Abstract
Background
Implementation of personalized medicine requires the accessibility of tumor molecular profiling in order to allow prioritization of appropriate targeted therapies for individual patients. Our aim was to study the role of comprehensive genomic profiling assays that may inform treatment recommendations for patients with solid tumors.
Materials and Methods
We performed a prospective study to evaluate the feasibility of application of the FoundationOne CDx panel—which detects substitutions, insertions and deletions, and copy number alterations in 324 genes, select gene rearrangements, and genomic signatures including microsatellite instability and tumor mutation burden (TMB)—to patients with advanced or recurrent solid tumors before its approval in Japan.
Results
A total of 181 samples were processed for genomic testing between September 2018 and June 2019, with data being successfully obtained for 175 of these samples, yielding a success rate of 96.7%. The median turnaround time was 41 days (range, 21–126 days). The most common known or likely pathogenic variants were TP53 mutations (n = 113), PIK3CA mutations (n = 33), APC mutations (n = 32), and KRAS mutations (n = 29). Among the 153 patients assessed for TMB, the median TMB was 4 mutations/Mb, and tumors with a high TMB (≥10 mutations/Mb) were more prevalent for lung cancer (11/32) than for other solid tumor types (9/121, Fisher's exact test p < .01). No clear trend toward increased efficacy for immune checkpoint inhibitor (ICI) monotherapy or ICI combination chemotherapy in patients with a high programmed cell death–ligand 1 tumor proportion score or a high TMB was apparent. Among the 174 patients found to harbor known or likely pathogenic actionable alterations, 24 individuals (14%) received matched targeted therapy.
Conclusion
The FoundationOne CDx assay was performed with formalin‐fixed, paraffin‐embedded tumor specimens with a success rate of >95%. Such testing may inform the matching of patients with cancer with investigational or approved targeted drugs.
Implications for Practice
This prospective cohort study was initiated to investigate the feasibility and utility of clinical application of FoundationOne CDx. A total of 181 samples were processed for genomic testing between September 2018 and June 2019, with data being successfully obtained for 175 of these samples, yielding a success rate of 96.7%, and 24 individuals (14%) received matched targeted therapy.
Keywords: Next‐generation sequencing, FoundationOne CDx, Solid tumors
Short abstract
Comprehensive genomic profiling assays can identify multiple actionable genetic alterations that may inform treatment recommendations for patients with solid tumors. This article evaluates the feasibility of the application of the FoundationOne CDx panel to patients with advanced or recurrent solid tumors.
Introduction
The increasing availability of targeted therapies for certain types of cancer with corresponding genetic alterations underscores the need to detect actionable driver mutations in patients with cancer. Next‐generation sequencing (NGS) has emerged as a key technology for simultaneous sequencing of multiple cancer predisposition genes with only small amounts of tissue. Since 2013, several institutions including the National Cancer Center (NCC) and academic hospitals in Japan have begun NGS‐based clinical sequencing for the purpose of matching investigational or approved drugs to patients with corresponding molecular alterations [1, 2, 3, 4]. Given that such screening was initially undertaken for research purposes, actionable gene alterations identified by NGS panels needed to be validated by other methods for enrollment of patients in clinical trials.
In December 2018, the Pharmaceuticals and Medical Devices Agency (PMDA) of Japan approved FoundationOne CDx and NCC Oncopanel as comprehensive genomic profiling (CGP) tests for all solid tumors [2]. FoundationOne CDx covers alterations of 324 genes known to drive cancer growth, and NCC Oncopanel captures mutations or amplifications of 114 genes including 12 gene fusions. Both tests thus provide potentially actionable information to help guide molecularly targeted therapy. Entrectinib was subsequently approved as the first tumor‐agnostic medicine in Japan. This drug targets NTRK gene fusions, which have been identified in a range of hard‐to‐treat solid tumor types, including pancreatic, thyroid, salivary gland, breast, colorectal, and lung cancer. Its approval was based on the promising results of the STARTRK‐2 trial, a phase II basket study of entrectinib for the treatment of patients with solid tumors positive for NTRK1, NTRK2, NTRK3, ROS1, or ALK fusions [5]. The difficulty of enriching NTRK fusion–positive patients with common tumors based on patient background such as smoking history and sex highlights the need for NGS panel testing in order to detect oncogenic drivers that occur at low frequency regardless of tumor type.
Before the approval of the two NGS panels in Japan, we initiated this prospective cohort study to investigate the feasibility and utility of clinical application of FoundationOne CDx, and we determined the proportion of tested patients who were treated with genotype‐directed targeted therapy and assessed their outcome.
Materials and Methods
Patients
Patients were eligible for genetic testing of their cancer if they had a cytologically or histologically confirmed diagnosis of an advanced or recurrent solid tumor and if both clinical information and a formalin‐fixed, paraffin‐embedded (FFPE) tissue block for genetic testing were available. It was desirable that ≥ 20% of the biopsy tissue area in the guide H&E slide be occupied by invasive cancer cells, as evaluated by a pathologist; however, samples with <20% of tumor cells were also submitted to Foundation Medicine (Cambridge, MA) for testing if no other alternative tissue was available. Patients also needed to be aged 20 years or older and to be candidates for chemotherapy. No restrictions on tumor histology, previous or subsequent treatment, performance status, or other factors were imposed. All patients provided written informed consent to the performance of genomic analysis, and the study was approved by the Institutional Review Board of Kindai University. The gene alterations identified by the FoundationOne CDx panel are categorized as (a) known or likely pathogenic variants in the Foundation Medicine database, which includes entries in the COSMIC database, or (b) variants of unknown significance (VUS), and they are addressed in the FoundationOne CDx Report. Treatment recommendations were determined after discussion by the multidisciplinary members of molecular tumor boards (MTBs). The proportion of patients who received matched targeted therapy includes those who received such therapy as a result of the FoundationOne CDx Report and those who had already received targeted therapy on the basis of standard diagnostic testing, the results of which were confirmed by the FoundationOne CDx panel. The primary endpoint of the study was to assess the feasibility of the application of FoundationOne CDx to Japanese patients with solid tumors before its approval by PMDA.
