Abstract
Context
Polygonatum sibiricum polysaccharide (PSP), derived from Polygonatum sibiricum Delar. ex Redoute (Liliaceae), is known to be able to delay the ageing process. However, the specific mechanisms underlying these effects are not clear.
Objective
To investigate the mechanisms underlying the effects of PSP treatment on brain ageing by the application of transcriptomic analysis.
Materials and methods
Forty Kunming mice were randomly divided into four groups (control, d-galactose, low-dose PSP, high-dose PSP). Mice were administered d-galactose (50 mg/kg, hypodermic injection) and PSP (200 or 400 mg/kg, intragastric administration) daily for 60 days. Behavioural responses were evaluated with the Morris water maze and the profiles of circRNA, miRNA, and mRNA, in the brains of experimental mice were investigated during the ageing process with and without PSP treatment.
Results
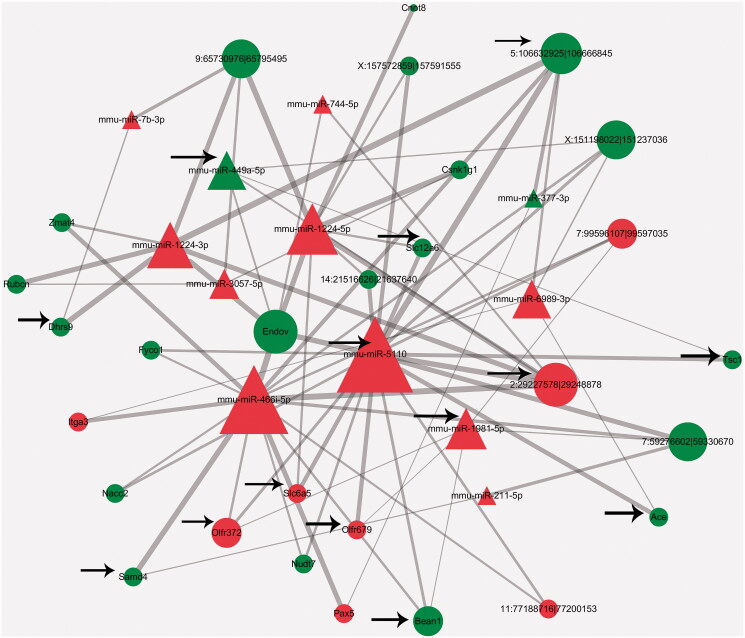
PSP improved cognitive function during brain ageing, as evidenced by a reduced escape latency time (p < 0.05) and an increase in the number of times mice crossed the platform (p < 0.05). A total of 37, 13, and 679, circRNAs, miRNAs, and mRNAs, respectively, were significantly altered by PSP treatment (as evidenced by a fold change ≥2 and p < 0.05). These dysregulated RNAs were closely associated with synaptic activity. PSP regulated regulate nine mRNAs (Slc6a5, Bean1, Ace, Samd4, Olfr679, Olfr372, Dhrs9, Tsc1, Slc12a6), three miRNAs (mmu-miR-5110, mmu-miR-449a-5p, mmu-miR-1981-5p), and two circRNAs (2:29227578|29248878 and 5:106632925|106666845) in the competing endogenous RNA (ceRNA) network.
Discussion and Conclusions
Our analyses showed that multiple circRNAs, miRNAs, and mRNAs responded to PSP treatment in mice experiencing brain ageing.
Keywords: Natural bioactive substances, anti-ageing, RNA sequencing, circRNA, ceRNA theory, synapse
Introduction
Ageing is a comprehensive and complex multifactorial process that refers to a gradual decline of physiological function in molecules, cells, tissues, and organs (López-Otín et al. 2013). Ageing has a particularly profound effect on the brain and the nervous system; this is because these systems incur the most severe damage caused by dysfunction in oxygen demand, regeneration ability, and endogenous antioxidant capacity (Mecocci et al. 2018). Brain ageing is also known to be related to the occurrence and development of Alzheimer’s disease, and other prevalent neurodegenerative disorders (Apple et al. 2017). Brain ageing has also been linked to a higher risk of several chronic diseases. Therefore, there is an urgent need to identify novel anti-ageing agents to delay brain ageing and improve the health of elderly individuals.
Drug discovery strategies from natural bioactive substances or components against neurodegeneration may represent a promising treatment with which to alleviate ageing-related brain injuries. Polygonatum sibiricum Delar. ex Redoute (Liliaceae) is a biologically active Chinese herb that has a long history of application for the invigoration of qi and the nourishment of yin. Polygonatum sibiricum polysaccharide (PSP) is the main active component of Polygonatum sibiricum Redouté and exhibits a range of biological functions, particularly with regards to anti-ageing effects (Zhao and Li 2015; Liu et al. 2018). A previous study demonstrated that PSP may play an important role in delaying the ageing process by regulating the Klotho/FGF23 endocrine axis, inhibiting oxidative stress, and by modulating the metabolism of calcium and phosphorus (Zheng 2020). Several recent reports have also reported that PSP acts directly on brain tissue during the anti-ageing process. Other research has shown that PSP intervention can improve the ultrastructure of the Cal region in the hippocampus of rats injected with Aβ1-42 (Yi et al. 2014). Telomerase activity was also activated in the brain and gonadal tissues of a mouse model of ageing when treated with PSP (Li et al. 2005). However, the specific effects of PSP on ageing in the brain have yet to be investigated in specific detail.
Micro-RNAs (miRNAs) are non-coding RNAs (ncRNAs) that are approximately 22 nucleotides in length; these miRNAs perform a repressive function on the expression of their target genes via miRNA response elements (Bartel 2004). Previous research based on RNA-Seq approaches found that miRNAs could represent good bioindicators for the investigation of ageing and age-related disease (Warnefors et al. 2014; Kumar et al. 2017). Circular RNAs (circRNAs) are an emerging class of ncRNA and represent a class of closed circRNA molecules that are enriched in neural tissues and exert a range of neural functions (Qu et al. 2015). CircRNAs have been demonstrated to play biological roles that are relevant to the ageing process in the nervous system (Knupp and Miura 2018). It is possible that circRNAs could associate with miRNAs via the competing endogenous RNA (ceRNA) network, in which circRNAs acting as miRNA sponges; these sponges can regulate splicing events and therefore modulate the expression of parental genes (Memczak et al. 2013). Interestingly, circRNA-associated-ceRNA networks have already been implicated in the onset of diseases that are related to ageing (Wang et al. 2018). Therefore, by analysing the ceRNA network, we were able to investigate the mechanisms underlying the restorative effect of PSP on age-related brain injuries.
Transcriptomics has proven to be an effective tool for unravelling the pharmacological mechanisms underlying specific physiological processes in various medical fields (Zhang et al. 2019). In the present study, we investigated the anti-ageing effect of PSP in a mouse model of brain ageing that was induced by d-galactose (d-gal). We were specifically interested in the effect of PSP on learning and memory. We also used RNA sequencing-based transcriptomics analysis to identify changes in the profiles of mRNA, miRNA, and circRNA, in order to establish a ceRNA network associated with the anti-ageing effects of PSP.
Material and methods
Animals and experimental design
Male Kunming mice, weighing 18–22 g, were provided by the Laboratory Animal Centre, Hunan Academy of Chinese Medicine (Changsha, China). All experimental procedures were conducted in accordance with institutional guidelines and were approved by the Institutional Animal Care and Use Committee in Hunan Academy of Chinese Medicine, Changsha, China. The mice were randomly divided into four groups (n = 10 for each group): (i) Control group, (ii) d-gal group (50 mg/kg/d for 60 days, i.h.), (iii) low-dose PSP (200 mg/kg/d for 60 days, i.g.) plus d-gal (i.h.), and high-dose PSP (400 mg/kg/d for 60 days, i.g.) plus d-gal. Behavioural tests were conducted after 60 consecutive days of modelling and administration. PSP was purchased from Ci Yuan Bio-Technology Co., Ltd (Shanxi, China) and was 98% pure (batch number: CY170510). The d-gal reagent was purchased from Kunming Nanjiang Pharmaceutical Co., Ltd (batch number: G19031406).
Morris water maze test
The Morris water maze test was carried out on days 54 to 60 the start of gavage treatment. The test was divided into two stages: navigation training and a probe trial. In the directional navigation test, we used a video analysis system to record the time it took mice to find a safe platform from the time they first entered the water (referred to as ‘escape latency’). The mice were trained four times a day for five consecutive days; we then carried out statistical analysis on escape latency data for each group of mice. After the directional navigation experiment, we removed the safety platform and allowed the mice to swim for 60 s. We then recorded the number of times each mouse crossed the location of the original platform and the time spent in each quadrant.
RNA extraction
Each mouse was sacrificed after behavioural analysis and specimens of brain tissue were collected. TRIzol Reagent (Invitrogen, Grand Island, NY, USA) was added to each brain tissue sample, ensuring that the volume of the tissue sample did not exceed 10% of the volume of TRIzol Reagent. The tissue sample was then homogenized. We then used a NanoDrop ND-1000 to determine the concentration of RNA; RNA integrity was assessed by standard denaturing agarose gel electrophoresis.
RNA sequencing analysis
Next, we constructed miRNA sequencing libraries in accordance with the protocol provided with the TruSeq® miRNA Sample Prep Kit v2 (Illumina, San Diego, CA, USA). Small RNA molecules were sequenced on a NovaSeq 6000 platform by Genergy Biotechnology Co., Ltd. (Shanghai, China). Unique small RNA sequences were mapped to reference sequences in the miRBase database by bowtie software. MiRDeep2 was then applied to predict novel miRNAs.
RNA-seq libraries for mRNAs and circRNAs were constructed in accordance with the guidelines provided with the TruSeq® strand RNA LT Sample Prep Kit (Illumina, San Diego, CA, USA). Sequencing was then performed on a NovaSeq 6000 platform. High-quality mRNA reads were aligned to the mouse reference genome (mm9) using SOAP aligner. The expression levels for each of the target genes were then normalized to reads per kilobase of exon model per million mapped reads (RPKM) so that we could compare mRNA levels between samples. For circRNA, sequencing data were analysed using CIRCexplorer2, an algorithm that can be used for de novo circRNA identification.
Gene ontology (GO) and KEGG pathway enrichment analyses
GO and KEGG pathway enrichment analyses are widely used to annotate gene function. In this study, we performed GO annotation and KEGG pathway analyses of differentially expressed (DE) mRNAs. We then used the Cluster Profiler package in R to identify the source genes of DE circRNAs and predict the target genes of the DE miRNAs. p < 0.05 was set as the cut-off criterion.
Construction of the ceRNA network
The miRanda software package (http://www.microrna.org/microrna/home.do) was used to predict miRNA-binding seed-sequence sites. We also identified miRNA response elements (MREs) that corresponded to the circRNAs and mRNAs. An overlap of the same miRNA-binding sites on both circRNA and mRNA indicated a potential circRNA–miRNA–mRNA network interaction. Based on the analysis of high-throughput sequencing data, we constructed a circRNA-related ceRNA network and visualized this network using Cytoscape software v.3.5.0 (San Diego, CA, USA).
Validation by real-time quantitative polymerase chain reaction (qPCR)
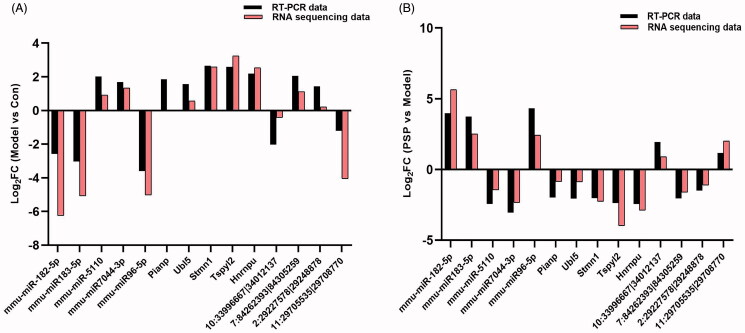
Quantitative real-time qPCR analysis was used to validate the results derived from RNA-Seq. qPCR was carried out with a LineGene FQD-96A Real-time PCR System (BIOER, China) in accordance with the manufacturer’s instructions. The expression levels of circRNA, mRNA, and miRNA, were normalized to those of β-actin, ACTB, and U6, respectively. Primer details are given in Table 1.
Table 1.
Primers designed for qRT-PCR validation of candidate miRNAs, mRNAs, and circRNAs.
| Names | Primer sequence (5′ to 3′) |
|---|---|
| Pianp | F: CACAGGCCATAGCACAGTCA R: TAGGCAACACGAGGGAGAGA |
| Ubl5 | F: CAACGACCGTCTCGGAAAGA R: ACCTTCCATCTGCTCACAGC |
| Stmn1 | F: AGGTGCTCCAGAAAGCCATC R: TTCTCTCGCAAGCGCTCC |
| Tspyl2 | F: CACTATTACCCCACCCTGAGC R: TGGCTCCTGCGATTATAGGC |
| Hnrnpu | F: CAGAGGTGGAATGCCCAACA R: CACAGCACATTCACATGGACA |
| 10:33996667|34012137 | F: CCTGACTACAATCCTGTTGCC R: TAAAGTGGAGTCTCATCAGGGT |
| 7:84262393|84305259 | F: CCGGCTGACCCTTCATCCTA R: TCCTTCTTTCACAGGGCCAA |
| 2:29227578|29248878 | F: GAGGATGGAGAGCGCCAAG R: TCAACTCTCACTTCATCCAGGTA |
| 11:29705535|29708770 | F: AAGGTGCCCTTATTGCTTCCAA R: AGAGGAGGGTATCACAGGCTC |
| mmu-miR-182-5p | F: GGCTTTGGCAATGGTAGAACTC RT: GTCGTATCGACTGCAGGGTCCGAGGTATTCGCAGTCGATACGACCGGTGT |
| mmu-miR-183-5p | F: GCGGCTATGGCACTGGTAGAA RT: GTCGTATCGACTGCAGGGTCCGAGGTATTCGCAGTCGATACGACAGTGAA |
| mmu-miR-5110 | F: CGGAGGAGGTAGAGGGTGGT RT: GTCGTATCGACTGCAGGGTCCGAGGTATTCGCAGTCGATACGACAATTCC |
| mmu-miR-7044-3p | F: CATGCAGCCCCGACCC RT: GTCGTATCGACTGCAGGGTCCGAGGTATTCGCAGTCGATACGACCTGTGA |
| mmu-miR-96-5p | F: CGGCTTTGGCACTAGCACATT RT: GTCGTATCGACTGCAGGGTCCGAGGTATTCGCAGTCGATACGACAGCAAA |
Statistical analysis
All data are expressed as mean ± standard deviation (SD). Statistical analysis involved the unpaired Student’s t-test for two comparisons or one-way analysis of variance (ANOVA) for repeated measures. p < 0.05 denoted a statistically significant difference. Sequencing data were processed by DEseq software. Genes and ncRNAs were defined as differentially expressed if p < 0.01 and |log2 (fold change)| >2.
Results
PSP improved the spatial memory ability of mice induced by d-gal
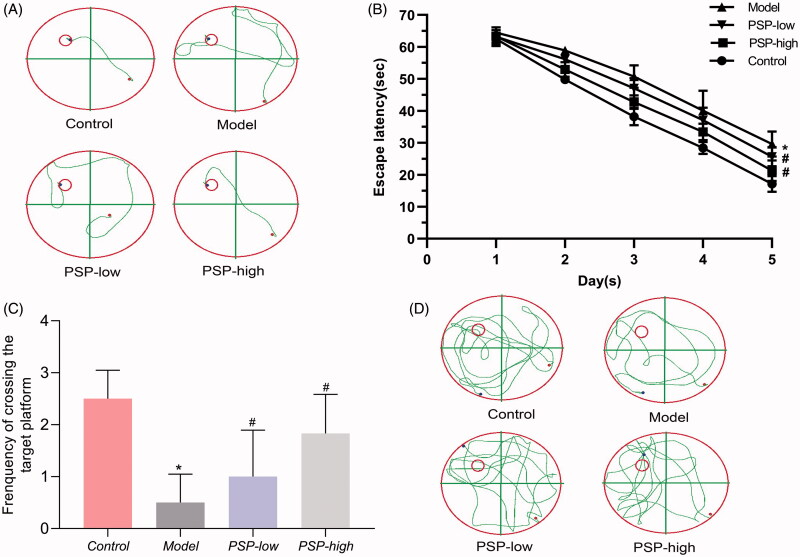
The d-gal animal model is internationally recognized and widely used to screen drugs and study the mechanisms involved with brain ageing (Zhang et al. 2005; Li et al. 2015). Compared to the Control group, the escape latency of mice in the Model group was significantly longer (p < 0.01). Furthermore, mice in the Model group had significantly fewer platform crossing times (p < 0.01), thus confirming that the mouse model of brain ageing had been successfully established. However, the escape latency in the PSP group was significantly shorter than that in the Model group (p < 0.05). Furthermore, PSP treatment significantly increased the number of platform crossings (p < 0.05) in mice experiencing brain ageing when compared with the Model group, thus suggesting the ameliorative effect of PSP on cognitive impairment in mice suffering from brain ageing induced by d-gal (Figure 1).
Figure 1.
Effects of PSP on spatial learning and memory deficiency in mice experiencing brain ageing. (A) Representative images of swimming paths undertaken on the fifth day of the spatial acquisition test, and (B) the time needed to reach the hidden platform. (C) Spatial memory function was tested by counting the number of times each mouse crossed the target platform within 90 s. (D) Representative images of swimming paths in the spatial probe trial. Data are expressed as the mean ± SD (n = 6 per group; escape latency was analysed by repeated measures analysis of variance (ANOVA); other data were analysed by one-way ANOVA followed by least significant difference tests). *p < 0.05, vs. Control group; #p < 0.05 vs. Model group.
Differentially expressed (DE) mRNAs and functional enrichment analysis
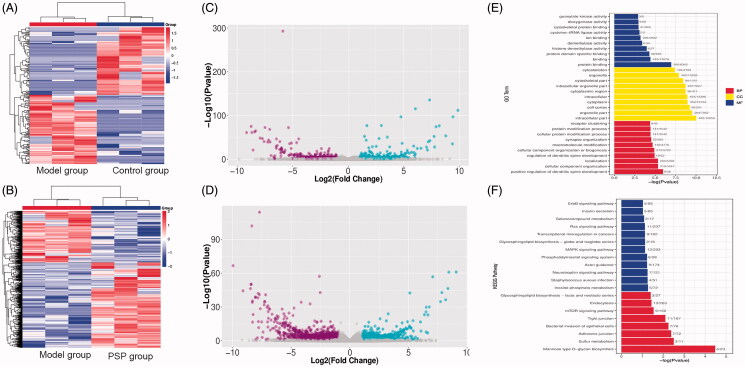
Next, we compared the gene profiles of the Model group to those of the Control group; this allowed us to identify a number of DE genes that we referred to as ‘ageing-related genes.’ These ageing-related genes were then compared to the gene profiles of the PSP group, and the genes that were regulated by PSP were considered as ‘PSP-related genes.’ Using p < 0.01 and |log 2 (fold change)| ≥ 2 as thresholds, we identified 2425 ageing-related mRNAs (1175 upregulated and 1250 downregulated) and 679 PSP-related mRNAs (Table 2). Compared with the Control group, 254 mRNAs were upregulated in the Model group. Interestingly, PSP intervention reversed these d-gal-induced alterations and yielded expression levels that were similar to those of the Control group. Similarly, compared with the Control group, 425 mRNAs were downregulated in the Model group; PSP treatment led to the upregulation of these mRNAs. Next, we created a heatmap; this revealed a distinct expression signature for mRNA in the Model group, Control group, and PSP group (Figure 2(A–B)). A volcano plot was then used to visualize the expression of DE genes in the ‘Model group vs. Control group’ and the ‘Model group vs. PSP group’ (Figure 2(C–D)). These results suggested that PSP treatment could reverse transcriptomic alterations induced by brain ageing to a significant extent.
