Abstract
From protein science, it is well understood that ordered folding and 3D structure mainly arises from balanced and noncovalent polar and nonpolar interactions, such as hydrogen bonding. Similarly, it is understood that single-chain polymer nanoparticles (SCNPs) will also compact and become more rigid with greater hydrophobicity and intrachain hydrogen bonding. Here, we couple high throughput photoinduced electron/energy transfer reversible addition-fragmentation chain-transfer (PET-RAFT) polymerization with high throughput small-angle X-ray scattering (SAXS) to characterize a large combinatorial library (>450) of several homopolymers, random heteropolymers, block copolymers, PEG-conjugated polymers, and other polymer-functionalized polymers. Coupling these two high throughput tools enables us to study the major influence(s) for compactness and flexibility in higher breadth than ever before possible. Not surprisingly, we found that many were either highly disordered in solution, in the case of a highly hydrophilic polymer, or insoluble if too hydrophobic. Remarkably, we also found a small group (9/457) of PEG-functionalized random heteropolymers and block copolymers that exhibited compactness and flexibility similar to that of bovine serum albumin (BSA) by dynamic light scattering (DLS), NMR, and SAXS. In general, we found that describing a rough association between compactness and flexibility parameters (Rg/Rh and Porod Exponent, respectively) with logP, a quantity that describes hydrophobicity, helps to demonstrate and predict material parameters that lead to SCNPs with greater compactness, rigidity, and stability. Future implementation of this combinatorial and high throughput approach for characterizing SCNPs will allow for the creation of detailed design parameters for well-defined macromolecular chemistry.
Graphical abstract
INTRODUCTION
Proteins are remarkable biopolymers that have evolved over millions of years to optimally perform several crucial functions such as transport, catalysis, and signaling. The primary structure or precise sequence of amino acids impacts the secondary and tertiary structures needed to properly perform a given task.1–4 Several proteins fold into specific 3D structures, giving rise to compact and ordered macromolecules, in order to carry out specific functions. Because proteins have been widely characterized, it is well understood that hydrophobic interactions are key to exceed the high configurational entropy associated with compaction.3, 5 In proteins, it has been theorized that about 30–80% hydrophobic character is required to form ordered structures.3 There is a fine balance between a highly hydrophilic structure that becomes extended due to its ability to interact with solvent and an extremely hydrophobic structure that aggregates out of solution. Hydrogen bonding is a secondary interaction that is also important for achieving the stable native state.6
The same types of interactions are considered for the design of single-chain polymer nanoparticles (SCNPs), which are single-chain polymers that fold by intramolecular cross-linking into collapsed polymers. SCNPs exhibit sizes on the order of proteins (< 20 nm in diameter, depending on the molecular weight).7–10 SCNPs are trending as a strong interest in polymer science with applications in drug delivery, biosensing, bioimaging, enzyme mimicry, and protein mimicry.7, 11–13 There is a need to control structural parameters such as flexibility and compactness in order to synthesize systems for these types of applications. In the realm of multivalent therapeutics, the binding affinity and selectivity of multivalent nanoparticles depends on compactness, flexibility, size, and ligand valency.14–16 While this has been shown for therapeutic biomolecules, it can be argued that these structural traits are important to control for other SCNP applications as well. Due to inefficiencies, much of the previous work in the SCNP space has focused on the synthesis of simple homopolymers such as poly(methyl acrylate) (PMA), polystyrene (PS), or poly(N-isopropryl acrylamide) (PNIPAM). This has prevented any in-depth experimental work to study SCNP compactness and flexibility.17 A rule-set has been established for understanding how SCNPs can be designed for compactness and greater ordering, mainly through molecular dynamics simulations. Three ways in which these structures become more ordered is through intrachain homocoupling, intrachain heterocoupling, and cross-linking-induced collapse.1, 18, 19 Similar to proteins, SCNPs require cross-linking in poor solvent conditions in order to achieve a high amount of compactness, as favorable solvent only induces local globule formation with substantial disorder.7, 13 In practice, the benefit of hydrophobicity to inducing order has been shown with various ABA triblock copolymers in which a hydrophobic core has enabled packed sphere formation.18, 20, 21 Additionally, supramolecular chemistry via hydrogen bonding aids in creating stable ordered structures.22–24
However, the major obstacles facing design of compact, ordered SCNPs are polydispersity and chain-to-chain compositional variations. These differences in molecular weight, solvent interactions, and random monomer compositions can limit order.12, 19 It is currently impossible to achieve sequence-level control of SCNPs, but with statistical control over monomer composition, it is possible to modulate blockiness and limit the coil-to-globule transition. Conversely, without this level of control, not only is there chain-to-chain variability but also intrachain variability, resulting in the formation of lower-ordered molten globules.25
In order to better understand how SCNPs can be designed to result in greater order and compactness, a combinatorial library of various polymers and functionalized polymers needs to be synthesized. However, traditional polymer synthesis techniques such as reversible addition-fragmentation chain-transfer polymerization (RAFT) and atom transfer radical polymerization (ATRP) tend to be inefficient due to requirements to degas and prevent oxygen from inhibiting the reaction.26 Over the past five years, three techniques have been established to enable synthesis of large combinatorial polymer libraries – enzyme-assisted RAFT (Enz-RAFT), air-tolerant ATRP, and photoinduced electron/energy transfer-RAFT (PET-RAFT).27–29 Enz-RAFT and air-tolerant ATRP utilize glucose oxidase which has the ability to degas the polymer reaction volume, enabling polymerizations in open-air conditions.27, 29, 30 Meanwhile, PET-RAFT incorporates zinc tetraphenylporphyrin (ZnTPP) to initiate the chain transfer agent (CTA) to begin polymerization and alter triplet oxygen to singlet oxygen that can be further trapped by DMSO.26, 28, 31–35 More recent work by the authors laid the groundwork for this paper which demonstrated the ability of PET-RAFT to synthesize a combinatorial library of linear, 3-arm, and 4-arm polymers functionalized with an acetylated azido-mannose via strain-promoted azide-alkyne cycloaddition (SPAAC) to screen for binding to concanavalin A.28
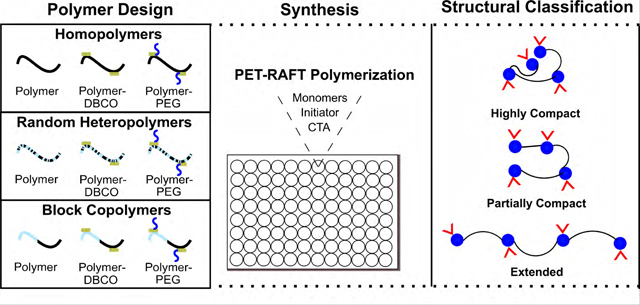
Here, we demonstrate a new addition to the SCNP structural design paradigm by coupling high throughput polymer synthesis with high throughput characterization. PET-RAFT was utilized as the SCNP synthesis platform as it is done in DMSO, which is amenable to several hydrophobic polymers needed for compactness.28, 33 Meanwhile, size-exclusion chromatography-multi-angle light scattering (SEC-MALS) enables rapid determination of molecular weight and dispersity (Ð). High throughput small-angle X-ray scattering (SAXS) at the National Synchrotron Light Source II Life Science X-ray Scattering (LiX) beamline36, 37 is also a unique tool that has aided in quantifying compactness and flexibility when used in parallel with dynamic light scattering (DLS) and diffusion ordered spectroscopy (DOSY) NMR. In this study, we synthesized over 450 diverse polymers with varied composition (homopolymers, random heteropolymers, and block copolymers), physicochemical characteristics (neutral, hydrophilic, hydrophobic, and charged), and a functionalization step (grafting of a pendant chain 2 kDa PEG-N3, 15 kDa PNIPAM-N3, or 2 kDa random heteropolymer HEA-PTMAEMA-nBA-N3) in order to survey this macromolecular landscape in high detail. The investigated SCNPs are not intramolecularly cross-linked but are self-interacting polymers whose dimensions and structural properties depend strongly on the nature of the solvent. Employing PET-RAFT and SAXS for synthesis and characterization of SCNPs has enabled us to efficiently create a diverse library of polymers with varying degrees of compactness and flexibility in a manner that has not been demonstrated before (Figure 1).
