods Cell Biol. 87, 169–185. 10.1016/S0091-679X(08)00209-4 [DOI] [PubMed] [Google Scholar]
- Ahn Y, Sanderson BW, Klein OD, Krumlauf R, 2010. Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development dev.054668. 10.1242/dev.054668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alappat S, Zhang ZY, Chen YP, 2003. Msx homeobox gene family and craniofacial development. Cell Res. 13, 429–442. 10.1038/sj.cr.7290185 [DOI] [PubMed] [Google Scholar]
- Barembaum M, Bronner ME, 2013. Identification and dissection of a key enhancer mediating cranial neural crest specific expression of transcription factor, Ets-1. Dev. Biol. 382, 567–575. 10.1016/j.ydbio.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braissant O, Wahli W, 1998. A Simplified In Situ Hybridization Protocol Using Non-radioactively Labeled Probes to Detect Abundant and Rare mRNAs on Tissue Sections. Biochemica 1. [Google Scholar]
- Brewer S, Feng W, Huang J, Sullivan S, Williams T, 2004. Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Dev. Biol. 267, 135–152. 10.1016/j.ydbio.2003.10.039 [DOI] [PubMed] [Google Scholar]
- Brewer S, Williams T, 2004. Loss of AP-2α impacts multiple aspects of ventral body wall development and closure. Developmental Biology 267, 399–417. 10.1016/j.ydbio.2003.11.021 [DOI] [PubMed] [Google Scholar]
- Chazaud C, Oulad-Abdelghani M, Bouillet P, Décimo D, Chambon P, Dollé P, 1996. AP-2.2, a novel gene related to AP-2, is expressed in the forebrain, limbs and face during mouse embryogenesis. Mechanisms of Development 54, 83–94. 10.1016/0925-4773(95)00463-7 [DOI] [PubMed] [Google Scholar]
- Chen Y, Bei M, Woo I, Satokata I, Maas R, 1996. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development 122, 3035. [DOI] [PubMed] [Google Scholar]
- Cho S-W, Lee H-A, Cai J, Lee M-J, Kim J-Y, Ohshima H, Jung H-S, 2007. The primary enamel knot determines the position of the first buccal cusp in developing mice molars. Differentiation 75, 441–451. 10.1111/j.1432-0436.2006.00153.x [DOI] [PubMed] [Google Scholar]
- Csanady A, Mosansky L, 2018. Skull morphometry and sexual size dimorphism in Mus musculus from Slovakia. North-Western Journal of Zoology 14, 102–106. [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP, 1998. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Current Biology 8, 1323–S2. 10.1016/S0960-9822(07)00562-3 [DOI] [PubMed] [Google Scholar]
- de Croze N, Maczkowiak F, Monsoro-Burq AH, 2011. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proceedings of the National Academy of Sciences 108, 155–160. 10.1073/pnas.1010740107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Fan C, Zhou J, Zhong Y, Liu R, Ren K, Hu X, Luo C, Xiao S, Wang Y, Feng D, Zhang J, 2006. GAS41 interacts with transcription factor AP-2beta and stimulates AP-2beta-mediated transactivation. Nucleic Acids Res. 34, 2570–2578. 10.1093/nar/gkl319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Luo C, Zhou J, Zhong Y, Hu X, Zhou F, Ren K, Gan L, He A, Zhu J, Gao X, Zhang J, 2009. The interaction of KCTD1 with transcription factor AP-2α inhibits its transactivation. Journal of Cellular Biochemistry 106, 285–295. 10.1002/jcb.22002 [DOI] [PubMed] [Google Scholar]
- Dressler S, Meyer-Marcotty P, Weisschuh N, Jablonski-Momeni A, Pieper K, Gramer G, Gramer E, 2010. Dental and Craniofacial Anomalies Associated with Axenfeld-Rieger Syndrome with PITX2 Mutation. Case Rep Med 2010. 10.1155/2010/621984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, Kato S, Sekine K, Ohuchi H, 2002. FGF10 maintains stem cell compartment in developing mouse incisors. Development 129, 1533–1541. [DOI] [PubMed] [Google Scholar]
- Harjunmaa E, Kallonen A, Voutilainen M, Hämäläinen K, Mikkola ML, Jernvall J, 2012. On the difficulty of increasing dental complexity. Nature 483, 324–327. 10.1038/nature10876 [DOI] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF, 2007. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 308B, 679–691. 10.1002/jez.b.21189 [DOI] [PubMed] [Google Scholar]