Table 2.
Dysregulated mRNAs associated with the anti-brain ageing effect of PSP.
| Transcript_ID | Symbol | Con | Mod | PSP | log2FoldChange |
Pvalue |
||
|---|---|---|---|---|---|---|---|---|
| Con_vs_Mod | Mod_vs_PSP | Con_vs_Mod | Mod_vs_PSP | |||||
| ENSMUST00000089161 | Tnfrsf13c | 2.896 | 0.000 | 3.085 | 2.896 | −3.085 | 0.001 | 0.000 |
| ENSMUST00000050709 | Dok7 | 5.681 | 4.226 | 5.458 | 1.455 | −1.232 | 0.007 | 0.001 |
| ENSMUST00000106464 | Ccdc163 | 3.874 | 0.000 | 5.163 | 3.874 | −5.163 | 0.001 | 0.000 |
| ENSMUST00000050228 | Tmem241 | 0.000 | 4.588 | 0.000 | −4.588 | 4.588 | 0.000 | 0.000 |
| ENSMUST00000113572 | Runx2 | 7.264 | 8.767 | 7.124 | −1.503 | 1.643 | 0.000 | 0.000 |
| ENSMUST00000121564 | Fam118b | 3.424 | 4.808 | 3.524 | −1.384 | 1.284 | 0.021 | 0.027 |
| ENSMUST00000151630 | Fam117b | 2.843 | 0.000 | 2.852 | 2.843 | −2.852 | 0.002 | 0.001 |
| ENSMUST00000167191 | Hs3st5 | 3.166 | 1.506 | 3.295 | 1.660 | −1.789 | 0.032 | 0.015 |
| ENSMUST00000019439 | Tmem129 | 9.154 | 7.625 | 8.971 | 1.529 | −1.346 | 0.012 | 0.042 |
| ENSMUST00000028880 | Slc20a1 | 6.181 | 8.633 | 6.944 | −2.452 | 1.689 | 0.026 | 0.000 |
| ENSMUST00000110267 | Sybu | 11.044 | 4.985 | 9.070 | 6.059 | −4.085 | 0.000 | 0.001 |
| ENSMUST00000169653 | Ankrd13d | 7.213 | 5.802 | 7.322 | 1.411 | −1.520 | 0.000 | 0.000 |
| ENSMUST00000089003 | Atp2b2 | 2.820 | 4.410 | 2.596 | −1.590 | 1.814 | 0.022 | 0.007 |
| ENSMUST00000023655 | Mx1 | 1.380 | 4.809 | 2.250 | −3.429 | 2.559 | 0.000 | 0.038 |
| ENSMUST00000103037 | Ush1g | 4.673 | 3.557 | 5.247 | 1.116 | −1.690 | 0.035 | 0.000 |
| ENSMUST00000164853 | Ccdc24 | 7.050 | 5.391 | 7.214 | 1.659 | −1.822 | 0.011 | 0.010 |
| ENSMUST00000126914 | Pgap2 | 3.198 | 1.378 | 3.386 | 1.820 | −2.008 | 0.013 | 0.003 |
| ENSMUST00000036300 | Col27a1 | 7.400 | 6.278 | 8.716 | 1.123 | −2.438 | 0.008 | 0.000 |
| ENSMUST00000169421 | Tmprss7 | 0.397 | 3.473 | 1.375 | −3.076 | 2.098 | 0.034 | 0.018 |
| ENSMUST00000179201 | Bsg | 6.943 | 4.359 | 8.507 | 2.584 | −4.148 | 0.001 | 0.000 |
| ENSMUST00000173341 | Gtf2i | 10.331 | 11.837 | 10.267 | −1.506 | 1.570 | 0.000 | 0.004 |
| ENSMUST00000119168 | Zfp740 | 3.804 | 0.000 | 5.294 | 3.804 | −5.294 | 0.000 | 0.000 |
| ENSMUST00000044616 | Ints8 | 7.647 | 5.398 | 8.788 | 2.249 | −3.389 | 0.000 | 0.000 |
| ENSMUST00000212404 | Spire2 | 1.069 | 3.159 | 5.773 | −2.090 | −2.615 | 0.049 | 0.027 |
| ENSMUST00000143089 | Rad54l | 0.613 | 3.775 | 2.533 | −3.162 | 1.242 | 0.000 | 0.030 |
| ENSMUST00000049655 | Tmem215 | 9.336 | 8.019 | 6.703 | 1.317 | 1.316 | 0.034 | 0.039 |
| ENSMUST00000089317 | Magi1 | 9.030 | 10.289 | 9.004 | −1.259 | 1.285 | 0.000 | 0.001 |
| ENSMUST00000028291 | Inpp5e | 5.419 | 3.414 | 5.763 | 2.006 | −2.350 | 0.019 | 0.006 |
| ENSMUST00000109134 | Zfp879 | 2.528 | 4.130 | 1.680 | −1.603 | 2.450 | 0.020 | 0.014 |
| ENSMUST00000134862 | Rnf44 | 6.929 | 5.278 | 6.694 | 1.652 | −1.416 | 0.007 | 0.008 |
| ENSMUST00000030858 | Fbxo6 | 2.623 | 6.664 | 4.897 | −4.041 | 1.768 | 0.003 | 0.005 |
| ENSMUST00000161769 | Hnrnpu | 8.561 | 10.751 | 8.308 | −2.190 | 2.443 | 0.008 | 0.000 |
| ENSMUST00000123479 | Kdm2b | 4.724 | 6.289 | 4.364 | −1.565 | 1.925 | 0.004 | 0.009 |
| ENSMUST00000198204 | Kcnt1 | 2.670 | 4.931 | 3.016 | −2.261 | 1.915 | 0.010 | 0.031 |
| ENSMUST00000102488 | Myo18a | 5.766 | 1.946 | 4.446 | 3.821 | −2.500 | 0.000 | 0.001 |
| ENSMUST00000183402 | Szt2 | 3.078 | 5.303 | 3.794 | −2.224 | 1.508 | 0.001 | 0.009 |
| ENSMUST00000053722 | Zfp607a | 4.114 | 2.795 | 4.073 | 1.318 | −1.277 | 0.022 | 0.034 |
| ENSMUST00000147938 | Tab2 | 4.825 | 6.131 | 7.981 | −1.306 | −1.850 | 0.001 | 0.001 |
| ENSMUST00000117929 | Tmcc3 | 4.656 | 2.849 | 5.018 | 1.807 | −2.169 | 0.008 | 0.003 |
| ENSMUST00000066601 | Hyou1 | 9.118 | 10.358 | 8.674 | −1.239 | 1.683 | 0.002 | 0.000 |
| ENSMUST00000113222 | Stxbp1 | 10.169 | 9.060 | 11.462 | 1.108 | −2.402 | 0.016 | 0.002 |
| ENSMUST00000113228 | Drp2 | 6.732 | 4.509 | 6.138 | 2.223 | −1.629 | 0.000 | 0.011 |
| ENSMUST00000135589 | Lpin2 | 2.458 | 4.631 | 3.322 | −2.173 | 1.310 | 0.030 | 0.037 |
| ENSMUST00000024639 | Map2 | 5.320 | 7.343 | 5.247 | −2.023 | 2.096 | 0.045 | 0.026 |
| ENSMUST00000076442 | Plec | 5.167 | 7.736 | 6.072 | −2.569 | 1.664 | 0.000 | 0.036 |
| ENSMUST00000061722 | Dlk2 | 3.005 | 6.320 | 3.275 | −3.316 | 3.045 | 0.002 | 0.001 |
| ENSMUST00000201588 | Medag | 4.550 | 2.955 | 4.187 | 1.595 | −1.232 | 0.001 | 0.014 |
| ENSMUST00000201583 | Tbc1d24 | 4.943 | 3.771 | 4.893 | 1.172 | −1.122 | 0.018 | 0.036 |
| ENSMUST00000145667 | Arl6ip4 | 4.969 | 6.035 | 4.808 | −1.067 | 1.228 | 0.000 | 0.000 |
| ENSMUST00000159415 | Fsd1l | 3.429 | 1.951 | 3.267 | 1.478 | −1.316 | 0.033 | 0.030 |
| ENSMUST00000215016 | Cx3cr1 | 4.244 | 7.929 | 4.604 | −3.685 | 3.325 | 0.000 | 0.000 |
| ENSMUST00000021544 | Plek2 | 4.636 | 3.225 | 4.890 | 1.411 | −1.665 | 0.008 | 0.000 |
| ENSMUST00000129010 | Nprl3 | 3.160 | 1.701 | 3.206 | 1.459 | −1.505 | 0.036 | 0.026 |
| ENSMUST00000148868 | Mpped1 | 5.797 | 6.850 | 4.844 | −1.053 | 2.006 | 0.001 | 0.001 |
| ENSMUST00000185177 | Rfx7 | 5.530 | 3.917 | 5.039 | 1.613 | −1.122 | 0.009 | 0.030 |
| ENSMUST00000180046 | Usp34 | 8.168 | 9.733 | 8.474 | −1.566 | 1.260 | 0.037 | 0.046 |
| ENSMUST00000196695 | Fubp1 | 7.705 | 5.913 | 7.450 | 1.792 | −1.538 | 0.035 | 0.006 |
| ENSMUST00000023465 | Top3b | 3.360 | 1.516 | 6.700 | 1.844 | −5.184 | 0.023 | 0.000 |
| ENSMUST00000211413 | Tulp2 | 0.000 | 2.038 | 0.000 | −2.038 | 2.038 | 0.036 | 0.021 |
| ENSMUST00000123203 | Scfd2 | 4.802 | 3.666 | 5.208 | 1.136 | −1.542 | 0.050 | 0.005 |
| ENSMUST00000137285 | Zfp644 | 6.557 | 5.354 | 6.717 | 1.203 | −1.363 | 0.003 | 0.000 |
| ENSMUST00000171151 | Washc3 | 2.371 | 4.283 | 2.402 | −1.912 | 1.882 | 0.009 | 0.004 |
| ENSMUST00000117994 | C330027C09Rik | 2.556 | 0.000 | 5.726 | 2.556 | −5.726 | 0.044 | 0.000 |
| ENSMUST00000220532 | Exoc2 | 5.604 | 6.715 | 5.614 | −1.110 | 1.100 | 0.017 | 0.034 |
| ENSMUST00000119316 | Ccnj | 7.932 | 5.262 | 8.081 | 2.670 | −2.819 | 0.003 | 0.001 |
| ENSMUST00000148162 | Flot2 | 8.595 | 6.725 | 8.382 | 1.870 | −1.658 | 0.000 | 0.000 |
| ENSMUST00000140359 | Kif3a | 0.000 | 2.723 | 0.000 | −2.723 | 2.723 | 0.050 | 0.026 |
| ENSMUST00000173989 | Rps6ka1 | 3.023 | 5.095 | 6.613 | −2.072 | −1.519 | 0.000 | 0.003 |
| ENSMUST00000191230 | Ilkap | 3.998 | 5.086 | 4.083 | −1.087 | 1.002 | 0.033 | 0.048 |
| ENSMUST00000187548 | Dnah7b | 4.385 | 3.141 | 5.733 | 1.245 | −2.592 | 0.041 | 0.000 |
| ENSMUST00000022398 | 1700011H14Rik | 3.107 | 0.540 | 3.171 | 2.567 | −2.631 | 0.009 | 0.004 |
| ENSMUST00000176374 | Pex12 | 2.695 | 0.000 | 2.710 | 2.695 | −2.710 | 0.042 | 0.034 |
| ENSMUST00000224676 | Arhgef33 | 0.000 | 2.140 | 3.481 | −2.140 | −1.341 | 0.027 | 0.044 |
| ENSMUST00000108238 | Yif1b | 7.476 | 6.319 | 7.659 | 1.157 | −1.340 | 0.005 | 0.001 |
| ENSMUST00000146291 | Crebzf | 3.911 | 0.000 | 3.710 | 3.911 | −3.710 | 0.019 | 0.012 |
| ENSMUST00000097666 | Cab39 | 4.829 | 2.983 | 4.866 | 1.847 | −1.883 | 0.021 | 0.028 |
| ENSMUST00000093829 | Megf11 | 7.114 | 5.973 | 7.014 | 1.141 | −1.042 | 0.000 | 0.008 |
| ENSMUST00000186807 | 2310035C23Rik | 6.499 | 4.018 | 6.464 | 2.481 | −2.446 | 0.035 | 0.040 |
| ENSMUST00000182003 | Anks1b | 3.321 | 4.853 | 2.148 | −1.532 | 2.705 | 0.006 | 0.000 |
| ENSMUST00000106727 | 0610037L13Rik | 0.000 | 2.800 | 0.000 | −2.800 | 2.800 | 0.005 | 0.002 |
| ENSMUST00000138547 | Ripor2 | 5.221 | 6.393 | 5.058 | −1.172 | 1.335 | 0.008 | 0.040 |
| ENSMUST00000110157 | Prokr2 | 0.611 | 3.076 | 0.654 | −2.464 | 2.421 | 0.017 | 0.023 |
| ENSMUST00000224182 | Hnrnpk | 3.250 | 1.368 | 3.141 | 1.881 | −1.773 | 0.024 | 0.018 |
| ENSMUST00000224455 | Emb | 4.776 | 5.892 | 7.753 | −1.116 | −1.861 | 0.027 | 0.021 |
| ENSMUST00000079719 | Tbx3 | 6.490 | 5.054 | 6.263 | 1.436 | −1.209 | 0.001 | 0.031 |
| ENSMUST00000165365 | Cd276 | 5.313 | 2.984 | 6.129 | 2.329 | −3.145 | 0.026 | 0.002 |
| ENSMUST00000173280 | H2-T22 | 3.382 | 0.000 | 6.312 | 3.382 | −6.312 | 0.024 | 0.000 |
| ENSMUST00000116388 | Rcor1 | 6.451 | 3.407 | 4.629 | 3.044 | −1.221 | 0.000 | 0.016 |
| ENSMUST00000199534 | Ldb2 | 4.819 | 2.555 | 5.140 | 2.264 | −2.584 | 0.023 | 0.002 |
| ENSMUST00000219554 | Myl6 | 6.794 | 3.364 | 6.543 | 3.431 | −3.179 | 0.000 | 0.000 |
| ENSMUST00000189871 | Ppp4r4 | 2.143 | 3.535 | 1.567 | −1.392 | 1.967 | 0.030 | 0.007 |
| ENSMUST00000113590 | Speg | 8.534 | 5.954 | 4.626 | 2.580 | 1.328 | 0.000 | 0.003 |
| ENSMUST00000058210 | Eps8 | 9.721 | 8.479 | 10.481 | 1.242 | −2.002 | 0.010 | 0.000 |
| ENSMUST00000214432 | Ncapd3 | 6.850 | 4.085 | 6.894 | 2.765 | −2.809 | 0.000 | 0.000 |
| ENSMUST00000114845 | Pfkfb3 | 1.026 | 4.719 | 0.638 | −3.693 | 4.081 | 0.001 | 0.000 |
| ENSMUST00000225845 | Zkscan4 | 3.827 | 0.000 | 3.330 | 3.827 | −3.330 | 0.030 | 0.015 |
| ENSMUST00000162242 | Impdh1 | 5.056 | 1.893 | 4.864 | 3.162 | −2.970 | 0.020 | 0.015 |
| ENSMUST00000171263 | Gbp7 | 6.787 | 5.436 | 7.245 | 1.351 | −1.809 | 0.001 | 0.000 |
| ENSMUST00000131390 | Mfsd9 | 1.815 | 5.920 | 1.356 | −4.105 | 4.564 | 0.000 | 0.000 |
| ENSMUST00000022551 | Rcbtb1 | 5.516 | 7.511 | 8.964 | −1.995 | −1.453 | 0.021 | 0.033 |
| ENSMUST00000110949 | Arhgap11a | 4.809 | 0.000 | 4.050 | 4.809 | −4.050 | 0.000 | 0.000 |
| ENSMUST00000063690 | Dhrs9 | 2.888 | 0.000 | 2.906 | 2.888 | −2.906 | 0.001 | 0.001 |
| ENSMUST00000077737 | Brd3 | 7.024 | 2.511 | 8.382 | 4.514 | −5.871 | 0.046 | 0.003 |
| ENSMUST00000022882 | Myo10 | 4.479 | 2.967 | 10.666 | 1.512 | −7.699 | 0.022 | 0.000 |
| ENSMUST00000044484 | Kdm6a | 7.843 | 6.412 | 8.811 | 1.431 | −2.399 | 0.000 | 0.000 |
| ENSMUST00000041175 | Ptger3 | 4.195 | 5.632 | 3.675 | −1.437 | 1.957 | 0.000 | 0.000 |
| ENSMUST00000114439 | Snx10 | 7.737 | 4.136 | 6.623 | 3.601 | −2.487 | 0.000 | 0.002 |
| ENSMUST00000095893 | Armt1 | 8.751 | 7.120 | 8.599 | 1.631 | −1.480 | 0.014 | 0.032 |
| ENSMUST00000075644 | Calcr | 6.433 | 2.341 | 6.188 | 4.092 | −3.847 | 0.029 | 0.036 |
| ENSMUST00000148313 | Tmc4 | 2.762 | 0.000 | 3.814 | 2.762 | −3.814 | 0.048 | 0.000 |
| ENSMUST00000105867 | Stmn1 | 5.629 | 8.281 | 6.252 | −2.652 | 2.029 | 0.000 | 0.000 |
| ENSMUST00000210312 | Gm45799 | 0.000 | 3.278 | 0.000 | −3.278 | 3.278 | 0.036 | 0.017 |
| ENSMUST00000127286 | Ndfip2 | 6.576 | 7.977 | 6.898 | −1.401 | 1.079 | 0.026 | 0.009 |
| ENSMUST00000081335 | Tgif2 | 5.372 | 3.721 | 5.335 | 1.651 | −1.614 | 0.000 | 0.005 |
| ENSMUST00000163324 | Fbxo34 | 3.088 | 9.177 | 6.669 | −6.089 | 2.508 | 0.029 | 0.003 |
| ENSMUST00000204481 | Ybx3 | 3.426 | 4.581 | 2.939 | −1.156 | 1.642 | 0.043 | 0.017 |
| ENSMUST00000113069 | Slc25a53 | 1.026 | 7.417 | 0.638 | −6.390 | 6.779 | 0.000 | 0.000 |
| ENSMUST00000133595 | Jmjd8 | 5.504 | 7.020 | 5.874 | −1.515 | 1.145 | 0.011 | 0.043 |
| ENSMUST00000179804 | Cldn12 | 2.761 | 5.030 | 3.085 | −2.269 | 1.945 | 0.007 | 0.005 |
| ENSMUST00000106128 | Adgrl2 | 10.700 | 7.484 | 9.072 | 3.216 | −1.589 | 0.000 | 0.011 |
| ENSMUST00000210988 | Mamstr | 2.678 | 5.486 | 3.984 | −2.808 | 1.502 | 0.000 | 0.035 |
| ENSMUST00000065330 | Clk3 | 6.371 | 8.003 | 6.799 | −1.631 | 1.204 | 0.001 | 0.020 |
| ENSMUST00000128187 | Hnrnpdl | 9.693 | 11.005 | 9.713 | −1.311 | 1.291 | 0.027 | 0.000 |
| ENSMUST00000111783 | Mapkapk5 | 3.762 | 0.986 | 2.814 | 2.777 | −1.828 | 0.005 | 0.019 |
| ENSMUST00000111785 | Ssfa2 | 9.541 | 7.345 | 10.966 | 2.195 | −3.621 | 0.003 | 0.000 |
| ENSMUST00000136114 | Plxna3 | 2.866 | 0.644 | 3.097 | 2.223 | −2.454 | 0.031 | 0.010 |
| ENSMUST00000203445 | Add2 | 5.633 | 3.838 | 4.999 | 1.796 | −1.162 | 0.000 | 0.017 |
| ENSMUST00000178982 | 43891 | 6.534 | 7.834 | 6.194 | −1.300 | 1.640 | 0.026 | 0.004 |
| ENSMUST00000093798 | Zbtb38 | 7.693 | 5.988 | 4.671 | 1.705 | 1.317 | 0.002 | 0.038 |
| ENSMUST00000225179 | Zcchc6 | 3.268 | 0.000 | 3.068 | 3.268 | −3.068 | 0.025 | 0.020 |
| ENSMUST00000173569 | Gne | 3.189 | 0.000 | 3.499 | 3.189 | −3.499 | 0.042 | 0.014 |
| ENSMUST00000149537 | Fastk | 3.888 | 1.693 | 3.981 | 2.195 | −2.288 | 0.033 | 0.014 |
| ENSMUST00000105500 | Snx3 | 0.000 | 3.077 | 0.000 | −3.077 | 3.077 | 0.030 | 0.013 |