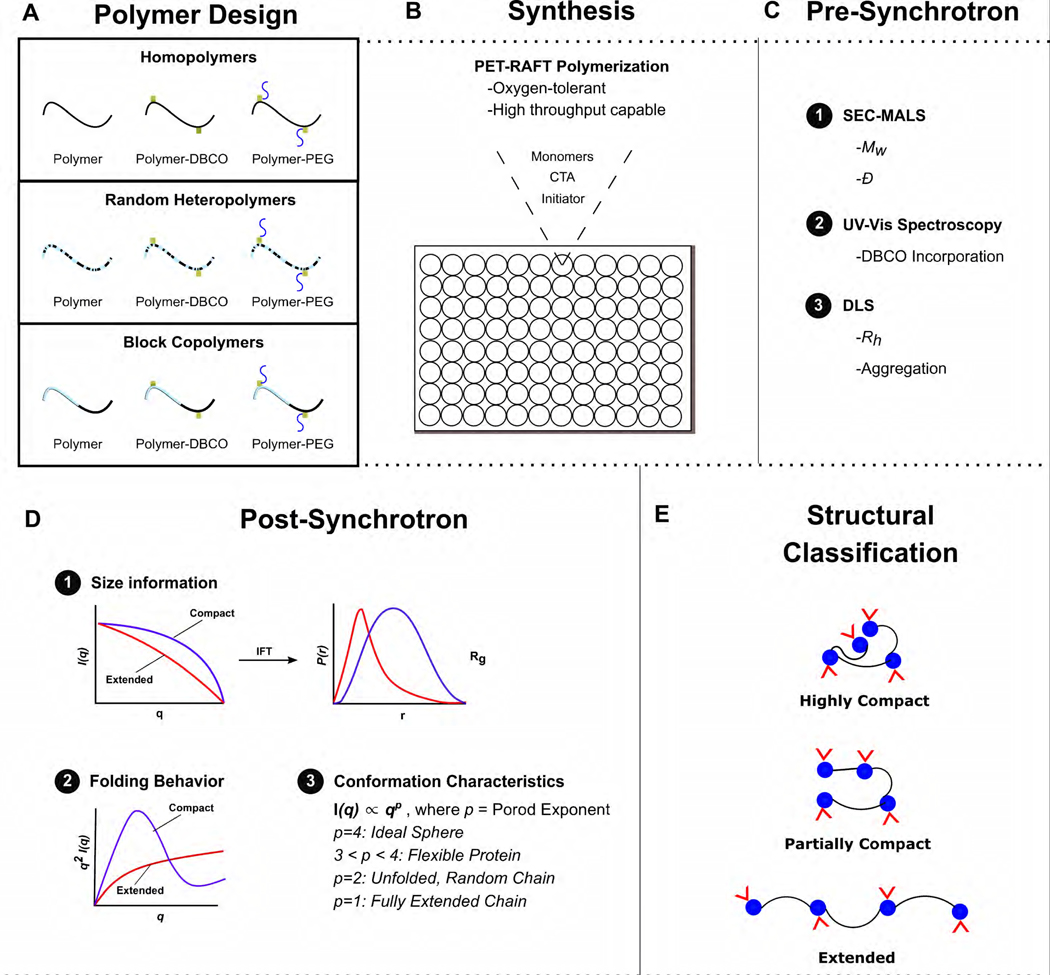
Figure 1. Experimental design schematic.
(A) The objective is to synthesize and characterize a variety of homopolymers, random heteropolymers, and block copolymers. (B) Using oxygen-tolerant polymerization, functional polymers can be efficiently synthesized in 96-well plates. (C) Further characterization was done by SEC-MALS, UV-Vis spectroscopy, and DLS to determine weight-average molecular weight (Mw), dispersity (Ð), and hydrodynamic radius (Rh). (D) By various SAXS data analysis techniques, radius of gyration (Rg) and Porod exponent can be quantified. (E) This enables the creation of a diverse polymer library with compact, partially compact, and extended structures along with varying degrees of flexibility.
EXPERIMENTAL
Materials.
Monomers were purchased from Sigma Aldrich, VWR, and Alfa Aesar and include 2-hydroxyethyl acrylate (HEA), 4-acryloylmorpholine (NAM), N-isopropylacrylamide (NIPAM), N,N-dimethyl acrylamide (DMA), 2-hydroxypropyl methacrylate (2-HPMA), 2-hydroxyethyl methacrylate (HEMA), methyl acrylate (MA), methyl methacrylate (MMA), n-butyl acrylate (nBA), [2-(methacryloyloxy)ethyl] trimethylammonium chloride solution (PTMAEMA), 2-methoxyethyl acrylate (mPEG acrylate), poly(ethylene glycol) monomethyl ether with Mn=350 Da and 500 Da (mPEG300, mPEG500), and acrylic acid N-hydroxysuccinimide ester (NHS acrylate). NHS acrylate was lightly sparged with argon before use in order to remove moisture and prevent hydrolysis.
The chain transfer agents (CTAs) utilized were 4-cyano-4-[(dodecylsulfanylthiocarbonyl)sulfanyl] pentanoic acid, 4-cyano-4-(phenylcarbonothioylthio) pentanoic acid, 2-(dodecylthiocarbonothioylthio) pentanoic acid, ethyl 2-(phenylcarbonothioylthio)-2-phenylacetate, and 2-(dodecylthiocarbonothioylthio)-2-methylpropionic acid 3-azido-1-propanol ester (for synthesizing HEA-PTMAEMA-nBA-N3), all purchased from Sigma Aldrich. The initiator was zinc tetraphenylporphyrin (ZnTPP) from Tokyo Chemical Industry Co., Ltd.
For post-polymerization functionalization, dibenzocyclooctyne-amine (DBCO-NH2) from Click Chemistry Tools, methoxypolyethylene glycol azide of Mn=2000 Da (PEG-N3), and PNIPAM azide of Mn=15,000 Da from Sigma Aldrich were purchased. Sephadex G-25 superfine resin (1000–5000 Da molecular weight cutoff range) from GE Healthcare Life Sciences was used to purify free DBCO-NH2, monomer, and initiator with 0.5 mL Zeba spin desalting columns (Thermo Fisher Scientific).
Polymerization Method.
Solutions of monomers (2 M) and CTA/initiator (50:0.5 mM) were made in DMSO, while NHS acrylate (2 M) was prepared in a DMSO/acetic acid mixture (1 mol equivalent acetic acid/NHS). Oxygen-tolerant PET-RAFT polymerizations were carried out in 96-well plates with a reaction volume of 200 μL. Plate sealing film was adhered onto the top of the plate to prevent evaporation or contamination and a 560 nm LED light was used to set off the polymerization (total reaction time was 15 hours).
Polymer Functionalization, Purification, and Preparation.
After polymerization was completed, DBCO-NH2 (1 mol equivalent/NHS) and dimethylaminopyridine (DMAP) (1 mol equivalent/NHS) were added to the polymer solution and left overnight (12 hours). Then spin purification was completed with Sephadex G-25 superfine resin (in DMSO) by packing and washing the resin at a centrifugation speed of 1000 g for 1 minute and purifying polymer and polymer-DBCO at 1000 g for 2 minutes. UV-Vis spectroscopy enabled quantification of percent DBCO incorporation, and PEG-N3, PNIPAM-N3, or HEA-PTMAEMA-nBA-N3 was then added (1 mol equivalent/DBCO). All samples were further dialyzed using Pierce 96-well Microdialysis Plates (Thermo Fisher Scientific) with 3500 Da molecular weight cutoff. Polymers and functionalized polymers were diluted 100x in water and lyophilized to a solid. This enabled absolute determination of concentration and buffer matching for SAXS experiments. Note: there were solubility concerns with most polymers functionalized with PNIPAM-N3 and HEA-PTMAEMA-nBA-N3 so this characterization data is not included in the paper.
logP Determination.
Log of the octanol-water partition coefficient (logP) was determined for each monomer type, DBCO-NH2, and PEG-N3 using the ChemAxon MarvinView logP plugin. Individual structures were created in ChemAxon MarvinSketch. The logP value was determined by an atomic hydrophobicity classification algorithm previously established,38 while this parameter has been used in the context of polymer science.39 For random heteropolymers and block copolymers, the weighted average of monomer and any other molecules (DBCO-NH2 or PEG-N3) was taken depending on the feed ratio (refer to Section S1.2 in Supporting Information).
SEC-MALS.
SEC was carried out with dimethylformamide (DMF) with LiBr as the eluent. A Phenomenex 5.0 μm guard column (50 × 7.5 mm) preceded two Phenomenex Phenogel columns (104 and 103 Å). In series with the columns were the Agilent 1200 Series differential refractive index (RI) detector, Agilent UV detector, and a Wyatt Technology miniDAWN TREOS MALS detector. Calibration of the SEC and normalization of the MALS detector was completed with Agilent PEO standards without correction. Polymers were prepared at 2 mg/mL in DMF and filtered with a 0.45 μm PTFE filter.
DLS.
DLS was completed on a Malvern Zetasizer Nano ZS with disposable polystyrene cuvettes. Polymers were prepared at 5 mg/mL in 99% PBS/1% DMSO from the lyophilized solid. A thermal equilibration time of 30 s was incorporated for each polymer at 25 °C. Diffusivity was measured by a size analysis protocol on the Zetasizer software, which was used to obtain hydrodynamic radius (Rh) by the Stokes-Einstein equation. The instrument had a 4 mW He-Ne laser with wavelength of 633 nm and scattering angle of 173°.
SAXS.
SAXS experiments were conducted at the National Synchrotron Light Source II (NSLS-II) at the Brookhaven National Lab (Upton, NY) beamline 16-ID for Life Science X-ray scattering (LiX). The instrument X-ray energy was 6.75 keV and employed three Pilatus 1M detectors and an autosampler for direct sample handling from PCR tubes. A secondary beam source was utilized along with refractive lenses for focusing. The q range used was 0.005–3.040 Å−1 (only 0.005–0.25 Å was taken as the small-angle region). Polymers at 5 mg/mL in 99% PBS/1% DMSO were exposed with 5-second exposures relative to buffer blanks. These five exposures were then averaged and the buffer scattering was subtracted.
SAXS Data Analysis.
