Abstract
Purpose
The pancreatic islet specific microRNA-375 (miR-375) is overexpressed in type 2 diabetes mellitus (T2DM) patients suppressing the glucose-induced insulin secretion. Thus, miR-375 may serve as a biomarker for the early prediction of T2DM among high-risk individuals. We conducted this clinical study to assess the significance of miR-375 among type 2 diabetes mellitus (T2DM) patients and their first-degree relatives.
Patients and Methods
We included 56 Han Chinese individuals (N: NGT = 21, T2DM = 10, FD-NGT =13 and FD-T2DM = 12) who received medical health check-ups from January 2018 to September 2018 at The Third Hospital of Yunnan Province, China. They were categorized as normal glucose tolerance (NGT), T2DM, first-degree relatives with normal glucose tolerance (FD-NGT) and first-degree relatives with T2DM (FD-T2DM). OGTT, C-peptide and Insulin tests were performed to confirm the diagnosis. The miR-375 levels were determined by Quantitative real-time RT-PCR (qRT-PCR).
Results
The OGTT test showed a significant difference in T2DM and FD-T2DM groups compared with NGT and FD-NGT (p< 0.05). Similar results were observed during C-peptide and insulin tests. Interestingly, the 2-hour insulin test showed FD-NGT group having a significantly higher mean ± standard error of (64.240 ± 12.775) compared to NGT (28.836 ± 10.875). Assessment of miR-375 expression levels in 4 groups showed a significant up-regulation in T2DM and FD-T2DM compared with NGT and FD-NGT groups. A slight increase in miRNA expression was observed in FD-NGT compared with NGT group but was not statistically significant.
Conclusion
The OGTT, C-peptide and insulin tests revealed a statistically significant difference in T2DM and FD-T2DM compared with NGT and FD-NGT groups. A significantly higher miR-375 expression was also observed in T2DM and FD-T2DM groups compared with NGT and FD-NGT and thus, miR-375 may serve as a stable biomarker for the early prediction of T2DM among high-risk individuals.
Keywords: C-peptide, insulin, microRNA-375, OGTT, type 2 diabetes mellitus
Introduction
MicroRNAs (miRNAs) are a class of small non-coding RNAs of 20–24 nucleotides which play an important role in regulating gene expression. Most of the miRNAs are transcribed from DNA sequences into primary miRNAs and processed into precursor miRNAs, and finally become mature miRNAs.1,2 In majority of cases, miRNAs interact with the 3′-untranslated region (UTR) sequences on target genes of target messenger RNAs (mRNAs) to induce mRNA degradation and translational repression.3 MiRNA can be found both intracellularly and extracellularly in fluids and in the circulation, such as in blood plasma4 making them a good candidate for blood-based biomarker development.5,6 Biomarkers are useful as they provide information on early detection of diseases, help in determining individuals at risk of developing complications or subtyping individuals for disease phenotypes. In addition, they may lead to better treatment strategies, personalized therapies, and improved outcome.7 The studies conducted previously showed the potential role of miRNAs as biomarkers in various diseased conditions like cancer, cardiovascular conditions and diabetes.8–10
In a recent study conducted by Alharbi et al, a strong genetic association between T2DM cases and controls was seen in the Saudi population with rs13266634 polymorphisms of the SLC30A8 gene.11 The association of single nucleotide polymorphisms (SNPs) was also seen in gestational diabetes mellitus (GDM) patients.12 Several studies indicated that miR-375 plays a direct role in insulin secretion13 and pancreatic islet development14 by targeting the pancreatic genes; insulin (INS), myotrophin (MTPN) and phosphoinositide-dependent protein kinase-1 (PDPK1). Many studies have confirmed the overexpression of miR-375 in type 2 diabetes mellitus (T2DM) patients.15,16 Analysis of miR-375 level in first degree relatives of T2DM patients may provide insights on importance of miR-375 as a potential biomarker for the early prediction of T2DM among high-risk individuals. However, till date, no clinical studies have been conducted assessing the level of miR-375 expression in first degree relatives of T2DM patients.