| ENSMUST00000037422 | Rnf44 | 7.400 | 5.172 | 6.682 | 2.228 | −1.510 | 0.034 | 0.036 |
| ENSMUST00000176263 | Ppp1r9a | 6.620 | 7.770 | 6.505 | −1.151 | 1.266 | 0.006 | 0.025 |
| ENSMUST00000144454 | Arrb2 | 5.954 | 4.713 | 6.091 | 1.241 | −1.378 | 0.000 | 0.000 |
| ENSMUST00000125399 | Rusc2 | 3.069 | 0.000 | 3.545 | 3.069 | −3.545 | 0.029 | 0.000 |
| ENSMUST00000170710 | Glod4 | 5.098 | 3.761 | 5.015 | 1.337 | −1.254 | 0.026 | 0.050 |
| ENSMUST00000137069 | Atat1 | 0.000 | 2.902 | 0.000 | −2.902 | 2.902 | 0.047 | 0.024 |
| ENSMUST00000180025 | Bhlhb9 | 6.544 | 4.457 | 6.145 | 2.087 | −1.688 | 0.003 | 0.009 |
| ENSMUST00000108173 | Rffl | 8.504 | 7.445 | 9.235 | 1.059 | −1.790 | 0.001 | 0.000 |
| ENSMUST00000140893 | Manba | 5.003 | 3.209 | 6.018 | 1.794 | −2.810 | 0.007 | 0.000 |
| ENSMUST00000124526 | Ahnak2 | 4.363 | 5.483 | 7.620 | −1.120 | −2.137 | 0.010 | 0.000 |
| ENSMUST00000035889 | Myo6 | 9.089 | 7.982 | 9.670 | 1.108 | −1.689 | 0.024 | 0.034 |
| ENSMUST00000146569 | Slx4 | 6.226 | 5.216 | 6.912 | 1.010 | −1.697 | 0.021 | 0.000 |
| ENSMUST00000169438 | Plec | 5.095 | 1.169 | 4.616 | 3.926 | −3.446 | 0.032 | 0.049 |
| ENSMUST00000114205 | Ablim2 | 4.691 | 3.387 | 5.333 | 1.304 | −1.946 | 0.020 | 0.001 |
| ENSMUST00000002298 | Ppm1j | 4.390 | 3.212 | 4.859 | 1.178 | −1.647 | 0.048 | 0.013 |
| ENSMUST00000174317 | Jmjd1c | 9.076 | 7.061 | 8.339 | 2.015 | −1.277 | 0.000 | 0.014 |
| ENSMUST00000180476 | Usp35 | 3.811 | 0.000 | 3.438 | 3.811 | −3.438 | 0.000 | 0.000 |
| ENSMUST00000151980 | Rdh12 | 3.736 | 0.663 | 5.123 | 3.074 | −4.460 | 0.014 | 0.000 |
| ENSMUST00000124922 | Afdn | 5.632 | 8.079 | 5.738 | −2.446 | 2.340 | 0.000 | 0.001 |
| ENSMUST00000112491 | Rps6ka3 | 5.850 | 4.059 | 5.286 | 1.791 | −1.227 | 0.001 | 0.019 |
| ENSMUST00000127303 | Cfap44 | 2.937 | 0.644 | 2.491 | 2.293 | −1.847 | 0.020 | 0.030 |
| ENSMUST00000144221 | Crebrf | 1.128 | 5.694 | 2.370 | −4.566 | 3.324 | 0.001 | 0.002 |
| ENSMUST00000224798 | Nr2f1 | 4.234 | 5.757 | 4.026 | −1.523 | 1.731 | 0.000 | 0.005 |
| ENSMUST00000120645 | Gsto2 | 4.340 | 2.527 | 4.633 | 1.813 | −2.105 | 0.015 | 0.004 |
| ENSMUST00000208394 | Gprc5b | 4.926 | 6.412 | 8.388 | −1.486 | −1.976 | 0.006 | 0.000 |
| ENSMUST00000126888 | Setd6 | 5.042 | 3.577 | 4.863 | 1.465 | −1.286 | 0.011 | 0.035 |
| ENSMUST00000216412 | Exosc7 | 6.588 | 2.165 | 7.703 | 4.423 | −5.538 | 0.000 | 0.000 |
| ENSMUST00000080394 | Sh3kbp1 | 0.000 | 3.861 | 1.310 | −3.861 | 2.551 | 0.002 | 0.010 |
| ENSMUST00000212046 | Elmo3 | 6.927 | 1.632 | 6.816 | 5.295 | −5.185 | 0.001 | 0.000 |
| ENSMUST00000219834 | Dgka | 1.268 | 3.884 | 1.649 | −2.616 | 2.235 | 0.038 | 0.018 |
| ENSMUST00000056517 | Gja10 | 0.000 | 1.979 | 0.000 | −1.979 | 1.979 | 0.039 | 0.023 |
| ENSMUST00000052368 | Kdm6a | 7.845 | 6.182 | 7.687 | 1.663 | −1.505 | 0.002 | 0.014 |
| ENSMUST00000143674 | Pde6d | 6.242 | 5.070 | 3.766 | 1.171 | 1.305 | 0.000 | 0.035 |
| ENSMUST00000133257 | Ggnbp1 | 5.509 | 7.011 | 5.586 | −1.502 | 1.426 | 0.012 | 0.016 |
| ENSMUST00000053950 | Lrrc28 | 6.986 | 8.515 | 6.264 | −1.528 | 2.250 | 0.000 | 0.000 |
| ENSMUST00000145066 | Trmt2b | 8.603 | 5.085 | 7.693 | 3.519 | −2.608 | 0.000 | 0.000 |
| ENSMUST00000136804 | Mrpl51 | 6.356 | 5.155 | 6.415 | 1.201 | −1.260 | 0.037 | 0.029 |
| ENSMUST00000213062 | Aprt | 1.026 | 2.806 | 0.963 | −1.780 | 1.843 | 0.034 | 0.017 |
| ENSMUST00000199592 | Elp6 | 7.085 | 4.917 | 5.969 | 2.169 | −1.053 | 0.000 | 0.040 |
| ENSMUST00000038522 | Slmap | 2.697 | 3.886 | 2.798 | −1.189 | 1.088 | 0.043 | 0.048 |
| ENSMUST00000114559 | Zfp467 | 2.878 | 0.000 | 2.565 | 2.878 | −2.565 | 0.044 | 0.034 |
| ENSMUST00000085786 | Card11 | 3.035 | 0.000 | 3.182 | 3.035 | −3.182 | 0.002 | 0.000 |
| ENSMUST00000220895 | Aaed1 | 7.058 | 0.000 | 6.957 | 7.058 | −6.957 | 0.000 | 0.000 |
| ENSMUST00000163281 | Skint7 | 0.000 | 3.241 | 0.000 | −3.241 | 3.241 | 0.001 | 0.000 |
| ENSMUST00000032479 | Pianp | 8.075 | 9.921 | 7.932 | −1.846 | 1.989 | 0.027 | 0.000 |
| ENSMUST00000064872 | Ahcyl2 | 6.697 | 4.046 | 5.881 | 2.651 | −1.835 | 0.001 | 0.008 |
| ENSMUST00000209203 | Chd9 | 9.069 | 7.290 | 8.575 | 1.779 | −1.285 | 0.004 | 0.032 |
| ENSMUST00000115101 | Nupl2 | 0.000 | 3.470 | 0.000 | −3.470 | 3.470 | 0.032 | 0.014 |
| ENSMUST00000091892 | Ctxn3 | 6.215 | 3.995 | 6.003 | 2.220 | −2.008 | 0.000 | 0.000 |
| ENSMUST00000209905 | Ebf3 | 2.516 | 4.386 | 6.214 | −1.870 | −1.828 | 0.008 | 0.009 |
| ENSMUST00000108707 | Ubc | 2.600 | 4.761 | 3.502 | −2.162 | 1.259 | 0.006 | 0.031 |
| ENSMUST00000139953 | Slc39a8 | 4.877 | 3.030 | 4.640 | 1.847 | −1.609 | 0.000 | 0.001 |
| ENSMUST00000195907 | Sema3a | 5.354 | 3.866 | 5.151 | 1.488 | −1.285 | 0.001 | 0.008 |
| ENSMUST00000098696 | Psd3 | 5.797 | 7.870 | 6.343 | −2.073 | 1.527 | 0.041 | 0.038 |
| ENSMUST00000163750 | Nedd8 | 4.412 | 2.811 | 5.239 | 1.601 | −2.428 | 0.023 | 0.001 |
| ENSMUST00000129290 | Fam117a | 2.331 | 0.000 | 2.323 | 2.331 | −2.323 | 0.012 | 0.007 |
| ENSMUST00000161523 | Tmem63a | 6.907 | 2.278 | 10.634 | 4.629 | −8.356 | 0.019 | 0.000 |
| ENSMUST00000204278 | Slc6a1 | 9.634 | 8.478 | 9.565 | 1.156 | −1.086 | 0.033 | 0.049 |
| ENSMUST00000162366 | Apbb2 | 6.479 | 4.712 | 3.147 | 1.767 | 1.565 | 0.004 | 0.005 |
| ENSMUST00000209537 | Pde2a | 4.150 | 5.379 | 2.592 | −1.228 | 2.787 | 0.022 | 0.001 |
| ENSMUST00000165881 | Mark2 | 2.741 | 0.644 | 2.700 | 2.097 | −2.056 | 0.047 | 0.032 |
| ENSMUST00000223611 | Nrn1 | 2.572 | 4.358 | 2.989 | −1.787 | 1.369 | 0.014 | 0.018 |
| ENSMUST00000176756 | Dnah11 | 4.345 | 0.993 | 4.764 | 3.352 | −3.771 | 0.001 | 0.000 |
| ENSMUST00000172670 | Ccdc9 | 4.587 | 3.119 | 4.370 | 1.467 | −1.251 | 0.002 | 0.009 |
| ENSMUST00000029707 | Gpatch4 | 2.703 | 5.226 | 2.221 | −2.523 | 3.005 | 0.005 | 0.002 |
| ENSMUST00000086167 | Nfrkb | 6.210 | 7.702 | 6.650 | −1.492 | 1.052 | 0.047 | 0.026 |
| ENSMUST00000219778 | Ascc1 | 3.076 | 4.690 | 3.563 | −1.614 | 1.127 | 0.006 | 0.040 |
| ENSMUST00000166724 | Hrh3 | 5.725 | 7.631 | 5.635 | −1.906 | 1.996 | 0.000 | 0.000 |
| ENSMUST00000217966 | Heatr5a | 1.341 | 3.987 | 5.478 | −2.646 | −1.491 | 0.000 | 0.000 |
| ENSMUST00000132274 | Rsad1 | 6.680 | 0.655 | 3.890 | 6.025 | −3.235 | 0.001 | 0.001 |
| ENSMUST00000225488 | Ntrk2 | 3.644 | 5.093 | 6.337 | −1.448 | −1.244 | 0.001 | 0.019 |
| ENSMUST00000129775 | Kyat3 | 3.884 | 1.704 | 3.732 | 2.180 | −2.028 | 0.009 | 0.007 |
| ENSMUST00000079777 | Pard3 | 0.000 | 3.448 | 0.289 | −3.448 | 3.159 | 0.031 | 0.013 |
| ENSMUST00000055014 | Skint7 | 1.471 | 4.871 | 1.862 | −3.400 | 3.009 | 0.000 | 0.001 |
| ENSMUST00000065111 | Usp45 | 7.905 | 6.299 | 8.529 | 1.606 | −2.230 | 0.013 | 0.001 |
| ENSMUST00000095775 | Setdb2 | 5.175 | 2.667 | 4.534 | 2.508 | −1.867 | 0.002 | 0.012 |
| ENSMUST00000124313 | Chaf1b | 2.511 | 0.000 | 2.988 | 2.511 | −2.988 | 0.008 | 0.001 |
| ENSMUST00000129335 | Numb | 0.000 | 3.593 | 0.000 | −3.593 | 3.593 | 0.029 | 0.012 |
| ENSMUST00000139003 | Pnkp | 5.066 | 3.502 | 4.854 | 1.564 | −1.353 | 0.003 | 0.004 |
| ENSMUST00000112394 | Csrnp3 | 4.840 | 6.150 | 5.142 | −1.310 | 1.007 | 0.000 | 0.006 |
| ENSMUST00000141927 | Efcab14 | 2.683 | 0.000 | 3.355 | 2.683 | −3.355 | 0.003 | 0.000 |
| ENSMUST00000126085 | Aamdc | 2.011 | 4.719 | 2.901 | −2.708 | 1.818 | 0.001 | 0.016 |
| ENSMUST00000198675 | Nhlh2 | 4.114 | 1.158 | 5.769 | 2.956 | −4.611 | 0.001 | 0.000 |
| ENSMUST00000113736 | Dlg3 | 5.751 | 8.014 | 6.095 | −2.262 | 1.918 | 0.008 | 0.008 |
| ENSMUST00000186315 | Usp40 | 6.001 | 7.957 | 6.456 | −1.955 | 1.500 | 0.016 | 0.002 |
| ENSMUST00000106220 | Macf1 | 7.714 | 9.280 | 7.743 | −1.567 | 1.537 | 0.000 | 0.001 |
| ENSMUST00000156444 | Zfp707 | 3.616 | 2.217 | 4.163 | 1.399 | −1.946 | 0.043 | 0.003 |
| ENSMUST00000156442 | Sec16a | 7.669 | 5.805 | 7.950 | 1.864 | −2.145 | 0.000 | 0.000 |
| ENSMUST00000147556 | Rtkn2 | 3.987 | 6.365 | 3.590 | −2.377 | 2.775 | 0.000 | 0.001 |
| ENSMUST00000048678 | Lss | 8.197 | 4.145 | 6.089 | 4.052 | −1.945 | 0.000 | 0.023 |
| ENSMUST00000166558 | Efemp2 | 2.950 | 0.000 | 2.395 | 2.950 | −2.395 | 0.002 | 0.006 |
| ENSMUST00000055281 | Skor1 | 2.230 | 3.594 | 6.538 | −1.363 | −2.944 | 0.034 | 0.000 |
| ENSMUST00000145622 | Zfp715 | 4.194 | 3.170 | 5.426 | 1.023 | −2.255 | 0.046 | 0.002 |
| ENSMUST00000203538 | Klhl5 | 6.404 | 5.283 | 6.832 | 1.121 | −1.549 | 0.002 | 0.003 |
| ENSMUST00000099742 | Cfap44 | 7.981 | 6.594 | 7.968 | 1.388 | −1.374 | 0.005 | 0.017 |
| ENSMUST00000131424 | Uggt2 | 4.287 | 2.242 | 4.690 | 2.045 | −2.448 | 0.046 | 0.008 |
| ENSMUST00000133896 | Slc11a1 | 2.612 | 0.000 | 2.328 | 2.612 | −2.328 | 0.005 | 0.008 |
| ENSMUST00000128706 | Tcf4 | 7.917 | 6.227 | 7.413 | 1.690 | −1.186 | 0.002 | 0.045 |
| ENSMUST00000211174 | Ankrd10 | 3.208 | 0.000 | 2.813 | 3.208 | −2.813 | 0.043 | 0.028 |
| ENSMUST00000178988 | Tmem205 | 4.231 | 0.931 | 3.836 | 3.300 | −2.904 | 0.010 | 0.031 |
| ENSMUST00000154718 | Reps1 | 2.293 | 3.902 | 1.909 | −1.609 | 1.993 | 0.026 | 0.001 |
| ENSMUST00000114100 | Sdccag3 | 3.231 | 0.000 | 3.069 | 3.231 | −3.069 | 0.034 | 0.023 |
| ENSMUST00000122945 | 44078 | 3.021 | 5.999 | 2.273 | −2.978 | 3.726 | 0.029 | 0.000 |
| ENSMUST00000143262 | Lrrc75a | 1.567 | 3.882 | 2.102 | −2.314 | 1.780 | 0.047 | 0.012 |
| ENSMUST00000214732 | Zfp651 | 3.956 | 1.840 | 4.446 | 2.116 | −2.606 | 0.049 | 0.013 |
| ENSMUST00000132517 | Abca7 | 8.436 | 5.958 | 9.478 | 2.478 | −3.520 | 0.010 | 0.000 |
| ENSMUST00000160620 | Arap2 | 9.418 | 6.553 | 7.556 | 2.866 | −1.003 | 0.000 | 0.026 |
| ENSMUST00000217197 | Gm1110 | 2.230 | 0.000 | 2.863 | 2.230 | −2.863 | 0.019 | 0.029 |
| ENSMUST00000167633 | Bean1 | 3.138 | 0.000 | 4.088 | 3.138 | −4.088 | 0.029 | 0.007 |
| ENSMUST00000212479 | Syde2 | 4.087 | 5.554 | 3.949 | −1.466 | 1.605 | 0.001 | 0.001 |
| ENSMUST00000006431 | Atp6v1b1 | 4.989 | 2.003 | 4.298 | 2.986 | −2.295 | 0.001 | 0.028 |
| ENSMUST00000048485 | Sln | 3.481 | 1.377 | 3.693 | 2.103 | −2.316 | 0.046 | 0.024 |
| ENSMUST00000123140 | Lrp8 | 4.427 | 8.085 | 4.750 | −3.658 | 3.335 | 0.004 | 0.000 |
| ENSMUST00000177840 | Cd151 | 7.394 | 6.120 | 8.358 | 1.274 | −2.238 | 0.005 | 0.000 |
| ENSMUST00000138091 | Ahcyl1 | 7.020 | 5.589 | 7.247 | 1.430 | −1.658 | 0.003 | 0.002 |
| ENSMUST00000068617 | Mettl27 | 0.000 | 2.776 | 0.000 | −2.776 | 2.776 | 0.010 | 0.004 |
| ENSMUST00000091420 | Skida1 | 3.501 | 7.791 | 5.321 | −4.289 | 2.469 | 0.000 | 0.015 |
| ENSMUST00000123594 | Otub1 | 0.613 | 4.015 | 1.567 | −3.403 | 2.448 | 0.000 | 0.000 |
| ENSMUST00000134105 | Slc2a1 | 4.878 | 2.774 | 5.156 | 2.104 | −2.382 | 0.002 | 0.000 |
| ENSMUST00000207252 | Crebzf | 6.366 | 4.601 | 5.734 | 1.764 | −1.133 | 0.004 | 0.045 |
| ENSMUST00000207879 | Supt5 | 7.658 | 5.677 | 9.073 | 1.981 | −3.396 | 0.024 | 0.000 |
| ENSMUST00000149972 | Nfat5 | 4.870 | 1.512 | 4.541 | 3.359 | −3.029 | 0.002 | 0.000 |
| ENSMUST00000165320 | Fiz1 | 4.853 | 3.254 | 5.193 | 1.599 | −1.939 | 0.023 | 0.007 |
| ENSMUST00000208131 | Myh14 | 3.223 | 0.000 | 4.033 | 3.223 | −4.033 | 0.001 | 0.000 |
| ENSMUST00000087133 | Exoc1 | 5.273 | 1.626 | 5.135 | 3.647 | −3.509 | 0.000 | 0.000 |
| ENSMUST00000110013 | Jak3 | 2.197 | 5.392 | 2.549 | −3.195 | 2.844 | 0.004 | 0.024 |
| ENSMUST00000111566 | Clip1 | 6.623 | 8.717 | 6.162 | −2.094 | 2.555 | 0.029 | 0.003 |
| ENSMUST00000220509 | Ppp2r5c | 7.300 | 3.326 | 5.073 | 3.974 | −1.747 | 0.000 | 0.015 |
| ENSMUST00000117460 | Tmcc3 | 7.888 | 6.057 | 8.562 | 1.831 | −2.505 | 0.003 | 0.000 |
| ENSMUST00000043429 | Nfyc | 6.958 | 8.069 | 6.807 | −1.112 | 1.262 | 0.001 | 0.006 |
| ENSMUST00000106494 | Pomgnt1 | 3.570 | 6.306 | 4.327 | −2.736 | 1.979 | 0.000 | 0.003 |
| ENSMUST00000114069 | Elmod3 | 4.610 | 1.414 | 4.729 | 3.196 | −3.315 | 0.002 | 0.000 |
| ENSMUST00000136721 | Ncln | 5.407 | 4.193 | 5.423 | 1.214 | −1.229 | 0.031 | 0.027 |
| ENSMUST00000062580 | Itsn2 | 8.126 | 5.605 | 6.955 | 2.521 | −1.350 | 0.000 | 0.000 |
| ENSMUST00000051390 | Zbtb45 | 8.726 | 5.951 | 7.269 | 2.775 | −1.318 | 0.000 | 0.003 |
| ENSMUST00000114882 | Rps10 | 6.118 | 4.729 | 6.976 | 1.389 | −2.247 | 0.025 | 0.000 |
| ENSMUST00000159959 | Hr | 2.769 | 0.342 | 2.345 | 2.427 | −2.003 | 0.006 | 0.023 |