From 1D SAXS intensity data size, compactness, and flexibility parameters and plots were obtained. BioXTAS RAW 1.5.0 was used to obtain radius of gyration (Rg) by Guinier analysis and Bayesian indirect Fourier transform (BIFT), while Kratky plots were used to determine a degree of compactness of the particle, i.e., the flexibility of the polymer chain.40, 41 The ATSAS 2.8.4 data analysis suite was utilized in RAW.42, 43
Porod analysis was carried out to quantify flexibility of the polymer chain. By following the deviations from Porod’s law, valid for particles with smooth surfaces:
| (1) |
where q is the momentum transfer or modulus of the scattering vector (q = 4π sin(θ) / λ), I(q)asym is the X-ray scattering intensity at high q, ρ is the average electron density of the scattered particles, ρ0 is the electron density of the buffer, and S is the total surface area. Deviations from this law that can be visualized by a Porod plot (Iq4 vs. q4) are used to quantify flexibility. These deviations were evaluated by calculating the scattering exponents in the equation:44–47
| (2) |
where Dm is the mass fractal dimension of the polymer chain. Dm captures the extent to which a linear chain (fractal dimension 1) fills a volume (fractal dimension 3), such that 1 ≤ Dm ≤ 3. Mass fractal values were quantified as the Porod exponent using the software package BIOISIS ScÅtter 3.0. First, the dataset was truncated to include only the hyperbolic Porod region. Then a linear range is set using a Porod-Debye plot (q4 I(q) vs. q4) in order to determine the Porod exponent.48, 49
NMR.
Translational diffusion of each polymer was measured by pulsed field gradient NMR experiments at 25 °C.50–52 NMR data were acquired using a Bruker AVANCE III 600 MHz spectrometer (Billerica, MA). Gradient strength was varied from 0.963–47.187 G/cm. For BSA, the total gradient time period was set to 2000 μs and the diffusion time was 0.4 s. For mPEG acrylate, the total gradient time period was set to 3500 μs and the diffusion time was 0.5 s. For RH 2-HPMA-nBA 10% PEG and BC 2-HPMA-MA 10% PEG, the total gradient time period was set to 1000 μs and the diffusion time was 0.15 s. The diffusion coefficient was derived for the methylene 1H peak of the polymers and the methyl 1H peak of BSA using the equation:
| (3) |
where D is the diffusion coefficient in m2/s, γ is the gyromagnetic radius of the observed nucleus, g is the gradient strength applied at each experimental point, Δ is the diffusion time, and δ is the total gradient time period. In order to account for differences in gradient length and diffusion time for each set of samples, this equation was re-expressed by taking the natural logarithm of both sides:
| (4) |
RESULTS AND DISCUSSION
First, polymers were synthesized by PET-RAFT and characterized by SEC-MALS and DLS (Table 1). SEC-MALS provided a molecular weight determination (weight-average molecular weight displayed throughout), while DLS and SAXS were used to quantify Rh and Rg, respectively. These polymers were homopolymers, random heteropolymers (RH), and block copolymers (BC) with nominal degree of polymerization of 400 and NHS acrylate/DBCO-NH2 incorporation of 10 mol%. DBCO-NH2 concentration was quantified by UV absorbance in Tables S3–S5 based on standard curve at 310 nm in Figure S4. For the definition and illustration of how the heteropolymers and block copolymers with 10% PEG are functionalized, refer to Section S1.1 and Figures S2–S3 in Supporting Information. Included are examples of the structures of RH 2-HPMA-nBA 10% PEG and BC 2-HPMA-MA 10% PEG. Molecular weight ranged from 20–70 kDa with Ð typically below 1.4. Note that molecular weight determined by MALS relied on dn/dc values, which were determined for each monomer individually and tabulated in Table S12. The overall sizes of each polymer are reported as Rg and Rh, ranging from about 7–20 nm and the logP is displayed for each polymer (lower value indicates hydrophilic while a higher value indicates hydrophobic). The proteins bovine serum albumin (BSA) and collagen type I are relatively more compact and extended, respectively.53, 54 In order to quantify the compactness of each polymer and account for molecular weight disparities, the Stokes ratio, or Rg/Rh, is used. A Stokes ratio of about 0.77 represents a compact or packed sphere, 1.00 amounts to a hollow sphere, and >1.00 constitutes an extended chain.55–58 Table 1 contains a representation of the most compact and extended polymers, but the structural information for other polymers studied along with the Flory scaling exponent (ν) can be viewed in Tables S6–S11.61 Additionally, confirmation of an increase in molecular weight due to PEG incorporation can be observed in Table S13 and Figures S12–S13.
Table 1.
Polymer Characterization Data by SEC-MALS, DLS, and SAXS
| # | Sample | logP | MWtheoretcal (Da) | MWGPC(Da) | MWMALS(Da) | ĐGPC | ĐMALS | Rg(nm) | Rh(nm) | Rg/Rh | Porod Exponent |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Collagen Type 1 | -- | 400,000 | -- | -- | -- | -- | 25.3 | 26.7 | 0.946 | 1.5 |
| 2 | NAM | −0.13 | 56,468 | 25,882 | 30,580 | 1.12 | 1.06 | 13.0 | 12.2 | 1.07 | 1.5 |
| 3 | mPEG acrylate | 0.33 | 52,056 | 20,097 | 27,140 | 1.14 | 1.06 | 19.3 | 13.9 | 1.38 | 1.4 |
| 4 | RH N1PAM-PTMAEMA | −0.042 | 49,046 | 41,395 | 36,480 | 1.40 | 1.04 | 23.5 | 25.4 | 0.92 | 1.5 |
| 5 | RH HEA-PTMAEMA | −0.18 | 50,140 | 36,566 | 42,020 | 1.28 | 1.10 | 15.0 | 12.4 | 1.22 | 1.5 |
| 6 | BSA | -- | 66,463 | -- | -- | -- | -- | 3.17 | 3.74 | 0.848 | 3.7 |
| 7 | RH 2-HPMA-nBA 10% PEG | 0.69 | 144,537 | 113,371 | 94,090 | 1.71 | 1.22 | 7.64 | 10.2 | 0.753 | 2.9 |
| 8 | BC 2-HPMA-MA 10% PEG | 0.53 | 118,618 | 112,472 | 94,830 | 1.71 | 1.19 | 8.45 | 10.2 | 0.827 | 2.4 |
The polymers with a more extended conformation with an Rg/Rh > 1.00 (mPEG acrylate, NAM, RH NIPAM-PTMAEMA, RH HEA-PTMAEMA) also display substantial flexibility with Porod exponent < 2. Meanwhile, compact polymers with an Rg/Rh < 0.90 (RH 2-HPMA-nBA 10% PEG and BC 2-HPMA-MA 10% PEG) display relatively greater rigidity (Porod exponent > 2.0). BSA has a larger Porod exponent when compared to these compact polymers, indicating that this protein is more rigid.
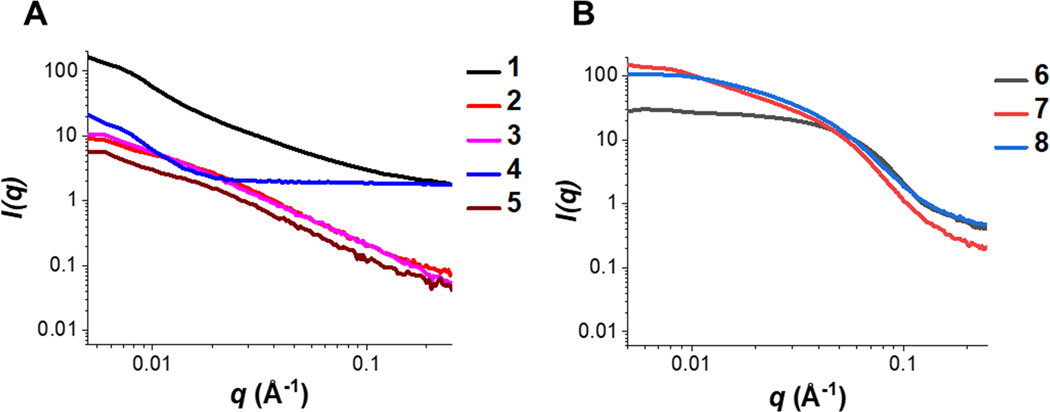
SAXS profiles are presented for extended polymers alongside collagen type I and compact polymers with BSA (Figure 2A–B). From these plots, a qualitative distinction between the two groups can be observed as the extended polymers (2–5) alongside collagen type I (1) exhibit a region at low q of linear decrease. Meanwhile, BSA (6) and compact polymers (7–8) exhibit a similar profile as BSA which is typical of structures with greater order.
Figure 2. SAXS intensity plots.
(A) SAXS intensity plots of collagen type I (1) with extended polymers NAM, mPEG acrylate, RH NIPAM-PTMAEMA, and RH HEA-PTMAEMA (2–5). (B) SAXS intensity plots of BSA (6) with compact polymers RH 2-HPMA-nBA 10% PEG and BC 2-HPMA-MA 10% PEG (7–8). Both groups of polymer scattering profiles overlap reasonably well with the respective proteins.