In the present study, we aimed to evaluate the differential expression of miR-375 in T2DM patients and their first-degree relatives with normal glucose tolerance and T2DM individuals by its isolation and characterization using quantitative real-time polymerase chain reaction (qRT-PCR).
Patients and Methods
Study Design
This was a single center cross-sectional study with a single time point data collection. The study was conducted at The Third Hospital of Yunnan Province, China after receiving approval from the Ethics Committee of the hospital (approval number: THYP0085XCNR) and was conducted in compliance to the Declaration of Helsinki. The written informed consent was obtained from all subjects prior to their enrollment.
Research Subjects
The study population consisted of Han Chinese individuals who received medical health check-ups from January 2018 to September 2018 in the Outpatient Department of Endocrinology, The Third Hospital of Yunnan Province, China. The diagnosis and recruitment of study subjects was based on American Diabetes Association (ADA) criteria.17 Subjects were divided into 4 groups as per the diagnosis. Group 1 included normal glucose tolerance subjects, Group 2 had T2DM patients, first degree relatives with normal glucose tolerance were considered as group 3 and group 4 included first degree relatives with T2DM. First degree relatives were considered as parents, children (≥18 years old) and siblings (same parents, ≥18 years old) of patients with T2DM. Judgment criteria were normal fasting blood glucose: 3.89–6.1mmol/L, diabetes mellitus: adults regardless of sex and age with fasting glucose >7.0mmol/l and OGTT (PG2 h) >11.1mmol/L, impaired fasting glucose (IFG): fasting glucose 6.1–7.0mmol/L and OGTT (PG2h) <7.8mmol/L, impaired glucose tolerance (IGT): fasting glucose <7.0mmol/L and OGTT (PG2h) ≥7.8–11.1mmol/L, hyperinsulinemia: fasting insulin ≥85 pmol/L, and homeostasis model of insulin resistance (HOMA-IR): >2.69. Individuals with emergency status of type 1 diabetes mellitus, malignant tumor, chronic infection, autoimmune diseases, cardiovascular disease, nephropathy or other chronic diseases which pose latent effect on miRNAs expression and who had previously been diagnosed with diabetes mellitus or had any history of medication for 6 months prior to the study were excluded.
All the diagnoses were confirmed by Oral Glucose Tolerance Test on the basis of fasting plasma glucose (FPG) and plasma glucose (PG) levels at 30 minutes, 1, 2, and, 3 hours. Among 56 individuals enrolled, 21 were healthy individuals with normal glucose tolerance (NGT), 10 were diagnosed with T2DM, first degree relatives of T2DM patients with normal glucose tolerance (FD-NGT) were 13 in number and 12 individuals were first degree relatives of T2DM patients with T2DM (FD-T2DM).
Laboratory Analyses
OGTT (Oral Glucose Tolerance Test)
A 3-hour OGTT (75 g of glucose) was performed in the laboratory department of The Third Hospital of Yunnan Province, China. The samples for plasma glucose (PG) were drawn at 0, 30 minutes, 1, 2 and, 3 hours Diagnosis was based on OGTT recommended by American Diabetes Association and evaluated as follows: Patients with (FPG<5.6 mmol/L and 2-hour PG<7.8 mmol/L) were considered as normal glucose tolerance (NGT) and those with (FPG 5.6–6.9 mmol/L and 2-hour PG 7.8–11.0 mmol/L) were considered as T2DM.17
C-Peptide Test
The Connecting peptide (C-peptide) is produced in equal amounts to insulin and is considered a measure of endogenous insulin secretion.18 Hence, C-peptide levels were analyzed using the automated Roche diagnostics (Manheim, Germany) E170 immuno-analyser (limit of detection 3.3 pmol/l, inter- and intra-assay coefficients of variation <4.5% and <3.3%, respectively). Fasting C–peptide level was: 1.1 to 4.4, 1 hour after meal it was 5 to 10 times the fasting level and 3 hours after meal it was back to the level of fasting.
