Abstract
The complete plastid genome of Chrysanthemum morifolium ‘Anhuishiliuye’, a Chinese traditional cultivar, was determined and analyzed in this work. It had a circular-mapping molecular with the length of 151,059 bp.The LSC and SSC of 82,857 bp and 18,294 bp were separated by two IRs of 24,954 bp. The chloroplast genome of C. morifolium ‘Anhuishiliuye’ contains 125 genes, including 83 protein-coding genes, 34 ribosomal RNA genes and 8 transfer RNA genes. Phylogenetic analysis showed that C. morifolium ‘Anhuishiliuye’ clustered together with other Chrysanthemum species. The data provided would be useful for elucidation of phylogenetics and evolution in Chrysanthemum cultivars.
Keywords: Complete plastid genome, Chrysanthemum, chloroplast, phylogenetics and evolution
Chrysanthemum (Chrysanthemum morifolium), in family Asteraceae, is one of the most popular and economically important floricultural crops all over the world, noted for ornamental, edible, tea and medicinal uses. It has approximately 1600 years of cultivation history, which was first cultivated in China and then successively introduced to Japan, Europe, and North American. So that cultivated chrysanthemums are highly heterozygous, whose genetic background are very complicated and coming from multiple species (Su et al. 2019). Understanding the plant chloroplast genetics is essential for identification of commercial cultivars and the determination of their purity. In this study, we reported and characterized the chloroplast genomes of a Chinese ancient cultivar, Chrysanthemum morifolium ‘Anhuishiliuye’, which is a big flower (disbud) potted variety. It is short daylight plant and the flowering period is from mid-November to early December in southern China. It is radial inflorescence, with tubular petals and light pink double flower.
By the cp genomic data, we reconstruct the phylogenetic tree of Asteraceae family to reveal the relationship and provide useful information for further study of Chrysanthemum cultivars. Fresh leaves of C. morifolium ‘Anhuishiliuye’ were collected from the mature plant in the green house of Zhongkai university of agriculture and engineering (22°40′18.69″N, 113°15′5.37″E) and stored in South China Botanical Garden, CAS. After DNA extraction, a library with the insertion size of 350 bp was constructed, and high-throughput DNA sequencing (pair-end 150 bp) was performed on an Illumina X10 platform. In total, 10 G raw reads were obtained for de novo assembly of cp genome in NOVOPlasty (Dierckxsens et al. 2017), and then annotated in Plann (Huang and Cronk 2015) and GeSeq using the default parameters. The complete genome sequence C. morifolium ‘Anhuishiliuye’ together with gene annotations were submitted to NCBI GenBank under the accession number MT976165.
The whole cp genome of C. morifolium ‘Anhuishiliuye’ was 151,060 bp in length, with two short inverted repeat (IRa and IRb) regions of 24,954 bp, a large single-copy (LSC) region of 82,858 bp, and a small single-copy (SSC) region of 18,294 bp. The overall GC content of cp genome is 37.45%. The cp genome of C. morifolium ‘Anhuishiliuye’ contained 125 genes, including 83 protein-coding genes, 34 rRNA genes, and 8 tRNA genes. Most of these genes did not contain intron, 16 genes contained one intron, and two genes (clpP and ycf3) contained two introns. All genes occurred as a single copy, except that 16 genes were duplicated in IR regions.
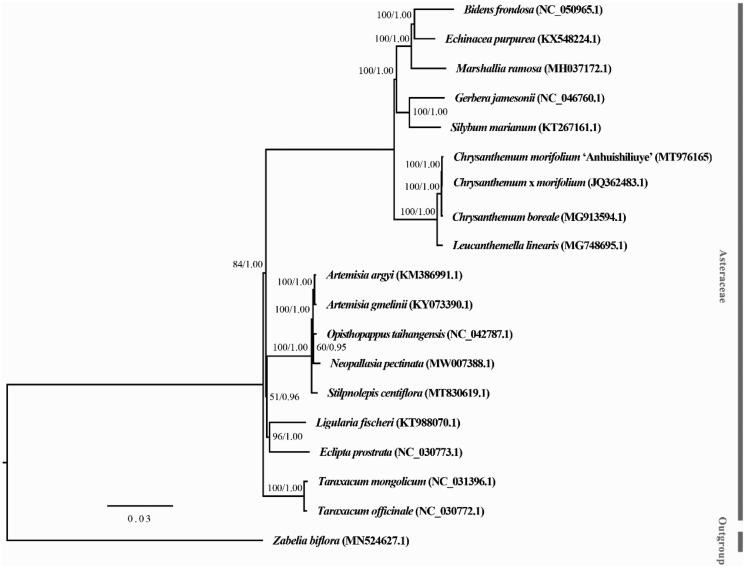
A phylogenetic analysis was carried out using cp genome sequences of C. morifolium ‘Anhuishiliuye’ and other 17 species within family Asteraceae and Zebelia biflora as outgroup (Figure 1). The sequences were aligned using MAFFT 7 (Katoh and Standley 2013). A maximum likelihood tree was constructed using RAxML 8.2.12 (Stamatakis 2014), and a Bayesian inference tree was constructed with MrBayes 3.2.7 (Ronquist et al. 2012). Phylogenetic analysis revealed that C. morifolium ‘Anhuishiliuye’ was grouped with species of genus Chrysanthemum, in consistent with the previous study in Chrysanthemum (Tyagi et al. 2019). In conclusion, this complete cp genome would establish a solid foundation for future genetic studies in C. morifolium and its relative species.
Figure 1.
Phylogeny of Chrysanthemum morifolium ‘Anhuishiliuye’, other Asteraceae species and Zebelia biflora. Phylogenetic tree was based on complete chloroplast genome with Maximum-likelihood (ML) and Bayesian (BI). Numbers above the branches are ML bootstrap/BI posterior probability.
Funding Statement
This work was supported by the National natural science foundation of China NFSC (32001244), the construction plan of Guangzhou science and technology basic conditions platform (201903010053), and the major technological innovation of Guangdong province of China (2020B020220009).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MT976165]. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682858 SRR13205719 and SAMN17012931, respectively.
References
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DI, Cronk QC.. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. . 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Jiang J, Zhang F, Liu Y, Ding L, Chen S, Chen F.. 2019. Current achievements and future prospects in the genetic breeding of chrysanthemum: a review. Hortic Res. 6:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Jung J-A, Kim JS, Kwon S-J, Won SY.. 2019. The complete chloroplast genome of an economic plant, Chrysanthemum morifolium ‘Baekma’. Mitochondrial DNA Part B. 4(2):3133–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MT976165]. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682858 SRR13205719 and SAMN17012931, respectively.