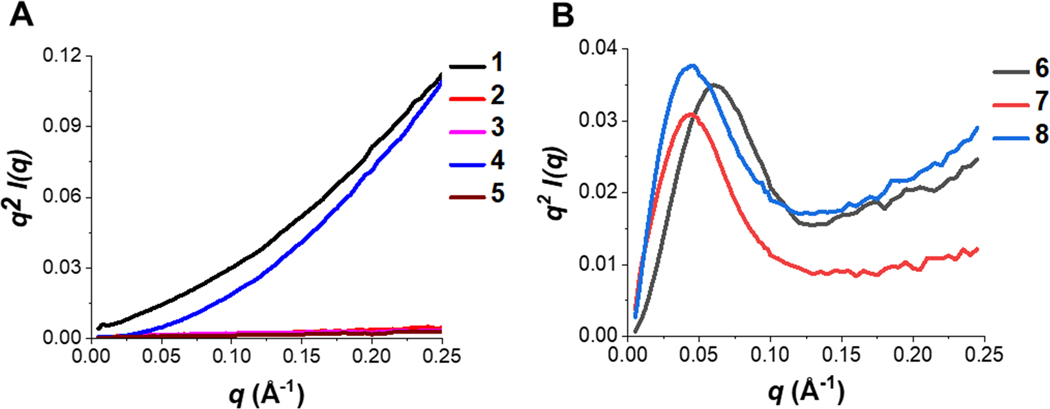
Kratky plots are presented for extended polymers (2–5) alongside collagen type I (1) and compact polymers (7–8) with BSA (6) (Figure 3A–B) to further support this data. The Kratky plots provide a qualitative way to compare compactness. The polymers that are similar in compactness to BSA are RH 2-HPMA-nBA 10% PEG (7) and BC 2-HPMA-MA 10% PEG (8), both of which contain a mixture of hydrophilic (2-HPMA and PEG) and hydrophobic (nBA/MA and DBCO) components.
Figure 3. Kratky plots for a qualitative assessment of compactness.
(A) Kratky plots of collagen type I (1) with extended polymers (2–5), as evidenced by the monotonically increasing profile. (B) Kratky plots of BSA (6) with compact polymers (7–8). Polymers 7–8 exhibit a peak at about q = 0.05 Å, signifying a compact structure.
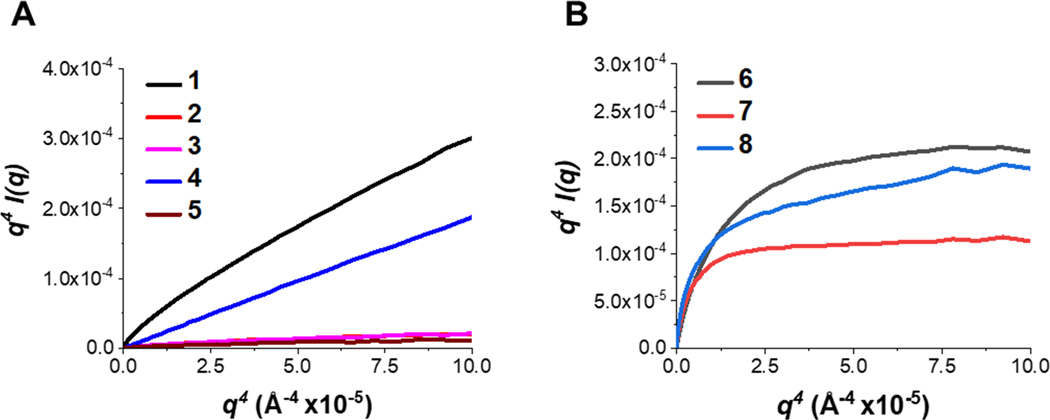
Porod plots further illustrate the differences in the conformation of the polymer chains (Figure 4). This curve increases monotonically for flexible, extended polymers (Figure 4A) while exhibiting an asymptotic Porod plateau for compact, folded polymers (Figure 4B). Collagen type I and similar polymers demonstrate an extended nature while BSA and its comparable polymers demonstrate a more compact conformation, as indicated by the behavior of the Porod-Debye plots.
Figure 4. Porod plots as a qualitative assessment of flexibility.
(A) Porod plots of collagen type I (1) with extended polymers (2–5). Polymers 2–5 are considered as flexible because they monotonically increase in the Porod q region. (B) Kratky plots of BSA (6) with compact polymers (7–8). Polymers 7–8 are considered as compact because they exhibit an asymptote in the Porod q region.
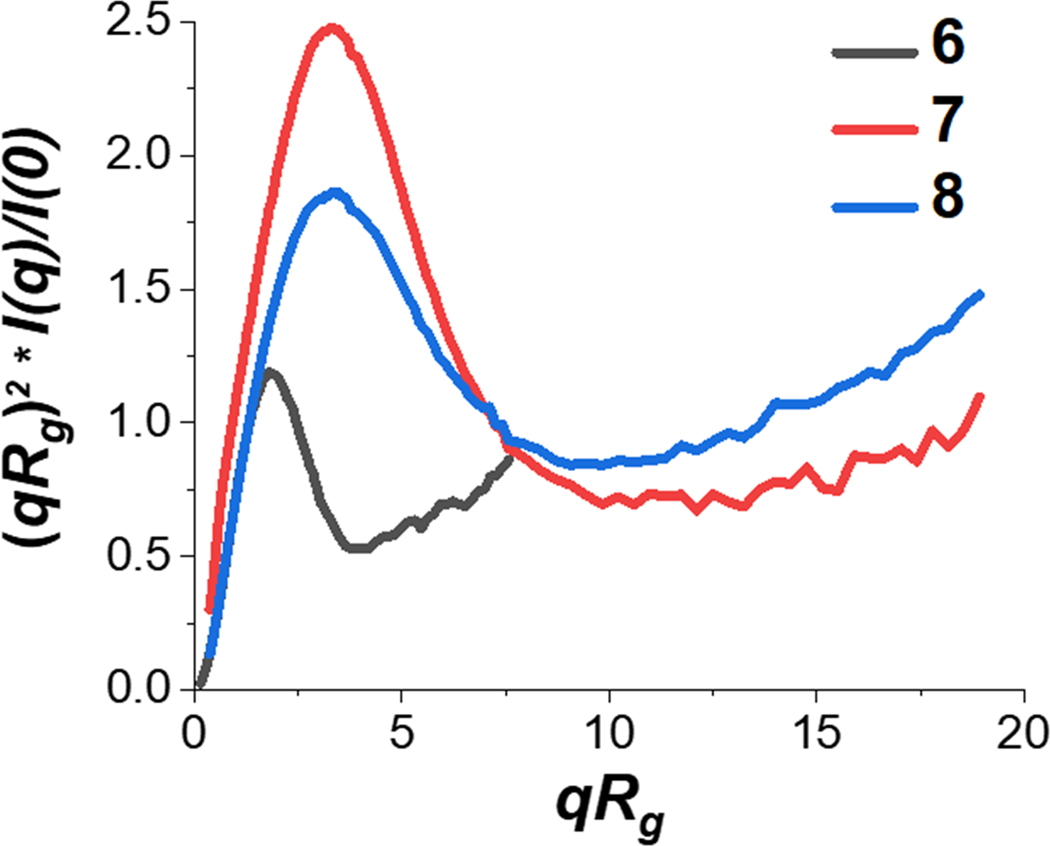
In further comparing the properties of BSA to other compact synthetic polymers, a normalized Kratky plot can be useful to highlight distinctions (Figure 5). In this normalized plot of (qRg)2 * I(q)/I(0) vs. qRg, the peak associated with BSA has a characteristic intensity of about 1.2 and qRg of about 2.5 for compact proteins. Both peaks associated with 6 and 7 demonstrate a slight shift in qRg and intensity, indicating that these polymers are more flexible compared to the compact protein.
Figure 5. Normalized Kratky plot.
Normalized Kratky plot for BSA (6) and compact polymers (7–8). BSA exhibits typical protein behavior with qRg of about 2.5 and intensity of about 1.2. The deviations of the 2 polymers indicates that they are more flexible than BSA.
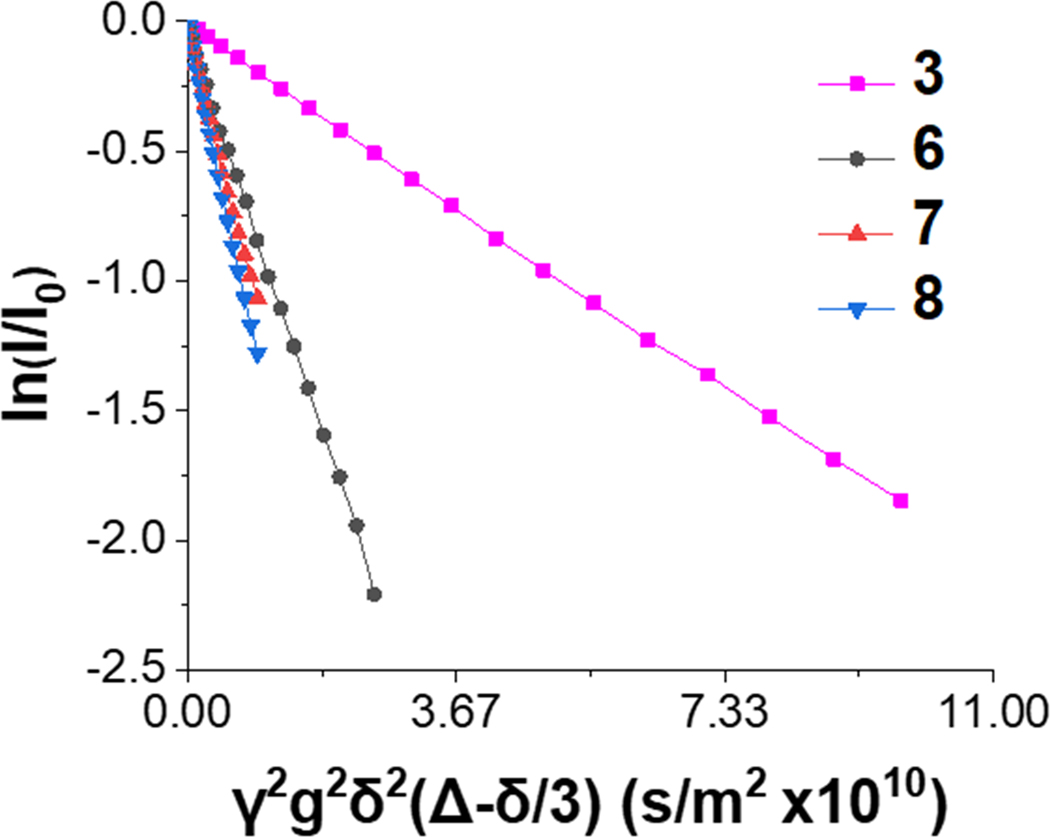
In order to confirm the compactness observed for the synthetic polymers 7 and 8, we conducted DOSY NMR experiments, which can be used to derive translational diffusion coefficients (D) in solution.59 By plotting the logarithmic normalization of the peak intensity vs. the experimental gradient strength, gradient time period, and diffusion time (Figure 6), the slope of the line is representative of the diffusion coefficient.50 We observe that the compact polymers have a diffusion coefficient similar to BSA, whereas the extended mPEG acrylate polymer exhibits much slower diffusion (Table 2). Further, the corresponding hydrodynamic radii were calculated by the Stokes-Einstein equation for a temperature of 25 °C and viscosity of 0.892 cP (Table 2). The Rh of extended mPEG acrylate was determined to be 12.54 nm, which contrasted with the Rh quantified for RH 2-HPMA-nBA 10% PEG (2.07 nm), BC 2-HPMA-MA 10% PEG (1.75 nm), and BSA (2.79 nm). DOSY NMR D summary and 1H-NMR can be viewed in Figures S7–S8.