FoundationOne CDx Testing and MTBs
FoundationOne CDx is a CGP platform that applies NGS to in vitro diagnostics with a hybrid capture‐based target enrichment approach and whole‐genome shotgun library construction in order to identify all four classes of somatic genomic alterations, including substitutions, insertions and deletions (indels), copy number alterations, and select rearrangements. The typical median depth of coverage is >500×. The FoundationOne CDx panel detects alterations in a total of 324 genes, including all coding exons of 309 cancer‐related genes, one promoter region, one noncoding RNA, and select intronic regions of 34 commonly rearranged genes, the coding exons of 21 of which are also included. FoundationOne CDx specimens are also simultaneously profiled for tumor mutation burden (TMB) as well as microsatellite instability (MSI) status (detailed information available at https://www.foundationmedicine.com/genomic-testing/foundation-one-cdx).
The complexity of the FoundationOne CDx Report delivered for each patient has highlighted the need for MTBs, also known as “expert panels” in Japan, which assess when molecular testing for tumor profiling is appropriate and address the opportunities for therapy with approved or investigational new drugs. MTBs consist of medical professionals including medical oncologists, a molecular biologist, pathologists, a geneticist, a genetic counselor, and a nurse navigator.
Immunohistochemical Staining of PD‐L1 in NSCLC
Assessment of the expression of programmed cell death–ligand 1 (PD‐L1) in advanced non‐small cell lung cancer (NSCLC) is now routine practice. The expression of PD‐L1 on the surface of tumor cells was evaluated by immunohistochemistry (IHC) as performed at SRL (Tokyo, Japan) with the use of a PD‐L1 IHC 22C3 pharmDx kit (Agilent Technologies, Santa Clara, CA) and a Dako Autostainer Link 48 platform (Dako, Carpinteria, CA). Staining intensity for PD‐L1 and the percentage of PD‐L1–positive tumor cells were determined for each sample by pathologists of the commercial vendor. The tumor proportion score (TPS) was calculated as the percentage of viable tumor cells showing partial or complete membrane staining (≥1+) relative to all viable tumor cells present in the sample.
Statistical Analysis
We conducted this observational study without determining sample size or power. Progression‐free survival (PFS) was calculated from the date of starting chemotherapy to the date of disease progression or death. The probability of survival as a function of time was estimated with the Kaplan‐Meier method. The relation between tumor type and TMB status was assessed with Fisher's exact test. A p value of <.05 was considered statistically significant. All statistical analysis was performed with GraphPad Prism software version 5.0 (GraphPad Software, San Diego, CA).
Results
Patient Characteristics
Between September 2018 and June 2019, written informed consent was obtained from 202 patients at Kindai University Hospital whose disease had already been cytologically or histologically proved (supplemental online Fig. 1). Eleven of these 202 patients were subsequently excluded from the study because they had insufficient tissue remaining for further genetic analysis on institutional pathological review (n = 9) or they had only a cytology specimen available (n = 2). Specimens from 191 patients were thus considered adequate for genomic testing and were shipped to Foundation Medicine. Pathological evaluation at Foundation Medicine revealed that an additional eight specimens contained an insufficient amount of tumor tissue for genomic testing, with an alternative specimen being available in one instance, and three samples yielded an insufficient amount of DNA. A total of 181 samples were therefore processed for genomic testing, 6 of which failed to complete the sequencing process. The remaining 175 samples yielded gene profiling data, making the success rate 96.7%. Among 175 samples available for gene profiling data, 60 were biopsy specimens and 4 were cell block specimens. Among those 64 small tumor samples, 58 tumors had ≥20% tumor cells, and the remaining 6 tumors had <20% tumor cell. Three of 6 patients (50%) whose tumor had <20% tumor cell were missing TMB data, whereas 5 of 58 patients (9%) whose tumor had ≥20% tumor cell were missing TMB data. Among six samples unavailable for genome profiling data, four were surgical specimens and the remaining two were biopsy specimens, and their tumor cells per the biopsy tissue area is 17% and 30%, respectively (supplemental online Table 1). The turnaround time, defined as the interval between the date informed consent was obtained and receipt of the NGS assay results by the treating physician after the MTB meeting, was 41 days (range, 21–126 days). The median turnaround time until providing the CGP results from attending physician to the patients was 11 days (range, 0–60 days), and 10 patients died before receiving CGP results.
The demographics of the 175 patients included in the study are shown in Table 1. Eighty‐seven patients (50%) were female, with the median age for all patients being 62 years (range, 19–83 years). One patient aged 19 years was allowed to enroll for ethical reasons. Most (90%) patients had a good Eastern Cooperative Oncology Group performance status (0 or 1) at the time of testing. The most common tumor types were lung cancer (23%), colorectal cancer (19%), breast cancer (13%), and head and neck cancer (7%). Sixty specimens (34%) were derived from tissue obtained at tumor biopsy, and 111 (63%) were from surgically resected tissue.
Table 1.