| ENSMUST00000110437 | Dtwd1 | 4.793 | 6.547 | 4.580 | −1.754 | 1.968 | 0.000 | 0.000 |
| ENSMUST00000176509 | Zbtb25 | 5.392 | 3.596 | 5.330 | 1.796 | −1.734 | 0.000 | 0.000 |
| ENSMUST00000039752 | Slc16a8 | 5.257 | 4.094 | 5.258 | 1.163 | −1.164 | 0.033 | 0.034 |
| ENSMUST00000152544 | Plk5 | 6.321 | 5.290 | 6.606 | 1.031 | −1.315 | 0.044 | 0.025 |
| ENSMUST00000201219 | Nat8f3 | 4.748 | 3.662 | 5.635 | 1.087 | −1.973 | 0.017 | 0.000 |
| ENSMUST00000033608 | Sytl4 | 5.079 | 3.077 | 4.722 | 2.002 | −1.645 | 0.002 | 0.020 |
| ENSMUST00000215324 | Olfr372 | 0.000 | 2.278 | 0.000 | −2.278 | 2.278 | 0.016 | 0.008 |
| ENSMUST00000161882 | Ubl5 | 4.207 | 5.776 | 3.717 | −1.569 | 2.060 | 0.002 | 0.000 |
| ENSMUST00000163421 | Axin1 | 8.800 | 6.591 | 8.773 | 2.209 | −2.182 | 0.000 | 0.000 |
| ENSMUST00000123384 | Angel2 | 6.848 | 5.171 | 6.595 | 1.677 | −1.424 | 0.003 | 0.011 |
| ENSMUST00000180636 | Slc35a5 | 3.489 | 5.460 | 2.816 | −1.971 | 2.644 | 0.007 | 0.001 |
| ENSMUST00000210358 | Isyna1 | 4.755 | 3.428 | 4.521 | 1.327 | −1.094 | 0.013 | 0.048 |
| ENSMUST00000110124 | Homer3 | 3.858 | 1.432 | 3.969 | 2.426 | −2.537 | 0.026 | 0.002 |
| ENSMUST00000169536 | Zfyve27 | 10.679 | 9.064 | 10.246 | 1.616 | −1.182 | 0.000 | 0.003 |
| ENSMUST00000045011 | Atrip | 4.230 | 7.404 | 5.683 | −3.174 | 1.722 | 0.000 | 0.003 |
| ENSMUST00000044204 | Tyw1 | 6.193 | 1.072 | 5.376 | 5.120 | −4.304 | 0.000 | 0.006 |
| ENSMUST00000102692 | Pemt | 1.113 | 3.936 | 0.638 | −2.823 | 3.298 | 0.001 | 0.000 |
| ENSMUST00000108690 | Sco1 | 2.953 | 4.820 | 3.336 | −1.867 | 1.484 | 0.003 | 0.039 |
| ENSMUST00000118810 | Mapk1ip1 | 5.624 | 3.973 | 6.284 | 1.651 | −2.311 | 0.000 | 0.000 |
| ENSMUST00000220218 | Dusp6 | 2.374 | 4.045 | 2.309 | −1.671 | 1.735 | 0.006 | 0.009 |
| ENSMUST00000139960 | Zfp740 | 6.075 | 4.873 | 6.204 | 1.202 | −1.331 | 0.026 | 0.001 |
| ENSMUST00000126638 | Spc25 | 2.813 | 0.342 | 2.245 | 2.471 | −1.903 | 0.006 | 0.034 |
| ENSMUST00000002145 | Hsf2bp | 4.455 | 2.903 | 4.282 | 1.552 | −1.378 | 0.004 | 0.007 |
| ENSMUST00000162637 | Reln | 5.735 | 4.265 | 6.382 | 1.470 | −2.117 | 0.007 | 0.000 |
| ENSMUST00000184503 | Gpr155 | 8.226 | 6.879 | 8.734 | 1.347 | −1.855 | 0.017 | 0.002 |
| ENSMUST00000152243 | Kctd9 | 5.620 | 4.192 | 6.162 | 1.429 | −1.970 | 0.011 | 0.000 |
| ENSMUST00000156420 | Ipo4 | 5.184 | 7.463 | 3.797 | −2.279 | 3.666 | 0.047 | 0.005 |
| ENSMUST00000201100 | Nsun7 | 5.774 | 4.667 | 6.183 | 1.106 | −1.515 | 0.000 | 0.001 |
| ENSMUST00000084325 | B4galt2 | 3.816 | 0.000 | 3.640 | 3.816 | −3.640 | 0.030 | 0.012 |
| ENSMUST00000020981 | Cenpo | 2.824 | 0.000 | 2.716 | 2.824 | −2.716 | 0.047 | 0.026 |
| ENSMUST00000217046 | Plekha7 | 6.090 | 7.206 | 5.530 | −1.116 | 1.676 | 0.001 | 0.000 |
| ENSMUST00000149575 | Spag8 | 0.000 | 3.071 | 0.000 | −3.071 | 3.071 | 0.003 | 0.001 |
| ENSMUST00000137699 | Lap3 | 3.857 | 5.296 | 3.500 | −1.438 | 1.795 | 0.006 | 0.003 |
| ENSMUST00000110232 | Adam33 | 1.734 | 4.021 | 0.000 | −2.287 | 4.021 | 0.024 | 0.000 |
| ENSMUST00000224385 | Nedd4l | 2.247 | 0.342 | 2.340 | 1.905 | −1.998 | 0.048 | 0.030 |
| ENSMUST00000163297 | Tmc3 | 3.530 | 0.000 | 5.492 | 3.530 | −5.492 | 0.033 | 0.000 |
| ENSMUST00000038369 | Cipc | 9.865 | 1.951 | 4.142 | 7.914 | −2.192 | 0.000 | 0.003 |
| ENSMUST00000155260 | Cmc2 | 5.463 | 4.162 | 5.530 | 1.300 | −1.368 | 0.001 | 0.019 |
| ENSMUST00000115098 | Kcnh2 | 0.759 | 6.438 | 7.830 | −5.679 | −1.392 | 0.000 | 0.005 |
| ENSMUST00000211431 | Trpm4 | 5.630 | 6.960 | 5.564 | −1.330 | 1.396 | 0.011 | 0.038 |
| ENSMUST00000178644 | Gm21969 | 4.940 | 3.544 | 5.875 | 1.396 | −2.331 | 0.013 | 0.000 |
| ENSMUST00000183241 | Rnasel | 3.583 | 0.000 | 5.634 | 3.583 | −5.634 | 0.021 | 0.000 |
| ENSMUST00000132123 | Ago3 | 3.829 | 2.512 | 3.889 | 1.318 | −1.377 | 0.028 | 0.020 |
| ENSMUST00000196090 | Lgi1 | 3.488 | 0.000 | 3.219 | 3.488 | −3.219 | 0.029 | 0.026 |
| ENSMUST00000126066 | Zbtb38 | 3.411 | 6.845 | 5.634 | −3.434 | 1.211 | 0.045 | 0.001 |
| ENSMUST00000027495 | 44076 | 7.256 | 8.760 | 7.174 | −1.503 | 1.585 | 0.009 | 0.003 |
| ENSMUST00000023441 | P2rx6 | 7.435 | 3.628 | 7.540 | 3.807 | −3.912 | 0.022 | 0.015 |
| ENSMUST00000129554 | Surf6 | 2.902 | 0.000 | 3.859 | 2.902 | −3.859 | 0.044 | 0.000 |
| ENSMUST00000096222 | Kat6b | 7.744 | 8.993 | 7.666 | −1.249 | 1.327 | 0.004 | 0.000 |
| ENSMUST00000109946 | Scrib | 8.472 | 6.619 | 8.105 | 1.853 | −1.486 | 0.000 | 0.000 |
| ENSMUST00000110132 | 1110034G24Rik | 4.710 | 3.363 | 5.112 | 1.347 | −1.749 | 0.023 | 0.004 |
| ENSMUST00000038959 | Psd3 | 4.254 | 2.557 | 4.598 | 1.697 | −2.040 | 0.010 | 0.001 |
| ENSMUST00000146003 | Mrpl48 | 6.039 | 7.915 | 5.505 | −1.875 | 2.410 | 0.001 | 0.000 |
| ENSMUST00000143685 | Fkbp5 | 3.618 | 1.516 | 3.902 | 2.103 | −2.386 | 0.018 | 0.003 |
| ENSMUST00000172728 | Fam92a | 4.685 | 3.567 | 4.938 | 1.118 | −1.371 | 0.040 | 0.006 |
| ENSMUST00000115443 | Met | 3.331 | 5.935 | 3.912 | −2.604 | 2.023 | 0.002 | 0.002 |
| ENSMUST00000018547 | Ggnbp2 | 9.278 | 7.995 | 9.206 | 1.282 | −1.211 | 0.000 | 0.000 |
| ENSMUST00000143867 | Slc35f6 | 3.274 | 0.000 | 2.723 | 3.274 | −2.723 | 0.028 | 0.026 |
| ENSMUST00000143864 | Trip4 | 0.000 | 4.650 | 1.841 | −4.650 | 2.809 | 0.000 | 0.003 |
| ENSMUST00000150258 | Sgpl1 | 6.009 | 7.546 | 6.306 | −1.537 | 1.240 | 0.023 | 0.037 |
| ENSMUST00000166735 | Papola | 4.496 | 5.868 | 4.842 | −1.372 | 1.026 | 0.020 | 0.042 |
| ENSMUST00000137822 | Rasa3 | 9.314 | 7.451 | 9.276 | 1.863 | −1.825 | 0.000 | 0.000 |
| ENSMUST00000030800 | Fastk | 2.527 | 5.315 | 2.357 | −2.788 | 2.958 | 0.004 | 0.003 |
| ENSMUST00000174779 | Csnk2b | 1.026 | 3.230 | 0.963 | −2.204 | 2.267 | 0.031 | 0.014 |
| ENSMUST00000127233 | Tcf7l2 | 7.483 | 5.683 | 7.752 | 1.799 | −2.068 | 0.045 | 0.020 |
| ENSMUST00000123440 | Rap1gap2 | 9.417 | 7.762 | 6.430 | 1.655 | 1.332 | 0.031 | 0.001 |
| ENSMUST00000199125 | Rilpl1 | 4.373 | 1.811 | 3.940 | 2.562 | −2.129 | 0.000 | 0.001 |
| ENSMUST00000105701 | Masp2 | 0.331 | 3.169 | 0.000 | −2.838 | 3.169 | 0.015 | 0.004 |
| ENSMUST00000107068 | E130114P18Rik | 0.695 | 3.945 | 0.674 | −3.250 | 3.271 | 0.000 | 0.000 |
| ENSMUST00000110506 | Sqor | 3.584 | 0.331 | 2.652 | 3.253 | −2.322 | 0.016 | 0.008 |
| ENSMUST00000200570 | Zzz3 | 5.183 | 0.000 | 5.078 | 5.183 | −5.078 | 0.000 | 0.000 |
| ENSMUST00000109997 | Zfp961 | 7.935 | 6.827 | 9.272 | 1.108 | −2.444 | 0.023 | 0.000 |
| ENSMUST00000222617 | Odc1 | 3.272 | 4.459 | 1.759 | −1.187 | 2.699 | 0.010 | 0.006 |
| ENSMUST00000088796 | Tex47 | 3.190 | 4.543 | 2.666 | −1.352 | 1.876 | 0.026 | 0.003 |
| ENSMUST00000156455 | Atf2 | 5.053 | 1.964 | 5.503 | 3.088 | −3.539 | 0.027 | 0.005 |
| ENSMUST00000109202 | Ifi47 | 3.744 | 0.986 | 3.294 | 2.758 | −2.308 | 0.002 | 0.002 |
| ENSMUST00000122066 | Zfp629 | 5.220 | 3.222 | 5.502 | 1.998 | −2.280 | 0.004 | 0.002 |
| ENSMUST00000113261 | Cntn4 | 6.036 | 7.280 | 5.645 | −1.244 | 1.635 | 0.016 | 0.011 |
| ENSMUST00000170538 | Eci2 | 0.000 | 3.488 | 0.943 | −3.488 | 2.545 | 0.041 | 0.029 |
| ENSMUST00000148702 | Uimc1 | 3.202 | 0.000 | 3.039 | 3.202 | −3.039 | 0.036 | 0.024 |
| ENSMUST00000167110 | Nfe2l1 | 4.342 | 1.516 | 3.130 | 2.826 | −1.614 | 0.002 | 0.030 |
| ENSMUST00000212716 | Enkd1 | 4.287 | 2.259 | 3.868 | 2.027 | −1.609 | 0.007 | 0.049 |
| ENSMUST00000187400 | Abi2 | 3.821 | 1.378 | 3.890 | 2.443 | −2.512 | 0.002 | 0.000 |
| ENSMUST00000177943 | Slc45a3 | 3.324 | 0.000 | 6.362 | 3.324 | −6.362 | 0.038 | 0.000 |
| ENSMUST00000029666 | Papss1 | 9.775 | 7.000 | 8.474 | 2.775 | −1.474 | 0.000 | 0.014 |
| ENSMUST00000022222 | Erbin | 1.026 | 3.028 | 0.963 | −2.001 | 2.064 | 0.025 | 0.012 |
| ENSMUST00000167096 | Agtpbp1 | 5.791 | 2.965 | 4.148 | 2.826 | −1.184 | 0.000 | 0.050 |
| ENSMUST00000223068 | Ispd | 5.296 | 0.331 | 4.069 | 4.965 | −3.738 | 0.000 | 0.010 |
| ENSMUST00000039165 | Golga1 | 4.925 | 6.119 | 7.625 | −1.194 | −1.506 | 0.031 | 0.010 |
| ENSMUST00000001963 | Ace | 9.300 | 7.967 | 10.447 | 1.333 | −2.480 | 0.007 | 0.000 |
| ENSMUST00000066545 | Shank3 | 7.184 | 8.216 | 6.908 | −1.032 | 1.308 | 0.024 | 0.039 |
| ENSMUST00000172578 | Khdc3 | 3.774 | 1.598 | 3.441 | 2.176 | −1.842 | 0.018 | 0.049 |
| ENSMUST00000064285 | Glcci1 | 10.489 | 7.730 | 9.740 | 2.759 | −2.010 | 0.006 | 0.040 |
| ENSMUST00000038193 | Wdr59 | 7.378 | 6.330 | 7.632 | 1.047 | −1.301 | 0.030 | 0.007 |
| ENSMUST00000161908 | Scyl3 | 3.947 | 6.819 | 4.701 | −2.872 | 2.117 | 0.002 | 0.010 |
| ENSMUST00000186242 | Ppil3 | 3.271 | 1.518 | 3.303 | 1.754 | −1.785 | 0.016 | 0.018 |
| ENSMUST00000084455 | Olfr61 | 3.150 | 4.240 | 3.054 | −1.090 | 1.186 | 0.034 | 0.020 |
| ENSMUST00000226068 | Myof | 3.580 | 2.194 | 5.175 | 1.387 | −2.981 | 0.035 | 0.000 |
| ENSMUST00000156053 | Btbd17 | 6.378 | 5.353 | 7.068 | 1.025 | −1.715 | 0.045 | 0.000 |
| ENSMUST00000021817 | Ippk | 4.258 | 5.309 | 4.173 | −1.051 | 1.136 | 0.023 | 0.045 |
| ENSMUST00000029448 | Sycp1 | 0.611 | 2.837 | 4.858 | −2.225 | −2.022 | 0.027 | 0.004 |
| ENSMUST00000133925 | Terf2 | 7.233 | 6.224 | 7.856 | 1.009 | −1.632 | 0.045 | 0.000 |
| ENSMUST00000029946 | Rragd | 0.397 | 2.554 | 0.638 | −2.157 | 1.916 | 0.041 | 0.025 |
| ENSMUST00000221379 | Rps6ka5 | 4.858 | 0.986 | 4.736 | 3.872 | −3.750 | 0.001 | 0.000 |
| ENSMUST00000130597 | Mavs | 2.699 | 1.372 | 2.751 | 1.327 | −1.379 | 0.049 | 0.034 |
| ENSMUST00000160143 | Rrnad1 | 3.663 | 1.706 | 3.160 | 1.957 | −1.454 | 0.002 | 0.032 |
| ENSMUST00000162170 | Pianp | 7.530 | 2.281 | 8.457 | 5.248 | −6.176 | 0.000 | 0.000 |
| ENSMUST00000140635 | Rabl6 | 4.685 | 3.541 | 5.380 | 1.143 | −1.839 | 0.039 | 0.000 |
| ENSMUST00000140632 | Ubl4a | 4.704 | 3.369 | 4.568 | 1.335 | −1.198 | 0.010 | 0.045 |
| ENSMUST00000125887 | Pde1a | 3.669 | 5.066 | 2.920 | −1.397 | 2.146 | 0.021 | 0.003 |
| ENSMUST00000032926 | Tmem219 | 2.651 | 0.000 | 4.685 | 2.651 | −4.685 | 0.042 | 0.000 |
| ENSMUST00000141305 | Ift122 | 4.784 | 7.336 | 5.010 | −2.552 | 2.326 | 0.001 | 0.000 |
| ENSMUST00000115593 | Deup1 | 3.664 | 0.788 | 3.619 | 2.876 | −2.831 | 0.010 | 0.015 |
| ENSMUST00000186017 | Nek7 | 8.854 | 7.807 | 8.992 | 1.047 | −1.185 | 0.038 | 0.027 |
| ENSMUST00000151296 | Osgepl1 | 6.673 | 5.373 | 6.659 | 1.300 | −1.286 | 0.002 | 0.000 |
| ENSMUST00000168649 | Xpo6 | 3.730 | 4.899 | 3.101 | −1.169 | 1.797 | 0.035 | 0.002 |
| ENSMUST00000114388 | Nsmf | 7.308 | 4.716 | 6.847 | 2.592 | −2.130 | 0.001 | 0.008 |
| ENSMUST00000058089 | Raly | 9.897 | 8.510 | 9.573 | 1.387 | −1.063 | 0.001 | 0.021 |
| ENSMUST00000140257 | Lsm14b | 2.992 | 0.000 | 2.648 | 2.992 | −2.648 | 0.038 | 0.034 |
| ENSMUST00000161648 | Mff | 0.000 | 2.936 | 0.000 | −2.936 | 2.936 | 0.049 | 0.025 |
| ENSMUST00000130658 | Zfp46 | 4.418 | 5.906 | 4.853 | −1.488 | 1.053 | 0.000 | 0.034 |
| ENSMUST00000150898 | Slc25a19 | 6.147 | 4.258 | 5.761 | 1.889 | −1.503 | 0.001 | 0.000 |
| ENSMUST00000100302 | Prkca | 2.683 | 4.429 | 2.629 | −1.745 | 1.800 | 0.001 | 0.001 |
| ENSMUST00000176983 | Wdr90 | 3.598 | 4.705 | 3.486 | −1.107 | 1.219 | 0.029 | 0.036 |
| ENSMUST00000029446 | Csde1 | 11.398 | 10.346 | 11.597 | 1.053 | −1.251 | 0.018 | 0.008 |
| ENSMUST00000199502 | Rasa4 | 2.472 | 3.803 | 4.923 | −1.331 | −1.120 | 0.023 | 0.020 |
| ENSMUST00000201094 | Tcf4 | 5.095 | 0.000 | 5.231 | 5.095 | −5.231 | 0.000 | 0.000 |
| ENSMUST00000187083 | Hivep2 | 8.934 | 10.610 | 8.374 | −1.676 | 2.235 | 0.000 | 0.000 |
| ENSMUST00000146642 | Homez | 0.000 | 2.362 | 0.674 | −2.362 | 1.688 | 0.039 | 0.046 |
| ENSMUST00000150606 | Pex11a | 4.102 | 2.612 | 4.208 | 1.490 | −1.596 | 0.035 | 0.013 |
| ENSMUST00000094154 | Serpinh1 | 9.021 | 7.710 | 8.850 | 1.311 | −1.140 | 0.006 | 0.035 |
| ENSMUST00000082121 | Glmn | 0.000 | 8.733 | 0.289 | −8.733 | 8.444 | 0.000 | 0.000 |
| ENSMUST00000179041 | Cep170b | 3.151 | 4.540 | 3.127 | −1.389 | 1.413 | 0.028 | 0.021 |
| ENSMUST00000101049 | Elac2 | 9.281 | 7.945 | 9.302 | 1.336 | −1.357 | 0.014 | 0.011 |
| ENSMUST00000192844 | Pfkfb3 | 8.749 | 6.867 | 8.808 | 1.882 | −1.942 | 0.000 | 0.000 |
| ENSMUST00000034046 | Acsl1 | 10.077 | 8.990 | 10.270 | 1.086 | −1.280 | 0.000 | 0.000 |
| ENSMUST00000103236 | Evi2a | 7.802 | 5.028 | 8.180 | 2.774 | −3.151 | 0.025 | 0.010 |