Insulin Test
Insulin was measured by chemiluminescent immunometric assay (Siemens Healthcare Diagnostics B.V., Breda, the Netherlands). The intra-assay variation was 6% at 47 pmol/L and 3% at 609 pmol/L. The inter-assay variation was 4% at 91 pmol/L and 6% at 120 pmol/L. The detection limit was 15 pmol/L. The fasting insulin level was 2.6 to 24.9 mIU/L, 1 hour after meal it was 5 to 10 times (13 to 249 mIU/L) the fasting level and 3 hours after meal it was back to the level of fasting.
RNA Isolation and Characterization
Peripheral blood was obtained by venipuncture and then coagulated at room temperature for 0.5–2 h. Following centrifugation at 3000g for 5 min, serum was collected and centrifuged again at 12,000g for 15 min to completely remove cell debris. It was then aliquoted and stored at −80°C until miRNA detection. Total RNA containing small RNA was extracted from 500 μL of serum using mirVana isolation kit (Ambion, Austin, USA) according to the manufacturer’s protocol. The final elution volume was 100 μL. The concentration of all RNA samples was quantified by NanoDrop 1000 (Nanodrop, Wilmington, USA), and 20 ng of serum RNA containing miRNA was reverse transcribed to cDNA using TaqMan MicroRNA Reverse Transcription Kit (Applied BioSystems, Foster, USA) and miRNA-specific primers provided by the manufacturer in an Applied BioSystems 9300 Thermocycler (Applied Biosystems, Foster, USA). All Complementary DNAs (cDNAs) were stored at −20°C until Quantitative real-time polymerase chain reaction (qRT-PCR) analysis. After 1:2 dilution, 4.5 μL was used as template in a 10 μL qPCR.
Quantitative Real-Time PCR (qRT-PCR)
Quantitative real-time RT-PCR (qRT-PCR) was performed to assess the levels of miR-375. Essential MicroRNA-specific data are presented in Table 1. Each reaction was performed in a final volume of 10μL containing 4.5μL cDNA, 5μL TaqMan 2 × Universal PCR Master Mix (No AmpErase) and 0.5μL TaqMan miRNA Assay (Applied BioSystems). The thermal cycle was set as start with 10 min template denaturation at 95°C, 40 cycles of denaturation at 95°C for 15 seconds and combined primer annealing/elongation at 60°C for 1 minute. Each sample was run in triplicate for analysis. As the internal control gene, non-coding small RNA RNU6B was used according to the Applied Biosystems Application Note. RNU6B has demonstrated both stable and abundant expression in different human tissues and organs. It is regarded as one of the control genes with the least variability for miRNAs assays and has been widely used in different fields including diabetic research.
Table 1.
Information About Detected miRNA
| Assay Name | Assay Type | AB Assay ID | miRBase Accession |
|---|---|---|---|
| hsa-miR-375 | Mature miRNA | 000564 | MIMAT0000728 |
| RNU6B | Control miR | 001093 | NR_002752 |
Statistical Analysis
Data were presented as means ± standard error. For qRT-PCR data, the difference of threshold cycle (Ct) between miR-375 and RNU6B (ΔCt) which was equivalent to the ratio of log2-transformed absolute copy numbers was employed to show the relative expression levels of miR-375. The one-way ANOVA followed by a post hoc multiple comparison test was used to compare the concentration of miR-375 among the four individual groups Statistical analysis was performed by using SPSS software. P value less than 0.05 was considered statistically significant.
Results
Baseline Demographics
A total of 56 Han Chinese individuals were enrolled, of which 21 of them had normal glucose tolerance (NGT), 10 were T2DM patients, 13 were individuals with normal glucose tolerance with first degree relatives having T2DM and 12 of them were T2DM patients with first degree relatives having T2DM (Table 2).
