Abstract
Background and objective
Lung ultrasound is an inherently user‐dependent modality that could benefit from quantitative image analysis. In this pilot study we evaluate the use of computer‐based pleural line (p‐line) ultrasound features in comparison to traditional lung texture (TLT) features to test the hypothesis that p‐line thickening and irregularity are highly suggestive of coronavirus disease 2019 (COVID‐19) and can be used to improve the disease diagnosis on lung ultrasound.
Methods
Twenty lung ultrasound images, including normal and COVID‐19 cases, were used for quantitative analysis. P‐lines were detected by a semiautomated segmentation method. Seven quantitative features describing thickness, margin morphology, and echo intensity were extracted. TLT lines were outlined, and texture features based on run‐length and gray‐level co‐occurrence matrix were extracted. The diagnostic performance of the 2 feature sets was measured and compared using receiver operating characteristics curve analysis. Observer agreements were evaluated by measuring interclass correlation coefficients (ICC) for each feature.
Results
Six of 7 p‐line features showed a significant difference between normal and COVID‐19 cases. Thickness of p‐lines was larger in COVID‐19 cases (6.27 ± 1.45 mm) compared to normal (1.00 ± 0.19 mm), P < 0.001. Among features describing p‐line margin morphology, projected intensity deviation showed the largest difference between COVID‐19 cases (4.08 ± 0.32) and normal (0.43 ± 0.06), P < 0.001. From the TLT line features, only 2 features, gray‐level non‐uniformity and run‐length non‐uniformity, showed a significant difference between normal cases (0.32 ± 0.06, 0.59 ± 0.06) and COVID‐19 (0.22 ± 0.02, 0.39 ± 0.05), P = 0.04, respectively. All features together for p‐line showed perfect sensitivity and specificity of 100; whereas, TLT features had a sensitivity of 90 and specificity of 70. Observer agreement for p‐lines (ICC = 0.65–0.85) was higher than for TLT features (ICC = 0.42–0.72).
Conclusion
P‐line features characterize COVID‐19 changes with high accuracy and outperform TLT features. Quantitative p‐line features are promising diagnostic tools in the interpretation of lung ultrasound images in the context of COVID‐19.
Keywords: coronavirus, COVID‐19, lung ultrasound, pleural line, point‐of‐care ultrasound (POCUS), quantitative features
1. INTRODUCTION
1.1. Background and importance
Coronavirus disease 2019 (COVID‐19) was declared a pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) after spreading to >180 countries by March 2020, 1 with >100 million cases confirmed and 2 million deaths. 2 Medical imaging plays a major role in COVID‐19 diagnosis and management. 3 The most common modality for imaging COVID‐19 patients is chest x‐ray, which detects presence of a local or bilateral patchy shadowing infiltrate, but it is known for low sensitivity, with findings absent in >40% of the cases. 4 Computed tomography (CT) scan has higher sensitivity and shows ground‐glass opacities 5 and, therefore, has been used in different therapeutic and triage strategies since the outbreak started. 6 The use of chest CT remains limited because of radiation exposure concerns and lack of availability in overextended healthcare facilities. 7 In the critically ill, the transport of unstable patients and exposure of infected patients also may outweigh the clinical benefit. There continues to be need for alternative imaging methods that enable quick, low‐cost, easy‐to‐use evaluation of COVID‐19 patients.
Lung ultrasound is a promising tool for lung monitoring in the intensive care unit (ICU), 8 , 9 particularly for assessing lung aeration. 10 , 11 , 12 A decade of clinical and physical studies demonstrates that lung ultrasound is able to detect interstitial lung disease, subpleural consolidations, and acute respiratory distress syndrome with different etiologies. With new COVID‐19, evolving evidence from clinical practice and several studies shows the usefulness of lung ultrasound for the management of pneumonia, from diagnosis to monitoring and follow‐up. 13 , 14 , 15 , 16 , 17 Characteristic ultrasound findings have been reported that can help physicians detect and stage the disease, tracking its progression. 13 The common patterns observed include irregular, thickened pleural lines, multiple B‐lines (vertical artifacts) ranging from focal to diffuse, which reflect the stage of inflammatory lung disease. These vertical artifacts of different shapes and lengths occur when the lung loses normal aeration but is not completely consolidated. These findings are usually bilateral with posterior basal predominance in the lungs.
Although there is a general consensus on lung ultrasound's usefulness for COVID‐19 pneumonia, 18 it is an inherently user‐dependent modality and, without proper training, could result in errors. 19 Limited experience with COVID‐19 further adds to the challenge of using lung ultrasound effectively. To overcome these limitations, a number of studies have evaluated quantitative methods for assessing lung ultrasound. 20 , 21 , 22 These studies assess the use of automated detection of the B‐line to provide critical visual information to clinicians in real time for diagnosis.
1.2. Goals of this investigation
In this study, we propose a comprehensive approach for detecting sonographic features, defining characteristics of pleural line (p‐line) and traditional lung texture (TLT) from B‐lines quantitatively. Thickening of the p‐line with irregular margin is highly suggestive of COVID‐19. 14 , 15 , 16 , 23 Furthermore, the presence of B‐lines influences the p‐line characteristics, as these patterns originate from the p‐line itself. A‐lines represent a more reflective p‐line, correlating with the brighter p‐line observed normally. The aim of this study is to demonstrate the proof of concept of using quantitative analysis of p‐lines for diagnosis and monitoring of COVID‐19 by ultrasound imaging. We evaluate the changes in p‐line features compared to the TLT features extracted from A‐ and B‐lines for differentiating COVID‐19 cases from normal individuals. The diagnostic performance and the observer variability of these features used individually or as a group were assessed for COVID‐19 diagnosis.