Characteristics of patients evaluated for genetic alterations (n = 175)
| Characteristic | Subset | No. of patients (%) |
|---|---|---|
| Median (range) age, years | 62 (19–83) | |
| Sex | Male | 88 (50) |
| Female | 87 (50) | |
| ECOG PS | 0 | 56 (32) |
| 1 | 102 (58) | |
| 2 | 14 (8) | |
| 3 | 3 (2) | |
| Type of cancer | Lung cancer | 41 (23) |
| Colorectal cancer | 33 (19) | |
| Breast cancer | 23 (13) | |
| Head and neck cancer | 12 (7) | |
| Esophageal carcinoma | 8 (5) | |
| Unknown primary cancer | 8 (5) | |
| Biliary cancer | 6 (3) | |
| Gastric cancer | 5 (3) | |
| Cervical cancer | 4 (2) | |
| Pancreatic cancer | 4 (2) | |
| Extramammary Paget disease | 4 (2) | |
| Thyroid carcinoma | 3 (2) | |
| Other | 24 (14) | |
| Lines of previous CTx | 0 | 39 (22) |
| 1 | 56 (32) | |
| ≥2 | 80 (46) | |
| Tissue source | Biopsy | 60 (34) |
| Cell block | 4 (2) | |
| Surgery | 111 (63) | |
Abbreviations: CTx, chemotherapy; ECOG PS, Eastern Cooperative Oncology Group performance status.
Prevalence of Genetic Alterations
A total of 1,999 gene alterations other than amplifications were detected, with a mean of 11.4 alterations per tumor, and 560 and 1,439 of the alterations were categorized as known or likely pathogenic variants or as VUS, respectively, by the Foundation Medicine database (Fig. 1A). Actionable fusions involving ALK or RET were detected in two patients each. The most common known or likely pathogenic alterations were in TP53 (n = 113), PIK3CA (n = 33), APC (n = 32), KRAS (n = 29), ARID1A (n = 21), MLL2 (n = 13), BRAF (n = 12), and CDKN2A (n = 10). A total of 599 gene amplifications were detected, with a mean of 3.4 amplifications per tumor, and 311 and 288 of the amplifications were categorized as known or likely pathogenic variants or as VUS, respectively (Fig. 1B). The most frequent known or likely pathogenic amplifications involved ERBB2 (n = 17), MYC (n = 15), FGF19 (n = 14), FGF3 (n = 14), FGF4 (n = 14), RAD21 (n = 13), and CCND1 (n = 12). In the present study, 35 cases (20%) included a potential germline mutation such as APC, RB1, or BRACA1/2. At the start of this study, no specific algorithms for potential germline mutation–detected tumor‐only genomic profiling had been established in Japan, and germline testing to confirm hereditary cancer had not been covered by insurance. None received genetic counseling and tested normal cells to confirm such hereditary mutations.
Figure 1.
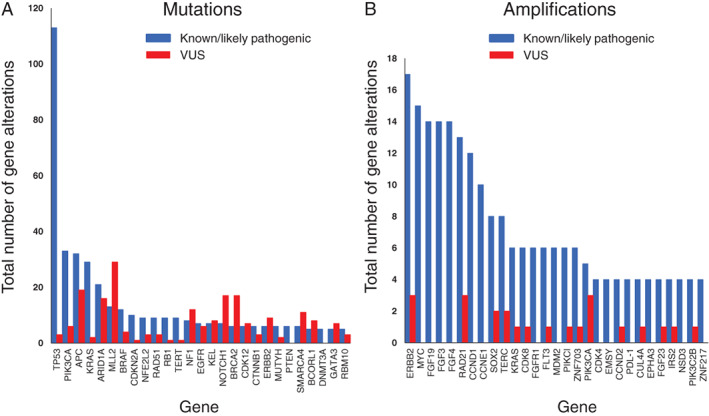
Distribution of the most frequent mutations (A) and amplifications (B) for the 175 patients with sequencing results. Genetic alterations are classified as known or likely pathogenic variants (blue) or as VUS (red).Abbreviation: VUS, variants of unknown significance.
Prevalence of MSI and TMB
Among the 175 patients assessed for MSI, 2 patients (1%) were categorized as MSI‐high, 150 patients (86%) as MSI‐stable, and the remaining 23 patients (13%) as “cannot be determined.” Among the 153 patients with TMB results available, the median TMB was 4 mutations/Mb, with a range of 0 to 122 mutations/Mb. The two MSI‐high cases included one patient with colorectal cancer and one with unknown primary cancer, with the TMB values for these two patients being 39 and 45 mutations/Mb, respectively. One colorectal cancer case classified as MSI‐stable with a high TMB (33 mutations/Mb) by FoundationOne CDx turned out to be categorized as MSI‐high by the MSI CDx test (FALCO). The distribution of TMB across different cancer types is shown in Figure 2A. The median TMB (mutations/Mb) was 6 for lung cancer and head and neck cancer; 5 for colorectal cancer and esophageal cancer; 4 for breast cancer, unknown primary cancer, and gastric cancer; 3 for biliary cancer; and 1 for pancreatic cancer. Classification of patients into two groups based on TMB value revealed that 20 and 133 had a high or low TMB (TMB of ≥10.0 or < 10.0 mutations/Mb), respectively. Tumors with a high TMB were more prevalent in lung cancer (11/32) compared with other solid tumors (9/121, Fisher's exact test p < .01).
Figure 2.