Figure 6. NMR diffusion summary.
Plot of logarithmic normalization of peak intensity vs. experimental gradient strength, gradient time period, and diffusion time. Slope intensity correlates with diffusion coefficient.
AGradient time period was 1750 μs while the diffusion time was 0.5 s. BGradient time period was 1000 μs while the diffusion time was 0.4 s. CGradient time period was 1000 μs while the diffusion time was 0.15 s. The error listed is the sample standard deviation.
Table 2.
Diffusion and NMR Hydrodynamic Size Data
| # | Sample | D (x10-11 m2/s) | Rh (nm) |
|---|---|---|---|
| 3 | mPEG acrylate | 1.95 ± 0.005A | 12.54 ± 0.030A |
| 6 | BSA | 8.78 ± 0.069B | 2.79 ± 0.022B |
| 7 | RH 2-HPMA-nBA 10% PEG | 11.8 ± 0.038C | 2.07 ± 0.007C |
| 8 | BC 2-HPMA-MA 10% PEG | 14.0 ± 0.015C | 1.75 ± 0.002C |
Gradient time period was 3500 μs while the diffusion time was 0.5 s.
Gradient time period was 2000 μs while the diffusion time was 0.4 s.
Gradient time period was 2000 μs while the diffusion time was 0.15 s.
The error listed is the sample standard deviation.
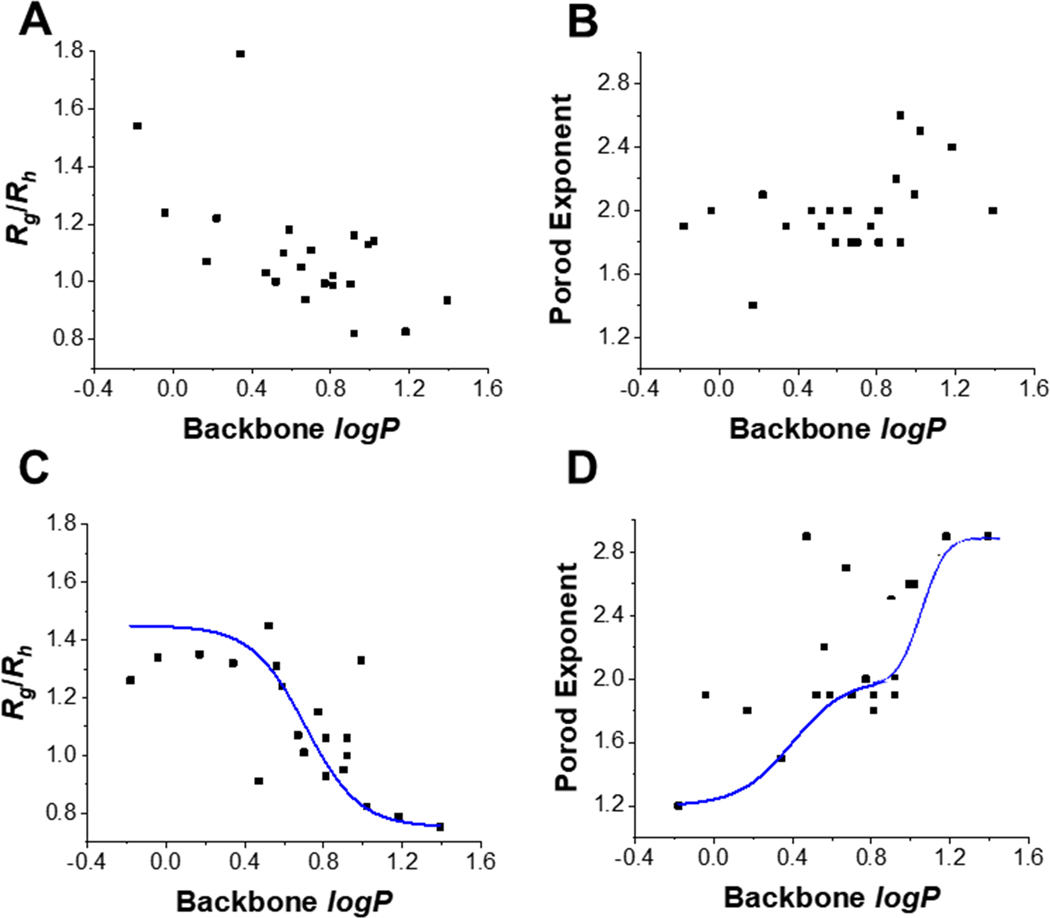
Further logP as a measure of hydrophobicity was plotted against compactness (Rg/Rh) and flexibility (Porod exponent) parameters to determine whether an association exists (Figure 7). This was done for block copolymers (Figures 7A–B) and for random heteropolymers (Figures 7C–D) in which all samples characterized were plotted to understand whether or not there is a global relationship that exists. Note that these figures take into account logP of the polymer backbone (only the monomeric composition was considered). Additional groups of logP plots can be found in Figures S9–S11. For Figures 7C–D, the blue curves indicate theoretical phase transitions that would describe the respective trends. This is supported by the literature but the exact model has not been defined here.60, 61
Figure 7. Relationship between hydrophobicity and compactness/flexibility for PEG- functionalized copolymers.
There is a roughly inverse relationship between hydrophobicity and Rg/Rh while there is a direct relationship between logP and Porod exponent. (A) Rg/Rh vs. backbone logP and (B) Porod exponent vs. backbone logP for all block copolymers functionalized with PEG. (C) Rg/Rh vs. backbone logP and (D) Porod exponent vs. backbone logP for all random heteropolymers functionalized with PEG. Theoretical two-phase and three-phase curve fits are included for (C) and (D), respectively.
The overall goal of this study was to utilize high throughput PET-RAFT and SAXS in order to explore the compositional landscape of synthetic polymers and demonstrate that polymers with varying degrees of flexibility and compactness can be synthesized. If this approach is used for bioactive molecules, a structure-activity relationship can be established for therapeutics in the space of tissue engineering and regenerative medicine. In the past, other groups have attempted to synthesize bioinspired polymers that are globular by creating polymers with varying amounts of hydrophobicity in the side groups.1, 62, 63 While polymers cannot be synthesized to closely resemble protein structure since they lack higher order secondary structure5, 6, they can still be synthesized to exhibit varying degrees of intermediate compactness and flexibility as evidenced by this work.
Biophysical characterization of polymers summarized in Table 1 was completed by GPC, MALS, DLS, and SAXS. We identified compact and extended polymeric nanomaterials from the entire polymer library by combining size information in Table 1 with qualitative compactness indications from Kratky plots in Figures 3A–B. However, in order to account for the differences in molecular weight between polymers, which would invariably lead to changes in size, the Stokes ratio (Rg/Rh) enabled normalization of polymers to quantify compactness with a single value. By comparing macromolecular hydrodynamic associations with geometric properties, compact structures can be differentiated from extended structures that may be random coils or rod-like in nature.
The two compact polymers we studied in more detail were RH 2-HPMA-nBA 10% PEG (7) and BC 2-HPMA-MA 10% PEG (8). The Kratky and normalized Kratky plots (Figures 3B and 5) indicate that, while the two polymers are compact, they are slightly more flexible when compared to BSA. This is further supported by the quantities displayed in Table 1, which demonstrates that the Stokes ratios of RH 2-HPMA-nBA 10% PEG and BC 2-HPMA-MA 10% PEG are 0.753 and 0.827, indicative of a compact structure. However, the Porod exponents of both polymers are markedly lower (2.9 and 2.4, respectively) when compared to BSA (3.7), confirming greater flexibility. This evidence indicates that these two polymers are at an intermediate level of compactness. In order to understand what is causing this, we listed biophysical quantities for polymer backbones that are extremely hydrophilic (low logP values of −0.042 and −0.18) (RH HEA-PTMAEMA and RH NIPAM-PTMAEMA) in Table 1. Both are extended and flexible as demonstrated by Rg/Rh > 1.00 and Porod exponent < 2. The polymers themselves are hydrophilic enough to remain extended in aqueous buffer, and we believe that the incorporation of hydrophobic DBCO-NH2 and subsequent functionalization of hydrophilic PEG-N3 for certain polymer compositions may result in the formation of a hydrophobic core and solvent-interacting PEGylated regions to promote compactness and solubility. This is supported by some experimental20 and MD simulation work13 that have proven the usefulness in designing SCNPs with both hydrophilic and hydrophobic components in order to synthesize complex structures.