2. METHODS
2.1. Study design and data acquisition
This multiple‐center, retrospective pilot study was conducted on 20 B‐mode ultrasound images that were used to evaluate the proposed quantitative analysis. Ten images were acquired from COVID‐19 patients and another independent 10 images were acquired from normal cases. Images were acquired by Dr. Yale Tung Chen, from the emergency department of Hospital Universitario La Paz, Madrid, Spain. Qualitative imaging findings and the results of diagnostic reverse transcription polymerase chain reaction tests were included with the images. The images were received for analysis anonymized and without patient‐related information. The patient consent for performing image analysis on anonymized images was exempted by the institutional review board. The images usually are acquired in a clip format that includes a large number of frames obtained during scanning of the area. Before quantitative analysis, each observer identifies and selects an image from the clip to choose the highest‐quality image for making quantitative measurements.
2.2. Interventions
2.2.1. Quantitative p‐line analysis and feature extraction
The quantitative analysis involving image segmentation and feature extraction was performed using software written in IDL (Interactive Data Language; version 8.5, L3Harris Geospatial, Boulder, Colorado, USA).
2.2.2. Semiautomated detection of p‐line
The p‐lines were segmented by a “wand” semiautomatic segmentation tool developed by the authors. This semiautomated segmentation approach is the simplest form of “region growing.” 24 Following selection of pixel seed by a user with a single mouse click, the algorithm automatically grows the region to include object pixels of similar grayscale within a tolerance range, ±10 gray levels by default. Only minimal input from the user is needed: to click and then validate the segmentation, making corrections in few cases where the segmented margin was not acceptable. Figure 1 shows examples of p‐line segmentation in images of COVID‐19 and normal cases.
FIGURE 1.

Examples of lung ultrasound images demonstrating the semiautomated detection method of pleural lines. The upper panel (A) shows a confirmed COVID‐19 case with pleural thickening and irregularity. Right panel A shows pleural line detected with the semiautomated segmentation. The lower panel (B) shows an example of a normal case where pleural line is outlined by semiautomated segmentation
2.2.3. P‐line feature extraction
Following the detection of p‐line, the software extracts quantitative features describing the depth (thickness), margin morphology, brightness, and heterogeneity. The features and their formulas are described in Appendix A, Table 1. In summary, the thickness parameters measure the change in horizontal depth of the p‐line. The margin morphology features, including tortuosity, projected intensity deviation (PID), and non‐linearity, measure irregularities of the p‐line margin shape. The last group of grayscale features is derived from the first‐order histogram to calculate the mean brightness and heterogeneity.
2.2.4. Traditional lung texture (TLT) feature extraction
In this study we introduce a predictive model, built on quantitative TLT features, that serves as an alternative to visual clinical assessment where an observer looks for A‐lines in normal lungs and B‐lines in COVID‐19 cases. Regions of interest (ROIs) defining lung areas showing A‐lines in normal cases and B‐lines in COVID‐19 cases were outlined manually by an expert user. Quantitative features were extracted from the lung images ROIs as grayscale first‐order statistics, and determined by run‐length and gray‐level co‐occurrence matrices (GLCM). 25 The 7 image features measured are described in Appendix 1. In brief, the TLT features computation indirectly represents the real texture of the tissue by the non‐deterministic properties that govern the distributions and relationships between the grey levels of the ultrasound image. The quantitative features measured demonstrate the changes in lung tissue texture, and should capture and quantify the prominence of A‐ and B‐lines used by human to discriminate lung ultrasound patterns found in COVID‐19 patients from the appearance of normal lung.
The Bottom Line
This study evaluates a novel quantitative approach for defining characteristics of the lung pleural lines, which performed better in the detection of COVID‐19 when compared with traditional qualitative analysis of lung ultrasound findings.
2.3. Measurements
Standard descriptive statistics were computed for the features extracted from the images: arithmetic mean and standard error. The 2‐tailed Student's t test of unequal variance was used to determine the statistical significance of the difference between 2 groups. P < 0.05 was considered significant.
2.4. Outcomes
2.4.1. Evaluation of the diagnostic performance of ultrasound features
The diagnostic performance for individual ultrasound features including area under the curve (AUC), as well as sensitivity and specificity at the Youden Index were calculated using MedCalc software (version 19.0.5, MedCalc Software Ltd., Ostend, Belgium). To assess the overall diagnostic performance of features as groups, features were normalized relative to their maximum and minimum values and assigned equal weights to calculate likelihood of COVID‐19. The averages of p‐line and TLT features after normalization were used independently to test the accuracy of each group for differentiating normal from COVID‐19 cases by receiver operating characteristics analysis.