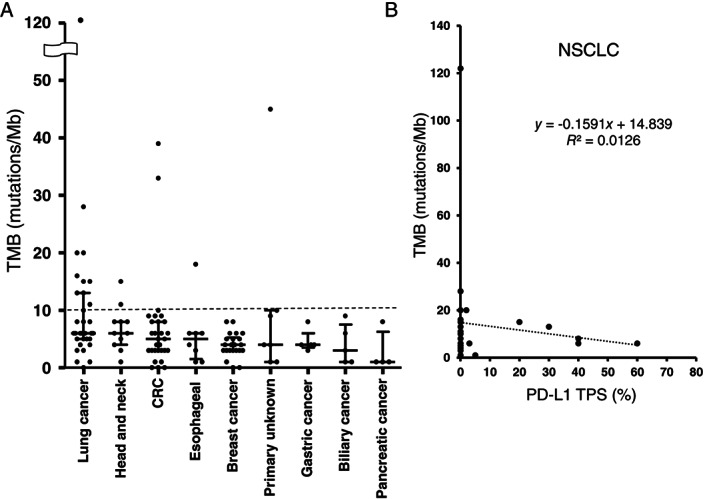
(A): Distribution of TMB according to tumor type. Bars indicate median and quartile values; the dashed line indicates the cutoff (10%) for high and low values. (B): Regression analysis for TMB and PD‐L1 TPS in patients with NSCLC (n = 26).Abbreviations: CRC, colorectal cancer; NSCLC, non‐small cell lung cancer; PD‐L1, programmed cell death–ligand 1; TMB, tumor mutation burden; TPS, tumor proportion score.
Relation Between PD‐L1 Expression and TMB in NSCLC
Given that PD‐L1 testing results are available for patients with NSCLC in routine clinical practice, we next evaluated the possible relation between PD‐L1 expression and TMB in such patients. Among the 41 patients with lung cancer who underwent FoundationOne testing, two patients were excluded because their tumors showed neuroendocrine features such as small cell lung cancer and carcinoid tumor. Another nine patients were excluded because the TMB value could not be determined, although information on clinically relevant genetic alterations and available clinical trials for these patients was available. A total of 26 patients with NCSLC had results available for both PD‐L1 TPS and TMB. No correlation was apparent between PD‐L1 TPS and TMB for these patients (R 2 = 0.0126; Fig. 2B).
Association Between PD‐L1 or TMB and ICI Efficacy
We next examined the relationship between PD‐L1 expression or TMB value and approved ICI efficacy across tumor types, including 21 patients with NSCLC, 8 with head and neck cancer, 4 with esophageal carcinoma, 2 with gastric cancer, and 1 with CRC, melanoma, and mesothelioma, respectively (Fig. 3). Among patients with NSCLC, 11 received immune checkpoint inhibitor (ICI) monotherapy and 10 received an ICI plus platinum‐based combination therapy. No clear trend toward increased efficacy for ICI monotherapy or ICI combination chemotherapy in patients with a high PD‐L1 TPS or a high TMB was apparent regardless of tumor type.
Figure 3.
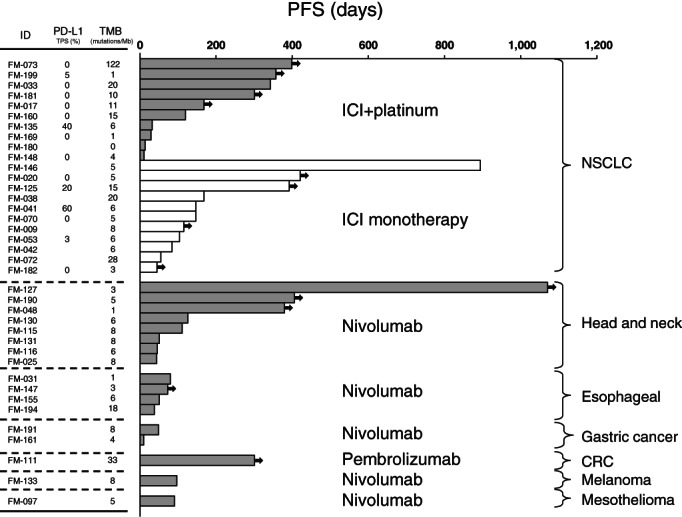
Swimmer plots for patients who received approved treatment with a programmed cell death–1 inhibitor either alone (ICI monotherapy) or together with a platinum‐based regimen (ICI + platinum). The length of each bar represents PFS, with the arrows indicating an ongoing response at data cutoff.Abbreviations: CRC, colorectal cancer; ICI, immune checkpoint inhibitor; NSCLC, non‐small cell lung cancer; PD‐L1, programmed cell death–ligand 1; PFS, progression‐free survival; TMB, tumor mutation burden.
Proportion of Patients Treated with Genotype‐Directed Therapy
To evaluate the clinical utility of the FoundationOne CDx panel, we determined the proportion of patients who were able to access matched targeted therapy. Among the 175 tested patients, 174 had at least one known or likely pathogenic gene alteration, and 24 of these patients (14%) received corresponding targeted therapy, including that with clinically approved (n = 11) or investigational (n = 11) agents, or both (n = 2; Table 2). Kaplan‐Meier analysis revealed that the median PFS for the patients who received matched targeted therapy was 12.1 months (95% confidence interval, 6.9–17.4 months; Fig. 4). Eight patients (33%) benefited from such targeted treatment for >1 year.
Table 2.