| ENSMUST00000135346 | Aspscr1 | 5.440 | 0.673 | 4.545 | 4.767 | −3.872 | 0.000 | 0.000 |
| ENSMUST00000043865 | Mpst | 3.832 | 2.033 | 5.771 | 1.799 | −3.738 | 0.019 | 0.000 |
| ENSMUST00000204182 | Rarres2 | 4.573 | 3.509 | 4.715 | 1.064 | −1.206 | 0.020 | 0.005 |
| ENSMUST00000207168 | Cars | 3.754 | 1.946 | 4.161 | 1.809 | −2.216 | 0.017 | 0.004 |
| ENSMUST00000071697 | Fxyd1 | 4.239 | 2.364 | 4.158 | 1.874 | −1.794 | 0.027 | 0.035 |
| ENSMUST00000160899 | B3gat1 | 7.206 | 4.797 | 7.239 | 2.409 | −2.443 | 0.000 | 0.000 |
| ENSMUST00000105839 | Usp48 | 6.243 | 4.496 | 5.996 | 1.747 | −1.500 | 0.010 | 0.018 |
| ENSMUST00000109731 | Cdk5rap1 | 5.720 | 2.819 | 4.874 | 2.901 | −2.055 | 0.010 | 0.031 |
| ENSMUST00000047075 | Setd1a | 8.795 | 5.346 | 7.673 | 3.449 | −2.327 | 0.000 | 0.000 |
| ENSMUST00000054449 | Plec | 8.869 | 5.080 | 8.668 | 3.789 | −3.588 | 0.012 | 0.001 |
| ENSMUST00000079056 | Arrb2 | 3.755 | 0.655 | 3.802 | 3.100 | −3.147 | 0.002 | 0.001 |
| ENSMUST00000112087 | Tnnt2 | 0.000 | 2.146 | 0.000 | −2.146 | 2.146 | 0.027 | 0.015 |
| ENSMUST00000213990 | Gnb5 | 7.190 | 3.929 | 6.721 | 3.261 | −2.792 | 0.000 | 0.023 |
| ENSMUST00000189542 | Nabp1 | 7.663 | 2.805 | 4.869 | 4.858 | −2.064 | 0.000 | 0.004 |
| ENSMUST00000143224 | Gramd2 | 6.278 | 7.463 | 6.093 | −1.185 | 1.370 | 0.015 | 0.006 |
| ENSMUST00000126123 | Optc | 3.631 | 0.540 | 3.776 | 3.091 | −3.236 | 0.001 | 0.000 |
| ENSMUST00000031845 | Dfna5 | 1.842 | 3.329 | 1.665 | −1.487 | 1.664 | 0.048 | 0.023 |
| ENSMUST00000210676 | Gab1 | 5.602 | 3.055 | 6.797 | 2.547 | −3.741 | 0.029 | 0.000 |
| ENSMUST00000166101 | Sptb | 10.109 | 7.509 | 10.113 | 2.600 | −2.603 | 0.000 | 0.000 |
| ENSMUST00000019447 | Psmc3ip | 3.554 | 5.089 | 3.651 | −1.535 | 1.438 | 0.030 | 0.002 |
| ENSMUST00000075180 | Baiap2 | 4.153 | 5.293 | 4.188 | −1.141 | 1.105 | 0.044 | 0.033 |
| ENSMUST00000185661 | Zfp672 | 7.882 | 0.741 | 8.445 | 7.141 | −7.704 | 0.000 | 0.000 |
| ENSMUST00000112634 | Galnt13 | 8.106 | 6.138 | 8.595 | 1.968 | −2.457 | 0.007 | 0.000 |
| ENSMUST00000193432 | Cog6 | 5.165 | 3.934 | 6.739 | 1.231 | −2.805 | 0.023 | 0.000 |
| ENSMUST00000056442 | Slc6a5 | 3.385 | 5.045 | 12.001 | −1.659 | −6.957 | 0.015 | 0.023 |
| ENSMUST00000053959 | Ints6 | 7.969 | 6.224 | 7.504 | 1.745 | −1.280 | 0.004 | 0.023 |
| ENSMUST00000225663 | Arhgef28 | 7.537 | 5.907 | 7.662 | 1.630 | −1.755 | 0.008 | 0.005 |
| ENSMUST00000087684 | Rgs12 | 4.907 | 8.100 | 3.735 | −3.192 | 4.365 | 0.000 | 0.010 |
| ENSMUST00000136375 | Rasal3 | 3.792 | 0.000 | 3.578 | 3.792 | −3.578 | 0.026 | 0.000 |
| ENSMUST00000062861 | Rabggta | 8.387 | 4.807 | 6.610 | 3.580 | −1.803 | 0.000 | 0.010 |
| ENSMUST00000134667 | Syvn1 | 4.491 | 1.506 | 6.144 | 2.985 | −4.638 | 0.014 | 0.000 |
| ENSMUST00000164877 | Rims1 | 4.824 | 2.684 | 4.527 | 2.140 | −1.843 | 0.006 | 0.006 |
| ENSMUST00000128668 | Fbxl21 | 4.868 | 3.770 | 4.908 | 1.098 | −1.138 | 0.038 | 0.032 |
| ENSMUST00000123277 | Slc35a2 | 3.579 | 0.000 | 2.611 | 3.579 | −2.611 | 0.000 | 0.035 |
| ENSMUST00000173114 | Csnk2b | 1.133 | 3.772 | 1.460 | −2.639 | 2.312 | 0.034 | 0.010 |
| ENSMUST00000058879 | Ntf5 | 2.516 | 0.000 | 2.977 | 2.516 | −2.977 | 0.009 | 0.000 |
| ENSMUST00000180196 | Slc39a8 | 3.596 | 0.342 | 3.502 | 3.254 | −3.159 | 0.033 | 0.014 |
| ENSMUST00000154043 | Numb | 0.298 | 3.548 | 0.000 | −3.250 | 3.548 | 0.026 | 0.012 |
| ENSMUST00000149685 | Tspyl2 | 6.814 | 9.395 | 7.024 | −2.581 | 2.371 | 0.004 | 0.013 |
| ENSMUST00000032132 | Efcc1 | 5.992 | 3.108 | 6.796 | 2.883 | −3.687 | 0.000 | 0.000 |
| ENSMUST00000115712 | Nrg2 | 4.944 | 3.428 | 5.212 | 1.516 | −1.784 | 0.016 | 0.005 |
| ENSMUST00000168428 | Znrf1 | 5.733 | 6.933 | 5.079 | −1.200 | 1.854 | 0.045 | 0.002 |
| ENSMUST00000177731 | Rabep1 | 3.096 | 6.252 | 3.283 | −3.156 | 2.969 | 0.000 | 0.000 |
| ENSMUST00000143141 | Arhgef16 | 4.191 | 0.000 | 4.717 | 4.191 | −4.717 | 0.000 | 0.000 |
| ENSMUST00000125946 | Gm42878 | 7.976 | 3.944 | 6.477 | 4.032 | −2.533 | 0.000 | 0.007 |
| ENSMUST00000126287 | Srcin1 | 0.000 | 4.140 | 0.000 | −4.140 | 4.140 | 0.000 | 0.000 |
| ENSMUST00000136640 | Fcho1 | 7.878 | 6.425 | 5.156 | 1.453 | 1.269 | 0.000 | 0.005 |
| ENSMUST00000043836 | Macrod2 | 7.039 | 2.711 | 4.985 | 4.328 | −2.274 | 0.000 | 0.013 |
| ENSMUST00000172697 | Mecom | 3.534 | 1.112 | 4.620 | 2.422 | −3.508 | 0.008 | 0.001 |
| ENSMUST00000085245 | Slc39a9 | 8.701 | 10.486 | 9.164 | −1.784 | 1.322 | 0.000 | 0.003 |
| ENSMUST00000187307 | Wdr89 | 1.897 | 3.356 | 1.050 | −1.458 | 2.306 | 0.047 | 0.041 |
| ENSMUST00000172330 | Pak3 | 10.944 | 7.820 | 10.732 | 3.125 | −2.913 | 0.002 | 0.001 |
| ENSMUST00000173689 | Jmjd1c | 9.721 | 4.331 | 8.017 | 5.390 | −3.686 | 0.007 | 0.022 |
| ENSMUST00000122327 | Fam160a2 | 5.303 | 3.959 | 5.117 | 1.343 | −1.158 | 0.017 | 0.035 |
| ENSMUST00000039021 | Ssx2ip | 9.876 | 0.000 | 9.956 | 9.876 | −9.956 | 0.000 | 0.000 |
| ENSMUST00000035237 | Usp4 | 8.121 | 5.669 | 8.651 | 2.452 | −2.982 | 0.006 | 0.001 |
| ENSMUST00000117692 | Rab27b | 5.426 | 2.876 | 4.587 | 2.549 | −1.710 | 0.000 | 0.008 |
| ENSMUST00000002395 | Rec8 | 0.000 | 3.280 | 5.304 | −3.280 | −2.024 | 0.000 | 0.001 |
| ENSMUST00000112197 | Kdm5b | 6.939 | 8.538 | 7.101 | −1.599 | 1.437 | 0.000 | 0.002 |
| ENSMUST00000170853 | Synj1 | 10.984 | 12.214 | 10.327 | −1.229 | 1.886 | 0.000 | 0.000 |
| ENSMUST00000173762 | Jmjd1c | 7.791 | 6.175 | 4.994 | 1.617 | 1.181 | 0.003 | 0.041 |
| ENSMUST00000041779 | Clec4a2 | 4.986 | 3.803 | 2.310 | 1.183 | 1.493 | 0.014 | 0.012 |
| ENSMUST00000201595 | Ubl3 | 7.333 | 8.648 | 7.206 | −1.316 | 1.442 | 0.015 | 0.006 |
| ENSMUST00000112923 | Ift122 | 2.912 | 6.264 | 3.521 | −3.352 | 2.743 | 0.000 | 0.001 |
| ENSMUST00000200380 | Crip2 | 2.146 | 5.257 | 3.632 | −3.112 | 1.625 | 0.001 | 0.003 |
| ENSMUST00000107115 | Ubtf | 7.742 | 6.573 | 7.602 | 1.169 | −1.029 | 0.015 | 0.027 |
| ENSMUST00000042009 | Dlg5 | 5.126 | 4.078 | 5.181 | 1.049 | −1.103 | 0.039 | 0.025 |
| ENSMUST00000027067 | Gsta3 | 5.229 | 0.876 | 4.995 | 4.353 | −4.118 | 0.002 | 0.001 |
| ENSMUST00000081393 | Nphp4 | 6.099 | 4.061 | 5.208 | 2.038 | −1.147 | 0.006 | 0.046 |
| ENSMUST00000128345 | Brinp3 | 7.279 | 4.132 | 5.144 | 3.147 | −1.012 | 0.027 | 0.045 |
| ENSMUST00000105914 | Ahdc1 | 1.934 | 3.983 | 1.510 | −2.048 | 2.472 | 0.018 | 0.029 |
| ENSMUST00000224693 | Nsd1 | 3.274 | 0.000 | 4.217 | 3.274 | −4.217 | 0.039 | 0.000 |
| ENSMUST00000196194 | Zfp697 | 6.015 | 7.119 | 5.567 | −1.104 | 1.551 | 0.005 | 0.000 |
| ENSMUST00000147104 | Tia1 | 3.915 | 2.756 | 4.696 | 1.160 | −1.941 | 0.047 | 0.004 |
| ENSMUST00000111143 | Lmo2 | 6.088 | 1.898 | 4.389 | 4.190 | −2.491 | 0.000 | 0.002 |
| ENSMUST00000125935 | Clip3 | 6.063 | 3.720 | 5.960 | 2.343 | −2.239 | 0.005 | 0.004 |
| ENSMUST00000060956 | Trappc1 | 2.862 | 0.000 | 3.002 | 2.862 | −3.002 | 0.050 | 0.019 |
| ENSMUST00000039153 | Cept1 | 3.517 | 2.278 | 3.304 | 1.239 | −1.026 | 0.044 | 0.048 |
| ENSMUST00000170928 | Med7 | 4.267 | 0.986 | 3.124 | 3.281 | −2.138 | 0.002 | 0.007 |
| ENSMUST00000140554 | Rita1 | 4.283 | 1.946 | 4.322 | 2.336 | −2.375 | 0.019 | 0.029 |
| ENSMUST00000204532 | Magi1 | 7.049 | 3.379 | 6.290 | 3.670 | −2.911 | 0.001 | 0.024 |
| ENSMUST00000224754 | Sec24c | 4.884 | 1.169 | 4.641 | 3.715 | −3.471 | 0.032 | 0.043 |
| ENSMUST00000137267 | Ydjc | 2.938 | 1.518 | 4.218 | 1.420 | −2.701 | 0.045 | 0.002 |
| ENSMUST00000066717 | Tcf4 | 4.646 | 2.527 | 4.198 | 2.119 | −1.671 | 0.008 | 0.021 |
| ENSMUST00000130116 | Sema3e | 6.030 | 7.952 | 5.766 | −1.922 | 2.186 | 0.007 | 0.006 |
| ENSMUST00000053666 | Slc12a6 | 6.995 | 4.400 | 5.526 | 2.595 | −1.126 | 0.000 | 0.028 |
| ENSMUST00000137563 | Atp5h | 8.178 | 6.322 | 8.183 | 1.856 | −1.861 | 0.019 | 0.021 |
| ENSMUST00000107201 | Cdc42se1 | 0.000 | 3.646 | 2.254 | −3.646 | 1.391 | 0.004 | 0.035 |
| ENSMUST00000107200 | Cdc42se1 | 6.911 | 5.125 | 6.410 | 1.786 | −1.285 | 0.000 | 0.010 |
| ENSMUST00000146588 | Tm9sf1 | 5.628 | 3.138 | 4.659 | 2.490 | −1.522 | 0.000 | 0.013 |
| ENSMUST00000091763 | Nme8 | 2.312 | 0.000 | 2.091 | 2.312 | −2.091 | 0.027 | 0.021 |
| ENSMUST00000114126 | Stx18 | 8.249 | 7.189 | 8.218 | 1.059 | −1.029 | 0.001 | 0.006 |
| ENSMUST00000195731 | Chst10 | 3.366 | 0.000 | 4.775 | 3.366 | −4.775 | 0.031 | 0.000 |
| ENSMUST00000122116 | Atg16l2 | 3.651 | 4.956 | 3.566 | −1.305 | 1.390 | 0.028 | 0.012 |
| ENSMUST00000145735 | Lamtor5 | 9.765 | 8.207 | 7.000 | 1.559 | 1.206 | 0.000 | 0.006 |
| ENSMUST00000124293 | Dlk1 | 4.827 | 3.749 | 5.203 | 1.078 | −1.454 | 0.047 | 0.005 |
| ENSMUST00000102911 | Slc44a1 | 9.991 | 8.484 | 10.370 | 1.507 | −1.886 | 0.000 | 0.000 |
| ENSMUST00000138282 | Zc3h7a | 7.805 | 4.072 | 6.284 | 3.733 | −2.211 | 0.000 | 0.000 |
| ENSMUST00000149074 | 1810043G02Rik | 4.509 | 3.260 | 4.930 | 1.249 | −1.670 | 0.034 | 0.007 |
| ENSMUST00000135944 | Pdlim2 | 2.428 | 0.000 | 2.072 | 2.428 | −2.072 | 0.010 | 0.027 |
| ENSMUST00000137543 | Samd4 | 9.819 | 8.056 | 10.356 | 1.763 | −2.300 | 0.000 | 0.000 |
| ENSMUST00000069325 | Dnaic2 | 5.967 | 4.074 | 6.252 | 1.893 | −2.179 | 0.000 | 0.000 |
| ENSMUST00000133019 | Trpv4 | 4.421 | 1.421 | 5.281 | 3.000 | −3.860 | 0.034 | 0.000 |
| ENSMUST00000117301 | Ddr1 | 0.000 | 3.331 | 1.865 | −3.331 | 1.465 | 0.008 | 0.035 |
| ENSMUST00000190785 | 2810474O19Rik | 2.198 | 5.566 | 8.132 | −3.368 | −2.567 | 0.004 | 0.007 |
| ENSMUST00000027800 | Tmem63a | 7.516 | 9.708 | 6.436 | −2.192 | 3.272 | 0.024 | 0.000 |
| ENSMUST00000149207 | Col6a4 | 1.572 | 3.846 | 0.000 | −2.274 | 3.846 | 0.007 | 0.000 |
| ENSMUST00000185692 | 2310035C23Rik | 7.998 | 3.290 | 5.524 | 4.708 | −2.234 | 0.008 | 0.037 |
| ENSMUST00000079322 | Heph | 3.689 | 2.086 | 6.842 | 1.603 | −4.756 | 0.014 | 0.000 |
| ENSMUST00000212989 | Slc27a1 | 5.106 | 2.616 | 4.585 | 2.490 | −1.969 | 0.000 | 0.000 |
| ENSMUST00000115026 | Cald1 | 8.622 | 7.021 | 9.284 | 1.601 | −2.262 | 0.002 | 0.000 |
| ENSMUST00000128381 | Haus6 | 4.856 | 5.926 | 4.893 | −1.070 | 1.034 | 0.017 | 0.014 |
| ENSMUST00000212161 | Spata33 | 3.873 | 0.000 | 2.791 | 3.873 | −2.791 | 0.000 | 0.027 |
| ENSMUST00000182903 | Baiap3 | 4.025 | 1.441 | 4.294 | 2.584 | −2.853 | 0.005 | 0.001 |
| ENSMUST00000086209 | Rnasel | 5.296 | 6.957 | 5.144 | −1.661 | 1.813 | 0.001 | 0.000 |
| ENSMUST00000220346 | Cep290 | 7.238 | 5.914 | 7.579 | 1.324 | −1.666 | 0.023 | 0.005 |
| ENSMUST00000098552 | Tmod2 | 12.398 | 14.537 | 12.595 | −2.139 | 1.941 | 0.000 | 0.000 |
| ENSMUST00000042227 | D11Wsu47e | 5.544 | 4.511 | 6.181 | 1.033 | −1.670 | 0.048 | 0.000 |
| ENSMUST00000108124 | Lingo2 | 4.730 | 3.330 | 6.034 | 1.400 | −2.704 | 0.049 | 0.001 |
| ENSMUST00000124725 | Pkp4 | 4.430 | 5.908 | 4.071 | −1.478 | 1.837 | 0.011 | 0.004 |
| ENSMUST00000187638 | Ccnyl1 | 3.578 | 1.041 | 2.781 | 2.536 | −1.740 | 0.002 | 0.050 |
| ENSMUST00000150780 | Acbd4 | 7.679 | 6.128 | 7.149 | 1.551 | −1.021 | 0.000 | 0.016 |
| ENSMUST00000074834 | Plec | 7.507 | 3.706 | 7.207 | 3.801 | −3.501 | 0.037 | 0.033 |
| ENSMUST00000107040 | Acbd4 | 7.435 | 5.212 | 6.657 | 2.224 | −1.446 | 0.000 | 0.021 |
| ENSMUST00000034692 | Olfm2 | 1.573 | 5.007 | 1.778 | −3.434 | 3.229 | 0.001 | 0.000 |
| ENSMUST00000087818 | Olfr262 | 0.000 | 2.179 | 0.000 | −2.179 | 2.179 | 0.027 | 0.015 |
| ENSMUST00000128338 | Btbd11 | 3.560 | 5.825 | 4.823 | −2.265 | 1.002 | 0.003 | 0.041 |
| ENSMUST00000155133 | Rars2 | 3.818 | 0.673 | 2.586 | 3.145 | −1.913 | 0.004 | 0.030 |
| ENSMUST00000165999 | Cyp4f17 | 7.821 | 6.340 | 7.719 | 1.482 | −1.379 | 0.000 | 0.001 |
| ENSMUST00000002007 | Siae | 6.510 | 3.371 | 2.158 | 3.138 | 1.213 | 0.041 | 0.041 |
| ENSMUST00000146201 | Fbxo22 | 8.794 | 7.007 | 8.637 | 1.787 | −1.630 | 0.001 | 0.001 |
| ENSMUST00000110849 | Rcor3 | 5.504 | 2.809 | 6.895 | 2.695 | −4.085 | 0.009 | 0.000 |
| ENSMUST00000171802 | Mcmdc2 | 0.000 | 3.094 | 0.000 | −3.094 | 3.094 | 0.049 | 0.026 |
| ENSMUST00000173669 | C4b | 5.750 | 3.805 | 5.298 | 1.946 | −1.494 | 0.000 | 0.001 |
| ENSMUST00000113645 | Phyhd1 | 3.110 | 1.176 | 3.438 | 1.934 | −2.262 | 0.046 | 0.017 |
| ENSMUST00000214328 | Olfr679 | 0.000 | 2.173 | 0.000 | −2.173 | 2.173 | 0.023 | 0.012 |