2.4.2. Assessment of observer agreement in feature analysis
To evaluate the reproducibility of the analysis, observer agreement for the same individual (intraobserver) and between 2 individuals (interobserver) were measured. Two physicians with >10 years of experience in ultrasound imaging analyzed the same image set. The observers were blinded to the diagnosis and to any other patient‐related information. Both observers performed the quantitative analysis, which includes selection of the seed for p‐line semiautomatic segmentation and outlining the ROI for the TLT analysis. No visual interpretation was made by the observers. To account for the variation related to selection of images, the analysis was repeated where one observer selected the images for analysis from a video clip. Intra‐ and interobserver observer agreement in feature analysis was measured by interclass correlation coefficient. 26 , 27
3. RESULTS
3.1. Characteristics of study subjects and images
Qualitative image findings were provided by an emergency physician experienced with lung ultrasound for both normal and COVID‐19 images. Normal images were described as having a thin, well‐defined p‐line with presence of A‐lines, whereas findings related to COVID‐19 patients included thick and irregular p‐lines with the presence of B‐lines. COVID‐19 images were from patients 8 to 12 days following COVID‐19 diagnosis. COVID‐19 patients showed mild to moderate symptoms. All had bilateral pneumonia with good oxygen saturation. Patient ages ranged from 30 to 60 years.
3.2. Main results
3.2.1. Quantitative p‐line features
From the 7 p‐line features extracted from lung ultrasound images, 6 showed statistically significant differences (P < 0.05) between normal and COVID‐19 cases. Figure 2 shows a comparison of the mean and standard errors for each feature of COVID‐19 cases to those of normal cases. The thickness of p‐lines was larger on average in COVID‐19 cases (6.27 ± 1.45 mm) compared to normal (1.00 ± 0.19 mm), P < 0.001. P‐line thickness variation was also larger on average in COVID‐19 cases, 2.86 ± 0.64 mm compared to 0.26 ± 0.07 mm, P < 0.001.
FIGURE 2.
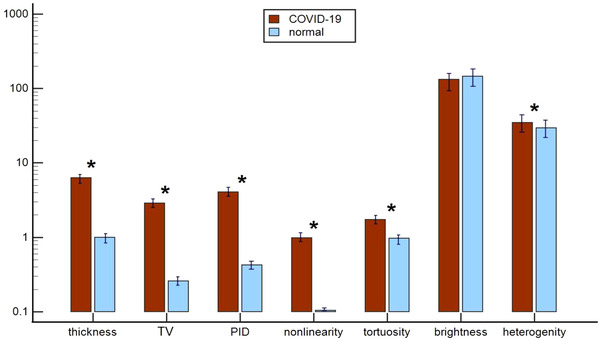
A comparison of computer‐based pleural line (p‐line) features of COVID‐19 and normal cases. Bars represent mean ± SE of each feature. Red bars represent COVID‐19 cases. Light blue bars represent normal cases. * indicates statistically significant difference (P < 0.05). Notably, 6 of 7 p‐line features showed statistically significant difference between the 2 groups. COVID‐19, coronavirus disease 2019; PID, projected intensity deviation; TV, thickness variation
Among features describing margin morphology, PID showed the largest difference between the 2 groups in COVID‐19 cases, 4.08 ± 0.32 compared to 0.43 ± 0.06 in normal cases, P < 0.001. Non‐linearity was also larger for the COVID‐19 cases, 0.98 ± 0.10 compared to 0.01 ± 0.01, P < 0.001. Margin tortuosity was about 2 times larger in COVID‐19 cases, 1.73 ± 0.09 compared to 0.97 ± 0.01, P < 0.001.
The mean p‐line brightness in normal p‐lines was higher in normal cases (145.98 ± 12.15) compared to COVID‐19 cases (132.61 ± 7.51) but was not significant (P = 0.14). On the other hand, p‐line heterogeneity increased significantly in COVID‐19 cases (34.67 ± 2.07) compared to normal cases (29.41 ± 2.87), P = 0.04.
3.2.2. The diagnostic performance of individual p‐line features
The sensitivity, specificity, and AUC for each p‐line feature are summarized in Table 1. Notably, p‐line margin morphology features showed the highest performances, with specificities ranging from 90 to 100 and sensitivities of 100. Similarly, thickness features showed high sensitivity values ranging from 80 to 90 and a specificity value of 100. On the contrary, p‐line brightness and heterogeneity had lower performance than other features, with sensitivity of 60 (Table 1).
TABLE 1.
The diagnostic performance for quantitative pleural line (p‐line) feature and TLT features showing AUC, sensitivity, and specificity for each feature
| P‐line features | AUC | Sensitivity | Specificity |
|---|---|---|---|
| Thickness (mm) | 0.91 | 80 | 100 |
| Thickness variation (mm) | 0.98 | 90 | 100 |
| Margin tortuosity | 1 | 100 | 100 |
| Projected intensity deviation | 1 | 100 | 100 |
| Non‐linearity | 0.97 | 100 | 90 |
| Brightness | 0.66 | 60 | 90 |
| Heterogeneity | 0.71 | 100 | 50 |
| TLT features | |||
| Echo intensity | 0.52 | 60 | 10 |
| Tissue heterogeneity | 0.64 | 80 | 50 |
| GLNU | 0.74 | 80 | 70 |
| RLNU | 0.75 | 90 | 70 |
| GLCM | 0.71 | 80 | 60 |
| Homogeneity | 0.69 | 40 | 100 |
| Entropy | 0.68 | 60 | 80 |
AUC, area under the curve; GLCM, gray‐level co‐occurrence matrix mean; GLNU, gray‐level non‐uniformity; RLNU, run‐length non‐uniformity; TLT, traditional lung texture
3.2.3. Traditional lung texture (TLT) features
Only 2 of the 7 features of TLT texture measurements showed a significant difference between COVID‐19 and normal cases (Figure 3). Run‐length matrix features including gray‐level non‐uniformity (0.32 ± 0.06) and run‐length non‐uniformity (0.59 ± 0.06) were both significantly higher in normal cases compared to those diagnosed with COVID‐19 (0.22 ± 0.02, 0.39 ± 0.05, P = 0.04), respectively.