Table 2.
Characteristic of the Study Subjects
| Characteristics | NGT | T2DM | FD-NGT | FD-T2DM |
|---|---|---|---|---|
| Gender (Male/Female) | 5/16 | 7/3 | 3/10 | 7/5 |
| Age (years) | 28.16 | 45.2 | 40.38 | 44.5 |
| BMI (kg/m2) | 21.49 | 22.84 | 22.66 | 23.09 |
| HbA1c (%) | 4.59 | 7.726 | 5.14 | 7.38 |
| TC (mmol/L) | 4.29 | 5.25 | 5.16 | 4.78 |
| TG (mmol/L) | 1.02 | 1.61 | 1.6 | 2.04 |
Abbreviations: BMI, body mass index; HbAlc, glycated hemoglobin; TG, triglycerides; TC, total cholesterol.
Clinical Data
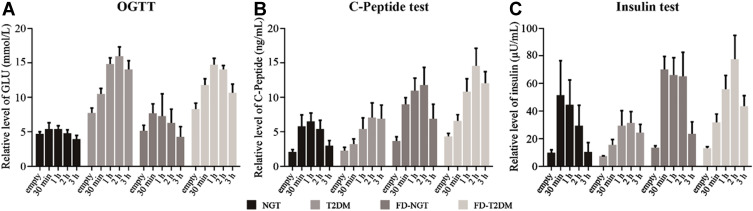
A significant difference was observed in T2DM and FD-T2DM groups compared to NGT and FD-NGT in terms of OGTT, C-peptide, and insulin test results. The 3-hour OGTT result showed a mean ± standard error of 3.844 ± 0.732 for NGT group whereas T2DM group showed a value of 13.810 ± 0.982 (p< 0.05). In the FD-T2DM group, the 3-hour value was observed as 10.583 ± 0.897 in comparison to NGT (3.844 ± 0.732) and FD-NGT (4.130 ± 0.861) showing statistical significance with p< 0.05 (Table 3, Figure 1A). Similar results were obtained from the C-peptide test. The C-peptide, mean ± standard error of 2.854 ± 1.351 was observed in NGT group after 3 hours whereas T2DM group showed a value of 6.875 ± 1.728 and was statistically significant with p< 0.05. FD-T2DM showed a value of 12.218 ± 1.728 and p< 0.05 when compared to NGT (2.854 ± 1.351), T2DM (6.875 ± 1.728) and FD-NGT (6.704 ± 1.590) (Table 4, Figure 1B). The 3-hour insulin test showed a mean ± standard error value of 9.802 ± 5.835, 23.543 ± 7.829, 22.523 ± 6.866, and 42.619 ± 7.147 in NGT, T2DM, FD-NGT, and FD-T2DM, respectively. A significantly higher value of p< 0.05 was observed in T2DM compared to NGT and in FD-T2DM also when compared to FD-NGT. Interestingly, FD-NGT showed a significantly higher value compared to NGT group (Table 5, Figure 1C).
Table 3.
Oral Glucose Tolerance Test
| Time for 75-g Oral Glucose | |||||
|---|---|---|---|---|---|
| Groups | FPG | 30 Min | 1 h | 2 h | 3 h |
| NGT | 4.541±0.490 | 5.293±0.543 | 4.763±0.677 | 4.649±0.595 | 3.844±0.732 |
| T2DM | 7.605±0.657* | 10.336±00.729* | 14.604±0.908* | 15.781±0.799* | 13.810±0.982* |
| FD-NGT | 4.928±0.576 | 7.547±0.639* | 7.193±0.796* | 6.150±0.701 | 4.130±0.861 |
| FD-T2DM | 8.141±0.600*Δ | 11.646±0.665*Δ | 14.550±0.829*Δ | 13.846±0.729*Δ | 10.583±0.897*Δ |
Notes: *p<0.05 in comparison to NGT, Δ p<0.05 in comparison to FD-NGT, Data are expressed as mean ± standard error.