- Hu X, Zhang S, Chen G, Lin C, Huang Z, Chen Y, Zhang Y, 2013. Expression of SHH signaling molecules in the developing human primary dentition. BMC Dev. Biol. 13, 11. 10.1186/1471-213X-13-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Hu X, Lin C, Chen S, Huang F, Zhang Y, 2014. Genome-wide analysis of gene expression in human embryonic tooth germ. J. Mol. Histol. 45, 609–617. 10.1007/s10735-014-9580-5 [DOI] [PubMed] [Google Scholar]
- Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I, 1994. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int. J. Dev. Biol. 38, 463–469. [PubMed] [Google Scholar]
- Kaufman MH, 1992. The Atlas of Mouse Development. Elsevier Science. [Google Scholar]
- Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR, 2008. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development 135, 377–385. 10.1242/dev.015081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RD, Javidan Y, Zhang T, Nelson S, Schilling TF, 2005. AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development 132, 3127–3138. 10.1242/dev.01879 [DOI] [PubMed] [Google Scholar]
- Kollar EJ, Baird GR, 1969. The influence of the dental papilla on the development of tooth shape in embryonic mouse tooth germs. Development 21, 131–148. [PubMed] [Google Scholar]
- Laugel-Haushalter V, Paschaki M, Thibault-Carpentier C, Dembelé D, Dollé P, Bloch-Zupan A, 2013. Molars and incisors: show your microarray IDs. BMC Res Notes 6, 113. 10.1186/1756-0500-6-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cornell RA, 2007. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Developmental Biology 304, 338–354. 10.1016/j.ydbio.2006.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden AG, 1988. Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development 103 Suppl, 155–169. [DOI] [PubMed] [Google Scholar]
- Marneros AG, 2020. AP-2B/KCTD1 Control Distal Nephron Differentiation and Protect against Renal Fibrosis. Developmental Cell 54, 348–366.e5. 10.1016/j.devcel.2020.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, 1990. Tissue patterning in the developing mouse limb. Int. J. Dev. Biol. 34, 323–336. 10.1387/ijdb.1702679 [DOI] [PubMed] [Google Scholar]
- Martino VB, Sabljic T, Deschamps P, Green RM, Akula M, Peacock E, Ball A, Williams T, West-Mays JA, 2016. Conditional deletion of AP-2β in mouse cranial neural crest results in anterior segment dysgenesis and early-onset glaucoma. Disease Models & Mechanisms 9, 849–861. 10.1242/dmm.025262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matalova E, Antonarakis GS, Sharpe PT, Tucker AS, 2005. Cell lineage of primary and secondary enamel knots. Dev. Dyn. 233, 754–759. 10.1002/dvdy.20396 [DOI] [PubMed] [Google Scholar]
- Metscher BD, 2009. MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev. Dyn. 238, 632–640. 10.1002/dvdy.21857 [DOI] [PubMed] [Google Scholar]
- Milunsky JM, Maher TA, Zhao G, Roberts AE, Stalker HJ, Zori RT, Burch MN, Clemens M, Mulliken JB, Smith R, Lin AE, 2008. TFAP2A Mutations Result in Branchio-Oculo-Facial Syndrome. The American Journal of Human Genetics 82, 1171–1177. 10.1016/j.ajhg.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina M, Kollar EJ, 1987. The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch. Oral Biol. 32, 123–127. [DOI] [PubMed] [Google Scholar]
- Moser M, Rüschoff J, Buettner R, 1997. Comparative analysis of AP-2 alpha and AP-2 beta gene expression during murine embryogenesis. Dev. Dyn. 208, 115–124. [DOI] [PubMed] [Google Scholar]
- Mostowska A, Kobielak A, Trzeciak WH, 2003. Molecular basis of non-syndromic tooth agenesis: mutations of MSX1 and PAX9 reflect their role in patterning human dentition. European Journal of Oral Sciences 111, 365–370. 10.1034/j.1600-0722.2003.00069.x [DOI] [PubMed] [Google Scholar]
- Nottoli T, Hagopian-Donaldson S, Zhang J, Perkins A, Williams T, 1998. AP-2-null cells disrupt morphogenesis of the eye, face, and limbs in chimeric mice. PNAS 95, 13714–13719. 10.1073/pnas.95.23.13714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Neubüser A, Kratochwil K, Balling R, 1998. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev 12, 2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pispa J, Jung HS, Jernvall J, Kettunen P, Mustonen T, Tabata MJ, Kere J, Thesleff I, 1999. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev. Biol. 216, 521–534. 10.1006/dbio.1999.9514 [DOI] [PubMed] [Google Scholar]