Characteristics of patients who received genotype‐matched therapy (n = 24)
| Type of cancer | Driver oncogene alteration | Targeted drug | No. of patients (%) |
|---|---|---|---|
| Lung cancer | EGFR Ex19del | Osimertinib | 2 (8) |
| EGFR L858R | Erlotinib | 1 (4) | |
| EGFR L861Q | Afatinib | 1 (4) | |
| EGFR Ex20ins | Investigational | 1 (4) | |
| HER2 amplification | Investigational | 1 (4) | |
| ALK rearrangement | Alectinib | 2 (8) | |
| Colorectal cancer | HER2 amplification | Investigational | 1 (4) |
| MSI‐high | Investigational | 1 (4) | |
|
TMB‐high/MSI‐stable (F1), MSI‐high (FALCO) |
Pembrolizumab | 1 (4) | |
| Gastric cancer | HER2 amplification | XP/HER | 1 (4) |
| Breast cancer | HER2 amplification | VNR/HER f/b Investigational | 1 (4) |
| HER2 amplification | T‐DM1 f/b Investigational | 1 (4) | |
| HER2 amplification | PER/HER/DTX | 1 (4) | |
| BRCA2 mutation | Olaparib | 1 (4) | |
| RET rearrangement | Investigational | 1 (4) | |
| Extramammary Paget disease | HER2 amplification | Investigational | 3 (13) |
| Unknow primary cancer | TMB‐high | Investigational | 1 (4) |
| BRAF mutation | Investigational | 1 (4) | |
| MSI‐high | Investigational | 1 (4) | |
| Cervical cancer | HER2 amplification | Investigational | 1 (4) |
Abbreviations: Ex19del, exon‐19 deletion; Ex20ins, exon‐20 insertion; F1, FoundationOne CDx; f/b, followed by; MSI, microsatellite instability; PER/HER/DTX, pertuzumab plus trastuzumab plus docetaxel; T‐DMI, trastuzumab emtansine; TMB, tumor mutation burden; VNR/HER, vinorelbine plus herceptin; XP/HER, capecitabine plus cisplatin plus herceptin.
Figure 4.
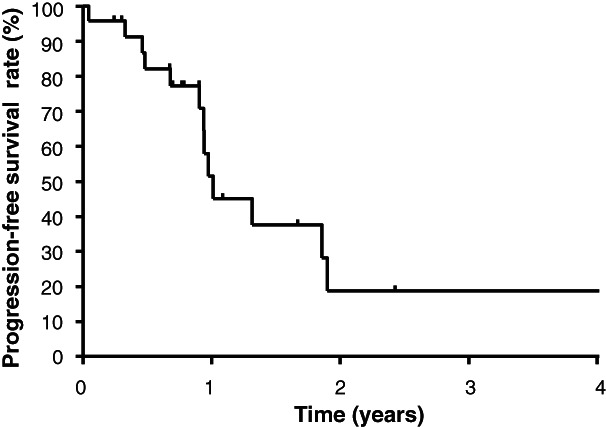
Progression‐free survival rate for patients who received genotype‐matched targeted therapy (n = 24).
Discussion
Clinical cancer research is undergoing a paradigm shift from a tumor‐specific treatment strategy to a tumor‐agnostic approach that targets specific genomic alterations or molecular features regardless of tumor site. In 2018, PMDA of Japan approved the ICI pembrolizumab for treatment of adult and pediatric patients with unresectable or metastatic solid tumors found to be either MSI‐high or mismatch repair–deficient [6]. In 2019, PMDA and subsequently the U.S. Food and Drug Administration approved entrectinib—a selective tyrosine kinase inhibitor that targets TRKA, TRKB, TRKC, ROS1, and ALK proteins—for patients with NTRK fusion–positive advanced or recurrent solid tumors. The clinical development of a RET inhibitor for RET‐altered solid tumors is currently underway in a global phase II study including patients in Japan. High‐throughput and multiplex genotyping tests are thus urgently required in daily clinical practice in order to take advantage of the development of such tumor‐agnostic drugs. We here investigated the feasibility and utility of the FoundationOne CDx panel, which was recently approved by PMDA for profiling of actionable mutations in solid tumors and which also provides an assessment of genomic signatures such as TMB and MSI.
The success rate of this NGS assay as performed with clinical FFPE samples including biopsy specimens (34%) was 96.7%, suggestive of the feasibility of such sequencing in clinical practice. The higher success rate observed in our study compared with a previous study [7] of FoundationOne CDx may be attributable to the fact that we evaluated tissue before shipment, even though the proportion of surgical specimens was similar in the two studies. On the other hand, the median turnaround time was 41 days (range, 21–126 days) in our study, which is not satisfactory for the clinical setting. Foundation Medicine announced that their turnaround time is 14 days on average from receipt of a sample that meets requirements. The long turnaround time in our study reflects the fact that it takes several days to ship tumor tissue from Japan to the U.S. as well as 1–2 weeks for the report to be returned to the referring physician after discussion by the MTB.
Most patients (99%) in the present study harbored known or likely pathogenic mutations, but only 24 of these individuals (14%) received matched targeted therapy, a proportion similar to that of a previous study with the NCC Oncopanel (13.4%) [2]. Our study enrolled patients with common cancer types such as lung cancer (n = 41) and breast cancer (n = 23). Most patients harboring known actionable driver alterations such as those with EGFR mutation–positive lung cancer or HER2‐positive breast cancer were not enrolled in the present study, given that such individuals already receive molecularly targeted therapy as a result of testing with rapid companion diagnostics such as the Cobas EGFR Mutation Test or HER2 fluorescence in situ hybridization. However, some patients with such alterations—EGFR‐mutated lung cancer (n = 4), ALK‐rearranged lung cancer (n = 2), HER2‐amplified breast cancer (n = 3), and HER2‐amplified gastric cancer (n = 1)—were included in this CGP analysis. The MATCH (Molecular Analysis for Therapy Choice) trial of the U.S. National Cancer Institute is a phase II basket study that was launched in August 2015 [8]. It will enroll ~1,000 patients with any solid tumor or lymphoma and assign them to treatment arms based on the molecular profile of their disease. A few tumor‐agnostic basket trails, such as for RET‐rearranged or ERBB2‐amplified tumor, are also ongoing in Japan. The RET inhibitor conducted in Japan is LOXO‐292 in RET‐fusion–positive cancers regardless of the tumor type. On the other hand, an example of the successful implementation is provided by the LURET study (UMIN000010095), a phase II study of vandetanib in patients with RET‐rearranged advanced NSCLC, showing a response rate of 47% [9]. There is an urgent need to develop novel tumor‐agnostic therapies, to guide patients to clinical trials evaluating drugs matched to identified molecular alterations, and to increase accessibility to such drugs at smaller regional cancer hospitals. The promising results in terms of PFS for patients who received matched targeted therapy in the present study should be interpreted with caution, however, given that the study was nonrandomized and observational in nature. Even the SHIVA trial, a randomized basket trial designed to evaluate whether matched targeted drugs could extend PFS compared with conventional chemotherapy in patients with advanced solid tumors, failed to show any improvement in survival or response with genome‐based targeted therapy [10].