FIGURE 3.

A comparison of computer‐based TLT features for COVID‐19 normal cases. Bars represent mean ± SE of each feature studied. Red bars represent COVID‐19 cases. Light blue bars represent normal cases. * indicates statistically difference (P < 0.05). COVID‐19, coronavirus disease 2019; GLCM, gray‐level co‐occurrence matrix mean; GLNU, gray‐level non‐uniformity; RLNU, run‐length non‐uniformity; TLT, traditional lung texture
TLT heterogeneity was also higher in normal cases (30.78 ± 3.91) compared to TLT heterogeneity observed in COVID‐19 cases (24 ± 0.07), P = 0.08. Conversely, mean echo intensity level was higher in COVID‐19 TLT (90.96 ± 13.06) compared to normal cases (85.40 ±8.78); however, this difference did not reach significance level, with P = 0.73.
Gray‐level co‐occurrence matrix (GLCM) features were all higher for COVID‐19 cases than for normal cases; however, none reached significance. GLCM mean in COVID‐19 patients was 2.98 ± 0.29 compared to 2.37 ± 0.23, P = 0.09. The other 2 GLCM features, entropy and homogeneity, were both higher in COVID‐19, 2.98 ± 0.29 and 2.78 ± 0.13, compared to normal cases, 2.37 ± 0.23 and 2.45 ± 0.18, P = 0.08 and P = 0.12, respectively.
3.2.4. The diagnostic performance of TLT features
Table 1 summarizes the sensitivity, specificity, and AUC for each TLT texture feature. Run‐length texture features showed the highest performance with sensitivity ranging between 80 and 90 and specificity of 70. TLT heterogeneity had a sensitivity of 80 and specificity of 60. Of the GLCM features, GLCM mean had the highest sensitivity (sensitivity = 80) and GLCM homogeneity was the most specific feature (specificity = 100). Echo intensity had the lowest performance among other features with a sensitivity of 60 and specificity of 10. Figure 4 shows an example of 2 cases that were diagnosed correctly by TLT analysis. The quantitative analysis showed that the case diagnosed with COVID‐19 has higher echogenicity and lower heterogeneity compared to an opposite pattern observed in the normal case.
FIGURE 4.
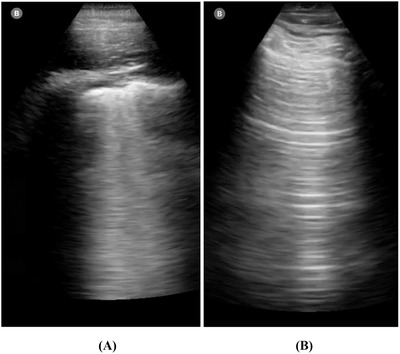
Two examples of cases that were diagnosed correctly by quantitative TLT analysis. The 2 cases showed different texture patterns. The COVID‐19 case (A) showed higher echogenicity and lower heterogeneity, RLNU, and GLNU. Although the normal case (B) showed lower echogenicity and higher heterogeneity, RLNU, and GLNU. COVID‐19, coronavirus disease 2019; GLNU, gray‐level non‐uniformity; RLNU, run length non‐uniformity; TLT, traditional lung texture
3.2.5. Comparison of the overall diagnostic performance of p‐line versus TLT features
Equally weighted p‐lines features when used together were better at differentiating normal cases from COVID‐19, achieving a perfect performance with AUC = 1.0 compared to AUC = 0.79 with equally weighted TLT features (Figure 5). P‐line features had perfect sensitivity of 100 and specificity of 100 in separating the 2 groups, whereas TLT features had a sensitivity of 90 and specificity of 70. The p‐line model showed a superior separation of the 2 groups compared to the TLT model, whose features were closely distributed and even overlapping (Figure 6). Figure 7 shows the cases that were incorrectly identified using TLT features, whereas p‐line features identified the cases correctly. Figure 7A is an example of a COVID‐19 case where the model built on quantitative p‐line features correctly diagnosed, but the model from TLT features was incorrect, diagnosing the case as normal. Similarly, in Figure 7B, a normal case was labeled incorrectly as COVID‐19, but p‐line features extracted by the computer correctly diagnosed the case. The cases that were misdiagnosed showed a TLT pattern that was overlapping between the two groups: COVID‐19 and normal cases.
FIGURE 5.

A comparison of the ROC curves for equally weighted p‐line features and TLT features showing outperformance of p‐line features in differentiating COVID‐19 from normal. AUC, area under the curve; COVID‐19, coronavirus disease 2019; p‐lines, pleural lines; ROC, receiver operating characteristics; sn, sensitivity; sp, specificity; TLT, traditional lung texture
FIGURE 6.
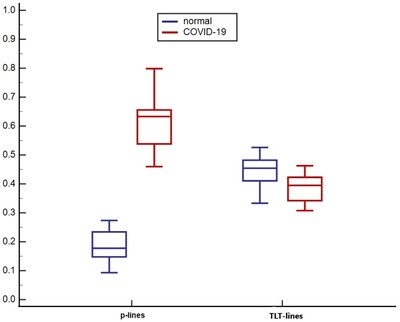
A box and whisker graph comparing the separation power of the 2 feature groups. P‐line features show a clear separation between normal and COVID‐9 cases whereas cases are closely distributed with TLT features. X‐axis represents equally weighted normalized features. COVID‐19, coronavirus disease 2019; p‐line, pleural line; TLT, traditional lung texture
FIGURE 7.