| ENSMUST00000137735 | Ctcf | 2.749 | 3.963 | 2.806 | −1.214 | 1.157 | 0.043 | 0.035 |
| ENSMUST00000185861 | Ly6e | 6.325 | 2.038 | 6.939 | 4.287 | −4.900 | 0.000 | 0.000 |
| ENSMUST00000138752 | Hdc | 3.228 | 0.680 | 3.919 | 2.548 | −3.240 | 0.037 | 0.000 |
| ENSMUST00000211743 | Trpm4 | 0.397 | 3.920 | 1.390 | −3.524 | 2.530 | 0.000 | 0.014 |
| ENSMUST00000133176 | B4galt7 | 2.775 | 0.000 | 3.105 | 2.775 | −3.105 | 0.037 | 0.018 |
| ENSMUST00000106923 | Rhog | 1.275 | 5.775 | 2.017 | −4.500 | 3.758 | 0.001 | 0.000 |
| ENSMUST00000108199 | Fut9 | 4.838 | 3.803 | 5.852 | 1.036 | −2.049 | 0.020 | 0.000 |
| ENSMUST00000177420 | Pcdh15 | 3.788 | 5.619 | 3.500 | −1.831 | 2.119 | 0.007 | 0.003 |
| ENSMUST00000019051 | Alox12e | 2.622 | 0.501 | 2.722 | 2.121 | −2.221 | 0.049 | 0.021 |
| ENSMUST00000143238 | 1500009C09Rik | 9.990 | 8.540 | 9.830 | 1.450 | −1.290 | 0.002 | 0.004 |
| ENSMUST00000042146 | Stx18 | 4.523 | 0.000 | 3.312 | 4.523 | −3.312 | 0.000 | 0.020 |
| ENSMUST00000112848 | Mapk10 | 4.548 | 5.837 | 3.563 | −1.289 | 2.275 | 0.022 | 0.001 |
| ENSMUST00000184560 | Nlrp6 | 0.000 | 3.118 | 0.000 | −3.118 | 3.118 | 0.040 | 0.020 |
| ENSMUST00000108429 | Rps19 | 7.779 | 0.000 | 7.264 | 7.779 | −7.264 | 0.000 | 0.000 |
| ENSMUST00000114877 | Parp9 | 6.318 | 3.075 | 5.270 | 3.243 | −2.196 | 0.000 | 0.005 |
| ENSMUST00000162541 | Wnk1 | 5.678 | 7.718 | 4.938 | −2.040 | 2.779 | 0.000 | 0.000 |
| ENSMUST00000123437 | Lmo2 | 5.598 | 3.935 | 5.000 | 1.664 | −1.065 | 0.000 | 0.023 |
| ENSMUST00000210599 | Cars2 | 5.474 | 1.961 | 4.309 | 3.513 | −2.348 | 0.000 | 0.009 |
| ENSMUST00000221694 | Tmem170b | 6.467 | 4.540 | 5.986 | 1.927 | −1.447 | 0.000 | 0.012 |
| ENSMUST00000029120 | Raly | 2.607 | 8.384 | 5.010 | −5.777 | 3.374 | 0.000 | 0.004 |
| ENSMUST00000029126 | Itch | 7.360 | 4.734 | 8.505 | 2.626 | −3.771 | 0.028 | 0.002 |
| ENSMUST00000196130 | Snx8 | 4.768 | 1.813 | 5.247 | 2.955 | −3.434 | 0.021 | 0.003 |
| ENSMUST00000065603 | Lrrc49 | 6.233 | 7.315 | 6.148 | −1.082 | 1.167 | 0.048 | 0.020 |
| ENSMUST00000204655 | C2cd5 | 4.185 | 5.219 | 3.663 | −1.034 | 1.557 | 0.041 | 0.012 |
| ENSMUST00000170771 | Frk | 2.726 | 0.000 | 3.136 | 2.726 | −3.136 | 0.041 | 0.018 |
| ENSMUST00000113870 | Tsc1 | 8.137 | 6.734 | 9.216 | 1.403 | −2.483 | 0.010 | 0.000 |
| ENSMUST00000103088 | Elmo2 | 9.184 | 0.986 | 3.332 | 8.198 | −2.346 | 0.000 | 0.017 |
| ENSMUST00000202991 | Ube2a | 0.000 | 3.593 | 0.000 | −3.593 | 3.593 | 0.000 | 0.000 |
| ENSMUST00000193899 | Pcdh11x | 7.327 | 6.108 | 8.261 | 1.219 | −2.154 | 0.016 | 0.000 |
| ENSMUST00000135424 | Hnrnpdl | 8.899 | 7.284 | 9.880 | 1.615 | −2.595 | 0.000 | 0.000 |
| ENSMUST00000182131 | Rnasel | 6.473 | 7.601 | 5.901 | −1.128 | 1.699 | 0.011 | 0.001 |
| ENSMUST00000155592 | Tfpt | 5.788 | 7.326 | 5.930 | −1.539 | 1.396 | 0.002 | 0.007 |
| ENSMUST00000176363 | Atp2c1 | 6.831 | 8.402 | 7.228 | −1.571 | 1.174 | 0.001 | 0.032 |
| ENSMUST00000072631 | Nkx2-9 | 0.526 | 2.744 | 4.350 | −2.218 | −1.606 | 0.028 | 0.014 |
| ENSMUST00000156357 | Edc4 | 5.082 | 6.432 | 4.385 | −1.351 | 2.047 | 0.024 | 0.000 |
| ENSMUST00000150862 | Dpy19l3 | 3.279 | 4.577 | 2.954 | −1.298 | 1.623 | 0.006 | 0.017 |
| ENSMUST00000178293 | Usp27x | 7.172 | 5.640 | 2.651 | 1.532 | 2.990 | 0.017 | 0.015 |
| ENSMUST00000197307 | Magi2 | 5.084 | 3.017 | 4.456 | 2.067 | −1.438 | 0.000 | 0.004 |
| ENSMUST00000027579 | Actr3 | 2.131 | 5.125 | 1.460 | −2.994 | 3.665 | 0.000 | 0.000 |
| ENSMUST00000138191 | Clu | 4.061 | 2.632 | 4.617 | 1.428 | −1.984 | 0.028 | 0.000 |
| ENSMUST00000098132 | Il11ra1 | 8.321 | 7.304 | 8.648 | 1.017 | −1.344 | 0.017 | 0.000 |
| ENSMUST00000145791 | Tcirg1 | 1.639 | 4.085 | 1.493 | −2.447 | 2.593 | 0.014 | 0.006 |
| ENSMUST00000133775 | Acad10 | 6.578 | 5.403 | 6.684 | 1.175 | −1.281 | 0.002 | 0.002 |
| ENSMUST00000111716 | Arpc3 | 3.780 | 4.835 | 3.384 | −1.055 | 1.450 | 0.033 | 0.012 |
| ENSMUST00000077146 | Dsg1a | 0.664 | 2.879 | 1.123 | −2.216 | 1.757 | 0.049 | 0.045 |
| ENSMUST00000068693 | Wdr86 | 4.658 | 0.663 | 3.469 | 3.995 | −2.806 | 0.000 | 0.015 |
| ENSMUST00000209061 | Zfp36 | 4.727 | 2.355 | 4.873 | 2.372 | −2.518 | 0.002 | 0.001 |
| ENSMUST00000209066 | Irf3 | 2.489 | 0.986 | 3.148 | 1.503 | −2.162 | 0.047 | 0.006 |
| ENSMUST00000196762 | Nme6 | 4.216 | 3.103 | 4.118 | 1.112 | −1.015 | 0.036 | 0.044 |
| ENSMUST00000043794 | Jakmip1 | 2.925 | 9.560 | 4.437 | −6.635 | 5.123 | 0.002 | 0.000 |
| ENSMUST00000109647 | Vstm2a | 6.268 | 4.844 | 6.017 | 1.424 | −1.173 | 0.007 | 0.023 |
| ENSMUST00000072481 | Sgip1 | 8.846 | 10.039 | 4.711 | −1.193 | 5.327 | 0.025 | 0.045 |
| ENSMUST00000135214 | Shank3 | 4.345 | 5.706 | 3.975 | −1.361 | 1.731 | 0.023 | 0.009 |
| ENSMUST00000132726 | Mepce | 2.950 | 0.331 | 3.406 | 2.619 | −3.075 | 0.003 | 0.000 |
| ENSMUST00000171738 | Slc25a53 | 7.214 | 0.000 | 7.063 | 7.214 | −7.063 | 0.000 | 0.000 |
| ENSMUST00000072521 | Adgrl3 | 7.400 | 5.987 | 7.010 | 1.413 | −1.023 | 0.003 | 0.026 |
| ENSMUST00000031797 | Ssmem1 | 2.838 | 3.971 | 2.953 | −1.133 | 1.017 | 0.030 | 0.042 |
| ENSMUST00000192666 | Mfsd7b | 4.830 | 0.000 | 2.853 | 4.830 | −2.853 | 0.000 | 0.026 |
| ENSMUST00000206976 | Mctp2 | 3.740 | 4.930 | 3.810 | −1.190 | 1.121 | 0.035 | 0.026 |
| ENSMUST00000169134 | Fbxl17 | 3.300 | 5.455 | 3.044 | −2.156 | 2.411 | 0.002 | 0.000 |
| ENSMUST00000162663 | Tyw5 | 3.356 | 4.933 | 3.218 | −1.577 | 1.715 | 0.011 | 0.005 |
| ENSMUST00000124745 | Dda1 | 8.804 | 7.454 | 8.510 | 1.349 | −1.055 | 0.007 | 0.050 |
| ENSMUST00000099084 | Zmynd8 | 3.297 | 7.930 | 3.230 | −4.633 | 4.700 | 0.015 | 0.006 |
| ENSMUST00000099296 | Bfsp1 | 0.872 | 4.585 | 0.838 | −3.713 | 3.747 | 0.005 | 0.000 |
| ENSMUST00000211301 | Trak1 | 2.821 | 5.592 | 2.895 | −2.771 | 2.697 | 0.042 | 0.023 |
| ENSMUST00000128256 | Mtmr3 | 9.864 | 4.541 | 9.905 | 5.323 | −5.364 | 0.000 | 0.000 |
| ENSMUST00000075869 | Fbxo40 | 3.006 | 4.209 | 6.307 | −1.203 | −2.098 | 0.040 | 0.005 |
| ENSMUST00000160098 | Tbc1d31 | 5.016 | 3.551 | 5.413 | 1.465 | −1.862 | 0.028 | 0.006 |
| ENSMUST00000081919 | Musk | 3.226 | 0.000 | 2.648 | 3.226 | −2.648 | 0.036 | 0.034 |
| ENSMUST00000186056 | Myrf | 8.043 | 6.501 | 7.789 | 1.542 | −1.288 | 0.001 | 0.009 |
| ENSMUST00000139266 | Sulf2 | 4.756 | 6.192 | 4.868 | −1.435 | 1.324 | 0.029 | 0.017 |
| ENSMUST00000147132 | Pcsk4 | 3.869 | 0.501 | 4.972 | 3.368 | −4.471 | 0.000 | 0.000 |
| ENSMUST00000111977 | Cfh | 3.149 | 1.613 | 3.912 | 1.536 | −2.299 | 0.044 | 0.006 |
| ENSMUST00000113800 | B3galt5 | 3.256 | 4.432 | 2.655 | −1.176 | 1.777 | 0.038 | 0.027 |
| ENSMUST00000118917 | Chdh | 3.785 | 4.932 | 2.547 | −1.147 | 2.385 | 0.039 | 0.007 |
| ENSMUST00000131462 | Plxnb1 | 4.340 | 3.106 | 4.548 | 1.233 | −1.442 | 0.023 | 0.003 |
| ENSMUST00000178615 | Rbm4 | 4.764 | 3.572 | 4.782 | 1.192 | −1.209 | 0.038 | 0.023 |
| ENSMUST00000172282 | Ptbp1 | 4.969 | 2.570 | 6.060 | 2.399 | −3.490 | 0.012 | 0.000 |
| ENSMUST00000173159 | Chd1 | 3.064 | 4.386 | 3.250 | −1.322 | 1.136 | 0.027 | 0.025 |
| ENSMUST00000152095 | 9330182L06Rik | 5.896 | 4.067 | 5.234 | 1.829 | −1.167 | 0.006 | 0.032 |
| ENSMUST00000100287 | Abca8a | 5.083 | 2.096 | 4.787 | 2.987 | −2.690 | 0.005 | 0.000 |
| ENSMUST00000064244 | Rexo4 | 1.541 | 4.468 | 1.908 | −2.926 | 2.559 | 0.000 | 0.000 |
| ENSMUST00000155491 | Bri3 | 0.000 | 2.581 | 0.517 | −2.581 | 2.064 | 0.006 | 0.034 |
| ENSMUST00000214640 | Commd4 | 1.444 | 3.654 | 1.465 | −2.210 | 2.189 | 0.002 | 0.001 |
| ENSMUST00000150029 | Reps1 | 5.907 | 4.148 | 5.775 | 1.759 | −1.627 | 0.034 | 0.049 |
| ENSMUST00000132689 | Ripor2 | 3.936 | 5.590 | 3.196 | −1.655 | 2.395 | 0.010 | 0.001 |
| ENSMUST00000082303 | Col15a1 | 3.513 | 0.000 | 2.657 | 3.513 | −2.657 | 0.001 | 0.041 |
| ENSMUST00000056338 | Zfp536 | 3.963 | 2.098 | 6.349 | 1.865 | −4.251 | 0.029 | 0.000 |
| ENSMUST00000132527 | Col9a3 | 2.495 | 5.164 | 0.325 | −2.668 | 4.839 | 0.000 | 0.000 |
| ENSMUST00000212414 | Kcnn1 | 0.000 | 2.616 | 0.468 | −2.616 | 2.147 | 0.013 | 0.032 |
| ENSMUST00000110371 | Acsl1 | 4.822 | 3.544 | 5.813 | 1.278 | −2.269 | 0.024 | 0.001 |
| ENSMUST00000165709 | Usp24 | 7.117 | 5.272 | 7.592 | 1.845 | −2.321 | 0.004 | 0.001 |
| ENSMUST00000137024 | Sirt3 | 5.150 | 3.624 | 5.926 | 1.526 | −2.302 | 0.009 | 0.000 |
| ENSMUST00000100411 | Pfkfb3 | 3.272 | 5.863 | 4.293 | −2.592 | 1.571 | 0.009 | 0.004 |
| ENSMUST00000202677 | Tex26 | 0.481 | 2.753 | 0.349 | −2.272 | 2.404 | 0.041 | 0.012 |
| ENSMUST00000151944 | Mfsd11 | 3.008 | 4.104 | 3.044 | −1.096 | 1.060 | 0.036 | 0.036 |
| ENSMUST00000120674 | Riox2 | 4.493 | 2.924 | 4.352 | 1.569 | −1.428 | 0.013 | 0.020 |
| ENSMUST00000160471 | Gpatch2 | 6.206 | 1.425 | 3.032 | 4.781 | −1.607 | 0.000 | 0.041 |
| ENSMUST00000009705 | Eng | 4.009 | 6.934 | 4.671 | −2.925 | 2.263 | 0.049 | 0.040 |
| ENSMUST00000045091 | Kirrel3 | 3.958 | 2.054 | 4.172 | 1.904 | −2.118 | 0.019 | 0.003 |
| ENSMUST00000046549 | Apobec2 | 3.694 | 2.243 | 3.550 | 1.451 | −1.307 | 0.016 | 0.036 |
| ENSMUST00000078021 | Glmn | 8.121 | 4.406 | 7.818 | 3.715 | −3.413 | 0.000 | 0.000 |
| ENSMUST00000111627 | Mobp | 4.522 | 3.318 | 5.593 | 1.203 | −2.275 | 0.039 | 0.000 |
| ENSMUST00000141425 | Rec8 | 2.433 | 0.000 | 3.317 | 2.433 | −3.317 | 0.013 | 0.000 |
| ENSMUST00000109266 | Zmynd8 | 9.867 | 3.816 | 10.110 | 6.051 | −6.294 | 0.000 | 0.000 |
| ENSMUST00000144625 | Cfap74 | 5.262 | 3.705 | 5.866 | 1.557 | −2.161 | 0.019 | 0.001 |
| ENSMUST00000142283 | Homez | 3.486 | 1.506 | 6.316 | 1.980 | −4.810 | 0.020 | 0.000 |
| ENSMUST00000150351 | Appl2 | 4.908 | 3.778 | 5.566 | 1.129 | −1.788 | 0.035 | 0.004 |
| ENSMUST00000211259 | Car11 | 4.758 | 6.166 | 5.121 | −1.408 | 1.045 | 0.007 | 0.031 |
| ENSMUST00000065573 | Gpatch2 | 3.001 | 6.545 | 3.896 | −3.544 | 2.649 | 0.044 | 0.021 |
| ENSMUST00000054564 | Pcolce | 3.209 | 0.342 | 5.026 | 2.867 | −4.684 | 0.036 | 0.000 |
| ENSMUST00000149871 | Irak1 | 2.757 | 0.313 | 4.495 | 2.444 | −4.182 | 0.046 | 0.000 |
| ENSMUST00000140490 | Steap3 | 0.000 | 3.767 | 0.000 | −3.767 | 3.767 | 0.027 | 0.011 |
| ENSMUST00000169736 | Pgghg | 6.469 | 3.687 | 6.436 | 2.782 | −2.749 | 0.001 | 0.000 |
| ENSMUST00000029795 | Rorc | 0.000 | 5.172 | 7.488 | −5.172 | −2.316 | 0.000 | 0.000 |
| ENSMUST00000144491 | Tmem44 | 6.909 | 9.480 | 7.047 | −2.571 | 2.433 | 0.000 | 0.000 |
| ENSMUST00000119339 | Ythdc1 | 4.671 | 1.174 | 4.738 | 3.497 | −3.564 | 0.002 | 0.000 |
| ENSMUST00000167063 | Mpp6 | 4.664 | 2.740 | 4.215 | 1.923 | −1.475 | 0.006 | 0.004 |
| ENSMUST00000096194 | Tmem2 | 7.834 | 4.161 | 7.027 | 3.673 | −2.866 | 0.001 | 0.008 |
| ENSMUST00000111106 | Psen2 | 1.583 | 3.212 | 1.343 | −1.629 | 1.869 | 0.036 | 0.013 |
| ENSMUST00000105466 | P4ha1 | 9.916 | 7.881 | 9.396 | 2.035 | −1.515 | 0.000 | 0.003 |
| ENSMUST00000125764 | Jkamp | 0.000 | 2.709 | 0.289 | −2.709 | 2.420 | 0.049 | 0.031 |
| ENSMUST00000144682 | 6430548M08Rik | 4.322 | 2.689 | 4.701 | 1.633 | −2.012 | 0.032 | 0.006 |
| ENSMUST00000049430 | Galnt18 | 5.219 | 6.594 | 4.492 | −1.375 | 2.102 | 0.013 | 0.002 |
| ENSMUST00000181402 | Emc8 | 6.112 | 7.204 | 8.401 | −1.092 | −1.197 | 0.039 | 0.046 |
| ENSMUST00000192068 | Prkar2a | 3.187 | 4.732 | 3.607 | −1.545 | 1.125 | 0.010 | 0.041 |
| ENSMUST00000023146 | Nubp1 | 7.291 | 5.370 | 6.928 | 1.920 | −1.557 | 0.000 | 0.002 |
| ENSMUST00000035844 | Josd2 | 3.979 | 6.494 | 5.176 | −2.515 | 1.318 | 0.000 | 0.020 |
| ENSMUST00000024791 | Trem2 | 6.450 | 4.107 | 6.036 | 2.344 | −1.929 | 0.000 | 0.002 |
| ENSMUST00000168122 | Sned1 | 4.786 | 3.648 | 5.170 | 1.137 | −1.522 | 0.025 | 0.001 |
| ENSMUST00000220845 | Zfand4 | 5.664 | 4.614 | 5.640 | 1.050 | −1.026 | 0.005 | 0.014 |
| ENSMUST00000168977 | Ylpm1 | 8.145 | 6.175 | 7.175 | 1.971 | −1.001 | 0.001 | 0.000 |
| ENSMUST00000164100 | Tmod2 | 8.858 | 6.568 | 8.791 | 2.290 | −2.223 | 0.039 | 0.027 |
| ENSMUST00000126603 | Lgals4 | 3.999 | 1.346 | 4.913 | 2.653 | −3.567 | 0.024 | 0.000 |
| ENSMUST00000001484 | Tbx21 | 3.126 | 0.000 | 2.137 | 3.126 | −2.137 | 0.041 | 0.015 |
| ENSMUST00000118390 | Synj1 | 10.688 | 9.094 | 10.835 | 1.594 | −1.741 | 0.009 | 0.011 |
Con: control; Mod: model; PSP: Polygonatum sibiricum polysaccharide.