The predictive role of TMB has been investigated largely in advanced NSCLC. CheckMate 568, a phase II trial that evaluated the efficacy and safety of the combination of the ICIs nivolumab and ipilimumab as first‐line treatment for advanced NSCLC, adopted the FoundationOne CDx assay and identified a TMB of ≥10 mutations/Mb as an effective cutoff for selection of patients most likely to benefit from this drug combination [11]. Moreover, the CheckMate 227 study met its coprimary endpoint of PFS for nivolumab plus ipilimumab compared with cytotoxic chemotherapy in patients with a TMB of ≥10 mutations/Mb, regardless of PD‐L1 expression status [12]. On the basis of these data, we selected a TMB of 10 mutations/Mb as the cutoff for TMB in the present study. We found that tumors with a high TMB were more frequent for lung cancer than for other solid tumor types, consistent with a previous study in which TMB was measured with other CGP panels targeting 315 genes across tumor types [13]. We also evaluated PD‐L1 expression by IHC with the Dako 22C3 pharmDx kit, which has been adopted as a biomarker to direct first‐line treatment, with a TPS of ≥50% having been established as the threshold for a positive result in KEYNOTE‐024 [14]. Although a high TMB has previously been found to be predictive of a favorable outcome for immunotherapy [11, 15], consistent with previous results [11, 16], we detected no clear relation between PD‐L1 expression and TMB in our study of specimens from patients with advanced or recurrent NSCLC—even though the method for determination of TMB and the antibody used to detect PD‐L1 differed between our and these previous studies. There are several reports investigating the correlation between TMB or PD‐L1 status and ICI efficacy. TMB or PD‐L1 may be a predictor of response for single‐agent immunotherapy, but it has failed to demonstrate the correlation with response when immunotherapy is combined with chemotherapy [17, 18, 19]. Because the efficacy of the ICI plus platinum‐based chemotherapy can be maintained independent of TMB or PD‐L1 status, evaluation of TMB or PD‐L1 is not required for making treatment decisions for patients eligible to receive combination therapy. Among ICI monotherapy–treated patients in our study, we could not demonstrate a superior clinical efficacy of ICIs in patients with NSCLC with high TMB or high PD‐L1 expression owing to small sample sizes.
Conclusion
We have demonstrated the successful application of the FoundationOne CDx assay to the performance of multiple genomic tests with a single small FFPE tumor specimen. In June 2019, two gene panels were approved with reimbursement: FoundationOne CDx Cancer Genomic Profile and OncoGuide NCC Oncopanel System. The fee for these products would be ¥560,000 (USD 5,100), and patients will have to pay 10%–30% of the total fees. It is an expensive test, but approximately 85% of patients do not benefit from panel testing. Such multiplex genomic testing will assist physicians in matching patients found to harbor actionable mutations with available targeted treatments or clinical trials of new targeted agents; however, there is a tremendous need to develop novel targeted agents matched with tumor molecular alteration.
Author Contributions
Conception/design: Masayuki Takeda
Collection and/or assembly of data: Takayuki Takahama, Kazuko Sakai, Shigeki Shimizu, Satomi Watanabe, Hisato Kawakami, Kaoru Tanaka, Chihiro Sato, Hidetoshi Hayashi, Yoshikane Nonagase, Kimio Yonesaka, Naoki Takegawa, Tatsuya Okuno, Takeshi Yoshida, Soichi Fumita, Shinichiro Suzuki, Koji Haratani
Data analysis and interpretation: Masayuki Takeda, Kazumasa Saigoh, Akihiko Ito, Tetsuya Mitsudomi, Hisashi Handa, Kazuya Fukuoka, Kazuhiko Nakagawa, Kazuto Nishio
Manuscript writing: Masayuki Takeda
Final approval of manuscript: Masayuki Takeda, Takayuki Takahama, Kazuko Sakai, Shigeki Shimizu, Satomi Watanabe, Hisato Kawakami, Kaoru Tanaka, Chihiro Sato, Hidetoshi Hayashi, Yoshikane Nonagase, Kimio Yonesaka, Naoki Takegawa, Tatsuya Okuno, Takeshi Yoshida, Soichi Fumita, Shinichiro Suzuki, Koji Haratani, Kazumasa Saigoh, Akihiko Ito, Tetsuya Mitsudomi, Hisashi Handa, Kazuya Fukuoka, Kazuhiko Nakagawa, Kazuto Nishio
Disclosures