A, an example of a COVID‐19 confirmed case showing pleural thickening and irregularity with presence of focal B‐lines. Quantitative pleural line features detected the case accurately as COVID‐19, whereas TLT features incorrectly identified the case as normal. B, an example of a normal lung image showing a thin and well‐defined pleural line and A‐lines. Quantitative pleural line features detected the case accurately as normal whereas TLT features incorrectly identified the case as COVID‐19. COVID‐19, coronavirus disease 2019; TLT, traditional lung texture
3.3. Observer agreement in feature analysis
3.3.1. Intraobserver agreement
The individual observer analyzed the same set of images 2 weeks following the first analysis. Interclass correlation coeffecients (ICCs) between the 2 analyses on average for p‐line features showed excellent agreement of ICC of 0.85 ± 0.09 (Table 2). ICC for individual p‐line features ranged from ICC of 0.72 (good agreement) for heterogeneity, to ICC of 0.95, excellent agreement, for non‐linearity.
TABLE 2.
Summary of the observer agreements in p‐line ultrasound features and TLT features using the same and different images of the subject
| P‐line features | Same image | Different images | ||
|---|---|---|---|---|
| Intraobserver ICC | Interobserver ICC | IntraobserverICC | InterobserverICC | |
| Thickness | 0.85 | 0.98 | 0.65 | 0.92 |
| Thickness variation | 0.76 | 0.94 | 0.68 | 0.85 |
| Tortuosity | 0.91 | 0.82 | 0.67 | 0.35 |
| Projected intensity deviation | 0.79 | 0.80 | 0.70 | 0.84 |
| Linearity | 0.95 | 0.63 | 0.82 | 0.89 |
| Brightness | 0.94 | 0.73 | 0.73 | 0.90 |
| Heterogeneity | 0.72 | 0.88 | 0.33 | 0.31 |
| Average ICC (±SE) | 0.85 ± 0.09 | 0.83 ± 0.12 | 0.71 ± 0.15 | 0.72 ± 0.27 |
| TLT features | ||||
| Echo intensity | 0.93 | 0.69 | 0.59 | 0.94 |
| Tissue heterogeneity | 0.82 | 0.56 | 0.33 | 0.66 |
| GLNU | 0.52 | 0.87 | 0.31 | 0.44 |
| RLNU | 0.55 | 0.61 | 0.41 | 0.40 |
| GLCM mean | 0.70 | 0.71 | 0.49 | 0.76 |
| Homogeneity | 0.67 | 0.67 | 0.54 | 0.45 |
| Entropy | 0.76 | 0.60 | 0.33 | 0.52 |
| Average ICC (±SE) | 0.71 ± 0.15 | 0.67 ± 0.10 | 0.45 ±0.11 | 0.59 ±0.20 |
GLCM, gray level co‐occurrence matrix mean; GLNU, gray‐level non‐uniformity; ICC, interclass correlation coefficients; p‐line, pleural line; RLNU, run‐length non‐uniformity; TLT, traditional lung texture
On the other hand, TLT features showed good agreement on average with ICC of 0.71 ± 0.15. ICC ranged from 0.52, fair agreement with run‐length non‐uniformity, to excellent agreement (ICC = 0.92) for echointensity (Table 2).
3.3.2. Interobserver agreement
High agreement levels were recorded between 2 observers who analyzed the same set of images (Table 2). The average ICC for p‐line features was 0.83 ± 0.12, excellent agreement. The feature that showed the highest agreement was p‐line thickness with ICC of 0.98, whereas p‐line brightness showed the lowest agreement (ICC = 0.63).
For TLT, overall agreement was good with ICC = 0.67 ± 0.10. Highest agreement in a feature was in GL non‐uniformity (ICC = 0.87); whereas, the lowest was seen in TLT heterogeneity with ICC = 0.56 (Table 2).
3.3.3. Observer agreement and the effect of image selection
Intra‐ and interobserver agreement were calculated for analyses using another set of images (Table 2). For both intra‐ and interobserver, ICCs for p‐line features on average were good (ICC of 0.65–0.71). On the other hand, the agreement for TLT features was lower, with ICC of 0.45–0.59, fair agreement.
4. LIMITATIONS
The study is not without limitations. One limitation is the small sample size. The motivation for the current pilot analysis is to test the use of computerized p‐line features for a potential role in diagnosis and monitoring of COVID‐19 by ultrasound. Future studies with larger sample sizes are needed to validate the proposed approach in a routine clinical setting, with a queue of COVID‐19 patients presenting at an emergency department or ICU. Future studies with larger sample size are also needed to reduce any potential selection bias. Another limitation is that the current analysis is used to differentiate 1 category of inflammatory diseases related to COVID‐19. In future, the methodology could be applied to a variety of lung inflammatory diseases. Ultrasound imaging is known to be user dependent where imaging presets, nature of the imaging device, and the training of the observer can influence diagnosis. Although quantitative analysis minimizes these effects, a broader study involving images acquired from multiple clinical devices with varying imaging presets used by different observers will generalize the use of quantitative p‐line and TLT for machine‐agnostic diagnosis.