Abbreviations: NGT, normal glucose tolerance; T2DM, type 2 diabetes mellitus; FD-NGT, in first-degree relatives of type 2 diabetes mellitus individuals with normal glucose tolerance; FD-T2DM, first-degree relatives of type 2 diabetes mellitus individuals with type 2 diabetes mellitus.
Figure 1.
Clinical data (A) OGTT (Oral Glucose Tolerance Test). (B) C-peptide test. (C) Insulin test.
Table 4.
The C-Peptide Test
| Time for Meal | |||||
|---|---|---|---|---|---|
| Groups | FPG | 30 Min | 1 h | 2 h | 3 h |
| NGT | 2.002±0.414 | 5.692±0.720 | 6.362±1.301 | 5.249±1.686 | 2.854±1.351 |
| T2DM | 2.241±0.529 | 3.495±0.921 | 5.598±1.665 | 7.505±2.157 | 6.875±1.728 |
| FD-NGT | 3.594±0.487* | 8.858±0.847* | 10.793±1.531* | 11.628±1.984* | 6.704±1.590* |
| FD-T2DM | 4.285±0.529*# | 6.383±0.921# | 10.722±1.665*# | 14.526±2.157*# | 12.218±1.728*#Δ |
Notes: *p<0.05 in comparison to NGT, Δ p<0.05 in comparison to FD-NGT, #p<0.05 in comparison toT2DM. Data are expressed as mean ± standard error.
Abbreviations: NGT, normal glucose tolerance; T2DM, type 2 diabetes mellitus; FD-NGT, first-degree relatives of type 2 diabetes mellitus individuals with normal glucose tolerance; FD-T2DM, first-degree relatives of type 2 diabetes mellitus individuals with type 2 diabetes mellitus.
Table 5.
The Insulin Test
| Time for Meal | |||||
|---|---|---|---|---|---|
| Groups | FPG | 30 Min | 1 h | 2 h | 3 h |
| NGT | 9.073±1.275 | 50.660±6.294 | 43.932±8.189 | 28.836±10.875 | 9.802±5.835 |
| T2DM | 6.540±1.711 | 14.789±8.444* | 28.961±10.987 | 30.773±14.566 | 23.543±7.829 |
| FD-NGT | 12.988±1.500 | 69.371±7.406 | 65.078±9.636 | 64.240±12.775* | 22.523±6.866 |
| FD-T2DM | 12.502±1.562# | 31.137±7.709Δ | 55.045±10.030Δ | 76.580±13.297*# | 42.619±7.147*Δ |
Notes: *p<0.05 in comparison to NGT, Δ p<0.05 in comparison to FD-NGT, #p<0.05 in comparison toT2DM. Data are expressed as mean ± standard error.
Abbreviations: NGT, normal glucose tolerance; T2DM, type 2 diabetes mellitus; FD-NGT, n first-degree relatives of type 2 diabetes mellitus individuals with normal glucose tolerance; FD-T2DM, first-degree relatives of type 2 diabetes mellitus individuals with type 2 diabetes mellitus.
These results also signify that individuals in FD-NGT group are at a higher risk of developing T2DM compared with individuals in NGT group.