In order to understand what may be causing compactness and rigidity, we attempted to determine whether or not logP plays a role as it is a measure of this hydrophobicity that is crucial for ordered SCNPs.38, 39 logP was determined for these complex polymers by taking the weighted average of monomeric components and accounting for the concentration of NHS acrylic acid, DBCO-NH2, and PEG-N3. logP is not often used for large macromolecules such as SCNPs so this is a method that can be improved upon. Overall relationships of logP for all of the polymers was shown in Figures S9–S11 while relationships between polymer backbone logP and compactness or flexibility are displayed in Figure 7. logP is shown from this work to have a stronger association in the random heteropolymer SCNPs as compared to block copolymers and seems to have an inverse relationship with Rg/Rh and positive association for Porod exponent. This confirms our previous intuitions that hydrophobic character is necessary to result in compact and rigid SCNPs.
Further differences are observed between block copolymers and random heteropolymers in terms of changes in Rg/Rh and Porod exponent with respect to logP. For random heteropolymers, there are two-phase and three-phase transitions displayed for the trends associated with Rg/Rh and Porod exponent, respectively (Figures 7C–D). These types of relationships are referred to in the literature for changes in Rg/Rh and the Flory scaling law during the coil-to-globule transition.60, 61 These types of structural parameters also tend to occur as a range of values rather than on the extremes as an “all or none” response (also supported by data in Figure 7 and Tables S6–S11). On the other hand, this phase transition is not as strong for block copolymers (Figures 7A–B). The importance of hydrophobicity, specifically due to hydrophobic interactions associated with the alkyl chain, has been demonstrated in amphiphilic random heteropolymers.64 It is not surprising that this is the case as proteins are random assortments of amino acids.3, 65 By creating a separate block of hydrophilic monomer (uneven distribution throughout the chain), we believe that some complexity is lost due to differences in self-association. For Rg/Rh, 60% of block copolymers are within the narrow range of 1.00–1.25, while only 27% of random heteropolymers fall within this range. Similarly, for Porod exponent, 68% of block copolymers are within the narrow range of 1.7–2.0 while just 45% of random heteropolymers are within this range.
We could also utilize this synthesis and characterization approach to study the effects of functionalization (in this case PEGylation) on the structure of SCNPs and how this varies between different groups. Observations can be made for SCNPs containing 90% of neutral component (HEA, NIPAM, 2-HPMA, and HEMA in Tables S8–S11). For example, PEGylation has a distinct effect on HEA polymers in contrast to NIPAM ones. For PEGylated HEA copolymers, the majority experience a larger size and greater rigidity while the majority of NIPAM PEGylated copolymers either experience a smaller size and greater rigidity or vice versa. This may be due to hydroxyl groups gaining greater solvent exposure in HEA copolymers due to PEG stabilization and thus engaging in more hydrogen bonding, but this mechanism would have to be studied more in-depth and with more data points. Likewise, differences in PEGylation between random heteropolymers and block copolymers can be examined. For 2-HPMA and NIPAM-based copolymers, the Porod exponent tends to decrease when comparing random heteropolymers to block copolymers. This indicates that the spacing associated with neutral components that can hydrogen bond (2-HPMA and NIPAM) along with charged or hydrophobic components is crucial. Surprisingly, HEA and HEMA-based polymers did not show a trend in Porod exponents between random and block copolymers. The major difference between these monomers and 2-HPMA is the alkyl chain substituent present (ethyl and propyl groups, respectively) so this may affect intra-chain cross-linking.
As displayed in Tables S9–S11, the only other polymers determined to have both compactness (Rg/Rh < 1.00) and rigidity (Porod exponent > 2) were RH 2-HPMA-MMA 10% PEG, RH 2-HPMA-MA 10% PEG, RH HEMA-nBA 10% PEG, BC HEMA-MMA 10% PEG, RH NIPAM-nBA 10% PEG, and BC NIPAM-nBA 10% PEG, and NIPAM-MMA. In an overwhelming majority (all except BC NIPAM-nBA) of these cases (including for polymers 7–8 in Table 1), there is 10% of the hydrophobic monomer (nBA, MA, or MMA) along with PEG functionalization. This gives the polymer complex hydrophobic character and induces self-interactions. Further, the neutral component of these polymers (2-HPMA, HEMA, or NIPAM) contains a hydroxyl or amine group, enabling non-covalent interactions to stabilize the SCNP. Most polymers were both extended and flexible while there was a small group (9 polymers) that exhibited either rigidity or compactness but not both. In the future, high throughput DLS (DynaPro Plate Reader, Wyatt Technology) can be utilized in order to measure Rh at various concentrations and in different solvent conditions. This would also improve the efficiency of our current characterization.
The greatest obstacles for polymer engineering as cited by Barron and colleagues is a lack of reproducibility and inability to design polymers in a sequence-specific manner.66 Our current system utilizing PET-RAFT for high throughput synthesis at the benchtop minimizes batch-to-batch variability and enables precise control of molecular weight.28 Even though we may not be able to achieve sequence-specific control, we can still maintain statistical control over polymer design. Others have demonstrated the ability to precisely control copolymer compositions for RAFT, ATRP, and nitroxide-mediated polymerization (NMP) using the Mayo-Lewis model and reactivity ratios of individual monomers.67, 68 By doing so, we could improve upon the order of hydrophobic random heteropolymers and block copolymers.25 Other possible directions may include conducting polymerizations in different solvents by using modified oxygen quenching techniques,32 further controlling polymer stereochemistry,69 and synthesizing polymers with non-linear geometries.28, 70 Overall, this work has demonstrated the ability to solve some challenges in the synthesis and design of SCNPs, namely incorporating techniques that can be scaled up, functionalizing SCNPs, and improving characterization.71 We believe that more complexity in SCNP design can be uncovered by utilizing a simulation-based approach to complement this work.
CONCLUSIONS
In conclusion, we demonstrated that our approach can be used to efficiently examine compactness and flexibility of SCNP combinatorial libraries. We were able to confirm that a rough correlation exists between hydrophobicity (logP) and the parameters for compactness and flexibility (Rg/Rh and Porod exponent) in studying polymers functionalized with PEG. We observed an inverse phase-transition relationship between logP and compactness along with a direct phase-transition relationship between logP and flexibility (much stronger for random heteropolymers). In the future, this type of approach when combined with molecular dynamics simulations can be utilized to further study structural parameters and devise more detailed design criteria for SCNPs.
Supplementary Material
ACKNOWLEDGMENT
R.U. acknowledges the support of the National Institute of General Medical Sciences of the National Institutes of Health under award number T32 GM008339. R.U. acknowledges Lin Yang, beamline scientist at NSLS-II beamline 16-ID for Life Science X-ray Scattering (LiX), for his assistance with conducting experiments at Brookhaven National Laboratory. The LiX beamline is part of the Life Science Biomedical Technology Research resource, jointly supported by the National Institute of Health (NIH), National Institute of General Medical Sciences under Grant P41 GM111244, and by the Department of Energy Office of Biological and Environmental Research under Grant KP1605010, with additional support from NIH Grant S10 OD012331. NSLS-II is a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. JK and SM were supported by “RESBIO” – the Integrated Resource for Polymeric Biomaterials (NIH Grant P41 EB001046). JB was supported by NIH grant R01 GM110577.
Funding Sources
This work was supported by a Busch Biomedical Award to A.J.G.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Additional characterization data, results, and schematics can be found in Supporting Information. The Supporting Information is available free of charge on the ACS Publications website.