5. DISCUSSION
Ultrasound is an ideal imaging tool for COVID‐19 diagnosis because of its high sensitivity, safety, portability, and affordability. 28 However, a significant disadvantage is that it is highly user dependent, and not all clinicians have training in performing lung ultrasound and reading the images. Operator experience may also affect specificity, becausee an expert will correlate different lung ultrasound patterns with different disease processes. 29 To overcome these problems, a number of studies have evaluated quantitative assessment methods 20 , 21 , 22 to help physicians in image interpretation. However, the focus of these studies is limited to developing semi‐quantitative scoring systems based on B‐line identification. There remains a need for an advanced and comprehensive approach that includes quantitative analysis of both the p‐line and TLT indicative of COVID‐19.
COVID‐19 is a pleural‐based disease, and when patients are infected by it, their pleura are thickened and inflamed. Pleural thickening is usually accompanied by tissue scarring, 30 , 31 often caused by acute inflammation of the pleura. In the normal lung, the p‐line appears as a thin curvilinear opaque lining 1–2 mm in thickness, completely continuous and well defined. It becomes gradually disrupted as the pathological condition worsens. This study focused on evaluating these pathological changes by ultrasound, which is ideally suited for imaging small structures. The idea central to the study is that significant pathological changes associated with COVID‐19 can be determined by quantitative analysis of pattern changes in the p‐line characteristics of lung ultrasound. A comparison between COVID‐19 and non‐COVID‐19 cases showed that it is possible to detect and characterize p‐line changes related to COVID‐19 with high accuracy using computer‐derived image features. The semiautomated segmentation tool detected the p‐line margins with fine details capturing the changes in margin morphology. Margin shape features including margin tortuosity, projected intensity deviation, and non‐linearity were significantly higher in COVID‐19 cases, correlating closely with the irregularity and changes in p‐line shape related to the inflammatory process reported in previous studies. Additionally, the mean thickness of p‐lines was measured as >6 times greater than p‐line thickness in normal cases, which is also consistent with qualitative clinical assessments of COVID‐19 cases on lung ultrasound. Notably, the p‐lines showed less mean brightness and higher heterogeneity in COVID‐19 compared to normal. These observed changes are consistent with the presence of inflammatory cells and edema in COVID‐19 cases, which cause the p‐line to lose its high intensity and uniform appearance seen in normal lungs.
The sensitivity and specificity for the individual p‐line features as well as the overall performance of all the features together were found to be high, thereby confirming their ability to differentiate normal and COVID‐19 cases with excellent accuracy. In particular, p‐line margin morphology and its thickness were more specific for COVID‐19 in comparison to brightness and heterogeneity. The high specificity of quantitative margin morphology features over other features could make these feature better suited for diagnostic models of COVID‐19.
The TLT features, extracted from A‐lines and B‐lines, were also able to detect COVID‐19‐related changes. However, these features, with AUC approaching 0.79, did not have a significant discriminatory power to diagnose the cases as that of p‐line features. The study results show that brighter and more homogenous areas are seen in COVID‐19 cases. With progression of disease, the lungs are filled with inflammatory cells as well as fluids that cause the ultrasound beam to be trapped between the inflammatory cells, producing the vertical artifacts called B‐lines. 12 , 13 These lines are brighter and more uniform compared to A‐lines seen in normal lungs. We were able to quantify these findings but did not observe a significant difference between the 2 groups, and it is unknown if the difference could become significant with more cases. Individual TLT features exhibited less sensitivity and specificity than p‐line features. One reason could be that B‐lines are imaging artifacts and their genesis remains unclear and multifactorial, correlating with various pathological conditions of the lung. In contrast, the p‐lines in lung ultrasound represent real physical structures that undergo acute inflammatory changes specific to COVID‐19. 32 Earlier studies also reported reduced blood flow in the p‐line by Doppler images in COVID‐19 patients compared to the increased flow seen in other types of viral pneumonia, 14 again because of the acute nature of the disease.
The high observer agreement indicates the consistency and reliability of the quantitative analyses. In all observer variability analyses, p‐line features were more stable and consistent compared to the TLT features, which had lower agreement between observations. A notable decrease in ICC of >0.15 was observed when different images were selected by the observer for the analysis, suggesting that >15% variation could result from the choice of images used for analysis. The effect of image selection has also been observed with B‐lines scoring, 33 where minute differences between images were found to significantly influence the scoring process and ultimately the final diagnosis. These findings underscore the importance of image selection in clinical settings.
In conclusion, we introduced a computer‐based system that captures p‐line pattern changes associated with COVID‐19 by quantitative analysis of lung ultrasound. Quantitative p‐line features showed high accuracy in detecting COVID‐19 cases compared to TLT features, which can be more uncertain. These results suggest that a comprehensive quantitative system that characterizes p‐lines would improve the diagnostic accuracy of COVID‐19 on lung ultrasound. However, future studies on a larger scale are needed before translating these techniques to clinical practice. In the current study, the automated methods were performed off‐line but these methods can be easily integrated into the operation of scanner for real‐time bedside assessment. Future larger studies will incorporate advanced machine learning methods to optimize the p‐line detection algorithm for robustness and automation. Given more cases, fully automated machine learning segmentation could be performed. This technology when implemented successfully in clinical practice will increase confidence in diagnosis, especially in low‐resource communities around the globe that lack experience in lung ultrasound.
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
AUTHOR CONTRIBUTIONS
LRS and CMS designed the project. LRS performed image, data, and statistical analysis. YTC provided the imaging data in addition to guidance in qualitative lung ultrasound findings. TWC designed the computer software and quantitative lung ultrasound features. KA performed the analysis for observer agreement. All of the authors contributed to the preparation and final review of the manuscript.