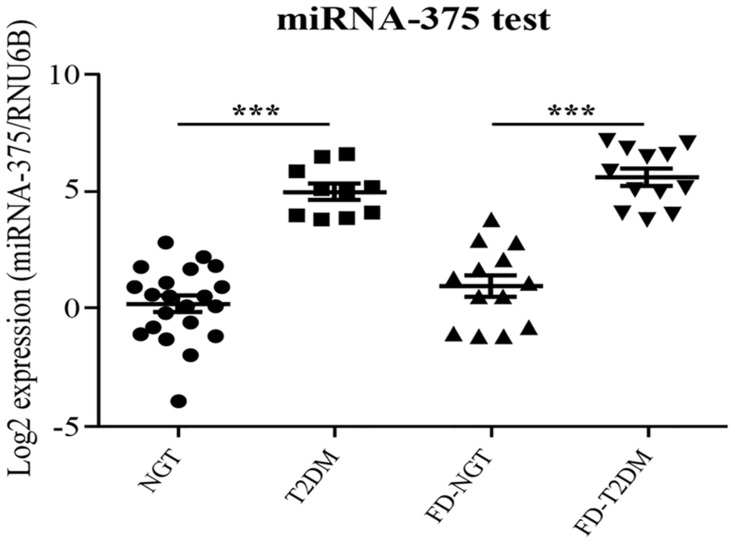
Expression Levels of miR-375 Were Significantly Elevated in T2DM and FD-T2DM
MiR-375 assessment by qRT-PCR showed significant up-regulation in T2DM group compared with NGT group (Figure 2). Similarly, a significant up-regulation was observed in FD-T2DM group compared with FD-NGT group. P value of <0.001 was observed in T2DM and FD-T2DM groups when compared with NGT and FD-NGT groups showing its statistical significance. The clinically significant elevation of miR-375 in FD-T2DM group suggests the potential role of miR-375 as a stable biomarker for the early prediction of T2DM among high-risk individuals. Interestingly, even though there was no significant difference between NGT and FD-NGT groups, we observed a slight increase in miRNA expression in FD-NGT compared with NGT group.
Figure 2.
Expression levels of miRNA-375.
Note: ***P value of <0.001 was observed in T2DM and FD-T2DM groups when compared with NGT and FD-NGT groups.
Discussion
The current study is the first clinical study to assess the differential expression of miR-375 in first degree relatives of T2DM patients in Han Chinese population. The results of the study revealed a significant difference in terms of OGTT, C-peptide and insulin test results in T2DM and FD-T2DM groups compared with NGT and FD-NGT. The OGTT test showed a significantly higher plasma glucose in T2DM compared with NGT and FD-T2DM compared with FD-NGT. The findings of our study were consistent with previously conducted clinical study which showed a significantly higher stepwise trends in FPG and 2-h PG from T2D-susceptible individuals with normal glucose tolerance (s-NGT) to pre-diabetes and the newly diagnosed T2D patients (n-T2D).15
The miR-375 level showed a significant up-regulation in T2DM compared with NGT group. A pilot cross-sectional study conducted to assess the miRNA levels in the circulation of subjects with diabetes showed similar results. The levels of miRNAs were significantly elevated in subjects with various forms of diabetes compared to healthy controls.19 A previous study on miR-375 up-regulation in T2DM patients showed its association with pancreatic islet amyloid formation and β-cell deficit. Owing to this, microRNA-375 may serve as a biomarker in the pathogenesis of type 2 diabetes related to islet amyloid deposition and β-cell dysfunction.20 Several studies have shown the potential role of miR-375 as a biomarker in diseases other than T2DM, such as osteosarcoma,21 non-alcoholic fatty liver disease,22 glioma cancer cells,23 colorectal cancer,24 oral premalignant lesions,25 non-small cell lung cancer26 etc. A meta-analysis conducted by Yan et al suggested that miR-375 profiling can be used as a screening test for cancers if the specific race and cancer is considered.27
Additionally, our study results revealed a significant up-regulation in FD-T2DM compared with FD-NGT group. Previous studies conducted on familial aggregation of T2DM showed a higher risk of developing diabetes and pre-diabetes progression in first degree relatives.28,29 The inclusion of first-degree relatives and assessing miRNA levels in them provides soundness to our study. A small sample size and consideration of only miR-375 in the analysis can be considered as the limitations of our study. Besides, this study was conducted only in Chinese population, and hence the results observed might not be generalizable. We observed higher values in FD-T2DM group when compared to T2DM group during C-peptide and insulin tests, further large multicenter studies are required to elucidate the possible mechanism for the observed result. In future studies, influences on circulating miRNA by gender, age and other factors (other diseases, medications, lifestyle factors) should be investigated. These studies will help to predict diabetes in the general population. Further, amassing data samples, standardizing miRNA detection technologies, and tracking and validating correlations with disease states will improve their predictive and diagnostic efficacy for developing strategies of therapeutic intervention.