REFERENCES
- (1).Pomposo JA Bioinspired single-chain polymer nanoparticles. Polym Int. [Online] 2014, 63 (4), 589–592. 10.1002/pi.4671 (September 11, 2019). [DOI] [Google Scholar]
- (2).Dill KA; MacCallum JL The protein-folding problem, 50 years on. Science [Online] 2012, 338 (6110), 1042–1046. 10.1126/science.1219021 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (3).Chan HS; Dill KA “Sequence space soup” of proteins and copolymers. J. Chem. Phys [Online] 1991, 95, 3775–3787. 10.1063/1.460828 (September 11, 2019). [DOI] [Google Scholar]
- (4).Lyon C; Prasher A; Hanlon A; Tuten B; Tooley C; Frank P; Berda E A Brief User’s Guide to Single-chain Nanoparticles. Polym. Chem [Online] 2015, 6 (2), 181–197. 10.1039/C4PY01217H (September 11, 2019). [DOI] [Google Scholar]
- (5).Dill KA Theory for the folding and stability of globular proteins. Biochemistry [Online] 1985, 24 (6), 1501–1509. 10.1021/bi00327a032 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (6).Dill KA Polymer principles and protein folding. Protein Sci. [Online] 1999, 8 (6), 1166–80. 10.1110/ps.8.6.1166 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Rabbel H; Breier P; Sommer J Swelling Behavior of Single-Chain Polymer Nanoparticles: Theory and Simulation. Macromolecules [Online] 2017, 50, 7410–7418. 10.1021/acs.macromol.7b01379 (September 11, 2019). [DOI] [Google Scholar]
- (8).Berda EB; Foster EJ; Meijer EW Toward Controlling Folding in Synthetic Polymers: Fabricating and Characterizing Supramolecular Single-Chain Nanoparticles. Macromolecules [Online] 2010, 43, 1430–1437. 10.1021/ma902393h (September 11, 2019). [DOI] [Google Scholar]
- (9).Li G; Tao F; Wang L; Li Y; Bai R A facile strategy for preparation of single-chain polymeric nanoparticles by intramolecular photo-crosslinking of azide polymers. Polymer [Online] 2014, 55 (16), 3696–3702. 10.1016/j.polymer.2014.05.064 (September 11, 2019). [DOI] [Google Scholar]
- (10).Pomposo JA Single-Chain Polymer Nanoparticles: Synthesis, Characterization, Simulations, and Applications. Wiley-VCH: Weinheim, Germany: 2017. [Google Scholar]
- (11).Perez-Baena I; Barroso-Bujans F; Gasser U; Arbe A; Moreno AJ; Colmenero J; Pomposo JA Endowing Single-Chain Polymer Nanoparticles with Enzyme-Mimetic Activity. ACS Macro Lett. [Online] 2013, 2, 775–779. 10.1021/mz4003744 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (12).Cole JP; Hanlon AM; Rodriguez KJ; Berda EB Protein-Like Structure and Activity in Synthetic Polymers. J. Polym. Sci., Part A: Polym. Chem [Online] 2017, 55, 191–206. 10.1002/pola.28378 (September 11, 2019). [DOI] [Google Scholar]
- (13).Lo Verso F; Pomposo JA; Colmenero J; Moreno AJ Simulation Guided Design of Globular Single-Chain Nanoparticles by Tuning the Solvent Quality. Soft Matter [Online] 2010, 11 (7), 1369–1375. 10.1039/C4SM02475C (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (14).Tjandra KC; Thordarson P Multivalency in Drug Delivery-When Is It Too Much of a Good Thing? Bioconjugate Chem. [Online] 2019, 30 (3), 503–514. 10.1021/acs.bioconjchem.8b00804 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (15).Mammen M; Choi SK; Whitesides GM Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem., Int. Ed. Engl [Online] 1998, 37 (20), 2755–2794. (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (16).Ehrlich PH Effect of Multivalency on the Specificity of Protein and Cell-Interactions. J. Theor. Biol [Online] 1979, 81 (1), 123–127. 10.1016/0022-5193(79)90085-7 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (17).Pomposo JA, Perez-Baena I, Lo Verso F, Moreno AJ, Arbe A, Colmenero J How Far are Single-Chain Polymer Nanoparticles in Solution from the Globular State? ACS Macro Lett. [Online] 2014, 3, 767–772. 10.1021/mz500354q (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (18).Lo Verso F; Pomposo JA; Colmenero J; Moreno AJ Multi-orthogonal folding of single polymer chains into soft nanoparticles. Soft Matter [Online] 2014, 10 (27), 4813–4821. 10.1039/C4SM00459K (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (19).Moreno AJ; Lo Verso F; Sanchez-Sanchez A; Arbe A; Colmenero J; Pomposo JA Advantages of Orthogonal Folding of Single Polymer Chains to Soft Nanoparticles. Macromolecules [Online] 2013, 46, 9748–9759. 10.1021/ma4021399 (September 11, 2019). [DOI] [Google Scholar]
- (20).Murthy NS; Zhang Z; Borsadia S; Kohn J Nanospheres with a smectic hydrophobic core and an amorphous PEG hydrophilic shell: structural changes and implications for drug delivery. Soft Matter [Online] 2018, 14 (8), 1327–1335. 10.1039/C7SM02472J (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Elacque E; Croom A; Lye DS; Weck M Coil-Helix and Sheet-Helix Block Copolymers via Macroinitiation from Telechelic ROMP Polymers. J. Polym. Sci., Part A: Polym. Chem [Online]. 2017, 55, 2991–2998. 10.1002/pola.28542 (September 11, 2019). [DOI] [Google Scholar]
- (22).Chen S; Rocher M; Ladavière C; Gérard JF; Lortie F, Bernard J AB/ABC/ABCD supramolecular block copolymers from Hamilton wedge and barbiturate-functionalized RAFT agents. Polym. Chem [Online] 2012, 3 (11), 3157–3165. 10.1039/C2PY20340E (September 11, 2019). [DOI] [Google Scholar]
- (23).Altintas O; Artar M; Ter Huurne G; Voets IK; Palmans ARA; Barner-Kowollik C; Meijer EW Design and Synthesis of Triblock Copolymers for Creating Complex Secondary Structures by Orthogonal Self-Assembly. Macromolecules [Online] 2015, 48, 8921–8932. 10.1021/acs.macromol.5b01990 (September 11, 2019). [DOI] [Google Scholar]
- (24).van Gorp JJ; Vekemans JA; Meijer EW Facile synthesis of a chiral polymeric helix; folding by intramolecular hydrogen bonding. Chem. Commun [Online] 2004, 60–61. 10.1039/B312407J (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (25).Rosales AM; Segalman RA; Zuckermann RN Polypeptoids: a model system to study the effect of monomer sequence on polymer properties and self-assembly. Soft Matter [Online] 2013, 9, 8400–8414. 10.1039/C3SM51421H (September 11, 2019). [DOI] [Google Scholar]
- (26).Yeow J; Chapman R; Gormley AJ; Boyer C Up in the air: oxygen tolerance in controlled/living radical polymerisation. Chem. Soc. Rev [Online] 2018, 47 (12), 4357–4387. 10.1039/C7CS00587C (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chapman R; Gormley AJ; Stenzel MH; Stevens MM Combinatorial Low-Volume Synthesis of Well-Defined Polymers by Enzyme Degassing. Angew. Chem., Int. Ed. Engl [Online] 2016, 55 (14), 4500–4503. 10.1002/anie.201600112 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (28).Gormley AJ; Yeow J; Ng G; Conway O; Boyer C; Chapman R An Oxygen-Tolerant PET-RAFT Polymerization for Screening Structure-Activity Relationships. Angew. Chem., Int. Ed. Engl [Online] 2018, 57 (6), 1557–1562. 10.1002/anie.201711044 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Enciso AE; Fu L; Russell AJ; Matyjaszewski KA A Breathing Atom-Transfer Radical Polymerization: Fully Oxygen-Tolerant Polymerization Inspired by Aerobic Respiration of Cells. Angew. Chem., Int. Ed. Engl [Online] 2018, 57 (4), 933–936. 10.1002/anie.201711105 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (30).Chapman R; Gormley AJ; Herpoldt K; Stevens MM Highly Controlled Open Vessel RAFT Polymerizations by Enzyme Degassing. Macromolecules [Online] 2014, 47, 8541–8547. 10.1021/ma5021209 (September 11, 2019). [DOI] [Google Scholar]
- (31).Oliver S; Zhao L; Gormley AJ; Chapman R; Boyer C Living in the Fast Lane-High Throughput Controlled/Living Radical Polymerization. Macromolecules [Online] 2018, 52 (1), 3–23. 10.1021/acs.macromol.8b01864 (September 11, 2019). [DOI] [Google Scholar]
- (32).Ng G; Yeow J; Xu JT; Boyer C Application of oxygen tolerant PET-RAFT to polymerization-induced self-assembly. Polym. Chem [Online] 2017, 8 (18), 2841–2851. 10.1039/C7PY00442G (September 11, 2019). [DOI] [Google Scholar]
- (33).Yeow J; Chapman R; Xu JT; Boyer C Oxygen tolerant photopolymerization for ultralow volumes. Polym. Chem [Online] 2017, 8 (34), 5012–5022. 10.1039/C7PY00007C (September 11, 2019). [DOI] [Google Scholar]
- (34).Xu J; Jung K; Atme A; Shanmugam S; Boyer C A robust and versatile photoinduced living polymerization of conjugated and unconjugated monomers and its oxygen tolerance. J. Am. Chem. Soc [Online] 2014, 136 (14), 5508–19. 10.1021/ja501745g (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (35).Ng G; Yeow J; Chapman R; Isahak N; Wolvetang E; Cooper-White JJ; Boyer C Pushing the Limits of High Throughput PET-RAFT Polymerization. Macromolecules [Online] 2018, 51 (19), 7600–7607. 10.1021/acs.macromol.8b01600 (September 11, 2019). [DOI] [Google Scholar]