Figure 2.
Distinct expression patterns of mRNAs and functional enrichment analyses. Heatmap of the expression profiles of significantly differentially expressed (DE) mRNAs between the Control group and Model group (A), and between the Model group and the PSP group (B). Volcano plot of the expression profiles of DE mRNAs between the Control group and Model group (C), and between the Model group and the PSP group (D). GO enrichment analysis of mRNAs that were altered by PSP treatment (E). The ordinate represents the GO term while the abscissa represents the − log10 (p-value). BP represents the biological process, CC represents the cellular component, and MF represents the molecular function. (F) KEGG enrichment analysis of mRNAs that were altered by PSP. The ordinate represents the KEGG pathway whilethe abscissa represents the − log10(p-value). PSP: Polygonatum sibiricum polysaccharide.
GO analysis of 679 PSP-related mRNAs identified several GO terms that were highly enriched (Figure 2(E)). The most enriched terms with regards to biological processes were positive regulation of dendritic spine development, cellular component organization, and localization. As for cellular components, the most enriched terms were the intracellular part, organelle part, and cell cortex. The most enriched molecular function terms were protein binding, binding, and protein domain–specific binding. As shown in Figure 2(E), KEGG analysis demonstrated that O-glycan biosynthesis, sulphur metabolism, and adherens junctions, may be closely associated with the ameliorative effect of PSP on cognitive impairment and brain ageing.
Functional analysis of differentially expressed circular RNAs
By comparing the Model group with the Control group, we identified 271 ageing-related circRNAs (137 upregulated and 134 downregulated). A total of 37 alterations of circRNA expression were reversed following PSP treatment (Table 3); in each case, expression levels returned to levels that were similar to those of the Control group. We also used heatmaps and volcano plots to determine the differential expression of circRNAs in the three groups (Figure 3(A–D)).
Table 3.
Dysregulated circRNAs associated with the anti-brain ageing effect of PSP.
| circRNA_ID | Con | Mod | PSP | log2FoldChange |
P value |
||
|---|---|---|---|---|---|---|---|
| Con_vs_Mod | Mod_vs_PSP | Con_vs_Mod | Mod_vs_PSP | ||||
| 11:21652226|21664385 | 2.4987 | 0.0000 | 1.5880 | 2.4987 | −1.5880 | 0.0079 | 0.0356 |
| 10:33996667|34012137 | 3.2867 | 1.2694 | 3.2118 | 2.0174 | −1.9425 | 0.0254 | 0.0351 |
| 2:29227578|29248878 | 2.8816 | 4.3125 | 2.8197 | −1.4309 | 1.4928 | 0.0277 | 0.0277 |
| 1:176874564|176919686 | 0.0000 | 2.2272 | 0.0000 | −2.2272 | 2.2272 | 0.0165 | 0.0157 |
| 11:29705535|29708770 | 5.2253 | 4.0147 | 5.1879 | 1.2106 | −1.1732 | 0.0068 | 0.0240 |
| 2:40919136|40928022 | 1.1005 | 3.0837 | 0.7580 | −1.9833 | 2.3258 | 0.0255 | 0.0325 |
| 4:106423460|106436855 | 0.0000 | 1.9829 | 0.0000 | −1.9829 | 1.9829 | 0.0290 | 0.0269 |
| 8:104517907|104527449 | 0.0000 | 1.8868 | 0.0000 | −1.8868 | 1.8868 | 0.0382 | 0.0354 |
| 4:44893533|44895989 | 2.2399 | 0.0000 | 2.0619 | 2.2399 | −2.0619 | 0.0157 | 0.0279 |
| 1:133753089|133753749 | 3.0811 | 4.2791 | 2.2187 | −1.1980 | 2.0604 | 0.0442 | 0.0011 |
| 15:81600908|81616863 | 1.9999 | 0.0000 | 2.0961 | 1.9999 | −2.0961 | 0.0280 | 0.0179 |
| 5:118077577|118095729 | 0.0000 | 1.5555 | 0.0000 | −1.5555 | 1.5555 | 0.0430 | 0.0392 |
| 10:105413217|105413787 | 2.6183 | 0.0000 | 2.0574 | 2.6183 | −2.0574 | 0.0050 | 0.0226 |
| 12:55659048|55677265 | 2.5505 | 0.0000 | 1.7102 | 2.5505 | −1.7102 | 0.0042 | 0.0353 |
| 3:88981233|89007877 | 2.0806 | 0.0000 | 1.8287 | 2.0806 | −1.8287 | 0.0271 | 0.0222 |
| 2:155432816|155440785 | 1.9271 | 0.0000 | 2.4661 | 1.9271 | −2.4661 | 0.0167 | 0.0097 |
| 5:118593333|118680971 | 0.4995 | 2.8826 | 0.0000 | −2.3831 | 2.8826 | 0.0191 | 0.0028 |
| 1:192116269|192130460 | 2.0510 | 0.0000 | 2.3057 | 2.0510 | −2.3057 | 0.0093 | 0.0203 |
| 9:110517505|110521364 | 0.0000 | 2.7886 | 0.0000 | −2.7886 | 2.7886 | 0.0038 | 0.0039 |
| 17:68455693|68463020 | 2.0998 | 0.0000 | 2.7630 | 2.0998 | −2.7630 | 0.0234 | 0.0028 |
| 11:104368680|104425293 | 1.7583 | 0.0000 | 2.3437 | 1.7583 | −2.3437 | 0.0255 | 0.0102 |
| 18:73648885|73678039 | 3.4036 | 1.7218 | 3.4239 | 1.6818 | −1.7021 | 0.0222 | 0.0309 |
| 5:25798446|25825215 | 1.1005 | 2.9398 | 0.0000 | −1.8393 | 2.9398 | 0.0405 | 0.0020 |
| 12:16558723|16560981 | 0.0000 | 1.6711 | 0.0000 | −1.6711 | 1.6711 | 0.0463 | 0.0422 |
| 5:107406032|107419879 | 0.0000 | 2.1166 | 0.0000 | −2.1166 | 2.1166 | 0.0233 | 0.0220 |
| 19:16707327|16725693 | 0.0000 | 2.9756 | 0.0000 | −2.9756 | 2.9756 | 0.0211 | 0.0180 |
| 15:99738665|99745946 | 0.7404 | 3.0031 | 1.0563 | −2.2627 | 1.9468 | 0.0429 | 0.0378 |
| 10:91158748|91166069 | 0.0000 | 1.7917 | 0.0000 | −1.7917 | 1.7917 | 0.0278 | 0.0376 |
| 6:83928653|83972283 | 1.8538 | 0.0000 | 2.0625 | 1.8538 | −2.0625 | 0.0156 | 0.0276 |
| 11:23354845|23364512 | 0.0000 | 1.9661 | 0.0000 | −1.9661 | 1.9661 | 0.0380 | 0.0349 |
| 10:109747194|109760528 | 0.4995 | 2.7214 | 0.0000 | −2.2219 | 2.7214 | 0.0324 | 0.0043 |
| 1:60437056|60453762 | 1.1015 | 3.0883 | 1.0563 | −1.9868 | 2.0320 | 0.0236 | 0.0240 |
| 16:52268382|52296984 | 2.4988 | 3.6868 | 0.5150 | −1.1880 | 3.1718 | 0.0468 | 0.0004 |
| 5:106632925|106666845 | 3.5873 | 1.9829 | 3.6412 | 1.6044 | −1.6583 | 0.0362 | 0.0409 |
| 7:84262393|84305259 | 0.0000 | 2.0461 | 0.0000 | −2.0461 | 2.0461 | 0.0115 | 0.0301 |
| 9:99262546|99263809 | 0.0000 | 1.6519 | 0.0000 | −1.6519 | 1.6519 | 0.0279 | 0.0483 |
| 13:83575535|83635469 | 0.0000 | 1.6695 | 0.0000 | −1.6695 | 1.6695 | 0.0474 | 0.0430 |
Con: control; Mod: model; PSP: Polygonatum sibiricum polysaccharide.
Figure 3.
Distinct expression patterns of circRNAs and functional enrichment analyses Heatmap of the expression profiles of significantly differentially expressed (DE) circRNAs between the Control group and the Model group (A), and between the Model group and the PSP group (B). Volcano plot of the expression profiles of DE circRNAs between the Control group and the Model group (C), and between the Model group and the PSP group (D). GO enrichment analysis of circRNAs that were altered by PSP treatment (E). The ordinate represents the GO term while the abscissa represents the − log10 (p-value). BP represents the biological process, CC represents the cellular component, and MF represents the molecular function. (F) KEGG enrichment analysis of circRNAs altered by PSP. The ordinate represents the KEGG pathway, while the abscissa represents the − log10 (p-value). PSP: Polygonatum sibiricum polysaccharide.
Subsequently, we performed GO and KEGG enrichment analysis based on the source genes of 37 PSP-related circRNAs (Figure 3(E–F)). GO analysis showed that the most enriched biological processes terms were anatomical structure development, synapse organization, and the regulation of cellular component biogenesis. The most enriched cellular component terms were synapse, neuron part, and dendritic shaft. The most enriched molecular function was protein domain–specific binding, PDZ domain binding, and calcium-transporting ATPase activity. KEGG analysis indicated that bacterial invasion of epithelial cells, sulphur metabolism, and adherens junctions were the most significantly enriched KEGG pathways related to the ameliorative effect of PSP on cognitive impairment and brain ageing.
Differentially expressed microRNAs and their targets
By comparing miRNA transcriptomes from the different groups of miRNA samples, we identified 38 ageing-related miRNAs (20 upregulated and 18 downregulated in the Model group), and 13 PSP-targeting miRNAs (Table 4). Next, we constructed heatmaps and a volcano plot (Figure 4(A–D)). We attempted to analyse the functions of these miRNAs by performing functional enrichment analyses on their target genes. By extracting consensus predictions using established miRNA target prediction tools, we obtained target genes for miRNAs with high levels of confidence.
Table 4.
Dysregulated miRNAs associated with the anti-brain ageing effect of PSP.
| Gene_ID | Con | Mod | PSP | log2FoldChange |
p Value |
||
|---|---|---|---|---|---|---|---|
| Con_vs_Mod | Mod_vs_PSP | Con_vs_Mod | Mod_vs_PSP | ||||
| mmu-miR-129b-5p | 5.4384 | 4.3598 | 5.4384 | 1.0785 | −1.0629 | 0.0150 | 0.0188 |
| mmu-miR-182-5p | 7.7668 | 10.3368 | 7.7668 | −2.5700 | 3.9824 | 0.0218 | 0.0000 |
| mmu-miR-183-5p | 7.1995 | 10.2440 | 7.1995 | −3.0445 | 3.7345 | 0.0161 | 0.0000 |
| mmu-miR-190a-5p | 5.2352 | 6.8532 | 5.2352 | −1.6180 | 1.8460 | 0.0095 | 0.0040 |
| mmu-miR-1981-5p | 10.2688 | 9.1380 | 10.2688 | 1.1309 | −1.2873 | 0.0057 | 0.0027 |
| mmu-miR-300-5p | 3.8334 | 5.0310 | 3.8334 | −1.1976 | 1.2587 | 0.0364 | 0.0235 |
| mmu-miR-32-5p | 6.8976 | 7.9036 | 6.8976 | −1.0060 | 1.8191 | 0.0481 | 0.0006 |
| mmu-miR-449a-5p | 4.7387 | 5.7892 | 4.7387 | −1.0505 | 1.0962 | 0.0248 | 0.0350 |
| mmu-miR-5100 | 4.0377 | 2.9478 | 4.0377 | 1.0899 | −2.0204 | 0.0309 | 0.0004 |
| mmu-miR-5110 | 2.4317 | 0.4057 | 2.4317 | 2.0260 | −2.4317 | 0.0114 | 0.0015 |
| mmu-miR-551b-5p | 4.2948 | 3.0900 | 4.2948 | 1.2048 | −1.2390 | 0.0406 | 0.0271 |
| mmu-miR-7044-3p | 3.0497 | 1.3562 | 3.0497 | 1.6934 | −3.0497 | 0.0454 | 0.0001 |
| mmu-miR-96-5p | 4.6470 | 8.2457 | 4.6470 | −3.5987 | 4.3200 | 0.0131 | 0.0000 |
Con: control; Mod: model; PSP: Polygonatum sibiricum polysaccharide.
Figure 4.
Distinct expression patterns of miRNAs and functional enrichment analyses Heatmap of significantly differentially expressed (DE) miRNAs between the Control group and the Model group (A), and between the Model group and the PSP group (B). Volcano plot of the expression profiles of DE miRNAs between the Control group and the Model group (C), and between the Model group and the PSP group (D). GO enrichment analysis of miRNAs altered by PSP treatment (E). The ordinate represents the GO term while the abscissa represents the − log10 (p-value). BP represents the biological process, CC represents the cellular component, and MF represents the molecular function. (F) KEGG enrichment analysis of miRNA altered by PSP: The ordinate represents the KEGG pathway while the abscissa represents the − log10(p value). PSP: Polygonatum sibiricum polysaccharide.
GO analysis of 13 PSP-related miRNAs identified several GO terms that were highly enriched (Figure 4(E)). The most enriched biological processes were system development, anatomical structure morphogenesis, and positive regulation of the biological process. The most enriched cellular component GO terms were intracellular, intracellular part, and cytoplasm. The most enriched molecular functions were protein binding, binding, and ion binding. A series of significantly enriched KEGG pathways were also detected (Figure 4(F)); of these pathways in cancer, the Wnt-signalling pathway, and axon guidance, were the most significant.
Validation of expression profiles
Real-time qPCR analysis is usually conducted to validate the findings obtained from RNA-Seq (Zhang et al. 2019). In the present study, a total of 14 dysregulated mRNAs, miRNAs, and circRNAs, were randomly selected to validate our microarray results by real-time qPCR. As shown in Figure 5, the expression levels determined by real-time qPCR were in agreement with the RNA-Seq results. Thus, all circRNAs, miRNAs, and mRNAs, were confirmed to be targets that were closely related to PSP treatment; these molecules were therefore included in all further analyses.
Figure 5.
Differential expression of mRNAs, miRNAs, and circRNAs, as validated by real-time quantitative polymerase chain reaction (q-PCR). The miRNA expression levels detected by RNA sequencing (mmu-miR-182-5p, mmu-miR-183-5p, mmu-miR-5110, mmu-miR-7044-3p, mmu-miR-96-5p), mRNAs (Pianp, Ubl5, Stmn1, Tspyl2, Hnrnpu), and circRNAs (10:33996667|34012137, 7:84262393|84305259, 2:29227578|29248878, 11:29705535|29708770) were consistent with q-PCR results. ‘PSP’ represents brain ageing model mice treated with PSP. ‘Control’ represents untreated mice. ‘Model’ represents brain ageing model mice. PSP: Polygonatum sibiricum polysaccharide.