To conclude, the present study demonstrated a significantly higher miR-375 expression in T2DM and FD-T2DM compared to NGT and FD-NGT groups, respectively, and thus, miR-375 may serve as a stable biomarker for the early prediction of T2DM among high-risk individuals.
Funding Statement
This study was supported by Yunnan Applied Basic Research Project-Youth Project (grant NO. 2017FD192).
Ethics Approval and Informed Consent
The ethics committee approval was obtained from the Third Hospital of Yunnan Province, China (approval number: THYP0085XCNR) and the study was conducted in compliance to the Declaration of Helsinki. The written informed consent was obtained from all subjects prior to their enrollment.
Disclosure
All authors declare that they have no competing financial interests and report no conflicts of interest in this work.
References
- 1.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402. doi: 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asahara H. Current status and strategy of microRNA research for cartilage development and osteoarthritis pathogenesis. J Bone Metab. 2016;23(3):121–127. doi: 10.11005/jbm.2016.23.3.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- 4.Zhu H, Fan G-C. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am J Cardiovasc Dis. 2011;1(2):138–149. [PMC free article] [PubMed] [Google Scholar]
- 5.Fransquet PD, Ryan J. Micro RNA as a potential blood-based epigenetic biomarker for alzheimer’s disease. Clin Biochem. 2018;58:5–14. doi: 10.1016/j.clinbiochem.2018.05.020 [DOI] [PubMed] [Google Scholar]
- 6.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seyhan AA. microRNAs with different functions and roles in disease development and as potential biomarkers of diabetes: progress and challenges. Mol Biosyst. 2015;11(5):1217–1234. doi: 10.1039/c5mb00064e [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Huang Z, Ni S, et al. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One. 2012;7(9):e44398. doi: 10.1371/journal.pone.0044398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wojciechowska A, Braniewska A, Kozar-Kamińska K. MicroRNA in cardiovascular biology and disease. Adv Clin Exp Med. 2017;26(5):865–874. doi: 10.17219/acem/62915 [DOI] [PubMed] [Google Scholar]
- 10.Tang X, Tang G, Ozcan S. Role of microRNAs in diabetes. Biochim Biophys Acta. 2008;1779(11):697–701. doi: 10.1016/j.bbagrm.2008.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alharbi KK, Abudawood M, Ali Khan I. Amino-acid amendment of arginine-325-tryptophan in rs13266634 genetic polymorphism studies of the SLC30A8 gene with type 2 diabetes-mellitus patients featuring a positive family history in the Saudi population. J King Saud Univ Sci. 2021;33(1):101258. doi: 10.1016/j.jksus.2020.101258 [DOI] [Google Scholar]
- 12.Khan IA, Jahan P, Hasan Q, Rao P. Genetic confirmation of T2DM meta-analysis variants studied in gestational diabetes mellitus in an Indian population. Diabetes Metab Syndr. 2019;13(1):688–694. doi: 10.1016/j.dsx.2018.11.035 [DOI] [PubMed] [Google Scholar]
- 13.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076 [DOI] [PubMed] [Google Scholar]
- 14.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RHA, Carrington JC. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5(8):e203. doi: 10.1371/journal.pbio.0050203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong L, Zhu J, Han W, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48(1):61–69. doi: 10.1007/s00592-010-0226-0 [DOI] [PubMed] [Google Scholar]