ACKNOWLEDGMENT
The authors thank Ms. Susan M. Schultz for her kind contribution to the study.
Biographies
Laith Sultan, MD, MPH, is a physician scientist and junior faculty in the Department of Radiology at the University of Pennsylvania School of Medicine.

Chandra M. Sehgal, PhD, is a research professor in the Department of Radiology and Directorof the Ultrasound Research Laboratory at the Hospital of the University of Pennsylvania.

1.
TABLE A1.
Description and formulas for pleural‐line (p‐line) features
| Features | Formulas | ||
|---|---|---|---|
|
Thickness and thickness variation (mm) For region R = {xi,yi},ranging over Nx horizontal values, the mean thickness is the mean of the y ranges taken through the region at each horizontal coordinate. |
|
||
|
Margin tortuosity (unitless) The perimeter of the lesion divided by the circumference of its best‐fit ellipse, also called elliptically normalized circumference. |
where P is the region's perimeter and C is the circumference of a best‐fit ellipse to the region. |
||
|
Projected intensity deviation (PID, gray level) Projected deviation measures the horizontal (transverse) irregularity in a region's gray level by computing the standard deviation of the depth‐projected mean intensity. Given image image Ix,y of dimensions Nx, Ny, it is given by the formula to the right. |
|
||
|
Non‐linearity (unitless) The probability that the points in a region R = {xi,yi} will lie on their linear regression fit a + bxi, where the regression minimizes χ2. This probability is the same as the probability of a t test with the 2 degrees of freedom (a,b) on the regression fit. |
where χ2 is the chi‐square of a linear fit, given by: |
||
|
Echo intensity mean (μ) and deviation (σ) of pleural line (gray level) Brightness and heterogeneity are first‐order histogram features: the mean brightness level of the segmented pleural lines and the measured variability in mean brightness, respectively. |
μ = , σ= where Ix,y is the intensity (grayscale) of the pleural line at coordinate [x,y]. |
TABLE A2.
Description and formulas for TLT‐line features
| Features | Formulas | |
|---|---|---|
|
Echo intensity mean (μ) and deviation (σ) (gray level) First order‐histogram features that describe the brightness of the tissue (mean) and the variability in brightness (standard deviation). |
μ = , σ= where Ix,y is the intensity (grayscale) of the echo line at coordinate [x,y]. |
|
|
Gray‐level non‐uniformity (unitless) This is a run‐length‐matrix (RLM) feature that measures disorderliness of homogeneous runs of gray along defined directions. The RLM p gives the length of homogeneous runs for each gray level, computed over 8 directions (vertical up and down, horizontal left and right, diagonals), so that element (i,j) is the number of homogeneous runs of j pixels with intensity i. |
where H is the total count of homogeneous runs in p, the run‐length matrix defined at left. |
|
|
Run‐length non‐uniformity (unitless) This is a run‐length matrix (RLM) feature that measures the disorderliness of the lengths of homogeneous runs, computed similarly to gray‐level non‐uniformity above, with the inner and outer sums switched. |
where H is the total count of homogeneous runs, and p is the RLM defined above. |
|
|
Gray‐level co‐occurrence matrix mean (unitless) The mean of the GLCM (p), the co‐occurrence matrix which records the counts of pixel intensity combinations occurring between neighboring pixels, computed over 8 directions (up and down, left and right, diagonals). Element (i,j) is the count of times a pixel of intensity i had a pixel of intensity j next to it in any direction at some scale. This is a measure of orderliness, and records whether small patterns repeat themselves. |
where p is the GLCM as defined at left. |
|
|
Gray level co‐occurrence matrix homogeneity of echo line (unitless) Also called the inverse difference moment, homogeneity increases if the region has less contrast. |
where p is the GLCM as defined above. |
|
|
Entropy (unitless) Larger entropy means the texture is more disordered. |
where p is the GLCM as defined above. |
Sultan LR, Chen YT, Cary TW, Ashi K, Sehgal CM. Quantitative pleural line characterization outperforms traditional lung texture ultrasound features in detection of COVID‐19. JACEP Open. 2021;2:e12418. 10.1002/emp2.12418.
Supervising Editor: Yiju Teresa Liu, MD, RDMS
Funding and support: By JACEP Open policy, all authors are required to disclose any and all commercial, financial, and other relationships in any way related to the subject of this article as per ICMJE conflict of interest guidelines (see www.icmje.org). The authors have stated that no such relationships exist.
REFERENCES
- 1. WHO. Coronavirus disease (COVID‐2019) situation reports. World Heal Organ. 2020.