- (36).Allaire M; Yang L Biomolecular solution X-ray scattering at the National Synchrotron Light Source. J. Synchrotron Rad [Online] 2011, 18 (1), 41–4. 10.1107/S0909049510036022 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).DiFabio J; Chodankar S; Pjerov S; Jakoncic J; Lucas M; Krywka C; Graziano V; Yang L The Life Science X-ray Scattering Beamline at NSLS-II. AIP Conf. Proc [Online] 2016, 1741, 1–5. 10.1063/1.4952872 (September 11, 2019). [DOI] [Google Scholar]
- (38).Vellarkad NV, Ghose AK, Revankar GR, Robins RK Atomic Physicochemical Parameters for Three Dimensional Structure Directed Quantitative Structure-Activity Relationships. 4. Additional Parameters for Hydrophobic and Dispersive Interactions and Their Application for an Automated Superposition of Certain Naturally Occurring Nucleoside Antibiotics. J. Chem. Inf. Comput. Sci [Online] 1989, 29 (3), 163–172. 10.1021/ci00063a006 (September 11, 2019). [DOI] [Google Scholar]
- (39).Richards SJ; Jones A; Tomas RMF; Gibson MI Photochemical “In-Air” Combinatorial Discovery of Antimicrobial Copolymers. Chem.--Eur. J [Online] 2018, 24 (52), 13758–13761. 10.1002/chem.201802594 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hopkins JB; Gillilan RE; Skou S BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr [Online] 2017, 50, 1545–1553. 10.1107/S1600576717011438 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Nielsen SS; Toft KN; Snakenborg D; Jeppesen MG; Jacobsen JK; Vestergaard B; Kutter JP; Arleth L BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J. Appl. Crystallogr [Online] 2009, 42, 959–964. 10.1107/S0021889809023863 (September 11, 2019). [DOI] [Google Scholar]
- (42).Franke D; Petoukhov MV; Konarev PV; Panjkovich A; Tuukkanen A; Mertens HDT; Kikhney AG; Hajizadeh NR; Franklin JM; Jeffries CM; Svergun DI ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr [Online] 2017, 50, 1212–1225. 10.1107/S0021889809023863 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Petoukhov MV; Franke D; Shkumatov AV; Tria G; Kikhney AG; Gajda M; Gorba C; Mertens HDT; Konarev PV; Svergun DI New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr [Online] 2012, 45, 342–350. 10.1107/S0021889812007662 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Murthy NS; Knox JR On Solué- Porod Plots of Protein X-Ray Scattering Data. J. Appl. Crystallogr [Online] 1977, 10, 137–140. 10.1107/S0021889877013120 (September 11, 2019). [DOI] [Google Scholar]
- (45).Martin JE; Hurd AJ Scattering from Fractals. J. Appl. Crystallogr [Online] 1987, 20, 61–78. 10.1107/S0021889887087107 (September 11, 2019). [DOI] [Google Scholar]
- (46).Besselink R; Stawski TM; Van Driessche AES; Benning LG Not just fractal surfaces, but surface fractal aggregates: Derivation of the expression for the structure factor and its applications. J. Chem. Phys [Online] 2016, 145 (21), 1–11. 10.1063/1.4960953 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (47).Teixeira J Small-Angle Scattering by Fractal Systems. J. Appl. Crystallogr [Online] 1988, 21, 781–785. 10.1107/S0021889888000263 (September 11, 2019). [DOI] [Google Scholar]
- (48).Rambo RP; Tainer JA Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers [Online] 2011, 95 (8), 559–571. 10.1002/bip.21638 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Hura GL; Menon AL; Hammel M; Rambo RP; Poole II FL.; Tsutakawa SE; Jenny FE Jr.; Classen S; Frankel KA; Hopkins RC; Yang S; Scott JW; Dillard BD; Adams MWW; Tainer JA Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods [Online] 2009, 6, 606–612. 10.1038/nmeth.1353 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Stejskal EO; Tanner JE Spin Diffusion Measurements: Spin Echoes in the Presence of a Time‐Dependent Field Gradient. J. Chem. Phys [Online] 1965, 42, 288–292. 10.1063/1.1695690 (September 11, 2019). [DOI] [Google Scholar]
- (51).Johnson CS Jr. Diffusion ordered nuclear magnetic resonance spectroscopy: principles and applications. Prog. Nucl. Magn. Reson. Spectrosc [Online] 1999, 34, 203–256. 10.1063/1.1695690 (September 11, 2019). [DOI] [Google Scholar]
- (52).Wu DH; Chen AD; Johnson CS Jr. An Improved Diffusion-Ordered Spectroscopy Experiment Incorporating Bipolar-Gradient Pulses. J. Magn. Reson., Series A [Online] 1995, 115 (2), 260–264. 10.1016/s0079-6565(99)00003–5 (September 11, 2019). [DOI] [Google Scholar]
- (53).Murthy NS; Knox JR Hydration of Proteins: SAXS Study of Native and Methoxy Polyethyleneglycol (mPEG)-Modified L-Asparaginase and Bovine Serum Albumin in mPEG Solutions. Biopolymers [Online] 2004, 74, 457–466. 10.1002/bip.20091 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (54).Oechsle AM; Wittmann X; Gibis M; Kohlus R; Weiss J Collagen entanglement influenced by the addition of acids. Eur. Polym. J [Online] 2014, 58, 144–156. 10.1016/j.eurpolymj.2014.06.015 (September 11, 2019). [DOI] [Google Scholar]
- (55).Voulgaris D; Tsitsilianis C Aggregation behavior of polystyrene/poly(acrylic acid) heteroarm star copolymers in 1,4-dioxane and aqueous media. Macromol. Chem. Phys [Online] 2001, 202 (17), 3284–3292. 10.1002/1521-3935(20011101)202:17<3284::AID-MACP3284>3.0.CO;2–0 (September 11, 2019). [DOI] [Google Scholar]
- (56).Roovers J; Martin JE The Hard-Sphere Model for Linear and Regular Star Polybutadienes. J. Polym. Sci., Part B: Polym. Phys [Online] 1989, 27, 2513–2524. 10.1002/polb.1989.090271209 (September 11, 2019). [DOI] [Google Scholar]
- (57).Ishizu K Architecture of Multi-Component Copolymer Brushes: Synthesis, Solution Properties and Applications for Nanodevices. Polym. J [Online] 2004, 36 (10), 775–792. 10.1295/polymj.36.77 (September 11, 2019). [DOI] [Google Scholar]
- (58).Nygaard M; Kragelund BB; Papaleo E; Lindorff-Larsen K An Efficient Method for Estimating the Hydrodynamic Radius of Disordered Protein Conformations. Biophys. J 2017, 113, 550–557. 10.1016/j.bpj.2017.06.042 (September 11, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Pagès G; Gilard V; Martino R; Malet-Martino M Pulsed-field gradient nuclear magnetic resonance measurements (PFG NMR) for diffusion ordered spectroscopy (DOSY) mapping. Analyst [Online] 2017, 142, 3771–3796. 10.1016/j.bpj.2017.06.042 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (60).Wu C; Zhou S First Observation of the Molten Globule State of a Single Homopolymer Chain. Phys. Rev. Lett [Online] 1996, 77 (14), 3053–3055. 10.1103/PhysRevLett.77.3053 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (61).Hong L; Lei J Scaling Law for the Radius of Gyration of Proteins and Its Dependence on Hydrophobicity. J. Polym. Sci., Part B: Polym. Phys [Online] 2009, 47, 207–214. 10.1002/polb.21634 (September 11, 2019). [DOI] [Google Scholar]
- (62).Hoogenboom R; Schlaad H Bioinspired Poly(2-oxazoline)s. Polymers [Online] 2011, 3, 467–488. 10.3390/polym3010467 (September 11, 2019). [DOI] [Google Scholar]
- (63).Patterson JP; Robin MP; Chassenieux C; Colombani O; O’Reilly RK. The analysis of solution self-assembled polymeric nanomaterials. Chem. Soc. Rev [Online] 2014, 43, 2412–2425. 10.1039/C3CS60454C (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (64).Terashima T; Sugita T; Fukae K; Sawamoto M Synthesis and Single-Chain Folding of Amphiphilic Random Copolymers in Water. Macromolecules [Online] 2014, 47 (2), 589–600. 10.1021/ma402355v (September 11, 2019). [DOI] [Google Scholar]
- (65).Kirshenbaum K; Zuckermann RN; Dill KA Designing polymers that mimic biomolecules. Curr. Opin. Struct. Biol [Online] 1999, 9 (4), 530–535. 10.1016/S0959-440X(99)80075-X (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (66).Barron AE; Zuckerman RN. Bioinspired polymeric materials: in-between proteins and plastics. Curr. Opin. Chem. Biol [Online] 1999, 3, 681–687. 10.1016/S1367-5931(99)00026-5 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (67).Smith AAA; Hall A; Wu V; Xu T Practical Prediction of Heteropolymer Composition and Drift. ACS Macro Lett. [Online] 2018, 8 (1), 36–40. 10.1021/acsmacrolett.8b00813 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (68).Ting JM; Navale TS; Bates FS; Reineke TM Precise Compositional Control and Systematic Preparation of Multimonomeric Statistical Copolymers. ACS Macro Lett. [Online] 2013, 2 (9), 770–774. 10.1021/mz4003112 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (69).Shanmugam S; Boyer C Stereo-, Temporal and Chemical Control through Photoactivation of Living Radical Polymerization: Synthesis of Block and Gradient Copolymers. J Am. Chem. Soc [Online] 2015, 137 (31), 9988–9999. 10.1021/jacs.5b05903 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (70).Ren JM; McKenzie TG; Fu Q; Wong EHH; Xu JT; An ZS; Shanmugam S; Davis TP; Boyer C; Qiao GG Star Polymers. Chemical Reviews [Online] 2016, 116 (12), 6743–6836. 10.1021/jacs.5b05903 (September 11, 2019). [DOI] [PubMed] [Google Scholar]
- (71).Hanlon AM; Lyon CK; Berda EB What Is Next in Single-Chain Nanoparticles? Macromolecules [Online] 2016, 49 (1), 2–14. 10.1021/acs.macromol.5b01456 (September 11, 2019). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.