- 16.Sun K, Chang X, Yin L, et al. Expression and DNA methylation status of microRNA-375 in patients with type 2 diabetes mellitus. Mol Med Rep. 2014;9(3):967–972. doi: 10.3892/mmr.2013.1872 [DOI] [PubMed] [Google Scholar]
- 17.Association AD. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes—2019. Diabetes Care. 2019;42(Supplement 1):S13–S28. doi: 10.2337/dc19-S002 [DOI] [PubMed] [Google Scholar]
- 18.Ludvigsson J. C-peptide in diabetes diagnosis and therapy. Front Biosci (Elite Ed). 2013;5:214–223. doi: 10.2741/e609 [DOI] [PubMed] [Google Scholar]
- 19.Seyhan AA, Nunez Lopez YO, Xie H, et al. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: a pilot cross-sectional study. Sci Rep. 2016;6(1). doi: 10.1038/srep31479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H, Guan J, Lee H-M, et al. Up-regulated pancreatic tissue microRNA-375 associates with human type 2 diabetes through beta-cell deficit and islet amyloid deposition. Pancreas. 2010;39(6):843–846. doi: 10.1097/MPA.0b013e3181d12613 [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Zhao X, Zhang Y-J, Fang G-W, Xue Y. MicroRNA-375 as a potential serum biomarker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J Int Med Res. 2018;46(3):975–983. doi: 10.1177/0300060517734114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei L, Zhou C, Yang X, Li L. Down-regulation of microRNA-375 regulates adipokines and inhibits inflammatory cytokines by targeting AdipoR2 in non-alcoholic fatty liver disease. Clin Exp Pharmacol Physiol. 2018;45(8):819–831. doi: 10.1111/1440-1681.12940 [DOI] [PubMed] [Google Scholar]
- 23.Zhang LX, Jin W, Zheng J, et al. MicroRNA-375 regulates proliferation and apoptosis of glioma cancer cells by inhibiting CTGF-EGFR signaling pathway. Bratisl Lek Listy. 2018;119(1):17–21. doi: 10.4149/BLL_2018_004 [DOI] [PubMed] [Google Scholar]
- 24.Alam KJ, Mo J-S, Han S-H, et al. MicroRNA 375 regulates proliferation and migration of colon cancer cells by suppressing the CTGF-EGFR signaling pathway. Int J Cancer. 2017;141(8):1614–1629. doi: 10.1002/ijc.30861 [DOI] [PubMed] [Google Scholar]
- 25.Harrandah AM, Fitzpatrick SG, Smith MH, Wang D, Cohen DM, Chan EKL. MicroRNA-375 as a biomarker for malignant transformation in oral lesions. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;122(6):743–752.e1. doi: 10.1016/j.oooo.2016.07.022 [DOI] [PubMed] [Google Scholar]
- 26.Chen L-J, Li X-Y, Zhao Y-Q, et al. Down-regulated microRNA-375 expression as a predictive biomarker in non-small cell lung cancer brain metastasis and its prognostic significance. Pathol Res Pract. 2017;213(8):882–888. doi: 10.1016/j.prp.2017.06.012 [DOI] [PubMed] [Google Scholar]
- 27.Yan J, She Q, Shen X, Zhang Y, Liu B, Zhang G. Potential role of MicroRNA-375 as biomarker in human cancers detection: a meta-analysis. Biomed Res Int. 2017;2017:1875843. doi: 10.1155/2017/1875843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran A, Snehalatha C, Satyavani K, Sivasankari S, Vijay V. Cosegregation of obesity with familial aggregation of type 2 diabetes mellitus. Diabetes Obes Metab. 2000;2(3):149–154. doi: 10.1046/j.1463-1326.2000.00067.x [DOI] [PubMed] [Google Scholar]
- 29.Li JKY, Ng MCY, So WY, et al. Phenotypic and genetic clustering of diabetes and metabolic syndrome in Chinese families with type 2 diabetes mellitus. Diabetes Metab Res Rev. 2006;22(1):46–52. doi: 10.1002/dmrr.577 [DOI] [PubMed] [Google Scholar]