- 2. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Inf Dis. 2020;20(5):533‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT‐PCR testing for coronavirus disease 2019 (COVID‐19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32‐E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guan W‐J, Ni Z‐Y, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID‐19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20(4):425‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang R, Ouyang H, Fu L, et al. CT features of SARS‐CoV‐2 pneumonia according to clinical presentation: a retrospective analysis of 120 consecutive patients from Wuhan city. Eur Radiol. 2020;30(8):4417‐4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tung‐Chen Y, Martí de Gracia M, Díez‐Tascón A, et al. Correlation between chest computed tomography and lung ultrasonography in patients with coronavirus disease 2019 (COVID‐19). Ultrasound Med Biol. 2020;46(11):2918‐2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bouhemad B, Mongodi S, Via G, Rouquette I. Ultrasound for “lung monitoring” of ventilated patients. Anesthesiology. 2015. [DOI] [PubMed] [Google Scholar]
- 9. Pesenti A, Musch G, Lichtenstein D, et al. Imaging in acute respiratory distress syndrome. Intensive Care Med United States. 2016;42(5):686‐698. [DOI] [PubMed] [Google Scholar]
- 10. Bouhemad B, Brisson H, Le‐Guen M, Arbelot C, Lu Q, Rouby J‐J. Bedside ultrasound assessment of positive end‐expiratory pressure‐induced lung recruitment. Am J Respir Crit Care Med United States. 2011;183(3):341‐347. [DOI] [PubMed] [Google Scholar]
- 11. Bouhemad B, Liu Z‐H, Arbelot C, et al. Ultrasound assessment of antibiotic‐induced pulmonary reaeration in ventilator‐associated pneumonia. Crit Care Med United States. 2010;38(1):84‐92. [DOI] [PubMed] [Google Scholar]
- 12. Soummer A, Perbet S, Brisson H, et al. Ultrasound assessment of lung aeration loss during a successful weaning trial predicts postextubation distress*. Crit Care Med United States. 2012;40(7):2064‐2072. [DOI] [PubMed] [Google Scholar]
- 13. Sultan LR, Sehgal CM. A review of early experience in lung ultrasound in the diagnosis and management of COVID‐19. Ultrasound in Medicine & Biology. 2020;46(9):2530–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Y, Wang S, Liu Y, et al. A preliminary study on the ultrasonic manifestations of peripulmonary lesions of non‐critical novel coronavirus pneumonia (COVID‐19). SSRN Electronic Journal. 10.2139/ssrn.3544750. [DOI] [Google Scholar]
- 15. Hui DS, I Azhar E, Madani TA, et al. The continuing 2019‐nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan. China Int J Infect Dis. 2020;91:264‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Inchingolo R, Smargiassi A, Moro F, et al. The diagnosis of pneumonia in a pregnant woman with COVID‐19 using maternal lung ultrasound. Am J Obstet Gynecol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalafat E, Yaprak E, Cinar G, et al. Lung ultrasound and computed tomographic findings in pregnant woman with COVID‐19. Ultrasound Obstet Gynecol. 2020;55(6):835‐837. [DOI] [PubMed] [Google Scholar]
- 18. Tung‐Chen Y. Lung ultrasound in the monitoring of COVID‐19 infection. Clin Med. 2020;20(4):e62‐e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinto A, Pinto F, Faggian A, et al. Sources of error in emergency ultrasonography. Crit Ultrasound J. 2013;5(Suppl 1):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moshavegh R, Hansen KL, Moller‐Sorensen H, Nielsen MB, Jensen JA. Automatic detection of B‐Lines in in vivo lung ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control. 2019;66(2):309‐317. [DOI] [PubMed] [Google Scholar]
- 21. Van Sloun RJG, Demi L. Localizing B‐lines in lung ultrasonography by weakly supervised deep learning, in‐vivo results. IEEE J Biomed Heal Informatics. 2020;24(4):957‐964. [DOI] [PubMed] [Google Scholar]
- 22. Correa M, Zimic M, Barrientos F, et al. Automatic classification of pediatric pneumonia based on lung ultrasound pattern recognition. PLoS One Public Library of Science. 2018;13(12):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Millington SJ, Koenig S, Mayo P, Volpicelli G. Lung ultrasound for patients with coronavirus disease 2019 pulmonary disease. Chest. 2021;159(1):205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tremeau A, Borel N. A region growing and merging algorithm to color segmentation. Pattern Recognit. 1997;30(7):1191‐1203. [Google Scholar]
- 25. Kairuddin WNHW, Mahmud WMHW. Texture feature analysis for different resolution level of kidney ultrasound images. IOP Conf Ser Mater Sci Eng. 2017;226:12136. [Google Scholar]
- 26. Johnson WD, Koch GG. Intraclass Correlation Coefficient. In: Lovric M. (eds) International Encyclopedia of Statistical Science. Berlin, Heidelberg: Springer, 2011. 10.1007/978-3-642-04898-2. [DOI] [Google Scholar]
- 27. Cicchetti D V. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol Assess. 1994;6(4):284‐290. [Google Scholar]
- 28. Trauer MM, Matthies A, Mani N, et al. The utility of lung ultrasound in COVID‐19: a systematic scoping review. Ultrasound. 2020;28(4):208–222. 10.1177/1742271x20950779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hankins A, Bang H, Walsh P. Point of care lung ultrasound is useful when screening for CoVid‐19 in emergency department patients. medRxiv Prepr Serv Heal Sci. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mongodi S, Bouhemad B, Orlando A, et al. Modified lung ultrasound score for assessing and monitoring pulmonary aeration Modifizierter Lungen‐US‐Score zur Bewertung und Überwachung der Belüftung der Lunge. Modif Lung Ultrasound Ultraschall Med. 2017. [DOI] [PubMed] [Google Scholar]
- 31. Onigbinde SO, Ojo AS, Fleary L, Hage R. Chest Computed tomography findings in COVID‐19 and influenza: a narrative review. Biomed Res Int. 2020;2020. 6928368. https://europepmc.org/articles/PMC7275219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. TMC deL, A F daSS, BR deL, ME deAB, J deAS. Mechanism of inflammatory response in associated comorbidities in COVID‐19. Diabetes Metab Syndr Clin Res Rev. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alfuraih AM. Point of care lung ultrasound in COVID‐19: hype or hope? BJR Open. 2020;2(1):20200027. [DOI] [PMC free article] [PubMed] [Google Scholar]