Prediction of circRNA–miRNA–mRNA interactions
Next, we constructed a circRNA-associated ceRNA network to reveal the novel inter-regulatory relationships between circRNAs, miRNAs, and mRNAs (Figure 6). Based on the differentially expressed RNAs between the Control group and the Model group, we constructed a circRNA-associated ceRNA network related to brain ageing (containing 19 mRNAs, 9 circRNAs, and 12 miRNAs). This network showed that by competitively binding with mmu-miR-5110, PSP can regulate two circRNAs (5:106632925|106666845, 2:29227578|29248878) and subsequently regulate nine mRNAs (Ace, Ubl5, Gab1, Cep170b, Synj1, Ccdc24, Bean1, Wdr89, Pianp). It is worth noting that PSP was able to regulate multiple RNAs (Slc6a5, Bean1, Ace, Samd4, Olfr679, Olfr372, Dhrs9, Tsc1, Slc12a6, mmu-miR-5110, mmu-miR-449a-5p, mmu-miR-1981-5p, 2:29227578|29248878, 5:106632925|106666845) in this ceRNA network, thus suggesting that circRNAs harbour miRNA response elements and play pivotal regulatory roles in the anti-ageing mechanism of PSP.
Figure 6.
A circRNA-miRNA-mRNA interaction network. The circle, square, and triangle represent mRNA, circRNA, and miRNA, respectively. Red and green represent upregulation and downregulation, respectively. The arrow represents RNAs that underwent changes in response to PSP treatment. PSP: Polygonatum sibiricum polysaccharide.
Discussion
Brain ageing is a process in which the brain gradually loses its ability to adapt to the environment as age increases. It has been established that the long-term systemic administration of high levels of d-gal induces behavioural and neurobiological changes that are similar to natural brain ageing (Pourmemar et al. 2017; Nam et al. 2019). In our study, the chronic injection of d-gal prolonged the escape latency and swimming distance of mice in an established behavioural test, thus indicating that the d-gal model is a well-established method for investigating anti-ageing pharmacological therapy. PSP is known to exhibit anti-ageing properties (Zheng 2020). In the present study, we found that the administration of PSP could reverse the dysfunction in learning and memory abilities caused by d-gal interference. Thus, as also reported by other groups, we documented evidence for the brain-protective effect of PSP on the d-gal-induced mouse model of ageing.
ncRNAs and ncRNA-regulatory processes are known to be important determinants in the pathogenesis of brain ageing and neurodegeneration (Esteller 2011; Szafranski et al. 2015). Thus, we focussed exclusively on ncRNA in order to elucidate the protective mechanisms of PSP on brain ageing. To the best of our knowledge, this is the first study to create a circRNA-associated ceRNA network and reveal regulator pathways relating to the protective effects of PSP on memory dysfunction in a mouse model of brain ageing induced by d-gal. We identified 37 DE circRNAs, 13 DE miRNAs, and 679 DE mRNAs, that were related to the protective function of PSP; these results were also validated by real-time qPCR. Next, we constructed a circRNA–miRNA–mRNA network. GO enrichment analysis and KEGG pathway analysis were also performed to functionally annotate the predicted target genes. Overall, these analyses provided clues to the mechanisms that underlie the ameliorative effect of PSP on the learning and memory abilities of mice experiencing brain ageing. This approach could shed new light on the prevention of neurodegenerative disorders.
First, we focussed on the DE coding genes. A total of 1175 upregulated and 1250 downregulated ageing-related mRNAs were detected. We also found that 679 mRNAs were potentially related to the protective effect of PSP against brain ageing. Earlier reports on the pathogenesis of ageing have already reported some of these RNAs. For example, the CD82-TRPM7-Numb signalling axis was previously shown to participate in the progression of age-related cognitive impairment. The upregulated expression of TRPM7 α-kinase cleavage and the phosphorylation of Numb were both associated with ageing and resulted in increased levels of Aβ secretion (Zhao et al. 2020). In another study, Carbajosa et al. (2018) investigated the impact of TREM2 deficiency using RNA-Seq and identified disruption in the gene networks related to endothelial cells that was more apparent in younger mice than aged mice. Interestingly, our findings indicate that PSP treatment can reduce the upregulated levels of TRPM2 and Numb caused by brain ageing. The SNX8, TSC1, and Sirt3, genes play important roles in ageing and age-associated diseases. A moderate increase in TSC1 expression is known to enhance overall health status (Zhang et al. 2017). An increase in SNX8 expression is also known to alleviate cognitive impairment in AD mice (Xie et al. 2019). The overexpression of Sirt3 in several tissues also suggested improved levels of protection against ROS-induced ageing (Tong et al. 2005; Qiu et al. 2010; Hallows et al. 2011). In the present study, we observed downregulated levels of SNX8, TSC1, and Sirt3, in the Dal-induced mouse when compared with controls. Interestingly, the expression levels of these genes were upregulated following PSP treatment. Current research is increasingly focussing on the role of synaptic organization in brain ageing. Interestingly, we also found that synapse-related genes were associated with mechanisms underlying the effects of PSP treatment. For example, abnormal expression levels of STXBP1 are known to impair synaptic neurotransmission; thus, STXBP1 has been identified as a new interventional target to protect the ageing brain (Lee et al. 2019). Other research has shown that Tcf4 is involved in synaptic plasticity in mature neurons, and that the functional loss of Tcf4 may contribute to neurological symptoms in Pitt–Hopkins syndrome (Crux et al. 2018). We observed downregulated expression levels of STXBP1 and upregulated expression levels of Tcf4, in the Dal-induced mouse compared with the Control group and showed that PSP treatment could reverse these changes.
Recent research on circRNAs has proved essential in revealing many of the molecular mechanisms that underlie the development and pathology of certain diseases, including ageing and age-related diseases (Knupp and Miura 2018). Drugs that target circRNAs potentially represent novel therapeutics for brain ageing in neurodegenerative diseases. In our research, we identified a total of 271 circRNAs that were related to brain ageing; of these, 38 circRNAs were found to play critical roles in the anti-brain ageing effects of PSP. GO analysis and KEGG analysis were performed to predict their function. It was evident that a significant number of GO terms were related to neuronal synapse (e.g., synapse, synapse organize, neuron to neuron synapse, and asymmetric synapse). Previous studies on brain tissue from a variety of mammalian species have concluded that the most consistent correlates with ageing are a reduced number of synaptic connections among neurons (Brunso-Bechtold et al. 2000; Peters et al. 2008; Soghomonian et al. 2010; VanGuilder et al. 2010) and cognitive decline (Dickson et al. 1995; VanGuilder et al. 2011). The balance between excitatory and inhibitory synaptic systems is a particularly hot topic in the field of brain ageing at present (Bishop et al. 2010; Deak & Sonntag 2012). Existing data, together with our present findings, indicate that pathological processes associated with synaptic dysfunction are most likely to lead to cognitive impairment in brain ageing. Further molecular and electrophysiological studies are now required to identify the specific molecular pathways that contribute to the specific effects of PSP at the synapse. It is also important that we identify the mechanisms that regulate learning and memory in brain ageing.
The regulation of brain ageing is associated with the differential expression of miRNAs, as revealed by previous studies involving RNA transcriptome analysis in the whole mouse brain (Eacker et al. 2011; Inukai et al. 2012). In the present study, a total of 38 miRNAs were associated with brain ageing. We also identified a limited number of miRNAs that play important roles in the anti-ageing effects of PSP. Previous research showed that the transfection of synthetic miR-182-5p and miR-183-5p mimics led to increased neurite outgrowth and the neuroprotection of dopaminergic neurons in vitro and in vivo, thus mimicking the effects of Glial cell line-derived neurotrophic factor (Roser et al. 2018). In the present study, we identified elevated levels of miR-182-5p and miR-183-5p in PSP-treated mice when compared with mice experiencing brain ageing. KEGG analysis further showed that the Wnt-signalling pathway is potentially a critical pathway and shows a strong association with the mechanism responsible for the ameliorative effects of PSP on brain ageing. Accumulating evidence now supports the hypothesis that dysfunctional Wnt signalling could be closely related to the pathogenesis of ageing-related brain diseases (Caricasole et al. 2004; Berwick and Harvey 2014; Smith-Geater et al. 2020). During the process of ageing, changes in the expression of Wnt-related molecules may lead to a reduction in the accumulation of presynaptic and postsynaptic proteins. Combined with other factors, this process will result in a decline in ability with regards to performing the morphometric adjustments required for neuronal plasticity, thus leading to cognitive decline in the elderly (Libro et al. 2016). These findings indicated that PSP potentially attenuates brain ageing by regulating Wnt signalling and synaptic activity.
Perturbations in ceRNA regulatory networks, including mRNAs, miRNAs, and circRNAs, are considered to play an essential role in the pathogenesis of ageing-related brain diseases (Wang et al. 2018). A circRNA-miRNA-mRNA regulatory network has already been constructed and used to elucidate the mechanisms underlying AD (Wang et al. 2018). Furthermore, the circDLGAP4/miR-134-5p/CREB-signalling pathway has been reported to exert functionality in the progression of PD (Feng et al. 2020). In the present study, we used ceRNA regulatory network analysis to generate a putative circRNA–miRNA–mRNA co-expression network during brain ageing. Moreover, we identified certain circRNAs that may act as sponges and play a role in the protective effect of PSP against brain ageing. We found that PSP-targeted circRNAs could competitively bind to mmu-miR-5110 and subsequently regulate the expression of nine mRNAs. These observations need to be investigated further.
Conclusions
We investigated whether PSP could improve learning and memory impairment in mice experiencing brain ageing induced by d-gal. We found that PSP could effectively ameliorate cognitive dysfunction during brain ageing. We also used high-throughput RNA sequencing to evaluate brain ageing-related mRNAs, circRNAs, and miRNAs, and identified the profiles of a range of RNAs that were dysregulated and targeted by PSP. We also constructed a circRNA–miRNA–mRNA network associated with brain ageing and PSP interventions. This network will help us to understand the transcriptional mechanisms associated with brain ageing and the pharmacological mechanisms of PSP. The corresponding roles and molecular mechanisms associated with these ncRNAs and mRNAs need to be investigated further.
Funding Statement
This work was financially supported by the National Natural Science Foundation of China [Grant numbers: 81873169 and 81603670] and the Hunan Provincial Natural Science Foundation of China [Grant numbers 2017JJ3459 and 2020JJ4803].
Author contributions
WP conceived the study. ZZ and WP drafted the manuscript and performed the analysis. Other authors helped to draft the manuscript and interpret the data. All authors read and approved the final manuscript.
Disclosure statement
The authors declare that there are no conflicts of interest in relation to this work.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Apple DM, Solano-Fonseca R, Kokovay E.. 2017. Neurogenesis in the aging brain. Biochem Pharmacol. 141:77–85. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 116(2):281–297. [DOI] [PubMed] [Google Scholar]
- Berwick DC, Harvey K.. 2014. The regulation and deregulation of Wnt signaling by PARK genes in health and disease. J Mol Cell Biol. 6(1):3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Lu T, Yankner BA.. 2010. Neural mechanisms of ageing and cognitive decline. Nature. 464(7288):529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunso-Bechtold JK, Linville MC, Sonntag WE.. 2000. Age-related synaptic changes in sensorimotor cortex of the Brown Norway X fischer 344 rat. Brain Res. 872(1-2):125–133. [DOI] [PubMed] [Google Scholar]
- Carbajosa G, Malki K, Lawless N, Wang H, Ryder JW, Wozniak E, Wood K, Mein CA, Dobson RJB, Collier DA, et al. 2018. Loss of Trem2 in microglia leads to widespread disruption of cell coexpression networks in mouse brain. Neurobiol Aging. 69:151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caricasole A, Copani A, Caraci F, Aronica E, Rozemuller AJ, Caruso A, Storto M, Gaviraghi G, Terstappen GC, Nicoletti F.. 2004. Induction of Dickkopf-1, a negative modulator of the Wnt pathway, is associated with neuronal degeneration in Alzheimer's brain. J Neurosci. 24(26):6021–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crux S, Herms J, Dorostkar MM.. 2018. Tcf4 regulates dendritic spine density and morphology in the adult brain. PLoS One. 13(6):e0199359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak F, Sonntag WE.. 2012. Aging, synaptic dysfunction, and insulin-like growth factor (IGF)−1. J Gerontol A Biol Sci Med Sci. 67(6):611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW, Crystal HA, Bevona C, Honer W, Vincent I, Davies P.. 1995. Correlations of synaptic and pathological markers with cognition of the elderly. Neurobiol Aging. 16(3):285–298; discussion 98-304. [DOI] [PubMed] [Google Scholar]
- Eacker SM, Keuss MJ, Berezikov E, Dawson VL, Dawson TM.. 2011. Neuronal activity regulates hippocampal miRNA expression. PLoS One. 6(10):e25068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. 2011. Non-coding RNAs in human disease. Nat Rev Genet. 12(12):861–874. [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang L, Wang S, Hong Q.. 2020. Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson's disease. Biochem Biophys Res Commun. 522(2):388–394. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devires MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, et al. 2011. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 41(2):139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai S, de Lencastre A, Turner M, Slack F.. 2012. Novel microRNAs differentially expressed during aging in the mouse brain. PLoS One. 7(7):e40028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knupp D, Miura P.. 2018. CircRNA accumulation: A new hallmark of aging? Mech Ageing Dev. 173:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Vijayan M, Bhatti JS, Reddy PH.. 2017. MicroRNAs as peripheral biomarkers in aging and age-related diseases. Prog Mol Biol Transl Sci. 146:47–94. [DOI] [PubMed] [Google Scholar]
- Lee YI, Kim YG, Pyeon HJ, Ahn JC, Logan S, Orock A, Joo KM, Lőrincz A, Deák F.. 2019. Dysregulation of the SNARE-binding protein Munc18-1 impairs BDNF secretion and synaptic neurotransmission: a novel interventional target to protect the aging brain. Geroscience. 41(2):109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Zhu Q, Lu YP, Zhao P, Feng ZB, Qian ZM, Zhu L.. 2015. Ligustilide prevents cognitive impairment and attenuates neurotoxicity in d-galactose induced aging mice brain. Brain Res. 1595:19–28. [DOI] [PubMed] [Google Scholar]
- Li YY, Deng HB, Wang R, Chen LL.. 2005. The change of telomerase activity in aging mice tissue and the interference of Plyonatum sibiricum. J. Clin. Res. 22:894–895. [Google Scholar]
- Libro R, Bramanti P, Mazzon E.. 2016. The role of the Wnt canonical signaling in neurodegenerative diseases. Life Sci. 158:78–88. [DOI] [PubMed] [Google Scholar]
- Liu N, Dong Z, Zhu X, Xu H, Zhao Z.. 2018. Characterization and protective effect of Polygonatum sibiricum polysaccharide against cyclophosphamide-induced immunosuppression in Balb/c mice. Int J Biol Macromol. 107(Pt A):796–802. [DOI] [PubMed] [Google Scholar]
- López-Otín C, Blasco M, Partridge L, Serrano M, Kroemer G.. 2013. The hallmarks of aging. Cell. 153(6):1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecocci P, Boccardi V, Cecchetti R, Bastiani P, Scamosci M, Ruggiero C, Baroni M.. 2018. A long journey into aging, brain aging, and Alzheimer's disease following the oxidative stress tracks. JAD. 62(3):1319–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- Nam SM, Seo M, Seo JS, Rhim H, Nahm SS, Cho IH, Chang BJ, Kim HJ, Choi SH, Nah SY.. 2019. Ascorbic acid mitigates d-galactose-induced brain aging by increasing hippocampal neurogenesis and improving memory function. Nutrients. 11(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Sethares C, Luebke JI.. 2008. Synapses are lost during aging in the primate prefrontal cortex. Neuroscience. 152(4):970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourmemar E, Majdi A, Haramshahi M, Talebi M, Karimi P, Sadigh-Eteghad S.. 2017. Intranasal cerebrolysin attenuates learning and memory impairments in d-galactose-induced senescence in mice. Exp Gerontol. 87(Pt A):16–22. [DOI] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D.. 2010. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 12(6):662–667. [DOI] [PubMed] [Google Scholar]
- Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H.. 2015. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 365(2):141–148. [DOI] [PubMed] [Google Scholar]
- Roser AE, Caldi Gomes L, Halder R, Jain G, Maass F, Tönges L, Tatenhorst L, Bähr M, Fischer A, Lingor P.. 2018. miR-182-5p and miR-183-5p act as GDNF mimics in dopaminergic midbrain neurons. Mol Ther Nucleic Acids. 11:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Geater C, Hernandez SJ, Lim RG, Adam M, Wu J, Stocksdale JT, Wassie BT, Gold MP, Wang KQ, Miramontes R, et al. 2020. Aberrant development corrected in adult-onset huntington's disease iPSC-derived neuronal cultures via WNT signaling modulation. Stem Cell Reports. 14(3):406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soghomonian JJ, Sethares C, Peters A.. 2010. Effects of age on axon terminals forming axosomatic and axodendritic inhibitory synapses in prefrontal cortex. Neuroscience. 168(1):74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski K, Abraham KJ, Mekhail K.. 2015. Non-coding RNA in neural function, disease, and aging. Front Genet. 6:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Fei W, Emily S, Qiang T.. 2005. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 280(14):13560–13567. [DOI] [PubMed] [Google Scholar]
- VanGuilder HD, Farley JA, Yan H, Van Kirk CA, Mitschelen M, Sonntag WE, Freeman WM.. 2011. Hippocampal dysregulation of synaptic plasticity-associated proteins with age-related cognitive decline. Neurobiol Dis. 43(1):201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGuilder HD, Yan H, Farley JA, Sonntag WE, Freeman WM.. 2010. Aging alters the expression of neurotransmission-regulating proteins in the hippocampal synaptoproteome. J Neurochem. 113(6):1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xu P, Chen B, Zhang Z, Zhang C, Zhan Q, Huang S, Xia ZA, Peng W.. 2018. Identifying circRNA-associated-ceRNA networks in the hippocampus of Aβ1-42-induced Alzheimer's disease-like rats using microarray analysis. Aging. 10(4):775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnefors M, Liechti A, Halbert J, Valloton D, Kaessmann H.. 2014. Conserved microRNA editing in mammalian evolution, development and disease. Genome Biol. 15(6):R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Niu M, Ji C, Huang TY, Zhang C, Tian Y, Shi Z, Wang C, Zhao Y, Luo H, et al. 2019. SNX8 enhances non-amyloidogenic APP trafficking and attenuates abeta accumulation and memory deficits in an AD mouse. Front Cell Neurosci. 13:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi YX, Wu SX, Ye MS, Zeng Y, Zhang P, Xie YQ.. 2014. Effect of Aβ (1-42) injection on hippocampus cells in rats and protective role of polygona-polysaccharose for Alzheimer’s disease. J. Cent. South. Univ. 39:344–348. [DOI] [PubMed] [Google Scholar]
- Zhang H-M, Diaz V, Walsh ME, Zhang Y.. 2017. Moderate lifelong overexpression of tuberous sclerosis complex 1 (TSC1) improves health and survival in mice. Sci Rep. 7(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li XK, Cui X, Zuo PP.. 2005. d-Galactose injured neurogenesis in the hippocampus of adult mice. Neurol Res. 27(5):552–556. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li B, Xu P, Yang B.. 2019. Integrated whole transcriptome profiling and bioinformatics analysis for revealing regulatory pathways associated with quercetin-induced apoptosis in HCT-116 cells. Front Pharmacol. 10:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li J.. 2015. Chemical constituents of the genus Polygonatum and their role in medicinal treatment. Nat Prod Comm. 10:683–688. [PubMed] [Google Scholar]
- Zhao Y, Kiss T, DelFavero J, Li L, Li X, Zheng L, Wang J, Jiang C, Shi J, Ungvari Z, et al. 2020. CD82-TRPM7-Numb signaling mediates age-related cognitive impairment. Geroscience. 42(2):595–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S. 2020. Protective effect of Polygonatum sibiricum polysaccharide on d-galactose-induced aging rats model. Sci Rep. 10(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.