Masayuki Takeda: Ono Pharmaceutical Co., Boehringer Ingelheim Japan Inc., Novartis Pharma K.K., AstraZeneca K.K., and Chugai Pharmaceutical Co. (H); Takayuki Takahama: AstraZeneca, Roche Diagnostics, and Boehringer Ingelheim (H); Kazuko Sakai: Roche Diagnostics, Bio‐Rad, SRL Diagnostics, AstraZeneca and Chugai Pharmaceutical Co., Ltd. (H); Hisato Kawakami: Diaiichi Sankyo (RF), Eli Lilly, Ono Pharmaceutical, Co., Ltd., Chugai Pharmaceutical, Co., Ltd., Taiho Pharmaceutical, Co., Ltd., MSD Pharmaceutical, Co., Ltd., Merck Serono, Takeda Pharmaceutical, Co., Ltd., AstraZeneca K.K., Yakult Pharmaceutical industry, Co., Ltd (H); Kaoru Tanaka: AstraZeneca, Merck Serono, Eisai, Bristol‐Myers Squibb, Ono Pharmaceutical, Merck Sharp & Dohme (H); Hidetoshi Hayashi: AstraZeneca K.K., Boehringer Ingelheim Japan Inc., Chugai Pharmaceutical Co. Ltd., Ono Pharmaceutical Co. Ltd. (RF), AstraZeneca K.K., Boehringer Ingelheim Japan Inc., Bristol‐Myers Squibb Co. Ltd., Chugai Pharmaceutical Co. Ltd., Eli Lilly Japan K.K., Kyorin Pharmaceutical Co. Ltd, Merck Biopharma Co., Ltd., MSD K.K., Novartis Pharmaceuticals K.K, Ono Pharmaceutical Co. Ltd., Pfizer Japan Inc., Shanghai Haihe Biopharm, Taiho Pharmaceutical Co. Ltd. (H); Koji Haratani: AstraZeneca K.K., MSD K.K. (RF), AstraZeneca K.K., MSD K.K., Pfizer Japan Inc., Ono Pharmaceutical Co. Ltd., As One Corporation, Bristol‐Myers Squibb Co. Ltd., Chugai Pharmaceutical Co. Ltd. (H); Kazuto Nishio: Otsuka Pharmaceutical Co., Ltd., Life Technologies Japan Ltd., Nippon Boehringer Ingelheim Co., Ltd., Eli Lilly Japan K.K., Ignyta,Inc., Astellas Pharma Inc. (RF), Otsuka Pharmaceutical Co., Ltd., Life Technologies Japan Ltd., Nippon Boehringer Ingelheim Co., Ltd., Eli Lilly Japan K.K., Ignyta, Inc., Astellas Pharma Inc., Chugai Pharmaceutical Co., Ltd., Eisai Co., Ltd., Pfizer Inc., Novartis Pharma K.K., MSD K.K., Ono Pharmaceutical Co. Ltd., Bristol‐Myers Squibb Company, SymBio Pharmaceuticals Limited., Solasia Pharma K.K., Yakult Honsha Co., Ltd., Roche Diagnostics K.K., AstraZeneca K.K., Sanofi K.K., Guardant Health Inc., Astellas Pharma Inc., Takeda Pharmaceutical Co. Ltd, Kobayashi Pharmaceutical Co. Ltd. (H); Tetsuya Mitsudomi: AstraZeneca, Chugai, Pfizer, Boehringer‐Ingelheim, Eli Lilly & Co., Amgen, Merck Sharp & Dohme, Bristol‐Myers Squibb, Ono, Novartis (C/A), Boehringer‐Ingelheim, AstraZeneca, Chugai, Eli Lilly & Co., Pfizer, Merck Sharp & Dohme, Daiichi‐Sankyo (RF), AstraZeneca, Chugai, Pfizer, Boehringer‐Ingelheim, Eli Lilly & Co., Merck Sharp & Dohme, Bristol‐Myers Squibb, Ono, Novartis (H); Kazuhiko Nakagawa: Takeda Pharmaceutical Co., Ltd., Kyorin Pharmaceutical Co., Ltd., Ono Pharmaceutical Co., Ltd., Pfizer Japan Inc., Eli Lilly & Co. Japan K.K (C/A), Merck Sharp & Dohme K.K., A2 Healthcare Corp., inVentiv Health Japan, Astellas Pharma Inc., Daiichi Sankyo Co., Ltd., Novartis Pharma K.K., AbbVie Inc., Quintiles Inc., IQVIA Services Japan K.K., ICON Japan K.K., Chugai Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., EP‐CRSU Co., Ltd., Gritstone Oncology, Linical Co., Ltd., Eli Lilly & Co. Japan K.K., Eisai Co., Ltd., Bristol‐Myers Squibb, Nippon Boehringer‐Ingelheim Co., Ltd., Taiho Pharmaceutical Co., Ltd., Pfizer Japan Inc., Parexel International Corp., SymBio Pharmaceuticals Limited, Ono Pharmaceutical Co., Ltd., Merck Serono Co., Ltd., Merck Biopharma Co., Ltd., AstraZeneca K.K., CMIC Shift Zero K.K., Kissei Pharmaceutical Co., Ltd., Kyowa Hakko Kirin Co., Ltd., EPS Corporation, Bayer Yakuhin, Ltd., Syneos Health, EPS International Co., Ltd., Pfizer R&D Japan G.K., Otsuka Pharmaceutical Co., Ltd. (RF), AstraZeneca K.K., Nichi‐Iko Pharmaceutical Co., Ltd., Astellas Pharma Inc., Takeda Pharmaceutical Co., Ltd., Merck Sharp & Dohme K.K., Taiho Pharmaceutical Co., Ltd., Ono Pharmaceutical Co., Ltd., Bristol‐Myers Squibb, Nippon Boehringer‐Ingelheim Co., Ltd., Eli Lilly & Co. Japan K.K., Novartis Pharma K.K., SymBio Pharmaceuticals Limited, Pfizer Japan Inc., Chugai Pharmaceutical Co., Ltd., Clinical Trial Co., Ltd., Nanzando Co., Ltd., Medicus Shuppan, Publishers Co., Ltd., Yodosha Co., Ltd., Care Net, Inc., Nikkei Business Publications, Inc., Reno Medical K.K., Daiichi Sankyo Co., Ltd., Kyorin Pharmaceutical Co., Ltd., Thermo Fisher Scientific K.K., Medical Review Co., Ltd., Yomiuri Telecasting Corporation, Roche Diagnostics K.K., Nippon Kayaku Co., Ltd., Bayer Yakuhin, Ltd., Merck Biopharma Co., Ltd., Medical Mobile Communications Co., Ltd., AbbVie Inc., 3H Clinical Trial Inc. (H). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Supplementary FIG 1 Patient flow.