- Rothstein M, Simoes-Costa M, 2020. Heterodimerization of TFAP2 pioneer factors drives epigenomic remodeling during neural crest specification. Genome Res. 30, 35–48. 10.1101/gr.249680.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoda M, Zhao F, Diaz GA, Burn J, Goodship J, Davidson HR, Pierpont MEM, Gelb BD, 2000. Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nature Genetics 25, 42–46. 10.1038/75578 [DOI] [PubMed] [Google Scholar]
- Satokata I, Maas R, 1994. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 6, 348–356. 10.1038/ng0494-348 [DOI] [PubMed] [Google Scholar]
- Schock EN, Struve JN, Chang C-F, Williams TJ, Snedeker J, Attia AC, Stottmann RW, Brugmann SA, 2017. A tissue-specific role for intraflagellar transport genes during craniofacial development. PLOS ONE 12, e0174206. 10.1371/journal.pone.0174206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ, 1996. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381, 235–238. 10.1038/381235a0 [DOI] [PubMed] [Google Scholar]
- Seberg HE, Van Otterloo E, Loftus SK, Liu H, Bonde G, Sompallae R, Gildea DE, Santana JF, Manak JR, Pavan WJ, Williams T, Cornell RA, 2017. TFAP2 paralogs regulate melanocyte differentiation in parallel with MITF. PLOS Genetics 13, e1006636. 10.1371/journal.pgen.1006636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanasubsinn N, Sittiwangkul R, Pongprot Y, Kawasaki K, Ohazama A, Sastraruji T, Kaewgahya M, Kantaputra PN, 2017. TFAP2B mutation and dental anomalies. J Hum Genet 62, 769–775. 10.1038/jhg.2017.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I, Keränen S, Jernvall J, 2001. Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation. Adv. Dent. Res. 15, 14–18. 10.1177/08959374010150010401 [DOI] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT, 1998. Transformation of Tooth Type Induced by Inhibition of BMP Signaling. Science 282, 1136–1138. 10.1126/science.282.5391.1136 [DOI] [PubMed] [Google Scholar]
- Tummers M, Thesleff I, 2003. Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development 130, 1049–1057. 10.1242/dev.00332 [DOI] [PubMed] [Google Scholar]
- Uchibe K, Shimizu H, Yokoyama S, Kuboki T, Asahara H, 2012. Identification of novel transcription-regulating genes expressed during murine molar development. Developmental Dynamics 241, 1217–1226. 10.1002/dvdy.23808 [DOI] [PubMed] [Google Scholar]
- Van Otterloo E, Li H, Jones KL, Williams T, 2018. AP-2α and AP-2β cooperatively orchestrate homeobox gene expression during branchial arch patterning. Development 145, dev157438. 10.1242/dev.157438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JT, 1968. Analysis of Dental Variation in Wild-caught California House Mice. The American Midland Naturalist 80, 360–380. 10.2307/2423531 [DOI] [Google Scholar]
- Wang F, Xiao J, Cong W, Li A, Song T, Wei F, Xu J, Zhang C, Fan Z, Wang S, 2014. Morphology and chronology of diphyodont dentition in miniature pigs, Sus Scrofa. Oral Diseases 20, 367–379. 10.1111/odi.12126 [DOI] [PubMed] [Google Scholar]
- Wang X, Pasolli HA, Williams T, Fuchs E, 2008. AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J Cell Biol 183, 37–48. 10.1083/jcb.200804030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T, Tjian R, 1991. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes & Development 5, 670–682. 10.1101/gad.5.4.670 [DOI] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, Williams T, 1996. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 381, 238–241. 10.1038/381238a0 [DOI] [PubMed] [Google Scholar]
- Zhang J, Williams T, 2003. Identification and regulation of tissue-specific cis-acting elements associated with the human AP-2alpha gene. Dev. Dyn. 228, 194–207. 10.1002/dvdy.10365 [DOI] [PubMed] [Google Scholar]
- Zhao F, Bosserhoff A-K, Buettner R, Moser M, 2011. A Heart-Hand Syndrome Gene: Tfap2b Plays a Critical Role in the Development and Remodeling of Mouse Ductus Arteriosus and Limb Patterning. PLOS ONE 6, e22908. 10.1371/journal.pone.0022908 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The μCT data from this study will be freely available to the public on FaceBase3 (https://www.facebase.org/).