Supplementary Table S1 Success rate of genome profiling and TMB according to tumor specimens
Acknowledgments
This study was supported in part by Japan Society for the Promotion of Science KAKENHI grant 19K07721.
Disclosures of potential conflicts of interest may be found at the end of this article.
Contributor Information
Masayuki Takeda, Email: takedamasa2004@yahoo.co.jp.
Kazuto Nishio, Email: knishio@med.kindai.ac.jp.
References
- 1. Takeda M, Sakai K, Terashima M et al. Clinical application of amplicon‐based next‐generation sequencing to therapeutic decision making in lung cancer. Ann Oncol 2015;26:2477–2482. [DOI] [PubMed] [Google Scholar]
- 2. Sunami K, Ichikawa H, Kubo T et al. Feasibility and utility of a panel testing for 114 cancer‐associated genes in a clinical setting: A hospital‐based study. Cancer Sci 2019;110:1480–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kenmotsu H, Serizawa M, Koh Y et al. Prospective genetic profiling of squamous cell lung cancer and adenosquamous carcinoma in Japanese patients by multitarget assays. BMC Cancer 2014;14:786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kou T, Kanai M, Yamamoto Y et al. Clinical sequencing using a next‐generation sequencing‐based multiplex gene assay in patients with advanced solid tumors. Cancer Sci 2017;108:1440–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doebele RC, Drilon A, Paz‐Ares L et al. Entrectinib in patients with advanced or metastatic NTRK fusion‐positive solid tumours: integrated analysis of three phase 1‐2 trials. Lancet Oncol 2020;21:271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marcus L, Lemery SJ, Keegan P et al. FDA approval summary: Pembrolizumab for the treatment of microsatellite instability‐high solid tumors. Clin Cancer Res 2019;25:3753–3758. [DOI] [PubMed] [Google Scholar]
- 7. De Falco V, Poliero L, Vitello PP et al. Feasibility of next‐generation sequencing in clinical practice: Results of a pilot study in the Department of Precision Medicine at the University of Campania 'Luigi Vanvitelli'. ESMO Open 2020;5:e000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flaherty KT, Gray RJ, Chen AP et al. Molecular landscape and actionable alterations in a genomically guided cancer clinical trial: National Cancer Institute Molecular Analysis for Therapy Choice (NCI‐MATCH). J Clin Oncol 2020;38:3883–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoh K, Seto T, Satouchi M et al. Vandetanib in patients with previously treated RET‐rearranged advanced non‐small‐cell lung cancer (LURET): An open‐label, multicentre phase 2 trial. Lancet Respir Med 2017;5:42–50. [DOI] [PubMed] [Google Scholar]
- 10. Le Tourneau C, Delord JP, Goncalves A et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open‐label, proof‐of‐concept, randomised, controlled phase 2 trial. Lancet Oncol 2015;16:1324–1334. [DOI] [PubMed] [Google Scholar]
- 11. Ready N, Hellmann MD, Awad MM et al. First‐line nivolumab plus ipilimumab in advanced non‐small‐cell lung cancer (CheckMate 568): Outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol 2019;37:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hellmann MD, Ciuleanu TE, Pluzanski A et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018;378:2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chalmers ZR, Connelly CF, Fabrizio D et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reck M, Rodriguez‐Abreu D, Robinson AG et al. Pembrolizumab versus chemotherapy for PD‐L1‐positive non‐small‐cell lung cancer. N Engl J Med 2016;375:1823–1833. [DOI] [PubMed] [Google Scholar]
- 15. Goodman AM, Kato S, Bazhenova L et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 2017;16:2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu H, Chen Z, Ballman KV et al. Correlation of PD‐L1 expression with tumor mutation burden and gene signatures for prognosis in early‐stage squamous cell lung carcinoma. J Thorac Oncol 2019;14:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paz‐Ares L, Luft A, Vicente D et al. Pembrolizumab plus chemotherapy for squamous non‐small‐cell lung cancer. N Engl J Med 2018;379:2040–2051. [DOI] [PubMed] [Google Scholar]
- 18. Gadgeel S, Rodriguez‐Abreu D, Speranza G et al. Updated analysis from KEYNOTE‐189: Pembrolizumab or placebo plus pemetrexed and platinum for previously untreated metastatic nonsquamous non‐small‐cell lung cancer. J Clin Oncol 2020;38:1505–1517. [DOI] [PubMed] [Google Scholar]
- 19. Carbone DP, Reck M, Paz‐Ares L et al. First‐line nivolumab in stage IV or recurrent non‐small‐cell lung cancer. N Engl J Med 2017;376:2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Supplementary FIG 1 Patient flow.
Supplementary Table S1 Success rate of genome profiling and TMB according to tumor specimens


